Significance
Terpenoids are the largest group of plant-specialized metabolites and include many valuable bioactive compounds, such as the blockbuster anticancer drugs vincristine and vinblastine, that are monoterpenoid indole alkaloids from the medicinal plant Catharanthus roseus (Madagascar periwinkle). A master regulator was discovered that activates the biosynthesis of the iridoids, the monoterpenoid precursors of vinblastine and vincristine, and the rate-limiting branch in their biosynthetic pathway. This master regulator can be used to boost production of iridoids and monoterpenoid indole alkaloids in C. roseus cell cultures and thus represents an interesting tool for the metabolic engineering of the sustainable production of these high-value compounds in cultures of the endogenous plant species.
Keywords: basic helix loop helix, Catharanthus roseus, jasmonate, Madagascar periwinkle, iridoids
Abstract
Plants make specialized bioactive metabolites to defend themselves against attackers. The conserved control mechanisms are based on transcriptional activation of the respective plant species-specific biosynthetic pathways by the phytohormone jasmonate. Knowledge of the transcription factors involved, particularly in terpenoid biosynthesis, remains fragmentary. By transcriptome analysis and functional screens in the medicinal plant Catharanthus roseus (Madagascar periwinkle), the unique source of the monoterpenoid indole alkaloid (MIA)-type anticancer drugs vincristine and vinblastine, we identified a jasmonate-regulated basic helix–loop–helix (bHLH) transcription factor from clade IVa inducing the monoterpenoid branch of the MIA pathway. The bHLH iridoid synthesis 1 (BIS1) transcription factor transactivated the expression of all of the genes encoding the enzymes that catalyze the sequential conversion of the ubiquitous terpenoid precursor geranyl diphosphate to the iridoid loganic acid. BIS1 acted in a complementary manner to the previously characterized ethylene response factor Octadecanoid derivative-Responsive Catharanthus APETALA2-domain 3 (ORCA3) that transactivates the expression of several genes encoding the enzymes catalyzing the conversion of loganic acid to the downstream MIAs. In contrast to ORCA3, overexpression of BIS1 was sufficient to boost production of high-value iridoids and MIAs in C. roseus suspension cell cultures. Hence, BIS1 might be a metabolic engineering tool to produce sustainably high-value MIAs in C. roseus plants or cultures.
Plants produce a vast amount of natural products, also called specialized metabolites, by which they can interact with their environment to ensure survival and reproductive success. These compounds often have bioactive properties that make them valuable for various pharmaceutical applications. The phytohormone jasmonate (JA) acts as an elicitor of the production of these bioactive metabolites and triggers a transcriptional reprogramming of plant metabolism, resulting in a concerted up-regulation of the expression of genes encoding enzymes involved in specific specialized metabolic pathways (1). The specialized defense metabolites are generally produced from primary metabolites, leading to precursor competition with cellular processes implicated in growth and development. For instance, terpenoid biosynthesis depends on the precursor isopentenyl diphosphate (IPP), which is produced either via the plastidial 2-C-methyl-d-erythritol 4-phosphate (MEP) or the cytosolic mevalonate (MVA) pathways (Fig. 1A). In the medicinal plant Catharanthus roseus (Madagascar periwinkle), the MEP pathway-derived IPP is one of the precursors of the monoterpenoid indole alkaloid (MIA) pathway that leads to the valuable anticancer molecules vincristine and vinblastine, of which C. roseus is the sole source (2–4), whereas the MVA-derived IPP is the precursor of the pentacyclic triterpenoid pathway (5, 6) (Fig. 1A). As IPP is also the precursor for the synthesis of gibberellins, brassinosteroids, phytosterols, carotenoids, and phytol, among others, the tight regulation of both IPP supply and consumption is crucial.
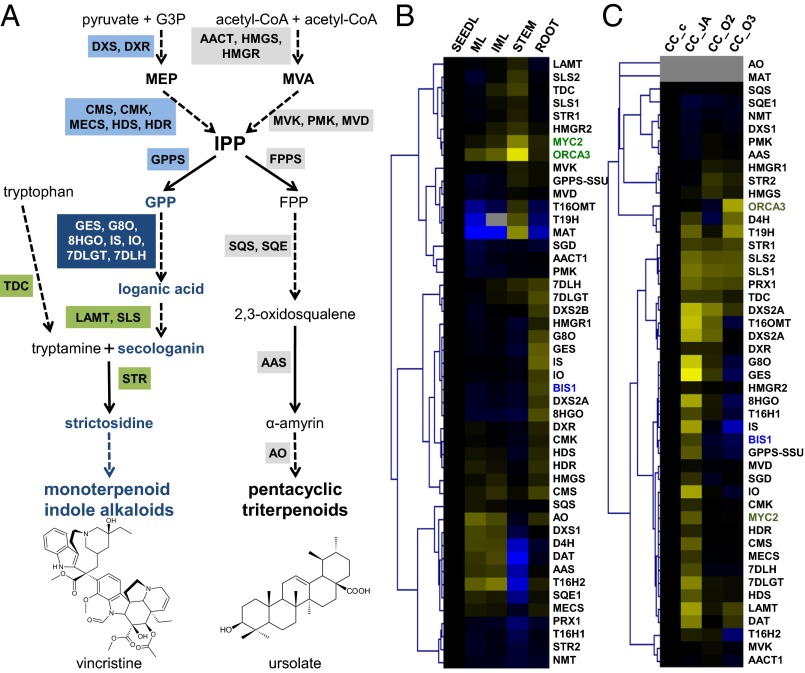
Fig. 1.
BIS1 is coexpressed with the iridoid pathway genes in C. roseus. (A) Pathways leading to the production of MIAs and triterpenoids in C. roseus. Genes activated by BIS1 and ORCA3 overexpression are boxed in blue and green, respectively. 7DLGT, 7-deoxyloganetic acid glucosyl transferase; 7DLH, 7-deoxyloganic acid hydroxylase; 8HGO, 8-hydroxygeraniol oxidoreductase; AAS, α-amyrin synthase; AO, amyrin oxidase; DXR, 1-deoxy-5-xylulose-5-phosphate reductase; FPP, farnesyl pyrophosphate; G3P, glyceraldehyde 3-phosphate; G8O, geraniol-8-oxidase; GES, geraniol synthase; GPP, geranyl diphosphate; HDS, (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; IPP, isopentenyl diphosphate; IO, iridoid oxidase; IS, iridoid synthase; LAMT, loganic acid O-methyltransferase; MECS, 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate synthase; MEP, 2-C-methyl-d-erythritol 4-phosphate; MVA, mevalonate; SLS, secologanin synthase; STR, strictosidine synthase; TDC, tryptophan decarboxylase. (B and C) Coexpression analysis of BIS1, ORCA3, MYC2, and the known MIA and triterpenoid pathway genes. The average linkage hierarchical clustering with Pearson correlation was used. FKPM values along with Caros and gene ID can be found in SI Appendix, Tables S1 and S2. Blue and yellow denote relative down-regulation and up-regulation, respectively. (B) Selected RNA-Seq data from the Medicinal Plant Genomics Resource (medicinalplantgenomics.msu.edu). Values were normalized to the seedling reads. IM, immature leaf; ML, mature leaf. (C) RNA-Seq from the Online Resource for Community Annotation of Eukaryotes (ORCAE) database from the SmartCell consortium (bioinformatics.psb.ugent.be/orcae/overview/Catro). Values were normalized to the control cell culture (CC_c). CC_JA, JA-treated cell culture; CC_O2 and CC_O3, cell culture overexpressing ORCA2 and ORCA3. Genes indicated in gray were not expressed in the cell culture.
In C. roseus, the MEP-derived production of the monoterpenoid, more specifically seco-iridoid, compound secologanin involves 10 enzymatic conversions starting from IPP, of which the eight genes upstream of loganic acid methyltransferase (LAMT) are tightly coexpressed (Fig. 1) (2, 3, 7). Secologanin is subsequently coupled to the indole compound tryptamine, forming the common alkaloid precursor strictosidine, from which all MIAs are derived (Fig. 1A). In contrast to the triterpenoid genes in C. roseus, genes involved in MIA production are JA inducible, clearly pointing to distinct regulatory circuits governing these two terpenoid pathways (Fig. 1) (8).
The concerted regulation of JA-induced genes is mediated by transcription factors (TFs) that are usually encoded by JA-inducible genes themselves (1). A combinatorial role for several TFs seems a plausible strategy for the control of biosynthetic pathways by JAs, but the identity of such TF arrays has so far remained elusive for most terpenoid pathways (1). The basic helix–loop–helix (bHLH) TF MYC2 has emerged as a central regulator in JA signaling cascades, including those leading to the biosynthesis of several classes of specialized metabolites (9). MYC2-type TFs have been shown to be involved in the regulation of terpene biosynthesis genes in Arabidopsis thaliana, Artemisia annua (sweet wormwood), and Solanum lycopersicum (tomato) (10–12). In C. roseus, MYC2 regulates the expression of the ethylene response factor (ERF) Octadecanoid derivative-Responsive Catharanthus APETALA2-domain 3 (ORCA3) (13), the JA-inducible regulatory TF that modulates the JA-induced expression of the genes of the indole branch of the pathway, strictosidine synthase (STR), and several steps downstream of strictosidine, thereby controlling part of the JA-responsive production of MIAs (14, 15). However, the iridoid genes upstream of LAMT, such as geraniol-8-oxidase (G8O), are not under the control of ORCA3 (Fig. 1) (14) but are clearly responsive to JA, as illustrated by transcriptome (8, 14) and G8O promoter (16) analysis, suggesting that several regulatory circuits might exist that drive divergent gene expression in the MIA pathway. These circuits probably involve both repressors and activators, such as the previously studied C. roseus G-box binding factors (GBFs), WRKY1, AT-hook, and zinc finger TFs (17–20), or TFs that interact with MYC2, for instance, as reported in Arabidopsis (21, 22).
As the accumulation of MIAs is low, leading to scarcity and high market prices, there is a general interest to boost the production of these compounds in planta. The use of TFs can be a powerful tool in such metabolic engineering programs. ORCA3 has been overexpressed in cell suspensions, hairy roots, and plants in attempts to increase MIA production (14, 23–25). However, because ORCA3 does not activate iridoid genes, MIA accumulation in these lines required either coexpression with G8O or feeding with iridoid precursors. Given the rate-limiting nature of the iridoid pathway in C. roseus cultures, we launched a screen, based on transcriptome analysis, to discover iridoid-regulating TFs.
Results
Identification of a pG8O-Transactivating TF.
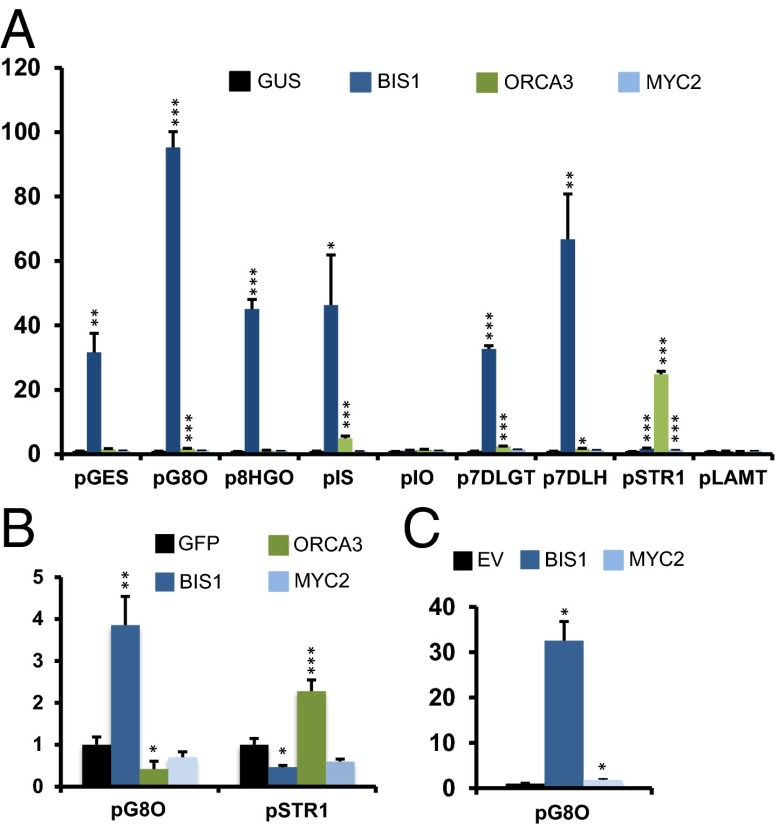
To identify regulators of the iridoid pathway branch, we set up a screen for transactivators of the G8O promoter (pG8O), the sequence of which had been determined previously (16). Candidate TFs were selected by mining recently generated RNA-Seq data (7, 8, 26) for TFs that showed coexpression with the iridoid genes upstream of LAMT. Essentially, TF encoding genes that showed expression patterns similar to G8O in the RNA-Seq data generated by the SmartCell consortium (bioinformatics.psb.ugent.be/orcae/overview/Catro), i.e., JA induced in both cell suspensions and seedlings but not induced by ORCA overexpression in cell suspensions, were selected for further analysis. Among them, a bHLH TF belonging to clade IVa of the family (27) (Fig. 1 B and C and SI Appendix, Fig. S1) was capable of transactivating pG8O 95.3-fold in a Nicotiana tabacum (tobacco) protoplast-based screen (Fig. 2A). The gene encoding this bHLH TF, designated “bHLH Iridoid Synthesis 1” (BIS1; Caros001862), responded to JA similarly to the genes encoding the enzymes involved in the iridoid and MIA pathways and those encoding known regulators, such as ORCA3 and MYC2 (Fig. 1C). Contrary to BIS1, the latter two transactivated pG8O only by 1.7-fold and 1.1-fold, respectively (Fig. 2A). Leaf disk assays with agroinfiltrated Nicotiana benthamiana plants confirmed the effect of BIS1 on pG8O (Fig. 2B). Finally, these results were corroborated by transient transactivation assays in bombarded C. roseus cells (Fig. 2C).
Fig. 2.
BIS1 transactivates iridoid genes in transient assays. (A) Transactivation in transfected N. tabacum protoplasts of iridoid and MIA promoters driving firefly luciferase (fLUC) expression by effector plasmids expressing BIS1, ORCA3, or MYC2. Values in the y axis are normalized fold-changes relative to protoplasts cotransfected with the reporter constructs and a pCaMV35S:GUS (GUS) control plasmid. For the normalization procedure, see SI Appendix, SI Materials and Methods. The error bars designate SE of the mean (n = 8). (B) Transactivation in agroinfiltrated N. benthamiana leaves of pG8O and pSTR1 driving fLUC expression by effector plasmids expressing BIS1, ORCA3, and MYC2. Values are normalized fold-changes relative to leaves coinfiltrated with the reporter constructs and a pCaMV35S:GFP (GFP) control plasmid. The error bars designate SE of the mean (n = 8). (C) Transactivation in bombarded C. roseus MP183L cells of pG8O driving GUS expression by effector plasmids expressing BIS1 and MYC2. Values are fold-changes relative to cells cobombarded with the reporter construct and a pCaMV35S:GFP (GFP) control plasmid. The error bars designate SE of the mean (n = 3). In all cases, statistical significance was determined by the Student’s t test (*P < 0.05, **P < 0.005, ***P < 0.0005).
BIS1 Specifically Transactivates the Iridoid Pathway Branch Upstream of LAMT.
Iridoid genes upstream of LAMT in C. roseus are highly coexpressed with G8O and thus might potentially be under a similar regulatory control. To test this hypothesis, we isolated the gene promoters of geraniol synthase (GES), 8-hydroxygeraniol oxidase (8HGO), iridoid synthase (IS), iridoid oxidase (IO), 7-deoxyloganetic acid glucosyltransferase (7DLGT), 7-deoxyloganic acid hydroxylase (7DLH), and LAMT from the recently revealed C. roseus genome sequence (28), as well as of the ORCA3-dependent gene STR1, and assessed them for transactivation by BIS1, MYC2, and ORCA3 (Fig. 2). In tobacco protoplasts, BIS1 was able to induce all iridoid promoters 31- to 95-fold, except for pIO (Fig. 2A). In contrast, ORCA3 only marginally transactivated the iridoid promoters, usually by less than twofold, except for pIS, which was induced 4.9-fold (Fig. 2A). No significant transactivation with MYC2 was observed for any of the iridoid promoters. As the minor effect of ORCA3 on these promoters did not correlate with increased expression of the corresponding genes in the ORCA3-overexpressing cell line (as determined by RNA-Seq analysis; Fig. 1C and SI Appendix, Table S2) (7), we did not further consider it. Conversely, the promoter of the known ORCA3 target gene STR1 was induced 24.7-fold by ORCA3 and only 1.3-fold by either BIS1 or MYC2 in tobacco protoplasts (Fig. 2A). The effect of ORCA3 on pSTR1 was validated in the N. benthamiana leaf assay, in which BIS1 or MYC2 slightly repressed the pSTR1-driven reporter expression (Fig. 2B). The LAMT promoter derivative used here could not be transactivated by any TF (Fig. 2B). Hence, the action range of BIS1 is limited to the iridoid branch of the MIA pathway (up to loganic acid synthesis) and is clearly distinct from that of ORCA3.
BIS1 Boosts MIA Production in C. roseus Cell Cultures.
In contrast to differentiated C. roseus plant tissues, the C. roseus cell suspension line MP183L accumulates only trace amounts of secologanin, strictosidine, or MIAs without exogenous supply of JA or loganin. In addition and in contrast to the ORCA3-dependent genes, G8O transcripts could not be detected in wild-type cells, illustrating the limiting nature of the seco-iridoid pathway in this cell line (14). The RNA-Seq data analysis confirmed the low transcript accumulation of all MEP and iridoid genes in nonelicited cells compared with that of the ORCA3-regulated genes, such as LAMT, SLS1, and STR1 (SI Appendix, Table S2). Overexpression of ORCA3 in this cell line increased the expression of LAMT, STR, and SLS, among other genes, but not of the iridoid genes upstream of LAMT (Fig. 1C) (7, 14). Accordingly, an increase in MIA accumulation in the ORCA3-overexpressing cells could only be achieved after addition of loganin to the cell culture medium (14).
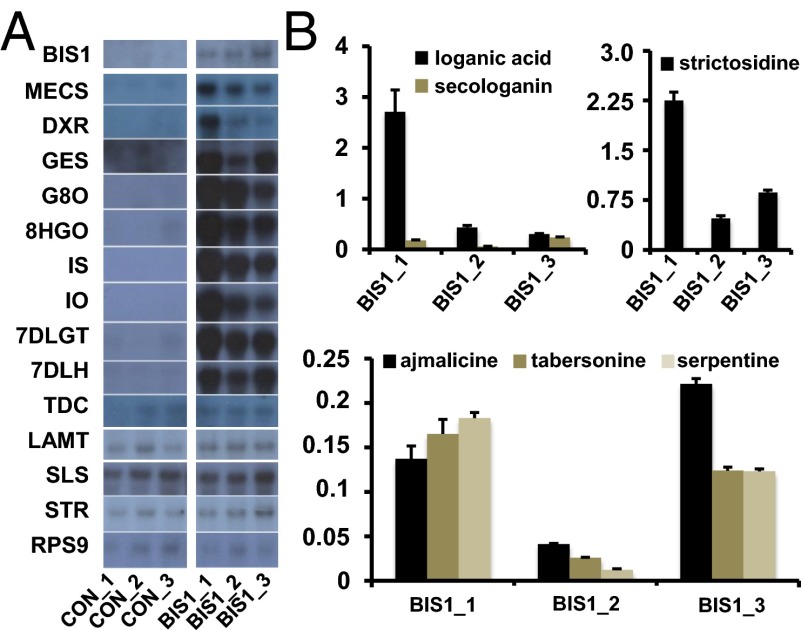
To evaluate the effect of BIS1 on MIA production and assess its value as a metabolic engineering tool, we generated three stable transgenic MP183L suspension cell lines constitutively overexpressing BIS1 (Fig. 3). Compared with control lines expressing the gene encoding the green fluorescent protein (GFP), BIS1 overexpression resulted in a high up-regulation of MEP genes and the iridoid genes upstream of LAMT in three independent BIS1 lines, whereas transcript accumulation of ORCA3-dependent genes remained unaltered (Fig. 3A). This up-regulation was accompanied by a strong effect on the metabolite level (Fig. 3B). In contrast to wild-type and control cells, the (seco)-iridoids loganic acid and secologanin accumulated in the BIS1-overexpressing cells. Likewise, we observed a strong accumulation of strictosidine, indicating that the tryptamine levels were not limiting, as well as of the downstream MIA compounds ajmalicine, serpentine, and tabersonine to levels previously unreported in untreated MP183L cells. In control cell lines, none of these MIA compounds accumulated to detectable levels. Together these data demonstrate that BIS1 overexpression is sufficient to boost MIA production in C. roseus MP183L cells.
Fig. 3.
BIS1 overexpression boosts MIA production in stably transformed C. roseus suspension cultures. (A) Expression analysis in transformed cells by RNA-blot hybridization analysis showing the effect of constitutive overexpression of BIS1 on selected biosynthetic genes in three independent cell suspension cultures of C. roseus. RPS9 was used as a reference gene. (B) Accumulation of (seco)-iridoid and MIA compounds in the BIS1-overexpressing cell lines. Metabolite levels are indicated in mg·g–1 dry weight. The error bars designate SE of the mean (n = 3). In control cell lines, no detectable levels of these compounds accumulated.
BIS1 Specifically Up-Regulates the Iridoid Genes in C. roseus Hairy Roots.
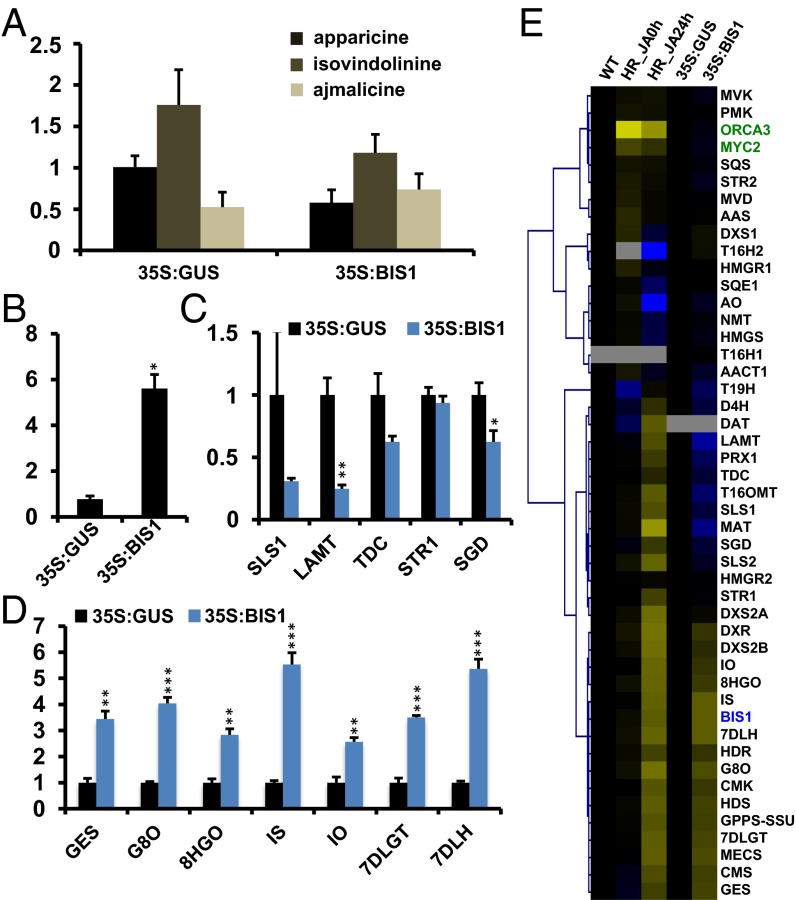
Overexpression of G8O had previously been reported to be sufficient to increase MIA accumulation in C. roseus hairy roots (24). Therefore, we also generated C. roseus hairy roots that overexpressed BIS1. In contrast to the cell suspensions, MIA accumulation did not increase in the transgenic hairy root lines (Fig. 4A), despite clear BIS1 overexpression (Fig. 4B).
Fig. 4.
Expression and metabolite profiling of BIS1-overexpressing C. roseus hairy roots. (A) Metabolite profiling on the BIS1-overexpressing and control (GUS) lines. MIA levels are indicated in mg·g–1 dry weight. The error bars designate SE of the mean (n = 3). (B) qPCR analysis showing BIS1 expression relative to the control lines set at 1. The error bars designate SE of the mean (n = 3). Statistical significance was determined by the Student’s t test (*P < 0.05, **P < 0.005, ***P < 0.0005). (C–E) Genome-wide expression profiling by RNA-Seq analysis. (C and D) Effect on expression of selected MIA (C) and (seco)-iridoid (D) genes indicated as fold induction relative to the control lines set at 1. The error bars designate SE of the mean (n = 3). (E) Average linkage hierarchical clustering of biosynthetic and TF genes. HR_JA0h and HR_JA24h, hairy roots treated with MeJA for 0 h and 24 h; WT, wild-type hairy roots (all obtained from bioinformatics.psb.ugent.be/orcae/overview/Catro). Values were normalized against the WT hairy roots and the pCaMV35S:GUS (35S:GUS) control hairy roots for the MeJA-treated and the pCaMV35S:BIS1 (35S:BIS1) samples, respectively. FKPM values along with Caros and gene ID can be found in SI Appendix, Table S2. Blue and yellow denote relative down-regulation and up-regulation, respectively.
Previously, MIA metabolite levels had not been found to increase significantly in ORCA3-overexpressing C. roseus hairy root lines either, not even after loganin and tryptophan feeding (23). This observation correlated not only with the anticipated lack of G8O induction but, unexpectedly, also with unaltered tryptophan decarboxylase (TDC) and repressed strictosidine-β-glucosidase (SGD) expression (23), two ORCA3-inducible genes in cell cultures (14).
To assess whether analogous flux limitations would occur in the BIS1-overexpressing roots, we carried out a transcriptome analysis by RNA-Seq. As in suspension cells, BIS1 overexpression up-regulated the expression of all iridoid genes upstream of LAMT (Fig. 4D) but not of the ORCA3 target gene STR1 (Fig. 4C). Overexpression of BIS1 also resulted in increased transcript accumulation of the genes encoding all MEP pathway enzymes and the geranyl diphosphate (GPP) synthase small subunit (Fig. 4E and SI Appendix, Table S2) but in a decreased expression of the ORCA3 target genes LAMT, secologanin synthase (SLS), TDC, and SGD (Fig. 4C and SI Appendix, Table S2). The latter effect might create a flux limit and might explain that BIS1 overexpression does not lead to increased MIA accumulation in hairy roots. Decreased expression of the ORCA3 target genes was not caused by decreased expression of ORCA2 or ORCA3, nor of any other known C. roseus TF encoding gene previously linked with regulation of the MIA pathway (SI Appendix, Fig. S2), and thus involves other, yet unknown, regulatory mechanisms.
It is conceivable that the increased MEP pathway activity may involve widespread reprogramming of terpenoid pathways dependent on MEP-derived IPP and GPP. To assess this, we mined the RNA-Seq data for genes encoding enzymes involved in MEP-dependent terpene biosynthesis, encompassing the monoterpenoid, diterpenoid (gibberellins), and carotenoid and phytyldiphosphate-reliant (chlorophyll and tocopherol) biosynthetic pathways. This analysis indicated that none of the above pathways was affected by BIS1 overexpression in hairy roots (SI Appendix, Fig. S3). In concordance with this observation, expression of the genes involved in these pathways was not affected by JA elicitation either (SI Appendix, Fig. S3).
C. roseus also produces pentacyclic triterpenoids, such as ursolate and oleanolate (5, 6), that are synthesized from MVA pathway-derived IPP. As BIS1 is JA inducible but the C. roseus triterpenoid pathway genes are not (8), we anticipated that the triterpenoid pathway genes would not be affected by BIS1 overexpression, as was indeed the case (Fig. 4E). Moreover, the triterpenoid precursor MVA pathway genes were not induced by BIS1 overexpression or by JA treatment (Fig. 4E) either, the latter in contrast to many other plant species (29). Together, this transcriptome analysis delineates the BIS1 specificity as a transcriptional activator of the iridoid pathway up to loganic acid.
Silencing of BIS1 Reduces MIA Accumulation in Planta.
Finally, we carried out a loss-of-function analysis of BIS1 in two different systems. First, virus-induced silencing (VIGS) was used to silence the expression of BIS1 in leaves of the C. roseus cultivar Little Bright Eyes. Unfortunately, despite several attempts, only a minor effect on the BIS1 transcript silencing and secologanin accumulation was obtained (SI Appendix, Fig. S4). No difference in downstream MIA products, such as strictosidine or vindoline, was observed. Second, we transformed C. roseus hairy roots with BIS1-targeting RNAi-silencing constructs. Two stably transformed root lines with significantly reduced BIS1 mRNA levels were obtained (SI Appendix, Fig. S4C). In these lines, MIAs accumulated at significantly decreased levels (SI Appendix, Fig. S4D). Together these results support a role for BIS1 in the regulation of the iridoid pathway in planta.
BIS1 Expression Is Enriched in IPAP-Containing Tissues.
The fragments per kilobase of exon per million fragments mapped (FPKM) values of BIS1 are too low to enable in situ hybridization signal detection (2). Therefore, to assess the cell specificity of BIS1 expression, we generated two sets of C. roseus tissues for quantitative PCR (qPCR) analysis. The first set was derived from leaves, from which we dissected the central vein as well as nearly veinless tissue, whereas the second set was derived from stems, from which we separated the epidermis from the rest of the stem tissue. qPCR analysis evidenced that the GES and G8O transcripts were enriched in the leaf central vein and peeled stem and absent from the stem epidermis (SI Appendix, Fig. S5), which is in agreement with their reported internal phloem-associated parenchyma (IPAP) localization (7, 30, 31). Conversely, SGD transcripts were enriched in the stem epidermis and veinless leaf and markedly less abundant in peeled stem and the leaf central vein (SI Appendix, Fig. S5), which is in agreement with its reported epidermal localization (32). BIS1 expression was enriched in the leaf central vein and peeled stem and largely absent from the stem epidermis (SI Appendix, Fig. S5), hence correlating with the expression of the IPAP-specific iridoid pathway genes. Furthermore, BIS1 expression was clearly distinct from that of the ORCA3 gene, which was also strongly expressed in the stem epidermis (SI Appendix, Fig. S5), thus further supporting its specificity.
Discussion
Identification of a bHLH-Type Transcriptional Regulator of MIA Production.
MIA production in the medicinal plant C. roseus requires tryptophan and IPP precursors that are shared with the primary metabolism as well as numerous other specialized metabolite pathways, such as of triterpenoids and sesquiterpenoids (5, 33). Therefore, biosynthesis of (seco)-iridoids and MIAs requires a tight transcriptional coordination of genes. Our understanding of this regulatory circuit remains limited to date, because, in general, only a few transcriptional regulators of the MEP (or MVA) as well as of the downstream terpenoid pathways have been identified (1, 34). Furthermore, MIA production in C. roseus involves a complex spatial organization, distributed in at least four cell types, and a marked and concerted elicitation of the pathway genes by JA treatment (2, 3). This complexity undoubtedly requires an array of TFs (1), many of which remain to be discovered.
The expression of several of these TFs may be JA inducible as part of the JA signaling cascade, as previously shown for ORCA3 and MYC2 (13, 15). This feature has turned out to be a powerful tool in the past to find pathway-regulating TFs in different plant species (1, 4, 35) and has been also successfully exploited here to identify the TF BIS1 as a regulator of the iridoid pathway. BIS1 belongs to clade IVa of the bHLH family (27), which is fundamentally different from the much larger MYC2-type TFs, because its members do not contain a known JAZ-interacting domain and thus seemingly lack a direct molecular connection to the currently known primary JA signaling module (1, 36). Therefore, determination of the exact hierarchical position of BIS1 in the JA signaling cascade will need further study.
BIS1 and ORCA3 Are Distinct Elements of the MIA Regulatory Circuit.
BIS1 specifically transactivates the promoters of iridoid genes up to 7DLH and induces their expression in transformed hairy root and cell suspension cultures. In addition, the MEP genes are also up-regulated in BIS1-overexpressing cultures, resulting in a coordinated activation of the iridoid and its precursor pathways. Such coordination of the expression of MEP precursor and terpenoid pathway genes has been reported previously and ensures the coupling of primary with secondary metabolism at the appropriate time and place (37) and is in agreement with the spatial coexpression of the MEP and iridoid genes in the IPAP cells (31).
To date, only one plant bHLH TF, phytochrome-interacting factor 5 (PIF5) from Arabidopsis, has been linked with the transcriptional activation of MEP pathway genes (38). Conversely, only one member of the bHLH clade IVa, NAI1 from Arabidopsis, has been characterized to some extent and has been reported to be involved in the formation of ER bodies, ER-derived organelles that have only been described in the plant order of Brassicales, where they are ubiquitous in roots and seedlings (39). However, NAI1 has not been linked with activation of MEP, terpenoid, or any (specialized) metabolite biosynthesis pathway yet. Because of the limited knowledge on TFs belonging to clade IVa and/or regulating MEP-dependent terpenoid biosynthesis (34), it is too early to speculate on the evolutionary specificity of BIS1.
The regulatory circuit of BIS1 differs markedly from that controlling the biosynthesis of the indole moiety and subsequent MIA pathway branches that are under control of ORCA3, which in turn is controlled by MYC2 (13, 14). Overall, the distinct BIS1 and ORCA3 circuits correlate well with the reported cell type-specific delineation of the pathway, with BIS1 targeting IPAP-specific genes. A 533-bp promoter sequence of the G8O gene had been isolated and shown to be sufficient for responsiveness to JA in C. roseus hairy roots (16). This promoter fragment contains several putative TF-binding sites that differ from those of the ORCA3-dependent gene promoters and was therefore postulated to be regulated by a different transcriptional cascade, as corroborated by our study. How and whether the different circuits interact remains to be resolved. BIS1, MYC2, ORCA3, and probably other, yet unknown, MIA-regulating TFs are coinduced by JA elicitation, possibly a prerequisite to guarantee optimal pathway functioning. Indeed, ORCA3-regulated genes are not up-regulated by BIS1, and some of them are even down-regulated. Conversely, in ORCA3-overexpressing cell cultures, BIS1 and several iridoid genes are down-regulated (Fig. 1C) (7). Likewise, the effect of ORCA3 overexpression on, for instance, SGD expression in cell suspension cultures (up) was found to be opposite to that in hairy root cultures (down) (23). The integration of the different circuits will need to be taken into account when engineering the production of MIA compounds in C. roseus cells.
BIS1 Boosts MIA Production Without Precursor Feeding.
Due to the low in planta abundance of the valuable MIAs and their precursors, several metabolic engineering and synthetic biology programs were launched with the aim to create sustainable MIA resources. In this regard, the recent reconstitutions of the pathway up to strictosidine in both tobacco (7) and yeast (40) represent important milestones. However, the use of transcriptional regulators to activate multiple steps is considered most promising to engineer MIA synthesis in the endogenous plant source. This belief has been fostered by the discovery of ORCA3 (14), the overexpression of which, however, was not sufficient to boost MIA production in C. roseus cultures.
In contrast to ORCA3, BIS1 overexpression boosted MIA production in C. roseus suspension cell cultures in the absence of exogenous precursors or JA elicitation. Although BIS1 “only” activated the pathway branch leading to loganic acid, its overexpression resulted in a dramatic increase, not only of loganic acid but also of downstream (seco)-iridoid and MIA products, such as secologanin, strictosidine, ajmalicine, serpentine, and tabersonine.
The biosynthetic route from strictosidine to tabersonine is largely unknown, but the seven-step biosynthesis pathway from tabersonine to vindoline has recently been fully elucidated and reassembled in yeast (41). Until now, production of vindoline in C. roseus suspension cultures seemed problematic, based on its complex cell-specific regulation. However, considering that the tabersonine-to-vindoline pathway can be assembled in a single yeast cell (41) and that the BIS1-overexpressing cells constitutively accumulate tabersonine, the latter cells may serve as a promising C. roseus-based platform for further metabolic engineering toward sustainable production of vindoline and, ultimately, vinblastine and vincristine. Successful engineering may depend not only on the full elucidation and reconstitution of the pathway at the enzyme level but also on the identification of additional regulatory TFs of the MIA or its precursor pathways. Availability of these TFs may enable the overexpression of a TF network that resembles the one activated by the plant under stress conditions and that may lead to high levels of the desired bioactive MIAs.
Materials and Methods
Plant Material.
C. roseus suspension cell line MP183L and cv. Würzburg hairy roots were used for overexpression and silencing experiments. C. roseus cv. Little Bright Eyes was used for the VIGS experiments and seedlings, treated with 1 mM of methyl JA (MeJA) for 24 h, for genomic DNA and RNA isolation.
Generation of DNA Constructs.
Constructs were made with the Gateway Technology (Invitrogen) or restriction enzyme digestion (SI Appendix). Primers used for cloning are listed in SI Appendix, Table S4.
Transient Transactivation Assays in Nicotiana spp.
Transactivation of C. roseus gene promoters by C. roseus TFs was assessed in transient expression assays in N. tabacum protoplasts, N. benthamiana leaves, and C. roseus cells (SI Appendix).
Generation and Profiling of Transformed C. roseus Cultures and Plants.
C. roseus suspension cells, hairy roots, and seedlings were stably and transiently transformed, and the subsequent metabolite and transcript profiling of the transformed tissues was carried out as described (SI Appendix).
Supplementary Material
Acknowledgments
We thank Siv Matomaa for technical assistance, Frederik Coppens for support with RNA-Seq, and Martine De Cock for help in preparing the manuscript. This work was supported by funding from the European Union Seventh Framework Program FP7/2007-2013 under Grant 222716-SMARTCELL, the Short-Term Scientific Missions (STSM) program from the European Union COST Action FA1006-PlantEngine, and the Research Foundation Flanders (Project G005212N). A.V.M. is indebted to the European Molecular Biology Organization for a long-term fellowship (EMBOCOFUND2010), the European Commission support from Marie Curie Actions (GA-2010-267154), and the STSM program from the European Union COST Action FA1006-PlantEngine; P.S. to the French Space Agency [Centre National d'Etudes Spatiales/Centre Spatial Guyanais (CNES/CSG)] for a scholarship; F.S. to the Swiss National Foundation for an Early Postdoc Mobility grant; R.P. to the John Innes Centre for a studentship; J.E. to the Advanced Human Capital Program (Becas Chile N°73140628-CONICYT) for a scholarship; and P.C.P. to the Directorate General of Higher Education of the Republic Indonesia for a scholarship. J.P. is a postdoctoral fellow of the Research Foundation-Flanders.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Short Read Archive (BioProject accession no. PRJNA276552) and the GenBank database [accession nos. KM409646 (BIS1), KP963953 (pGES), KP963954 (p8HGO), KP963955 (pIO), KP963956 (pIS), KP963957 (p7DLH), KP963958 (p7DLGT), and KP963959 (pLAMT)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504951112/-/DCSupplemental.
References
- 1.De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012;17(6):349–359. doi: 10.1016/j.tplants.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Courdavault V, et al. A look inside an alkaloid multisite plant: The Catharanthus logistics. Curr Opin Plant Biol. 2014;19(6):43–50. doi: 10.1016/j.pbi.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 3.De Luca V, Salim V, Thamm A, Masada SA, Yu F. Making iridoids/secoiridoids and monoterpenoid indole alkaloids: Progress on pathway elucidation. Curr Opin Plant Biol. 2014;19(6):35–42. doi: 10.1016/j.pbi.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Dugé de Bernonville T, et al. Phytochemical genomics of the Madagascar periwinkle: Unravelling the last twists of the alkaloid engine. Phytochemistry. 2015;113:9–23. doi: 10.1016/j.phytochem.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Murata J, Roepke J, Gordon H, De Luca V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell. 2008;20(3):524–542. doi: 10.1105/tpc.107.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L, et al. Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus. Planta. 2012;236(5):1571–1581. doi: 10.1007/s00425-012-1712-0. [DOI] [PubMed] [Google Scholar]
- 7.Miettinen K, et al. The seco-iridoid pathway from Catharanthus roseus. Nat Commun. 2014;5:3606. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Moerkercke A, et al. CathaCyc, a metabolic pathway database built from Catharanthus roseus RNA-Seq data. Plant Cell Physiol. 2013;54(5):673–685. doi: 10.1093/pcp/pct039. [DOI] [PubMed] [Google Scholar]
- 9.Kazan K, Manners JM. MYC2: The master in action. Mol Plant. 2013;6(3):686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 10.Hong G-J, Xue X-Y, Mao Y-B, Wang L-J, Chen X-Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24(6):2635–2648. doi: 10.1105/tpc.112.098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji Y, et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014;55(9):1592–1604. doi: 10.1093/pcp/pcu090. [DOI] [PubMed] [Google Scholar]
- 12.Spyropoulou EA, Haring MA, Schuurink RC. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics. 2014;15:402. doi: 10.1186/1471-2164-15-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, et al. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011;67(1):61–71. doi: 10.1111/j.1365-313X.2011.04575.x. [DOI] [PubMed] [Google Scholar]
- 14.van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289(5477):295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- 15.van der Fits L, Memelink J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 2001;25(1):43–53. doi: 10.1046/j.1365-313x.2001.00932.x. [DOI] [PubMed] [Google Scholar]
- 16.Suttipanta N, et al. Promoter analysis of the Catharanthus roseus geraniol 10-hydroxylase gene involved in terpenoid indole alkaloid biosynthesis. Biochim Biophys Acta. 2007;1769(2):139–148. doi: 10.1016/j.bbaexp.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Suttipanta N, et al. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011;157(4):2081–2093. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sibéril Y, et al. Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol Biol. 2001;45(4):477–488. doi: 10.1023/a:1010650906695. [DOI] [PubMed] [Google Scholar]
- 19.Pauw B, et al. Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J Biol Chem. 2004;279(51):52940–52948. doi: 10.1074/jbc.M404391200. [DOI] [PubMed] [Google Scholar]
- 20.Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J. Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 2007;144(3):1680–1689. doi: 10.1104/pp.107.096115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweizer F, et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013;25(8):3117–3132. doi: 10.1105/tpc.113.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peebles CAM, Hughes EH, Shanks JV, San K-Y. Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab Eng. 2009;11(2):76–86. doi: 10.1016/j.ymben.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Wang C-T, Liu H, Gao X-S, Zhang H-X. Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep. 2010;29(8):887–894. doi: 10.1007/s00299-010-0874-0. [DOI] [PubMed] [Google Scholar]
- 25.Pan Q, et al. Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics. PLoS ONE. 2012;7(8):e43038. doi: 10.1371/journal.pone.0043038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Góngora-Castillo E, et al. Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS ONE. 2012;7(12):e52506. doi: 10.1371/journal.pone.0052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 28.Kellner F, et al. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015;82(4):680–692. doi: 10.1111/tpj.12827. [DOI] [PubMed] [Google Scholar]
- 29.Pollier J, et al. The protein quality control system manages plant defence compound synthesis. Nature. 2013;504(7478):148–152. doi: 10.1038/nature12685. [DOI] [PubMed] [Google Scholar]
- 30.Simkin AJ, et al. Characterization of the plastidial geraniol synthase from Madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry. 2013;85:36–43. doi: 10.1016/j.phytochem.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Burlat V, Oudin A, Courtois M, Rideau M, St-Pierre B. Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 2004;38(1):131–141. doi: 10.1111/j.1365-313X.2004.02030.x. [DOI] [PubMed] [Google Scholar]
- 32.Guirimand G, et al. Strictosidine activation in Apocynaceae: Towards a “nuclear time bomb”? BMC Plant Biol. 2010;10:182. doi: 10.1186/1471-2229-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Pinho PG, et al. Volatile composition of Catharanthus roseus (L.) G. Don using solid-phase microextraction and gas chromatography/mass spectrometry. J Pharm Biomed Anal. 2009;49(3):674–685. doi: 10.1016/j.jpba.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 34.Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 35.Goossens A. It is easy to get huge candidate gene lists for plant metabolism now, but how to get beyond? Mol Plant. 2015;8(1):2–5. doi: 10.1016/j.molp.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Pauwels L, Goossens A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–3100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudareva N, et al. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci USA. 2005;102(3):933–938. doi: 10.1073/pnas.0407360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mannen K, et al. Coordinated transcriptional regulation of isopentenyl diphosphate biosynthetic pathway enzymes in plastids by phytochrome-interacting factor 5. Biochem Biophys Res Commun. 2014;443(2):768–774. doi: 10.1016/j.bbrc.2013.12.040. [DOI] [PubMed] [Google Scholar]
- 39.Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I. NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell. 2004;16(6):1536–1549. doi: 10.1105/tpc.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown S, Clastre M, Courdavault V, O’Connor SE. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc Natl Acad Sci USA. 2015;112(11):3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu Y, et al. Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc Natl Acad Sci USA. 2015;112(19):6224–6229. doi: 10.1073/pnas.1501821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.