Significance
The development of Clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated genes (CAS)–based technology for targeted genome editing has revolutionized molecular biology approaches, but significant and outstanding gaps exist for applications in bacteria, the native hosts of these adaptive immune systems. This study shows that CRISPR-Cas systems can be directed to target and delete genomic islands that are flanked by insertion-sequence elements and devoid of essential genes. Naturally occurring minor subpopulations harboring deletions in genomic islands were identified and readily isolated using CRISPR-Cas screening. Promising applications of this approach can define minimal bacterial genomes, determine essential genes, and characterize genetically heterogeneous bacterial populations.
Keywords: CRISPR, lactic acid bacteria, transposons, IS-elements, Cas9
Abstract
Genomic analysis of Streptococcus thermophilus revealed that mobile genetic elements (MGEs) likely contributed to gene acquisition and loss during evolutionary adaptation to milk. Clustered regularly interspaced short palindromic repeats–CRISPR-associated genes (CRISPR-Cas), the adaptive immune system in bacteria, limits genetic diversity by targeting MGEs including bacteriophages, transposons, and plasmids. CRISPR-Cas systems are widespread in streptococci, suggesting that the interplay between CRISPR-Cas systems and MGEs is one of the driving forces governing genome homeostasis in this genus. To investigate the genetic outcomes resulting from CRISPR-Cas targeting of integrated MGEs, in silico prediction revealed four genomic islands without essential genes in lengths from 8 to 102 kbp, totaling 7% of the genome. In this study, the endogenous CRISPR3 type II system was programmed to target the four islands independently through plasmid-based expression of engineered CRISPR arrays. Targeting lacZ within the largest 102-kbp genomic island was lethal to wild-type cells and resulted in a reduction of up to 2.5-log in the surviving population. Genotyping of Lac− survivors revealed variable deletion events between the flanking insertion-sequence elements, all resulting in elimination of the Lac-encoding island. Chimeric insertion sequence footprints were observed at the deletion junctions after targeting all of the four genomic islands, suggesting a common mechanism of deletion via recombination between flanking insertion sequences. These results established that self-targeting CRISPR-Cas systems may direct significant evolution of bacterial genomes on a population level, influencing genome homeostasis and remodeling.
Mobile genetic elements (MGEs) present bacteria with continuous challenges to genomic stability, promoting evolution through horizontal gene transfer. The term “MGE” encompasses plasmids, bacteriophages, transposable elements, genomic islands, and many other specialized genetic elements (1). MGEs encompass genes conferring high rates of dissemination, adaptive advantages to the host, and genomic stability, leading to their nearly universal presence in bacterial genomes. To cope with the permanent threat of predatory bacteriophages and selfish genetic elements, bacteria have evolved both innate and adaptive immune systems targeting exogenous genetic elements. Innate immunity includes cell-wall modification, restriction/modification systems, and abortive phage infection (2). Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated genes (Cas) are an adaptive immune system targeted against invasive genetic elements in bacteria (3). CRISPR-Cas–mediated immunity relies on distinct molecular processes, categorized as acquisition, expression, and interference (3). Acquisition occurs via molecular sampling of foreign genetic elements, from which short sequences, termed “spacers,” are integrated in a polarized fashion into the CRISPR array (4). Expression of CRISPR arrays is constitutive and inducible by promoter elements within the preceding leader sequence (5, 6). Interference results from a corresponding transcript that is processed selectively at each repeat sequence, forming CRISPR RNAs (crRNAs) that guide Cas proteins for sequence-specific recognition and cleavage of target DNA complementary to the spacer (7). CRISPR-Cas technology has applications in strain typing and detection (8–10), exploitation of natural/engineered immunity against mobile genetic elements (11), programmable genome editing in diverse backgrounds (12), transcriptional control (13, 14), and manipulation of microbial populations in defined consortia (15).
Although sequence features corresponding to CRISPR arrays were described previously in multiple organisms (16, 17), Streptococcus thermophilus was the first microbe in which the roles of specific cas genes and CRISPR-array components were elucidated (4). S. thermophilus is a nonpathogenic, thermophilic Gram-positive bacterium used as a starter culture that catabolizes lactose to lactic acid in the syntrophic production of yogurt and various cheeses (18). S. thermophilus encodes up to four CRISPR-Cas systems, two of which (CRISPR1 and CRISPR3) are classified as type II-A systems that are innately active in both acquisition and interference (4, 19). Accordingly, genomic analysis of S. thermophilus and its bacteriophages established a likely mechanism for phage/DNA protection in CRISPR-Cas systems. Investigation of CRISPR-Cas systems in S. thermophilus led to bioinformatic analysis of spacer origin (4, 20), discovery of the proto-spacer adjacent motif (PAM) sequences (19, 21), understanding of phage–host dynamics (22, 23), demonstration of Cas9 endonuclease activity (7, 24, 25), and, recently, determination of the transactivating crRNA tracrRNA structural motifs governing function and orthogonality of type II systems (26). Genomic analysis of S. thermophilus revealed evolutionary adaptation to milk through the loss of carbohydrate catabolism and virulence genes found in pathogenic streptococci (18). S. thermophilus also underwent significant acquisition of niche-related genes, such as those encoding including cold-shock proteins, copper resistance proteins, proteinases, bacteriocins, and lactose catabolism proteins (18). Insertion sequences (ISs) are highly prevalent in S. thermophilus genomes and contribute to genetic heterogeneity among strains by facilitating dissemination of islands associated with dairy adaptation genes (18). The concomitant presence of MGEs and functional CRISPR-Cas systems in S. thermophilus suggests that genome homeostasis is governed at least in part by the interplay of these dynamic forces. Thus, S. thermophilus constitutes an ideal host for investigating the genetic outcomes of CRISPR-Cas targeting of genomic islands.
CRISPR-Cas systems recently have been the subject of intense research in genome editing applications (12), but the evolutionary roles of most endogenous microbial systems remain unknown (27). Even less is known concerning evolutionary outcomes of housing active CRISPR-Cas systems beyond the prevention of foreign DNA uptake (7), spacer acquisition events (4), and mutation caused by chromosomal self-targeting (28–32). Thus, we sought to determine the outcomes of targeting integrated MGEs with endogenous type II CRISPR-Cas systems. Four islands were identified in S. thermophilus LMD-9, with lengths ranging from 8 to 102 kbp and totaling ∼132 kbp, or 7% of the genome. To target genomic islands, plasmid-based expressions of engineered CRISPR arrays with self-targeting spacers were transformed into S. thermophilus LMD-9. Collectively, our results elucidate fundamental genetic outcomes of self-targeting events and show that CRISPR-Cas systems can direct genome evolution at the bacterial population level.
Results
Identification of Expendable Genomic Regions.
In silico prediction of mobile and expendable loci for CRISPR-Cas targeting was performed on the basis of (i) the location, orientation, and nucleotide identity of IS elements, and (ii) the location of essential ORFs. In Bacillus subtilis, 271 essential ORFs were identified by determining the lethality of genome-wide gene knockouts (33). The S. thermophilus genome was queried for homologs to each essential gene from B. subtilis using the BLASTp search tool under the default scoring matrix for amino acid sequences. Homologs to ∼239 essential ORFs were identified in S. thermophilus, all of which were chromosomally encoded (Table S1). Proteins involved in conserved cellular processes including DNA replication/homeostasis, translation machinery, and core metabolic pathways were readily identified. No homologs corresponding to cytochrome biosynthesis/respiration were observed, in accordance with the metabolic profile of fermentative bacteria. Each putative essential ORF was mapped to the reference genome using SnapGene software, facilitating visualization of their location and distribution in S. thermophilus LMD-9 (Fig. 1).
Fig. 1.

Map of essential genes, insertion sequences, and genomic islands showing the location and distribution of putative essential ORFs (red), insertion sequences (gray), and putative genomic islands (blue). Type II CRISPR-Cas loci in S. thermophilus LMD-9 are shown in black.
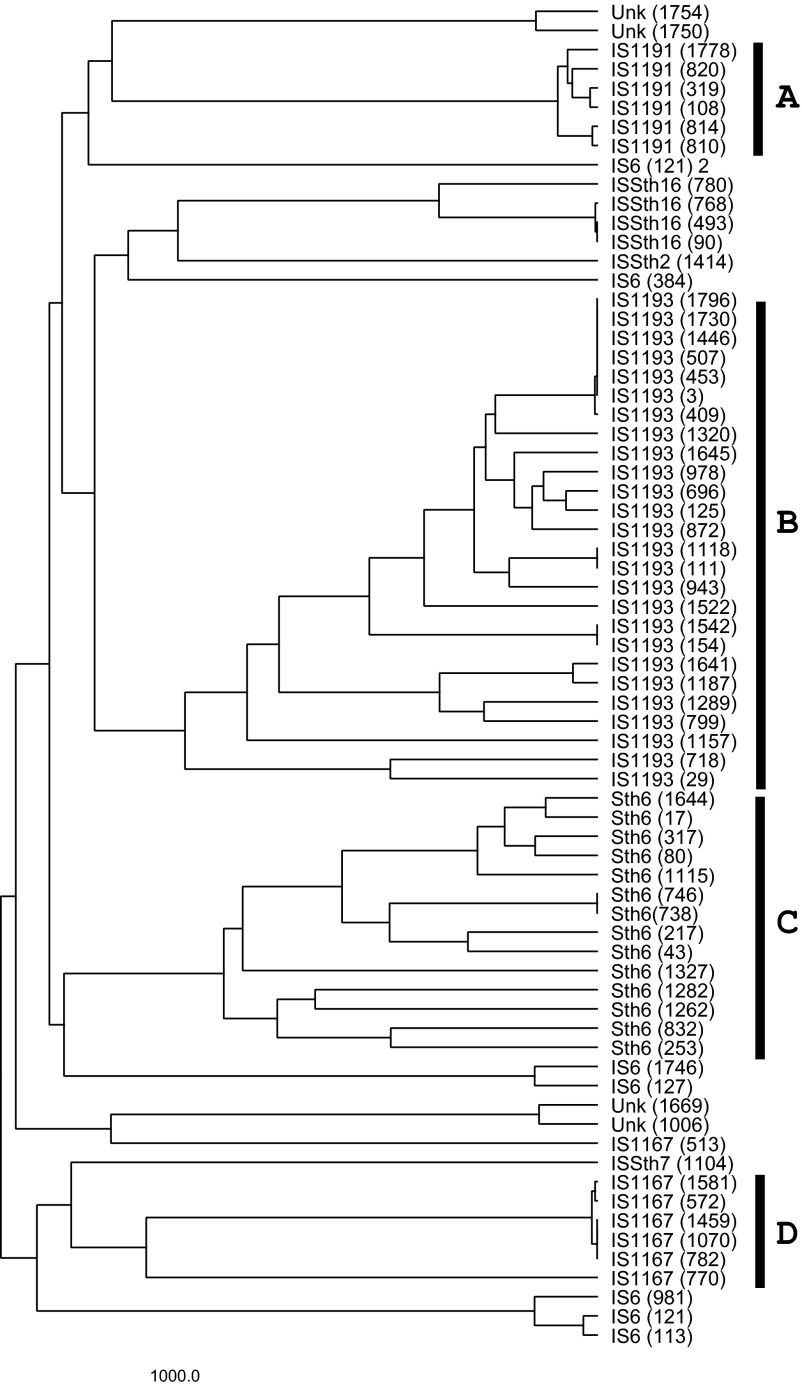
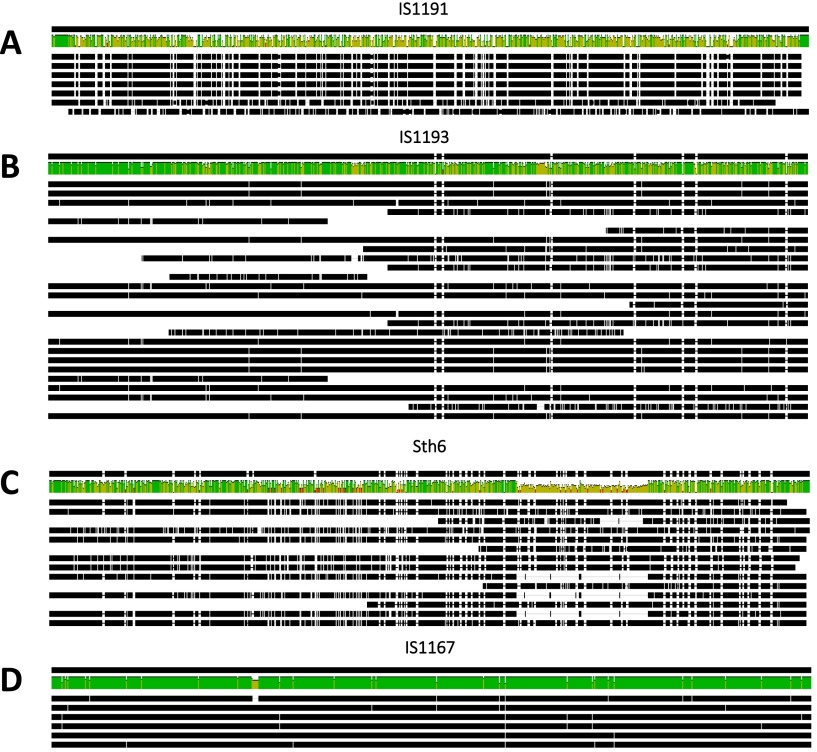
IS elements within the S. thermophilus genome were grouped by aligning transposon coding sequences using Geneious software (Fig. S1). Family designations were determined according to BLAST analysis within the IS element database (https://www-is.biotoul.fr//). To predict the potential for recombination-mediated excision of chromosomal segments, the relative locations of related IS elements were mapped to the S. thermophilus genome (Fig. 1). The IS1193 and Sth6 families of IS elements appeared most frequently in the genome and are commonly found in Streptococcus pneumoniae and Streptococcus mutans (34). IS1191 elements were not frequent but exhibited nearly perfect identity between the copies identified in the genome (Figs. S1A and S2A). Despite the prevalence of IS1193 elements, many of these loci were shown to be small fragments that exhibited some polymorphism and degeneracy, but several copies with a high level of sequence identity were present also (Figs. S1B and S2B). The Sth6 family exhibited considerable polymorphism and high degeneracy, with some copies harboring significant internal deletions (Figs. S1C and S2C). IS1167 elements were well conserved (Figs. 1D and S2D). Based on the conservation of length and sequence of the IS1167 and IS1191 elements of S. thermophilus and their relative proximity to milk adaptation genes, we postulate that these conserved/high-fidelity transposons were acquired in the genome recently.
Fig. S1.
Dendrogram of transposon coding sequences distributed throughout the genome of S. thermophilus LMD-9. Alignment was created using the Geneious Software. Family designations were assigned using https://www-is.biotoul.fr A, IS1191 elements; B, IS1193 elements; C, Sth6 transposons; D, IS1167 elements.
Fig. S2.
Geneious alignments of transposon coding sequences for each major IS family found in the S. thermophilus LMD-9 genome. Different families exhibited varying levels of conservation of length and nucleotide identity. (A) IS1191 elements exhibited high identity. (B) IS1193 had high-fidelity copies but exhibited the greatest intrafamily diversity in length. (C) Sth6 transposons were highly polymorphic and apparently degenerate because of internal deletions in some of the copies. (D) IS1167 elements had fewer copies but maintained high fidelity in length and identity.
By combining the location of predicted essential ORFs and highly similar IS elements, expendable islands were identified (Fig. 1 and Table 1). The first island contained an operon unique to S. thermophilus LMD-9, encoding a putative ATP-dependent oligonucleotide transport system with unknown specificity (Fig. S3A) (35). The second harbors the cell-envelope proteinase PrtS, which contributes to the fast-acidification phenotype of S. thermophilus (Fig. S3B) (36). Notably, although prtS is not ubiquitous in S. thermophilus genomes, it has been demonstrated that the genomic island encoding prtS is transferable between strains using natural competence (36). The third island contains a putative ATP-dependent copper efflux protein and is present in every sequenced S. thermophilus strain (Fig. S3C). The fourth island is the largest by far in terms of length, at 102 kbp, and gene content, with 102 predicted ORFs including the lac operon (Fig. S3D). This island is found in all strains of S. thermophilus, but the specific gene content and length vary among strains. To determine the outcome of targeting a large genomic island with both endogenous type II systems, repeat-spacer arrays were generated for the lacZ coding sequence (Fig. S3D) and were cloned into pORI28 (Table S2). The fourth island was selected for CRISPR-Cas targeting because of its size, ubiquity in S. thermophilus strains, and the ability to screen for lacZ mutations on the basis of a β-galactosidase–negative phenotype.
Table 1.
Genomic island characteristics and CRISPR targets
| GEI | ORF region | Length, bp | IS family | Target gene | CRISPR system (PAM) | Spacer | PAM |
| 1 | STER_139-STER_148 | 8,490 | IS6 | Oligopeptide transporters | 3 (NGGNG) | GGTGGGCTGGATGTTTTATCTCGTGTTATC | TGGGG |
| 2 | STER_840-STER_848 | 11,932 | ISSth1/ IS1167 | Proteinase prtS | 3 (NGGNG) | GCGTGTATTCTCAGACCTCAAAGCTACAAC | AGGCG |
| 3 | STER_881-STER_888 | 9,891 | IS1191 | Copper efflux | 3 (NGGNG) | TTAGCAGCTGAAACGATTGATTGAGCAATC | GGGTG |
| 4 | STER_1277-STER_1380 | 102,087 | IS1193 | lac operon; 102 ORFs | 1 (NNAGAAW) | ATTAAGAGATTGTCTTAACTTCATCTCCCCT | TCAGAAA |
| 3 (NGGNG) | TACAGCAAGCTGGTTGGAAGACCAAGACTTC | TGGAG | |||||
| 3 (NGGNG) | AGTATTTTGAATCTCTTGAAGAATTTTCTGA | AGGGG |
GEI, genomic island.
Fig. S3.
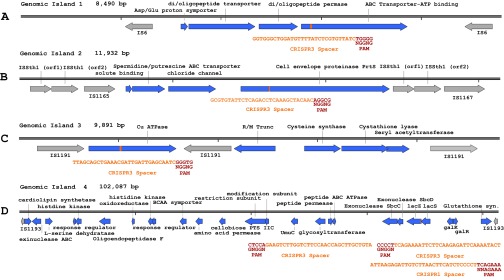
(A) Genomic island 1: 8,490 bp, flanked by IS6 elements, encodes the target ATP-binding ABC oligopeptide transport gene. (B) Genomic island 2: 11,932 bp, flanked by ISSth1 elements, contains the target gene cell-envelope proteinase prtS. (C) Genomic island 3: 9,891 bp, flanked by IS1191 elements, encodes the target ATPase copper-efflux gene. (D) Genomic island 4: 102,087 bp, flanked by IS1193 elements, encodes lacZ, the target for three independent spacers.
CRISPR-Cas Targeting of lacZ Selects for Large Deletion Events.
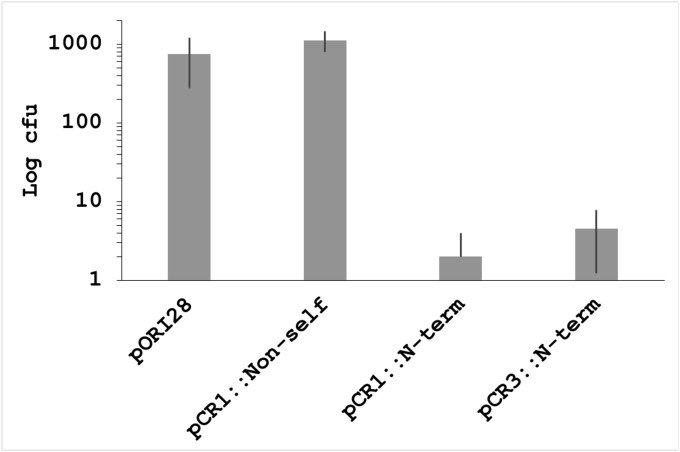
In type II systems, Cas9 interrogates DNA and binds reversibly to PAM sequences with activation of Cas9 at the target occurring via formation of the tracrRNA::crRNA duplex (37), ultimately resulting in dsDNA cleavage (Fig. S4 A and B) (25). Transformation with plasmids eliciting chromosomal self-targeting by CRISPR-Cas systems appeared cytotoxic as measured by the relative reduction in surviving transformants compared with non–self-targeting plasmids (15, 29). Targeting the lacZ gene in S. thermophilus resulted in an ∼2.5-log reduction in recovered transformants (Fig. 2), approaching the limits of transformation efficiency. Double-stranded DNA breaks (DSBs) constitute a significant threat to the survival of organisms. The corresponding repair pathways often require end resection to repair blunt-ended DNA. Cas9-effected endonucleolysis further exacerbates the pressure for mutations caused by DSBs to occur, because restoration of the target locus to the wild type does not circumvent subsequent CRISPR targeting. Identification of spacer origins within lactic acid bacteria revealed that 22% of spacers exhibit complementarity to self and that the corresponding genomic loci were altered, likely facilitating the survival of naturally occurring self-targeting events (28).
Fig. S4.
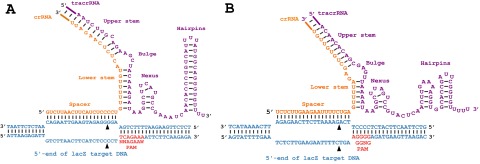
Structural basis for and apparent cytotoxicity of DNA targeting by CRISPR-Cas9. Spacer sequences for CRISPR1 (A) and CRISPR3 (B) for targeting lacZ. Cas9 interrogates DNA and binds reversibly to PAM sequences with stabilization of Cas9 at the target occurring via formation of the tracrRNA::crRNA duplex. Activation of the Cas9 causes simultaneous cleavage of each strand by the RuvC and HNH domains, as denoted by black triangles.
Fig. 2.
Transformants recovered after electroporation of control and self-targeting plasmids. Bars show average clones ± SD screened across independent transformation experiments (n = 4) for each of the plasmids tested. N-term, N-terminal.
To determine if the target locus was mutated in response to Cas9-induced cleavage, transformants first were screened for loss of β-galactosidase activity. Clones deficient in activity were genotyped at the lacZ locus. No mutations caused by classical or alternative end joining and no spontaneous SNPs were observed in any of the clones sequenced. The absence of SNPs may be attributed to a low transformation efficiency compounded by a low incidence of point mutations, and the absence of Ku and ligase IV homologs correlated with an absence of nonhomologous end joining (38). PCR screening indicated that wild-type lacZ was not present, but the PCR amplicons did not correspond to the native lacZ locus; rather, an IS element-flanked sequence at another genomic locus was amplified. To investigate the genotype responsible for the loss of β-galactosidase activity, single-molecule real-time sequencing was performed on two clones, one generated from CRISPR3 targeting the 5′ end of lacZ and one generated from CRISPR3 targeting the sequence encoding the ion-binding pocket necessary for β-galactosidase catalysis (Fig. 3 A and B). This sequencing strategy was used for its long read length to circumvent difficulty in reliably mapping reads to the proper locus because of the high number of IS elements in the genome (35). Reads were mapped to the reference genome sequence using Geneious software and revealed the absence of a large segment (∼102 kbp) encoding the lacZ ORF (Fig. 3 A and B). Both sequenced strains confirmed the reproducibility of the large deletion boundaries and showed that the deletion occurred independently of the lacZ spacer sequence used for targeting. However, the sequencing data did not reliably display the precise junctions of the deletion.
Fig. 3.
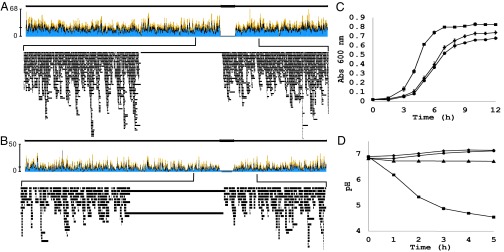
Genome sequencing and phenotypic analysis of Lac− clones. (A and B) Sequence data revealed an absence of the chromosomal segment encoding lacZ in two mutants independently created by targeting the 5′ end (A) and cation-binding residue (B) coding sequences of lacZ using the CRISPR3 system. The size of the deletions ranged from 101,865 to 102,146 bp in length, constituting ∼5.5% of the genome of S. thermophilus. (C) Growth of large-deletion strains generated by CR3 spacer 1 (circles) and CR3 spacer 2 (diamonds) compared with wild type (squares) in semisynthetic Elliker medium represented as mean ± SD. Shown is OD at 600 nm of three independent biological replicates. (D) Acidification capacity of wild-type S. thermophilus (squares) and large-deletion strains (circles/diamonds) in skim milk relative to an uninoculated control (triangles).
The 102-kbp segments deleted constitute ∼5.5% of the 1.86-Mbp genome of S. thermophilus. The region contained 102 putative ORFs (STER_1278–1379), encoding ATP-binding cassette (ABC) transporters, two-component regulatory systems, bacteriocin synthesis genes, phage-related genes, lactose catabolism genes, and several cryptic genes with no annotated function (35). The effect of the deletion on growth phenotype was assessed in broth culture by measuring OD at 600 nm over time (Fig. 3C). The deletion clones appeared to have a longer lag phase and lower final OD (P < 0.01) and exhibited a significantly longer generation time during log phase (average of 103 min compared with 62 min for the wild type; P < 0.001). Although the deletion derivatives have 5.5% less of the genome to replicate per generation and expend no resources in transcription or translation of the eliminated ORFs, no apparent increase in fitness was observed relative to the wild type. β-Galactosidase activity is a hallmark feature for industrial applications of lactic acid bacteria and is essential for preservation of food systems through acidification. The capacity of lacZ-deficient S. thermophilus strains to acidify milk therefore was assessed by monitoring pH (Fig. 4D). Predictably, the deletion strain failed to acidify milk over the course of the experiment, in sharp contrast to the rapid acidification phenotype observed in the wild type.
Fig. 4.
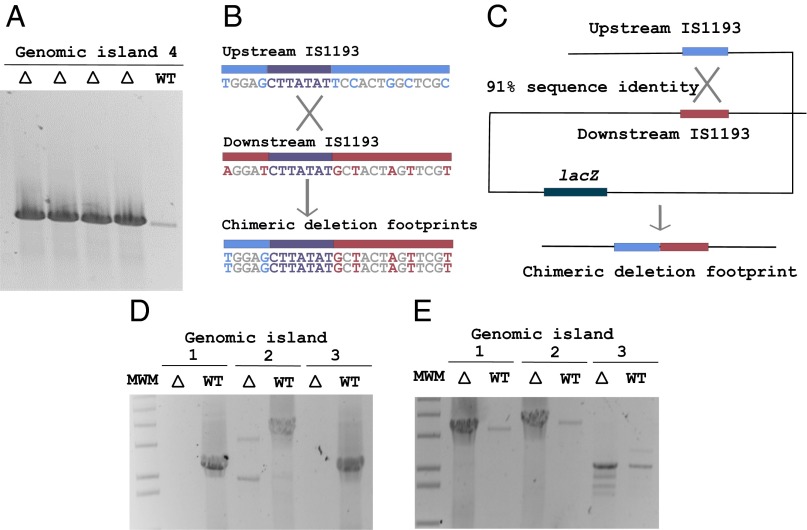
Depiction of recombination events between ISs. (A) Large-deletion amplicons yielded by PCR analysis of gDNA recovered from transformants. Lanes denoted Δ were amplified from gDNA of Lac− clones. (B) Sequences of predicted recombination sites were determined by mapping SNPs corresponding to upstream (blue) or downstream (red) IS elements. (C) Schematic of ISs predicted to recombine during chromosomal deletion of the island encoding lacZ. (D) Amplicons generated from primers flanking genomic islands 1, 2, and 3 to confirm deletions. (E) Amplicons generated from internal primers to confirm the absence of wild-type sequences in each CRISPR-induced deletion culture. Lanes denoted Δ were amplified from genomic DNA of clones recovered after CRISPR-Cas–mediated targeting.
Genomic Deletions Occur Through Recombination Between Homologous IS Elements.
To investigate the mechanism of deletion, the nucleotide sequences flanking the segment were determined. The only homologous sequences observed at the junctions were two truncated IS1193 insertion sequences exhibiting 91% nucleotide sequence identity globally over 727 bp. Accordingly, a primer pair flanking the two IS elements was designed to amplify genomic DNA of surviving clones exhibiting the deletion. Each of the deletion strains exhibited a strong band of the predicted size (∼1.2 kb) and confirmed the large genomic deletion event (Fig. 4A). Interestingly, a faint amplicon corresponding to the chromosomal deletion was observed in the wild type, indicating that this region may excise naturally from the genome at a low rate within wild-type populations. Sequencing of the junction amplicon was performed for 20 clones generated by chromosomal self-targeting by CRISPR3. Genotyping of the locus revealed the presence of one chimeric IS element in each clone and furthermore revealed the transition from the upstream element to the downstream sequence within the chimera for each clone (Fig. 4B). The size of deletions observed ranged from 101,865 to 102,146 bp. The exact locus of transition was variable but was nonrandom within the clones, implying the potential bias of the deletion mechanism. S. thermophilus harbors typical recombination machinery encoded as RecA (STER_0077), AddAB homologs functioning as dual ATP-dependent DNA exonucleases (STER_1681 and STER_1682), and a helicase (STER_1742) of the RecD family. The high nucleotide identity between the flanking IS elements and the capacity for S. thermophilus to carry out site-specific recombination (4) confirms the potential for RecA-mediated recombination to mediate excision of the genomic segment (Fig. 4C).
It next was hypothesized that CRISPR-Cas targeting could facilitate isolation of deletions for each locus with the same genetic architecture. Thus, three CRISPR3 repeat-spacer arrays, one targeting the oligonucleotide transporter in the first locus, one targeting prtS from the second locus, and one targeting the ATPase copper efflux gene from the third locus, were generated and cloned into pORI28 (Table S2). To screen for deletions, primers flanking the IS elements at each locus were designed to amplify each deletion junction (Fig. 4D). The absence of wild-type loci also was confirmed in each case by designing internal primers for each genomic island (Fig. 4E). After transformations with the targeting plasmids, deletions at each locus were isolated, and the absence of wild type was confirmed. Sequencing of the deletion junction amplicons confirmed that a single chimeric IS element footprint remained, indicating a common mechanism for deletion at each locus. Interestingly, primers flanking the IS elements also amplified from wild-type gDNA, further suggesting that population heterogeneity that naturally occurred at each locus resulted from spontaneous genomic deletions. These results imply that sequence-specific Cas9 cleavage selects for the variants lacking protospacer and PAM combinations necessary for targeting. Thus, spontaneous genomic deletions can be isolated using CRISPR-Cas targeting as a strong selection for microbial variants that already have lost those genomic islands.
Discussion
In this study, native type IIA systems harbored in S. thermophilus were repurposed for defining spontaneous deletions of large genomic islands. By independently targeting four islands in S. thermophilus, stable mutants collectively lacking a total of 7% of the genome were generated. Characterization of the deletion junctions suggested that an IS-dependent recombination mechanism contributes to population heterogeneity and revealed deletion events ranging from 8 to 102 kbp. Precise mapping of the chimeric IS elements indicated that natural recombination events are likely to be responsible for the large chromosomal deletions in S. thermophilus and potentially could be exploited for targeted genome editing. Recent landmark studies have highlighted the potential for CRISPR-Cas–induced chromosomal deletions and rearrangements in bacteria (29, 30). Jiang et al. (29) first reported that sequence-specific Cas9 cleavage selects for preexisting variants lacking protospacer and PAM combinations necessary for targeting in S. pneumoniae. Similarly, Vercoe et al. (30) demonstrated that chromosomal targeting by a type-IF CRISPR-Cas system caused elimination of a horizontally acquired pathogenicity island in Pectobacterium atrosepticum (30). The concept of sequence-based removal of specific genotypes was developed further as a tool for manipulation of microbial consortia via CRISPR-Cas targeting, resulting in directed genome evolution at the population level (15). In accordance with previous work, our results demonstrate that wild-type clones were removed from the population, but mutants without CRISPR-Cas–targeted features survived. Thus, adaptive islands were identified and validated, showing that precise targeting by an endogenous Cas9 can be exploited for isolating large deletion variants in mixed populations.
Genome evolution of bacteria occurs through horizontal gene transfer, intrinsic mutation, and genome restructuring. Genome sequencing and comparative analysis of S. thermophilus strains have revealed significant genome decay but also indicate that adaptation to nutrient-rich food environments occurred through niche-specific gene acquisition (18, 35). The presence of MGEs including integrative and conjugative elements, prophages, and IS elements in S. thermophilus genomes is indicative of rapid evolution to a dairy environment (39, 40). Mobile genetic features facilitate gene acquisition and, conversely, inactivation or loss of nonessential sequences. Consequently, MGEs confer genomic plasticity as a means of increasing fitness or changing ecological lifestyles. Our results strongly indicate that CRISPR-Cas targeting of these elements may influence chromosomal rearrangements and homeostasis. This finding is in contrast to experiments targeting essential features, which resulted in the selection of variants with inactivated CRISPR-Cas machinery (41). Mutation of essential ORFs is not a viable avenue for circumvention of CRISPR-Cas targeting, and thus only those clones with inactivated CRISPR-Cas systems remain. By design, targeting genetic elements predicted to be hypervariable and expendable demonstrated that variants with altered loci were viable, maintaining active CRISPR-Cas systems during self-targeting events.
Despite the nearly ubiquitous distribution of IS elements in bacterial genomes, they remain an enigmatic genetic entity, largely because of their diversity and plasticity in function (34). Our results suggest it is possible to predict recombination between related IS elements by analyzing their location, orientation, and sequence conservation (Figs. S1 and S2). CRISPR-Cas targeting then can be used to validate population heterogeneity empirically at each predicted locus and simultaneously to increase the recovery of low-incidence mutants. The high prevalence of MGEs in lactic acid bacteria, and especially S. thermophilus, is in accordance with their role in speciation of these hyper-adapted bacteria through genome evolution (39, 40). Moreover, recovery of genomic deletion mutants using CRISPR-Cas targeting could facilitate phenotypic characterization of genes with unknown function. Mutants exhibiting the deletion of the 102-kb island encoding the lac operon had significantly increased generation times relative to the wild type and achieved a lower final OD. With 102 predicted ORFs therein, it is likely that additional phenotypes are affected, and many of the genes do not have annotated functions. CRISPR-Cas targeting allows direct assessment of how island-encoded genes contribute to adaption to grow in milk; this understanding is important, given the industrial relevance of niche-specific genes such as prtS. Moreover, it is in the natural genomic and ecological context of these horizontally acquired traits, because they likely were acquired as discrete islands. These results establish avenues for the application of self-targeting CRISPR-Cas9 systems in bacteria to investigate transposition, DNA repair mechanisms, and genome plasticity.
CRISPR-Cas systems generally limit genetic diversity through interference with genetic elements, but acquired MGEs also can provide adaptive advantages to host bacteria. Thus, the benefit of maintaining genomically integrated MGEs despite CRISPR-Cas targeting is an important driver of genome homeostasis. Collectively, our results establish that in silico prediction of GEIs can be coupled with CRISPR-Cas targeting to isolate clones exhibiting large genomic deletions. Chimeric insertion sequence footprints at each deletion junction indicated a common mechanism of deletion for all four islands. The high prevalence of self-targeting spacers exhibiting identity to genomic loci, combined with experimental demonstrations of genomic alterations, suggests that CRISPR-Cas self-targeting may contribute significantly to genome evolution of bacteria (28, 30). Collectively, studies on CRISPR-Cas–induced large deletions substantiate this approach as a rapid and effective means to assess the essentiality and functionality of gene clusters devoid of annotation and to define minimal bacterial genomes based on chromosomal deletions occurring through transposable elements.
Materials and Methods
Bacterial Strains.
All bacterial strains are listed in Table S2. Bacterial cultures were cryopreserved in an appropriate growth medium with 25% glycerol (vol/vol) and stored at −80 °C. S. thermophilus was propagated in Elliker medium (Difco) supplemented with 1% beef extract (wt/vol) and 1.9% (wt/vol) β-glycerolphosphate (Sigma) broth under static aerobic conditions at 37 °C or on solid medium with 1.5% (wt/vol) agar (Difco), incubated anaerobically at 37 °C for 48 h. Concentrations of 2 µg/mL of erythromycin and 5 µg/mL of chloramphenicol (Sigma) were used for plasmid selection in S. thermophilus, when appropriate. Escherichia coli EC1000 was propagated aerobically in Luria–Bertani (Difco) broth at 37 °C or on brain-heart infusion solid medium (Difco) supplemented with 1.5% agar. Antibiotic selection of E. coli was maintained with 40 µg/mL kanamycin and 150 µg/mL of erythromycin for recombinant E. coli, when appropriate. Screening of S. thermophilus derivatives for β-galactosidase activity was assessed qualitatively by supplementing a synthetic Elliker medium with 1% lactose, 1.5% agar, and 0.04% bromo-cresol purple as a pH indicator.
DNA Isolation and Cloning.
All kits, enzymes, and reagents were used according to the manufacturers' instructions. DNA purification and cloning were performed as described previously (42). Plasmids with lacZ targeting arrays were constructed with each consisting sequentially of the (i) native leader sequence specific to CRISPR1 or (ii) CRISPR3 native repeats specific to CRISPR 1 or (iii) CRISPR 3 spacer sequence specific to the 5′ end of lacZ, another native repeat. To engineer each plasmid, the sequence features listed above were ordered as extended oligomers (Table S3), combined using splicing by overlap extension PCR (42), and cloned into pORI28 (Table S3).
Selection and Design of CRISPR Spacers.
Putative protospacers were constrained by first defining the location of all putative PAM sequences in the sense and antisense strands of lacZ. Within the 3,081-nt gene, there were 22 CRISPR1 (AGAAW) and 39 CRISPR3 (GGNG) PAM sites that were identical to their bioinformatically derived consensus sequences (21). After potential spacers were identified, the complete proto-spacer, seed, and PAM sequence were subjected to BLAST analysis against the genome of S. thermophilus LMD-9 to prevent additional targeting of nonspecific loci. The spacers for CRISPR1 and CRISPR3 were disparate in sequence and corresponding PAM sites but were designed to target the 5′ end of lacZ, resulting in predicted cleavage sites residing 6 nt apart. The leader sequences, repeats, and spacers on each plasmid represented orthogonal features unique to CRISPR1 or CRISPR3, respectively. To assess target locus-dependent mutations, an additional CRISPR3 plasmid was created with a spacer to the metal cation-binding residue essential for β-galactosidase activity. A CRISPR1 array plasmid containing a nonself spacer was used as a control to quantify lethality of self-targeting.
Transformation.
Plasmids were electroporated into competent S. thermophilus containing the temperature-sensitive helper plasmid pTRK669. An overnight culture of S. thermophilus was inoculated at 1% (vol/vol) into 50 mL of Elliker medium supplemented with 1% beef extract, 1.9% β-glycerophosphate, and chloramphenicol selection. When the culture achieved an OD600 at nm of 0.3, penicillin G was added to achieve a final concentration of 10 µg/mL. Cells were harvested by centrifugation and washed three times in 10 mL cold electroporation buffer (1 M sucrose and 3.5 mM MgCl2). The cells were concentrated 100-fold in electroporation buffer, and 40 µL of the suspension was aliquoted into 0.1-mm electroporation cuvettes. Each suspension was combined with 700 ng of plasmid. Electroporation conditions were set at 2,500 V, 25 µF capacitance, and 200 Ω resistance. Time constants were recorded and ranged from 4.4 to 4.6 ms. The suspensions were combined immediately with 950 µL of recovery medium and were incubated for 8 h at 37 °C. Cell suspensions were plated on selective medium, and electroporation cuvettes were washed with medium to ensure recovery of cells.
Growth and Activity Assessment.
Cultures were preconditioned for growth assays by subculturing for 12 generations in a semisynthetic Elliker medium with glucose as the sole carbohydrate source. Fresh medium was inoculated with an overnight culture at 1% (vol/vol) and incubated at 37 °C statically. OD600 was monitored hourly until the cultures achieved stationary phase. Acidification of milk was assessed by inoculating skim milk with an overnight culture to a level of 108 cfu/mL and incubation at 42 °C. The pH was subsequently monitored using a Mettler Toledo Seven Easy pH meter and Accumet probe. Skim milk was acquired from the North Carolina State University dairy plant and was pasteurized for 30 min at 80 °C.
Supplementary Material
Acknowledgments
We thank Allie Briner, Madelyn Shoup, Chase Beisel, Yong Jun Goh, Sarah O'Flaherty, and Brant Johnson for helpful discussions, and the North Carolina State University (NCSU) Dairy Plant for providing skim milk. This work was supported by NCSU start-up funds, the North Carolina Agriculture Foundation, and DuPont Nutrition and Health. K.S. was supported in part by the 2013–2014 Dannon Probiotics Fellow Program of The Dannon Company, Inc. during the completion of this work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508525112/-/DCSupplemental.
References
- 1.Darmon E, Leach DR. Bacterial genome instability. Microbiol Mol Biol Rev. 2014;78(1):1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Brouns SJJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young JC, et al. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS ONE. 2012;7(5):e38077. doi: 10.1371/journal.pone.0038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 8.Groenen PM, Bunschoten AE, van Soolingen D, van Embden JD. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10(5):1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 9.Yin S, et al. The evolutionary divergence of Shiga toxin-producing Escherichia coli is reflected in clustered regularly interspaced short palindromic repeat (CRISPR) spacer composition. Appl Environ Microbiol. 2013;79(18):5710–5720. doi: 10.1128/AEM.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu F, et al. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol. 2011;77(6):1946–1956. doi: 10.1128/AEM.02625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrangou R, Horvath P. CRISPR: New horizons in phage resistance and strain identification. Annu Rev Food Sci Technol. 2012;3:143–162. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 12.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41(15):7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomaa AA, et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2014;5(1):e00928–e13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 18.Bolotin A, et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol. 2004;22(12):1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath P, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(Pt 8):2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 21.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paez-Espino D, et al. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- 23.Sun CL, et al. Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol. 2013;15(2):463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- 24.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39(21):9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briner AE, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56(2):333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 27.Bondy-Denomy J, Davidson AR. To acquire or resist: The complex biological effects of CRISPR-Cas systems. Trends Microbiol. 2014;22(4):218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Horvath P, et al. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol. 2009;131(1):62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 29.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013a;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vercoe RB, et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9(4):e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh JH, van Pijkeren JP. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42(17):e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selle K, Barrangou R. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015;23(4):225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi K, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100(8):4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62(3):725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goh YJ, Goin C, O’Flaherty S, Altermann E, Hutkins R. Specialized adaptation of a lactic acid bacterium to the milk environment: The comparative genomics of Streptococcus thermophilus LMD-9. Microb Cell Fact. 2011;10(Suppl 1):S22. doi: 10.1186/1475-2859-10-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dandoy D, et al. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb Cell Fact. 2011;10(Suppl 1):S21. doi: 10.1186/1475-2859-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11(8):1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 2007;189(4):1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103(42):15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang W, et al. Dealing with the evolutionary downside of CRISPR immunity: Bacteria and beneficial plasmids. PLoS Genet. 2013;9(9):e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goh YJ, et al. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2009;75(10):3093–3105. doi: 10.1128/AEM.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.