Significance
During the past 30 years, the methyl transfer community has attempted to find the molecular origin of the methyltransferases’ catalytic power. This report describes a combination of experimental and computational studies of enzymatic methyl transfer catalyzed by catechol-O-methyltransferase and its mutants at position Tyr68. The results show structural and dynamical differences between WT and mutants, as well as a role for substrate ionization in the generation of active site compaction. For the first time, to our knowledge, we are able to show a trend in donor–acceptor distance in the ground state that can be correlated with catalytic efficiency. This work provides an important step forward and a clear new direction for understanding enzymatic methyl transfer.
Keywords: compaction, COMT, donor–acceptor distance, methyltransferase, QM/MM
Abstract
Enzymatic methyl transfer, catalyzed by catechol-O-methyltransferase (COMT), is investigated using binding isotope effects (BIEs), time-resolved fluorescence lifetimes, Stokes shifts, and extended graphics processing unit (GPU)-based quantum mechanics/molecular mechanics (QM/MM) approaches. The WT enzyme is compared with mutants at Tyr68, a conserved residue that is located behind the reactive sulfur of cofactor. Small (>1) BIEs are observed for an S-adenosylmethionine (AdoMet)-binary and abortive ternary complex containing 8-hydroxyquinoline, and contrast with previously reported inverse (<1) kinetic isotope effects (KIEs). Extended GPU-based computational studies of a ternary complex containing catecholate show a clear trend in ground state structures, from noncanonical bond lengths for WT toward solution values with mutants. Structural and dynamical differences that are sensitive to Tyr68 have also been detected using time-resolved Stokes shift measurements and molecular dynamics. These experimental and computational results are discussed in the context of active site compaction that requires an ionization of substrate within the enzyme ternary complex.
Methyltransferases are widely distributed in nature, playing critical roles in metabolic transformations, natural product biosynthesis (1, 2), and cellular regulation via the reversible methylation of proteins and nucleic acids (3–5). The ability to understand the origin of catalysis within this class of reactions is crucial to any efforts at protein redesign (6, 7) or inhibition (5, 8, 9). Catechol-O-methyltransferase (COMT), a drug target for a number of neurological diseases, emerged early as a prototype for mechanistic investigations (8, 10, 11). A compelling and unusual feature of COMT catalysis is the presence of a large inverse kinetic isotope effect (KIE) during transfer of the methyl group from the cofactor S-adenosylmethionine (AdoMet) to catechol acceptor. Comparison of the enzymatic behavior with a model reaction in solution led to the proposal of a role for transition state compression in enzymatic rate acceleration (12–18).
Over the past decade, there has been a major shift in focus away from a historical interpretation of enzyme catalysis within the context of static 3D protein structures. Increasingly, protein motions are seen as integral to protein function at every level, from ligand binding to allosteric control and enzyme catalysis (19–22). Models for catalysis that depend on such protein motions have been particularly important in the area of C–H activation, where the transfer of hydrogen by tunneling mechanisms implicates a critical dependence of the reaction rate on barrier width, and not just barrier height (23, 24). Achievement of the tunneling-ready state requires a transient sampling of enzymatic ground states that achieves a reduced distance between the H-donor and acceptor (25). This feature raises the question of whether the reported inverse KIE in COMT could result from ground state interactions that bring about catalytically relevant changes in the reactants’ geometry and distance (26).
The human COMT functions to inactivate the neurotransmitters dopamine, norepinephrine, and epinephrine via methylation of a catechol oxygen (8). This physiologically important enzyme has been subjected to extensive structural and kinetic investigation, and operates via an ordered binding mechanism in which the addition of AdoMet is followed by binding of Mg2+ and then substrate (SI Appendix, Fig. S1). The chemical reaction proceeds via SN2 attack of an oxygen of the substrate on the methyl group of AdoMet, resulting in methylated catechol and S-adenosyl-l-homocysteine as products (SI Appendix, Fig. S2). Numerous computational studies have attempted to understand the origin of the experimentally measured inverse KIE (15–18, 27–33). Using the available X-ray structures for the soluble form of COMT, Lau and Bruice (16) applied classical molecular dynamics (MD) to identify a small subset of ground state structures that bring the donor and acceptor atoms closer than van der Waals distances. Subsequent ab initio studies of a 22-heavy atom model of the COMT active site that included three truncated amino acid side chains (Met40, Tyr68, and Asp141), AdoMet, and catechol led to the proposal of greater contributions from ground state than transition state interactions (29). Semiempirical quantum mechanics/molecular mechanics (QM/MM) studies of the COMT transition state structure, either alone or in combination with ensemble averaging (31), have consistently been unable to detect any reduction in the methyl group of AdoMet to catechol oxygen distance (C···O) relative to the solution reaction. Although the latter studies were able to rationalize the inverse KIEs in COMT by invoking an increase in the reaction coordinate frequency, they were unable to provide a physical model for the origins of catalysis (17, 18).
This laboratory introduced the use of site-specific mutagenesis to examine the extent to which a conserved Tyr at position 68, residing behind the sulfur of the methyl donor of AdoMet in COMT and other methyl transfer enzymes, would control active site properties (34). Kinetic data for WT COMT and five variants indicated a 103-fold reduction in rate that correlates with an increase in the magnitude of the inverse secondary KIE toward the solution value of unity. The data obtained (for second-order rate constants) were unable to distinguish ground state from transition state effects and, further, could not provide a molecular mechanism for the large rate differentials. In a recent computational study of COMT and its Tyr68 mutant (Y68A), the empirical valence bond methodology developed by Warshel and Weiss (35) was also unable to provide a physical basis for the observed trends in KIEs (33), dismissing these effects as “circular interpretations of experimental results.”
In this study, we present experimental measurements of the impact of Tyr68 mutants on binding isotope effects (BIEs) for formation of the binary complex of COMT with AdoMet that are accompanied by graphics processing unit (GPU)-accelerated QM studies on a catalytically relevant ternary complex (36). Additional probes of protein structural and dynamics include time-resolved fluorescence lifetimes and Stokes shifts and >80-ns MD simulations for WT, Y68F, and Y68A. The aggregate results implicate a critical role for Tyr68 in achieving the catalytically optimal alignment of the methyl donor (AdoMet) in relation to its ionized (catecholate) acceptor.
Results
BIEs.
The experimental measurement of equilibrium BIEs was accomplished in the context of a kinetically competent formation of a complex between AdoMet, Mg2+, and COMT, which is followed by the binding of catechol to form the catalytic complex (SI Appendix, Fig. S1). The previously reported kcat/Km KIEs involved reaction of a fully tritiated methyl donor, [3H3]-AdoMet, with unlabeled substrate (dopamine) at a concentration below the Km value for dopamine (34). In this work, we repeated the KIE measurements for WT COMT at a range of dopamine concentrations (0.5–3 Km of dopamine), and the results are given in SI Appendix, Table S1. Although the tritium KIE may begin to rise slightly toward unity at the highest dopamine concentration as substrate begins to saturate the enzyme and suppress the competitive KIE (37), the former conclusion of a secondary KIE ≤0.8 is validated in the present study, providing a robust frame of reference for the experimental BIEs (Table 1). As a control, we also extended the measurement of the primary 14C KIE, originally determined for WT, to the mutants, finding values in the range of 6–14% (SI Appendix, Table S2). These data are consistent with a single rate-determining methyl transfer step in all cases under the second-order kinetic condition of kcat/Km.
Table 1.
Experimental equilibrium BIEs and secondary KIEs for the recombinant human COMT and its mutants
| COMT | Kd(AdoMet),* μM | Secondary KIE,† kCH3/kCT3 | Binary BIE,‡ CH3/CT3 | Ternary BIE,‡ CH3/CT3 |
| 108V(WT) | 19 ± 1 | 0.791 ± 0.012 | 1.015 ± 0.011 | 1.117 ± 0.010 |
| Y68F | 155 ± 10 | 0.822 ± 0.021 | 1.008 ± 0.021 | 1.124 ± 0.114 |
| Y68A | 186 ± 4 | 0.850 ± 0.012 | 1.008 ± 0.024 | 1.058 ± 0.026 |
| V108M | 22 ± 1 | 0.784 ± 0.014 | 1.017 ± 0.009 | 1.136 ± 0.014 |
| V108M/Y68F | 163 ± 9 | 0.822 ± 0.007 | 1.039 ± 0.027 | 1.133 ± 0.025 |
| V108M/Y68A | 189 ± 9 | 0.863 ± 0.019 | 1.010 ± 0.008 | 1.070 ± 0.004 |
Fitted values from SI Appendix, Eq. S3 and their SDs.
Data from ref. 34.
Binary BIEs were determined by a competitive method using the mixture of [methyl-3H] AdoMet and [carboxyl-14C] AdoMet. The ternary BIEs were determined by a competitive method using the mixture of [methyl-3H] AdoMet and [carboxyl-14C] AdoMet in the presence of the 8-hydroxyquinoline. Average values from (at a minimum) triplicate measurements and their SDs are shown.
From Table 1, it is clear that the binary tritium BIEs are all normal (i.e., in the opposite direction from the KIEs). Further, within experimental error, there is no difference among the binary BIEs following mutation on Tyr68. Considering that the binary complex must bind a catechol substrate to accomplish the reaction, the BIEs were repeated in the presence of the well-characterized competitive inhibitor 3,5-dinitrocatechol (8). However, this potent inhibitor of COMT was found to prevent a full equilibration between the enzyme-bound AdoMet and free AdoMet, a prerequisite for the detection of reliable BIEs involving the cofactor. After screening a variety of inhibitors, 8-hydroxyquinoline (38) was found to be suitable for the BIE measurements (Table 1). Surprisingly, the values for the BIEs within this ground state ternary complex of COMT are all normal, ranging from 6–14%. These results indicate that the experimentally available BIEs with 8-hydroxyquinoline are unable to capture the changes in force constants reported upon in the previously described KIEs (34).
Time-Resolved Fluorescence Lifetimes and Stokes Shifts.
Time-dependent fluorescence spectroscopy offers an independent means of examining ground state interactions that may affect the reactivity of COMT and its Tyr68 variants. Two Trp residues are present in WT COMT. Trp143 gates the entrance of the binding pocket of AdoMet and interacts with the Ado ring of cofactor, whereas Trp38 resides near the catechol site (Fig. 1A). The single-Trp construct, W143in, in which Trp38 has been replaced by Phe, was chosen for further study using variants involving the insertion of either Phe or Ala at position 68. The kinetic behavior of the W143in series is similar to the WT series with regard to its sensitivity to mutation at Tyr68 (SI Appendix, Table S3), making these constructs suitable candidates for fluorescence lifetime and Stokes shifts studies. The lifetime data for apoenzyme indicate little difference among the Tyr68 series (SI Appendix, Fig. S3A). The addition of AdoMet, however, leads to an impact of Tyr68 (SI Appendix, Fig. S3B). Analysis of the data indicates multiexponential decay (SI Appendix, Fig. S4), in which a third transient (τ1) appears with a shorter lifetime at the expense of the longest lived transient (τ3). This property generates a smaller overall lifetime <τ> for W143in than either W143in/Y68F or W143in/Y68A (SI Appendix, Table S4). Time-resolved dynamic Stokes shifts parameters were further reconstructed from the time-correlated single-photon counting technique (39) (SI Appendix, Figs. S5 and S6), and are summarized in Table 2. It can be seen that upon addition of AdoMet, an increase in the magnitude of the Stokes shifts (Δν) parallels a decrease in relaxation lifetime (τ) in all cases. The magnitude of the impact of cofactor is especially significant for WT, which shows a sixfold increase in Δν (from 114–724 cm−1) (Table 2). The data indicate an active site that has become more structured upon AdoMet binding (increased Δν), generating an environment that enhances the relaxation rate of the excited state dipole at Trp143 (decreased τ). Significantly, mutations at Tyr68, ca. 7 Å away, perturb the environment in a direction that brings Δν closer to the apoenzyme.
Fig. 1.
(A) Active site of human WT COMT complex with catechol and AdoMet showing Y68 and two Trps (W38 and W143) (derived from Protein Data Bank ID code 3BWM). (B) Proposed reaction coordination. The dominant ground state (GS) structure shows catechol in its ring-protonated form. Although not indicated, Lys144 is proposed to function as a general base in the formation of the catecholate intermediate (28). As illustrated, the catechol is fully deprotonated before methyl transfer. Mg2+ is presented as a gray sphere. E, COMT; R1(R2)-S+-CH3, AdoMet.
Table 2.
Experimental parameters for time-resolved fluorescence Stokes shifts
| COMT | Kd(AdoMet),* μM | τapo, ns | Δνapo, cm−1 | τholo, ns | Δνholo, cm−1 |
| W143in | 18 ± 1 | 1.64 ± 0.01 | 114 | 0.77 ± 0.03 | 724 |
| W143in/Y68F | 40 ± 2 | 1.48 ± 0.01 | 76 | 0.52 ± 0.05 | 154 |
| W143in/Y68A | 51 ± 3 | 1.81 ± 0.01 | 72 | 0.42 ± 0.03 | 148 |
τapo and τholo refer to decay times for apoenzyme and binary AdoMet–enzyme complex Stokes shifts, respectively. The data represent average values from triplicate measurements and their SDs. Δνapo and Δνhoho refer to total red shift in Stokes measurements for apoenzyme and holoenzyme (with AdoMet), respectively.
Fitted values from SI Appendix, Eq. S3 and their SDs.
Extended GPU-Based ab Initio Computations of the COMT–AdoMet–Catecholate Complex.
The investigation of ground state ternary complexes of COMT presents a major challenge to the goals of this study. Although BIEs for a ternary complex have been measured using 8-hydroxyquinoline to mimic the substrate, this complex is inherently distinct from the reactive ternary complex. Computational studies on COMT are not limited in this manner and allow us to estimate ground state structures that arise from productive complexes containing both cofactor and activated (i.e., deprotonated) substrate. Advances in GPU-accelerated quantum chemistry make it possible to study enzymes straightforwardly using a QM/MM scheme (40–43) in which the QM region is on the order of 1,000 atoms (44). The computational cost of this procedure is comparable to the computational cost for a calculation of only tens of atoms in the QM region on traditional central processing unit (CPU) processors. Recent GPU-based studies of WT COMT have focused on identifying convergence of both charge and internuclear distances with increasing QM region size, showing that the system converges at ca. 26–30 protein residues (36). A QM region that extends to within a 5-Å radius of the AdoMet and catecholate reactants is thus needed to produce a stable solution. These studies, which focus on the enthalpically most favored ground state distribution for WT COMT, indicate a shortened bond distance of 2.87 Å for the AdoMet methyl and catecholate oxygen (36), as anticipated from the available crystal structures (cf. SI Appendix, Table S5). For the present work, the GPU methodology has been extended to Y68F and Y68A, again using a range-corrected ωPBEh density-functional with a 6–31G basis set for the large QM region that is combined with a force field description of the remainder of the solvated protein.
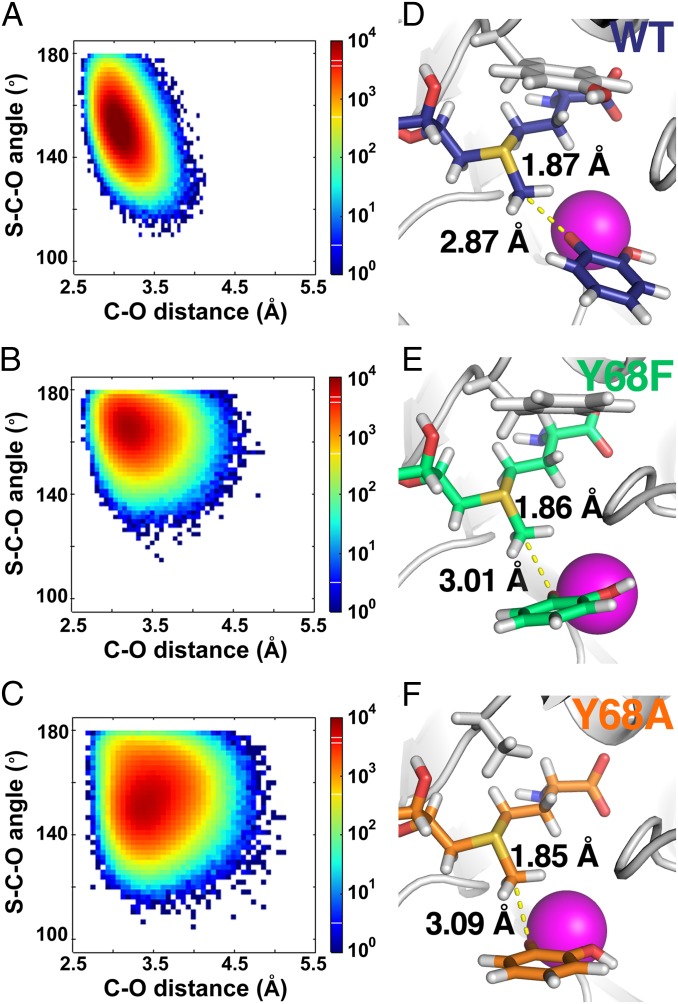
The studies were initiated via >80-ns MD simulations (Fig. 2 A–C), and representative structures at median C···O distances from these MD runs were selected for in-depth QM/MM analysis. After optimization of the structures of Y68F and Y68A using the expansive QM/MM approach (36), the methyl carbon-to-catecholate oxygen distance is found to increase relative to WT, to 3.01 Å (Y68F) and 3.09 Å (Y68A) (Table 3). At the same time, an elongation of the S+–CH3 bond seen in WT undergoes a small reduction from 1.87 Å (WT) to 1.85 Å (Y68A). Significantly, the computed changes in orientation and distances, illustrated in Fig. 2 D–F, indicate an overall shift in the sulfur-to-oxygen distance from 4.65 Å for WT enzyme to 4.86 Å (Y68F) and 4.93 Å (Y68A) (Table 3). This analysis shows that mutation at Tyr68 not only leads to changes in individual bond lengths (sulfur to carbon and C···O) but also to more expanded overall sulfur-to-oxygen distances (S+···C···O) that approach the estimated van der Waals value of 4.94 Å.
Fig. 2.
Results from computations on the complex of COMT, AdoMet, Mg2+, and the anion of 2-hydroxyphenol. (A–C) Results from classical MD simulations. The distribution of both the S–C–O angle and the C–O distance increase in the order WT < Y68F < Y68A. (D–F) Results from the expanded ab initio analysis on WT, Y68F, and Y68A, respectively. The methyl donor-to-acceptor distance increases from the value in WT (2.87 Å) to 3.01 and 3.09 Å in Y68F and Y68A, respectively. At the same time, the bond length between the carbon of the methyl group and sulfur in the cofactor AdoMet decreases from 1.87 Å for WT to 1.86 Å and 1.85 Å for Y68F and Y68A, respectively.
Table 3.
Structural characteristics of the computed ternary complexes in WT COMT, COMT mutants and in solution
| COMT | d(+S–C), Å | d(C···O), Å | θ (+S···C···O), ° | d(+S–C···O),* Å | d(+S···O),† Å |
| WT (108V)‡ | 1.87 | 2.87 | 156 | 4.74 | 4.65 |
| Y68F | 1.86 | 3.01 | 167 | 4.87 | 4.86 |
| Y68A | 1.85 | 3.09 | 174 | 4.94 | 4.93 |
| Solution | 1.82 | 3.12¶ | 180 | 4.94 | 4.94 |
S, C, and O refer to the sulfur in AdoMet, the transferred methyl carbon, and catechol oxygen, respectively.
This distance is the sum of the d(+S–C) and d(C···O), rather than the actual distance between sulfur and oxygen.
Actual distance between sulfur and oxygen.
Data from ref. 36.
This value is presented as van der Waals distance of carbon (1.72 Å) and oxygen (1.40 Å).
Discussion
One initially surprising finding from this work was that the BIEs for both binary (E–AdoMet) and ternary (E–AdoMet–inhibitor) complexes of COMT are close to unity or slightly elevated, in contrast to the inverse (<1) KIEs reported earlier for WT and a series of Tyr68 mutants (34). As summarized in Table 1, BIEs for binary complex are all quite small and are ≤1% within 1 SD. It can be reasonably concluded that little of interest is occurring within the E–AdoMet complex with regard to changes in force constant at the transferred methyl group. Clearly, the ability of the AdoMet to bind to COMT is affected to some degree within the Tyr68 mutants of WT ( 10-fold; Table 1) and of W143in (threefold or less; Table 2). Similarly, the fluorescence lifetimes and size of the Stokes shift at Trp143 are sensitive to the presence of both AdoMet and mutation at Tyr68. We conclude that the presence of AdoMet has multiple effects on the WT, including an increase in structure surrounding Trp143 that is also accompanied by increased local dynamics (Table 2 and SI Appendix, Table S4). The data reveal communication between Tyr68 and Trp143 in the binary complex, with mutations at Tyr68, decreasing the magnitude of the Stokes shift (Table 2) as well as the relative contribution of the most rapid fluorescence lifetime (SI Appendix, Table S4). In aggregate, these results indicate a remote role for Tyr68 in altering the region of the protein contacting the Ado ring of the bound AdoMet, without any direct effect on the methyl group itself.
The fact that the BIEs for abortive ternary complexes of COMT are uniformly larger than in the binary complexes (Table 1) indicates that an interaction is taking place that alters (reduces) the vibrational frequencies at the reactive methyl group in the presence of the inhibitor 8-hydroxyquinoline. Although it would have been useful to measure fluorescence lifetimes and Stokes shifts for ternary complexes, this measurement was found to be precluded by the extensive quenching of W143in fluorescence by bound inhibitor. One possible explanation for BIEs >1 is a change in local electrostatics that fails to be accompanied by any overall decrease in the distance between the methyl donor (S+) and acceptor (represented by the hydroxyl group of 8-hydroxyquinoline). We note that in a related methyl transfer system, SET7/9, X-ray crystallography and NMR of the binary complex have led to the proposal of ground state hydrogen bonding between the C–H of the methyl group in AdoMet and electron-accepting protein side chain(s) (45, 46). A role for such putative C–H···O bonding in reducing vibrational frequencies at the transferred methyl group would be consistent with BIEs larger than unity, with the caveat that the elevated BIE data presented herein are for a different enzyme family and only occur in the presence of the relatively weak binding inhibitor 8-hydroxyquinoline.
The X-ray structures of abortive ternary complexes of COMT with substrate-like catechol inhibitors provide an alternative window into the properties of ternary complexes. In a large number of published structures, a surprisingly short C···O (methyl-to-catechol oxygen) bond has been detected, less than the expected van der Waals distance in all cases (SI Appendix, Table S5). On inspection, it is seen that all of the interrogated structures contain inhibitors with electron withdrawing that will lower the pKa of one of the catechol-OH group (SI Appendix, Figs. S7 and S8), likely resulting in the binding of the anionic form of inhibitor. This type of pKa alteration is absent in free catechol and cannot occur in the one inhibitor, 8-hydroxyquinoline, found to be suitable for BIE measurements. We considered that the reduced C···O bond distances in the X-ray structures are a direct result of pKa reductions in inhibitor, and that short donor–acceptor distances require prior ionization at one of the ring hydroxyl groups of inhibitor.
For the above reasons, GPU-based computation of the enzyme ternary complex has been particularly insightful regarding the nature of the catalytically competent ground state ternary complex in which substrate can be assigned to its catecholate form. The findings (Table 3) indicate a clear-cut ground state perturbation that includes (i) a slightly elongated S+–C bond, (ii) a decreased C···O bond, and (iii) an overall reduced S+···O distance. Each of these properties is affected to differing degrees by mutation at Tyr68, with the computed distances in the least catalytic Y68A approximating the computed distances expected for unperturbed molecules in solution. Further results, to be published separately, indicate that binding of catechol, rather than catecholate, eliminates the perturbed bond lengths and trends with Y68X (Table 3). It is of considerable interest and importance that the ability to detect active site compaction in WT enzyme, together with its sensitivity to Tyr68 mutants, is only possible when a very large region of COMT (five- to 10-fold larger than normally used) is included in the QM region of the QM/MM calculations.
The precise structural features that could give rise to intersubstrate compaction in WT COMT could, in principle, be revealed from comparative X-ray structures of binary and abortive ternary complexes of COMT. A comparative structure for a ternary complex formed with dintro-catechol as a substrate analog does show a reduced distances between the methyl group of AdoMet and active site side chain (at Asp141) and main chain carbonyl (at Asp141 and Met40) (SI Appendix, Fig. S9 and Table S6); as noted above, the lowered pKa value of the dinitro-catechol (pKa = 3.32; SI Appendix, Fig. S8) makes it likely that this particular structure contains catecholate in its ionized form. There were no further analyses of structural changes at more remote residues, and, in any case, the method reported reflects static protein structures. Our initial classical MD studies of the COMT–AdoMet–catecholate complex (Fig. 2 A–C and SI Appendix, Fig. S10) show distinctive differences in the shape and distribution of ground state configurations for WT in relation to the Y68 variants, indicating how changes in ground state dynamics may contribute to the altered properties of the Y68X series.
A key question in the present study has been the interrelationship of binding to kinetic effects in COMT. To explore the possibility that factors other than a reduction in donor–acceptor distances were contributing to the kinetic effects, density functional theory methods were used to compute the equilibrium isotope effect for the transfer of a methyl group from a sulfonium ion to an oxygen nucleophile:
Focusing on tritium effects, we estimate the equilibrium isotope effect, T(Keq), to be 0.83 (SI Appendix, Table S7), a value quite close to the KIE reported for WT COMT. This result raised the question of whether the magnitude of T(kcat/Km) could arise solely from force constant changes occurring upon transfer of a methyl group from a sulfonium ion to an oxyanion center. Fortunately, the trends in the KIEs observed for WT and Y68X allow us to examine this premise. The key observation is that the reduction in rate upon mutation at Tyr68 is accompanied by an increase in the magnitude of the secondary KIE toward unity. A structure-reactivity correlation would predict a later, not earlier, transition state for the less reactive variants of COMT and, hence, accompanying more inverse KIEs. This prediction is in direct opposition to the experimental observations, leading us to rule out early vs. late transition states as the origin of the trends in the measured KIEs for WT and Y68F and Y68A. This analysis corroborates the validity of a focus on active site compaction that is initiated via substrate ionization as the origin of both the magnitude and trends in the secondary KIEs and rates for COMT (cf. ref. 47).
In the context of the aggregate experimental and computational data available for COMT, we now present a working model for its catalysis. As shown from fluorescence data, the initial formation of the binary complex with AdoMet produces an increase in both structure and dynamics in the region of the protein surrounding Trp143, and, furthermore, these features are influenced by the Tyr68 that is ca. 7 Å away. At the same time, mutants at Tyr68 have no apparent influence on the bending and stretching force constants at the more proximal methyl group of AdoMet in the binary complex (Table 1). The addition of the inhibitor 8-hydroxyquinoline to form an abortive ternary complex introduces new interactions that lead to a reduction in the force constants at the methyl group experimental BIEs (>1) that are affected by the mutation at Tyr68 and may reflect hydrogen binding between the methyl group of AdoMet and protein side chains (45, 46) or a small increase in the S+–CH3 bond length that has not been compensated for by a reduction of the C···O bond distance. Importantly, the elevated pKa of the 8-hydroxyquinoline yields a ground state ternary structure in which the ring hydroxyl group is unionized, which we propose maintains the methyl donor and acceptor at their van der Waals distance. This behavior contrasts with the properties of ternary complexes that implicate shortened donor–acceptor distances, detected either in X-ray structures of ternary complexes of COMT with low pKa catechol-type inhibitors (SI Appendix, Table S5) or via our QM/MM computations that involve the ionized form of substrate (Fig. 2). Importantly, an impact of position 68 on the vibrational frequencies of the transferred methyl group arises within ternary complexes, with a change in direction for both BIEs (Table 1) and KIEs (34) toward unity.
The following sequence of force constant changes at the methyl group of AdoMet is thus proposed (Fig. 1B): (i) Upon formation of binary complex, no significant alteration in the methyl group takes place; (ii) formation of a neutral substrate or inhibitor ternary complex produces a reduction in force constant that may be due to some elongation of the S+–CH3 bond (BIEs >1); and (iii) a subsequent ionization of the bound catechol leads to a activated ground state structure that lies between the dominant ground state and transition state. The latter feature produces a significant reduction in donor–acceptor distance that is a primary source of increased force constants at the methyl group, contributing to the KIEs <1 (4). The magnitude of the latter is highly dependent on the proximal Tyr68 as well as residues distributed throughout the core of the COMT structure, as dictated by the large number of atoms required to reach convergence in the QM/MM computations. The placement of the activated complex between ground state and transition state structures for COMT satisfies the experimental observations of inverse KIEs on both kcat and kcat/Km (14).
In the context of the mechanism of Fig. 1B, it will be of future value to obtain X-ray structures of COMT that are mutated at Tyr68, as well as structures that contain an expanded range of inhibitors with different pKa values. Because all of the AdoMet-dependent methyltransferases share an equivalent AdoMet-binding fold to COMT, it will also be of considerable interest to determine the degree to which the catalytic strategies presented herein are conserved among this large family of enzymes that includes protein, DNA, and RNA methyltransferases.
Regarding the ongoing quest for a set of basic physical principles that govern enzyme catalysis, this study has offered the opportunity to examine the degree to which enzymatic methyl transfer reactions display properties similar to the extensively characterized C–H activation reactions. The central role of H-nuclear tunneling in the latter case indicates that catalysis requires a close approach between the H-donor and acceptor, estimated to be reduced by 0.3–0.6 Å from donor–acceptor van der Waals distances. For native and optimized enzymes, reduced distances are attributed to a stochastic search among protein ground states that leads to short, tunneling-ready donor–acceptor configurations (25). Significantly, the present work also uncovers a ground state compaction in an activated, substrate-ionized ground state that correlates with very high turnover rates for COMT (compare Fig. 2 D–F) (34). Although these computed geometries represent static, energy-minimized states, protein conformational sampling is also predicted to play a role in the reaction (16, 28, 29). Looking to the future, the further development of GPU-based computational methods may be expected to enable conformational sampling while maintaining the greatly expanded QM regions of protein shown to be critical to the properties of COMT (36) (Fig. 2). This synergy of experimental and computational methods provides a powerful platform for advancing our understanding of biological methyl transfer.
Materials and Methods
Expression and Purification of COMT.
The human soluble form of COMT and mutants in the Novagen pET22b(+) vector were transformed and expressed in Escherichia coli BL21 (DE3) cells overnight. Purification was taken on a nickel-nitrilotriacetic acid metal affinity column and polished by size exclusion chromatography.
BIE Measurement.
Competitive binding of [methyl-3H] AdoMet and [carboxyl-14C] AdoMet was performed using the ultrafiltration method as detailed in SI Appendix.
KIE Measurement.
KIE was measured by competitive methods using [methyl-14C]-AdoMet and [phenyl-2,5,6-3H]-dopamine for primary KIE, and [methyl-3H] AdoMet and [8-14C] dopamine for secondary KIE as described previously (34).
Computation Method.
Large-scale QM/MM simulations of WT and mutants COMT were carried out with the TeraChem/AMBER12 interface. The QM description is obtained at the ωPBEh/6–31g level, whereas the remainder of the protein in solvating water is described with an AMBER ff03 force field description. Equilibrium isotope effect calculations were carried out using the TeraChem package and Gaussian 09 (Gaussian).
Supplementary Material
Acknowledgments
We thank Vern L. Schramm (Albert Einstein College of Medicine of Yeshiva University) for sharing the ultrafiltration apparatus used in measurements of BIEs and Hui Zhu for helpful discussion. This work was supported by NIH Grants GM025765 and GM039296 (to J.P.K.) and by the Department of Defense (Office of the Director of Defense Research and Engineering) through a National Security Science and Engineering Faculty Fellowship (to T.J.M.). H.J.K. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506792112/-/DCSupplemental.
References
- 1.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29(10):1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 2.Struck A-W, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem. 2012;13(18):2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: Histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27(8):396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 5.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8(9):724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 6.Klimasauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25(3):99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Luo M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem Biol. 2012;7(3):443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Männistö PT, Kaakkola S. Catechol-O-methyltransferase (COMT): Biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999;51(4):593–628. [PubMed] [Google Scholar]
- 9.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97(20):1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 10.Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: Relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004;29(11):1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- 11.Marsala SZ, Gioulis M, Ceravolo R, Tinazzi M. A systematic review of catechol-0-methyltransferase inhibitors: Efficacy and safety in clinical practice. Clin Neuropharmacol. 2012;35(4):185–190. doi: 10.1097/WNF.0b013e31825c034a. [DOI] [PubMed] [Google Scholar]
- 12.Mihel I, Knipe JO, Coward JK, Schowen RL. Alpha-deuterium isotope effects and transition-state structure in an intra-molecular model system for methyl-transfer enzymes. J Am Chem Soc. 1979;101(15):4349–4351. [Google Scholar]
- 13.Gray CH, Coward JK, Schowen KB, Schowen RL. Alpha-deuterium and c-13 isotope effects for a simple, inter-molecular sulfur-to-oxygen methyl-transfer reaction - transition-state structures and isotope effects in transmethylation and transalkylation. J Am Chem Soc. 1979;101(15):4351–4358. [Google Scholar]
- 14.Hegazi MF, Borchardt RT, Schowen RL. Alpha-deuterium and c-13 isotope effects for methyl transfer catalyzed by catechol o-methyltransferase–SN2-like transition-state. J Am Chem Soc. 1979;101(15):4359–4365. doi: 10.1021/ja00426a079. [DOI] [PubMed] [Google Scholar]
- 15.Williams IH. Theoretical modeling of compression effects in enzymic methyl transfer. J Am Chem Soc. 1984;106(23):7206–7212. [Google Scholar]
- 16.Lau EY, Bruice TC. Importance of correlated motions in forming highly reactive near attack conformations in catechol O-methyltransferase. J Am Chem Soc. 1998;120(48):12387–12394. [Google Scholar]
- 17.Ruggiero GD, Williams IH, Roca M, Moliner V, Tuñón I. QM/MM determination of kinetic isotope effects for COMT-catalyzed methyl transfer does not support compression hypothesis. J Am Chem Soc. 2004;126(28):8634–8635. doi: 10.1021/ja048055e. [DOI] [PubMed] [Google Scholar]
- 18.Kanaan N, Ruiz Pernía JJ, Williams IH. QM/MM simulations for methyl transfer in solution and catalysed by COMT: Ensemble-averaging of kinetic isotope effects. Chem Commun (Camb) 2008;(46):6114–6116. doi: 10.1039/b814212b. [DOI] [PubMed] [Google Scholar]
- 19.Nagel ZD, Klinman JP. A 21st century revisionist’s view at a turning point in enzymology. Nat Chem Biol. 2009;5(8):543–550. doi: 10.1038/nchembio.204. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz SD, Schramm VL. Enzymatic transition states and dynamic motion in barrier crossing. Nat Chem Biol. 2009;5(8):551–558. doi: 10.1038/nchembio.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay S, Scrutton NS. Good vibrations in enzyme-catalysed reactions. Nat Chem. 2012;4(3):161–168. doi: 10.1038/nchem.1223. [DOI] [PubMed] [Google Scholar]
- 22.Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham’s razor to enzyme dynamics and catalysis. Nat Chem. 2012;4(3):169–176. doi: 10.1038/nchem.1244. [DOI] [PubMed] [Google Scholar]
- 23.Nagel ZD, Klinman JP. Tunneling and dynamics in enzymatic hydride transfer. Chem Rev. 2006;106(8):3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 24.Hu S, et al. Extremely elevated room-temperature kinetic isotope effects quantify the critical role of barrier width in enzymatic C-H activation. J Am Chem Soc. 2014;136(23):8157–8160. doi: 10.1021/ja502726s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinman JP. Importance of protein dynamics during enzymatic C-H bond cleavage catalysis. Biochemistry. 2013;52(12):2068–2077. doi: 10.1021/bi301504m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klinman JP. Looking in new directions for the origins of enzymatic rate accelerations. In: Wüthrich K, Wilson IA, Hilvert D, Wolan DW, Wit AD, editors. New Chemistry and New Opportunities from the Expanding Protein Universe. World Scientific; Singapore: 2014. pp. 64–69. [Google Scholar]
- 27.Rodgers J, Femec DA, Schowen RL. Isotopic mapping of transition-state structural features associated with enzymic catalysis of methyl transfer. J Am Chem Soc. 1982;104(12):3263–3268. [Google Scholar]
- 28.Zheng YJ, Bruice TC. A theoretical examination of the factors controlling the catalytic efficiency of a transmethylation enzyme: Catechol O-methyltransferase. J Am Chem Soc. 1997;119(35):8137–8145. [Google Scholar]
- 29.Kahn K, Bruice TC. Transition-state and ground-state structures and their interaction with the active-site residues in catechol O-methyltransferase. J Am Chem Soc. 2000;122(1):46–51. [Google Scholar]
- 30.Kuhn B, Kollman PA. QM-FE and molecular dynamics calculations on catechol O-methyltransferase: Free energy of activation in the enzyme and in aqueous solution and regioselectivity of the enzyme-catalyzed reaction. J Am Chem Soc. 2000;122(11):2586–2596. [Google Scholar]
- 31.Roca M, et al. Theoretical modeling of enzyme catalytic power: Analysis of “cratic” and electrostatic factors in catechol O-methyltransferase. J Am Chem Soc. 2003;125(25):7726–7737. doi: 10.1021/ja0299497. [DOI] [PubMed] [Google Scholar]
- 32.García-Meseguer R, Zinovjev K, Roca M, Ruiz-Pernía JJ, Tuñón I. Linking electrostatic effects and protein motions in enzymatic catalysis. A theoretical analysis of catechol o-methyltransferase. J Phys Chem B. 2015;119(3):873–882. doi: 10.1021/jp505746x. [DOI] [PubMed] [Google Scholar]
- 33.Lameira J, Bora RP, Chu ZT, Warshel A. Methyltransferases do not work by compression, cratic, or desolvation effects, but by electrostatic preorganization. Proteins. 2015;83(2):318–330. doi: 10.1002/prot.24717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Klinman JP. Enzymatic methyl transfer: Role of an active site residue in generating active site compaction that correlates with catalytic efficiency. J Am Chem Soc. 2011;133(43):17134–17137. doi: 10.1021/ja207467d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warshel A, Weiss RM. An empirical valence bond approach for comparing reactions in solutions and in enzymes. J Am Chem Soc. 1980;102(20):6218–6226. [Google Scholar]
- 36.Kulik HJ, Zhang J, Klinman JP, Martínez TJ. How large should the QM region be in QM/MM calculations? The case of catechol O-methyltransferase 2015 doi: 10.1021/acs.jpcb.6b07814. . arXiv:1505.05730v1 [q-bio.BM] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinman JP, Humphries H, Voet JG. Deduction of kinetic mechanism in multisubstrate enzyme reactions from tritium isotope effects. Application to dopamine beta-hydroxylase. J Biol Chem. 1980;255(24):11648–11651. [PubMed] [Google Scholar]
- 38.Borchard RT. Catechol o-methyltransferase .2. in-vitro inhibition by substituted 8-hydroxyquinolines. J Med Chem. 1973;16(4):382–387. doi: 10.1021/jm00262a016. [DOI] [PubMed] [Google Scholar]
- 39.Meadows CW, Ou R, Klinman JP. Picosecond-resolved fluorescent probes at functionally distinct tryptophans within a thermophilic alcohol dehydrogenase: Relationship of temperature-dependent changes in fluorescence to catalysis. J Phys Chem B. 2014;118(23):6049–6061. doi: 10.1021/jp500825x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monard G, Merz KM. Combined quantum mechanical/molecular mechanical methodologies applied to biomolecular systems. Acc Chem Res. 1999;32(10):904–911. [Google Scholar]
- 41.Gao J, Truhlar DG. Quantum mechanical methods for enzyme kinetics. Annu Rev Phys Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- 42.Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl. 2009;48(7):1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 43.Ranaghan KE, Mulholland AJ. Investigations of enzyme-catalysed reactions with combined quantum mechanics/molecular mechanics (QM/MM) methods. Int Rev Phys Chem. 2010;29(1):65–133. [Google Scholar]
- 44.Liao R-Z, Thiel W. Convergence in the QM-only and QM/MM modeling of enzymatic reactions: A case study for acetylene hydratase. J Comput Chem. 2013;34(27):2389–2397. doi: 10.1002/jcc.23403. [DOI] [PubMed] [Google Scholar]
- 45.Horowitz S, Yesselman JD, Al-Hashimi HM, Trievel RC. Direct evidence for methyl group coordination by carbon-oxygen hydrogen bonds in the lysine methyltransferase SET7/9. J Biol Chem. 2011;286(21):18658–18663. doi: 10.1074/jbc.M111.232876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horowitz S, et al. Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases. J Am Chem Soc. 2013;135(41):15536–15548. doi: 10.1021/ja407140k. [DOI] [PubMed] [Google Scholar]
- 47.Jencks WJ. Catalysis in Chemistry and Enzymology. Dover; Toronto: 1987. pp. 282–287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.