Abstract
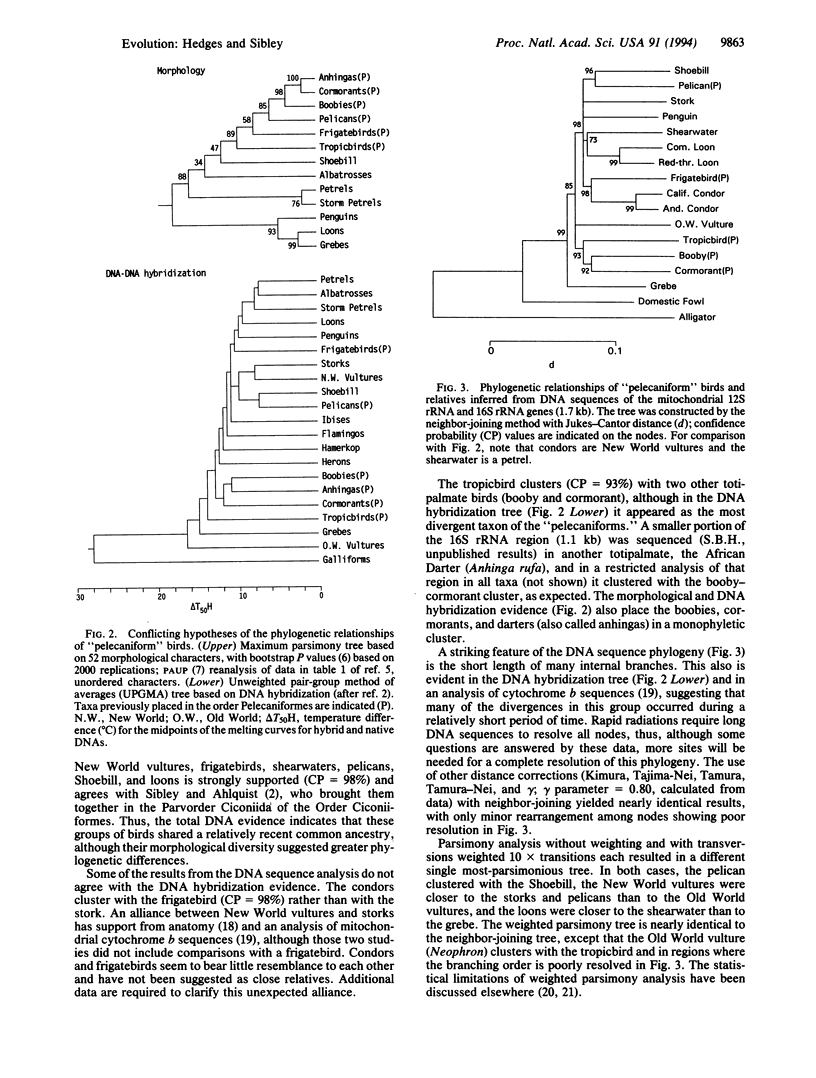
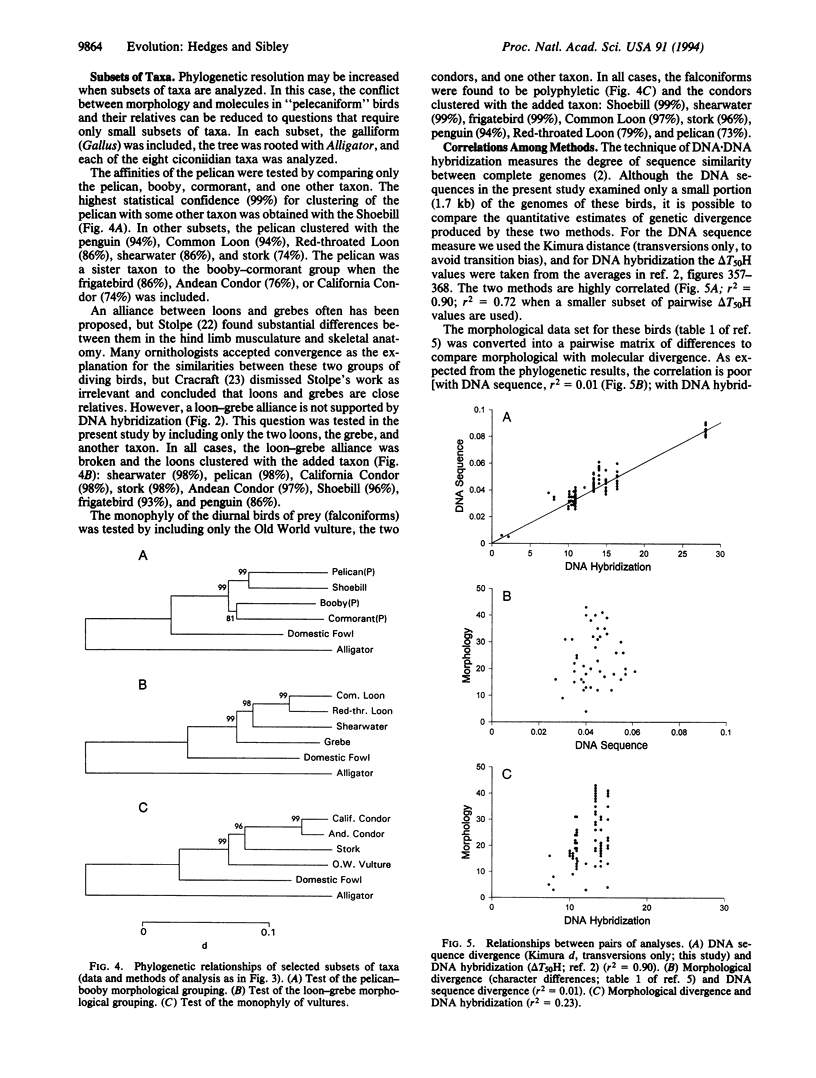
The traditional avian Order Pelecaniformes is composed of birds with all four toes connected by a web. This "totipalmate" condition is found in ca. 66 living species: 8 pelicans (Pelecanus), 9 boobies and gannets (Sula, Papasula, Morus), ca. 37 cormorants (Phalacrocorax), 4 anhingas or darters (Anhinga), 5 frigatebirds (Fregata), and 3 tropicbirds (Phaethon). Several additional characters are shared by these genera, and their monophyly has been assumed since the beginning of modern zoological nomenclature. Most ornithologists classify these genera as an order, although tropicbirds have been viewed as related to terns, and frigatebirds as relatives of the petrels and albatrosses. DNA.DNA hybridization data indicated that the pelicans are most closely related to the Shoebill (Balaeniceps rex), a stork-like bird that lives in the swamps of central Africa; the boobies, gannets, cormorants, and anhingas form a closely related cluster; the tropicbirds are not closely related to the other taxa; and the frigatebirds are closest to the penguins, loons, petrels, shearwaters, and albatrosses (Procellarioidea). Most of these results are corroborated by DNA sequences of the 12S and 16S rRNA mitochondrial genes, and they provide another example of incongruence between classifications derived from morphological versus genetic traits.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Nelson W. S., Sibley C. G. DNA sequence support for a close phylogenetic relationship between some storks and New World vultures. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5173–5177. doi: 10.1073/pnas.91.11.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabot E. L., Beckenbach A. T. Simultaneous editing of multiple nucleic acid and protein sequences with ESEE. Comput Appl Biosci. 1989 Jul;5(3):233–234. doi: 10.1093/bioinformatics/5.3.233. [DOI] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Nucleotide sequence and evolution of coding and noncoding regions of a quail mitochondrial genome. J Mol Evol. 1991 Feb;32(2):153–161. doi: 10.1007/BF02515387. [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Bezy R. L. Phylogeny of xantusiid lizards: concern for data and analysis. Mol Phylogenet Evol. 1993 Mar;2(1):76–87. doi: 10.1006/mpev.1993.1008. [DOI] [PubMed] [Google Scholar]
- Hedges S. B. Molecular evidence for the origin of birds. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2621–2624. doi: 10.1073/pnas.91.7.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994 Apr;10(2):189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]




