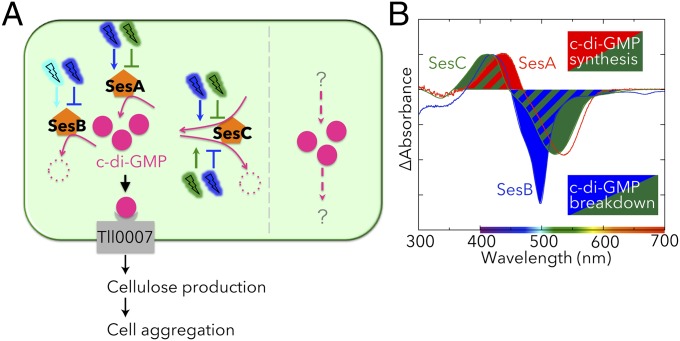
Significance
Cyanobacteria have sophisticated photosensory systems to adapt to ambient-light conditions to improve oxygenic photosynthesis efficiency. Their genomes contain many genes encoding cyanobacteriochromes (CBCRs), which are the photoreceptors of light-signaling pathways. Although the photochemical properties of many CBCRs have been characterized, whether and how multiple photoreceptors work together are unknown. Herein we describe how three CBCRs work together in a light color-sensitive manner to regulate cyanobacterial cell aggregation. The three CBCRs have distinguishable, but congruent, light color-dependent c-di-GMP synthetic and/or degrading activities. Ours is the first report, to our knowledge, concerning synchronization of distinctive CBCR activities, which emphasizes the underlying need for CBCR photoreceptors with diverse activities.
Keywords: photoreceptors, signal transduction, light sensing, second messenger, sessility
Abstract
Cyanobacteriochromes (CBCRs) are cyanobacterial photoreceptors that have diverse spectral properties and domain compositions. Although large numbers of CBCR genes exist in cyanobacterial genomes, no studies have assessed whether multiple CBCRs work together. We recently showed that the diguanylate cyclase (DGC) activity of the CBCR SesA from Thermosynechococcus elongatus is activated by blue-light irradiation and that, when irradiated, SesA, via its product cyclic dimeric GMP (c-di-GMP), induces aggregation of Thermosynechococcus vulcanus cells at a temperature that is suboptimum for single-cell viability. For this report, we first characterize the photobiochemical properties of two additional CBCRs, SesB and SesC. Blue/teal light-responsive SesB has only c-di-GMP phosphodiesterase (PDE) activity, which is up-regulated by teal light and GTP. Blue/green light-responsive SesC has DGC and PDE activities. Its DGC activity is enhanced by blue light, whereas its PDE activity is enhanced by green light. A ΔsesB mutant cannot suppress cell aggregation under teal-green light. A ΔsesC mutant shows a less sensitive cell-aggregation response to ambient light. ΔsesA/ΔsesB/ΔsesC shows partial cell aggregation, which is accompanied by the loss of color dependency, implying that a nonphotoresponsive DGC(s) producing c-di-GMP can also induce the aggregation. The results suggest that SesB enhances the light color dependency of cell aggregation by degrading c-di-GMP, is particularly effective under teal light, and, therefore, seems to counteract the induction of cell aggregation by SesA. In addition, SesC seems to improve signaling specificity as an auxiliary backup to SesA/SesB activities. The coordinated action of these three CBCRs highlights why so many different CBCRs exist.
Cyanobacteria are photoautotrophic prokaryotes that carry out oxygenic photosynthesis. Light exposure is essential for their nutritionally independent growth; therefore, cyanobacteria have a very large and photochemically diverse number of photosensory systems that respond to a broad spectrum of light. When exposed to far-red light, certain cyanobacteria express far-red light–absorbing chlorophylls, namely chlorophyll d and f, to optimize photosynthesis (1). During complementary chromatic acclimation, certain cyanobacteria alter their antenna pigment and protein compositions in response to the ambient red/green-light ratio (2, 3). The ability of cyanobacteria to move toward or away from light (phototaxis) is also a light color-dependent process and is usually controlled by UV/blue- or green/red-light exposure (4–7). Conversely, to date, cell aggregation has been shown to be dependent on only blue-light exposure (8).
Photoreceptors are the proteins that sense ambient light for acclimation. Cyanobacteria contain photoreceptors, denoted cyanobacteriochromes (CBCRs) (9, 10), that contain a covalently bound linear tetrapyrrole (bilin) chromophore in their GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domains. CBCRs respond to a wide spectral range of light from the near UV to the far red (3, 11–14). CBCRs reversibly convert between two photo-induced states according to their bilin chromophore isomerization between its C15-Z and C15-E states (10). Although much is known about the mechanisms of CBCR photoconversions, why CBCRs possess such diverse photochemical properties has not been elucidated.
How CBCRs transduce signals to downstream components also remains mostly unexplored, except for signaling mechanisms of the histidine kinase-type CBCRs CcaS and RcaE (2, 3, 15). The phototaxis regulator Cph2 and the cell-aggregation regulator SesA produce the second-messenger bis(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) upon irradiation of blue light (5, 8). C-di-GMP is universally found in bacteria, including cyanobacteria, and generally induces sessile multicellular lifestyles and represses motility in these organisms (16). It seems that light is a major factor regulating c-di-GMP signaling (5, 8, 17–19). The GGDEF domain usually synthesizes c-di-GMP via its diguanylate cyclase (DGC) activity (20), whereas the EAL and HD-GYP domains usually degrade c-di-GMP via their phosphodiesterase (PDE) activities (21, 22).
A common feature of CBCR-GAF and c-di-GMP synthesis/degradation domains (GGDEF/EAL/HD-GYP) is their large number in bacterial and the majority of freshwater cyanobacterial genomes, especially compared with the numbers of other types of photoreceptors, such as LOV or BLUF. For example, in Synechocystis sp. PCC 6803, there are seven CBCR genes, whereas there is only one each for LOV and BLUF (23). Regarding c-di-GMP domains, there are 29 Escherichia coli, 41 Pseudomonas aeruginosa PAO1, and 28 Synechocystis sp. PCC 6803 genes encoding these domains (16, 19). Many of the c-di-GMP synthesis/degradation proteins have been shown to be specifically involved in a particular response pathway, owing to, for example, transcriptional regulation, protein–protein interaction(s), and variation in the binding affinity of c-di-GMP receptors for c-di-GMP (16). Conversely, little is known about how CBCRs work together and why so many CBCRs are needed in cyanobacteria.
The CBCR SesA from the thermophilic cyanobacterium Thermosynechococcus elongatus has a CBCR-GAF domain activated by blue-light irradiation, and disruption of Thermosynechococcus vulcanus sesA has been shown to inhibit cell aggregation (8). The cellulose synthase T. vulcanus Tll0007, which has also been shown to be essential for cell aggregation (24), contains a c-di-GMP–binding domain (25, 26) and may be the downstream acceptor for SesA-produced c-di-GMP. In addition, although Thermosynechococcus spp. genomes possess five CBCR genes, two of the other four CBCRs are homologs of participants in pathways not involved in cell aggregation in other cyanobacteria (4, 27) and, of the 10 c-di-GMP synthesis/degradation domain proteins, only SesA (Tlr0924), SesB (Tlr1999), and SesC (Tlr0911) contain a photosensory domain. The functions of SesB and SesC to our knowledge had not been characterized before this report, but the presence of a CBCR-GAF domain in these two proteins implied that they might also be involved in the light-regulated cell aggregation.
For this study, we first characterized the photobiochemical properties of SesA, SesB, and SesC from the thermophilic cyanobacteria T. vulcanus and T. elongatus that had been expressed in and purified from a cyanobacterial expression system, and then investigated the effects of disruption of sesA, sesB, and sesC separately and in combination with the temperature-sensitive aggregation of T. vulcanus.
We found coordinated regulation of cell aggregation by SesA, SesB, and SesC via c-di-GMP signaling, which partially explains why multiple CBCR and c-di-GMP synthesis/degradation proteins are present in cyanobacteria.
Results
Photobiochemical Properties of SesA.
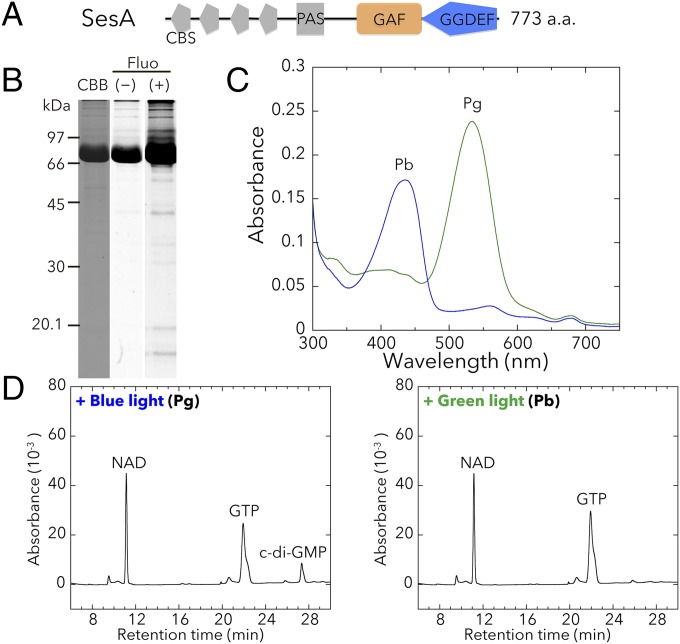
Previously, we reported the physiological role of T. vulcanus sesA together with the photobiochemical properties of SesA protein from the closely related T. elongatus (8). Here, we prepared the full-length T. vulcanus SesA holoprotein (∼90.5 kDa) from a cyanobacterial expression system and confirmed that it is indeed a blue light-induced DGC (Fig. 1 and Fig. S1A).
Fig. 1.
Photobiochemical properties of T. vulcanus SesA. The full-length SesA holoprotein was prepared in and purified from the cyanobacterial expression system. (A) Domain composition of SesA (Tlr0924) deduced by SMART (smart.embl-heidelberg.de). CBS, cystathionine beta synthase; PAS, Per/ARNT/Sim. (B) SDS/PAGE of SesA after Coomassie brilliant blue (CBB) staining and fluorescence (Fluo) in the gel before (–) and after (+) Zn2+ addition. (C) Absorption spectra of native SesA Pb (blue line) after irradiation with green light, and SesA Pg (green line) after irradiation with blue light. (D) HPLC chromatograms assessing SesA DGC activity (GTP→c-di-GMP). NAD served as the internal control. Reaction mixtures, including 100 μM GTP, were incubated for 5 min under blue light (Left) or green light (Right).
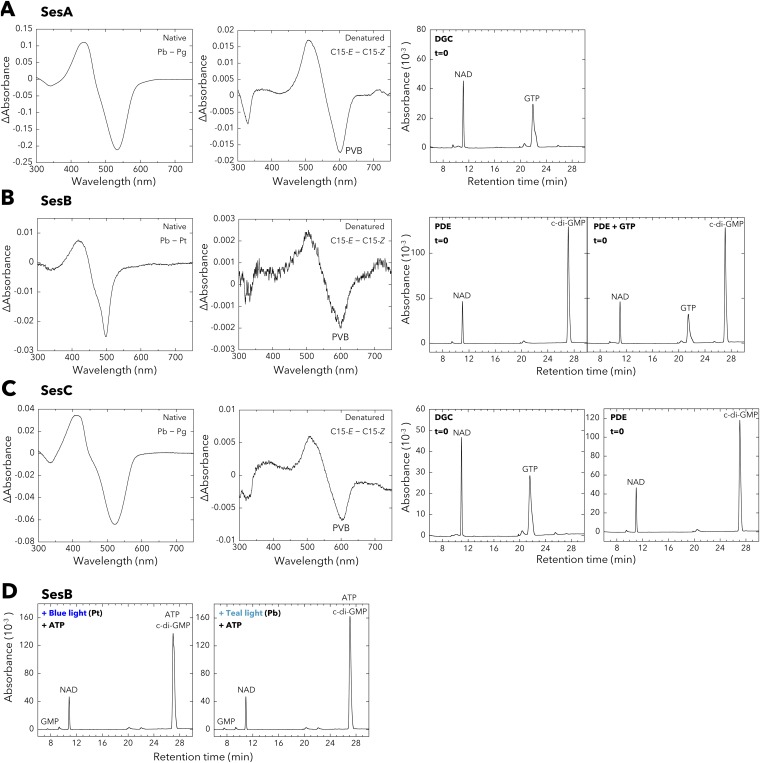
Fig. S1.
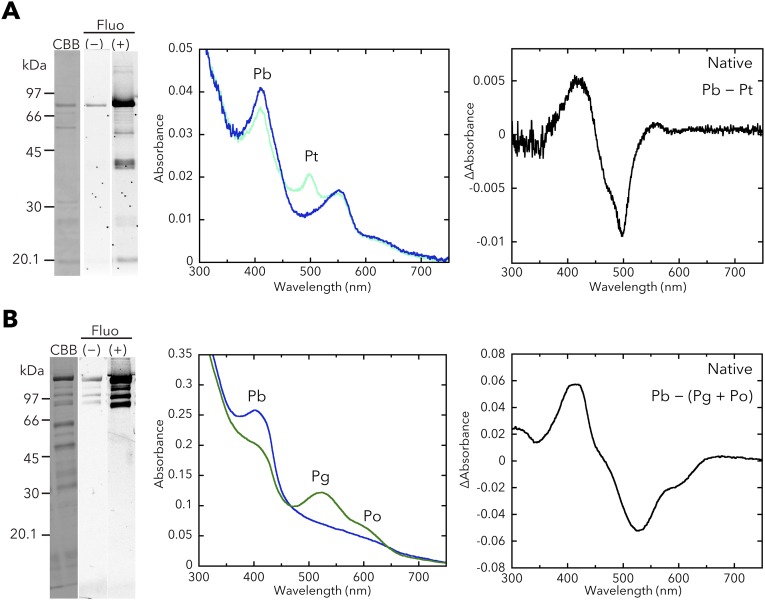
Properties of the full-length T. vulcanus SesA, SesB, and SesC holoproteins expressed in and purified from the cyanobacterial expression system. (A, Left) Difference spectrum of the absorption spectra of native SesA Pb and Pg. (Middle) White light-induced difference spectrum of the chromophore (C15-E – C15-Z) for the urea/acid-denatured SesA. (Right) Control HPLC chromatogram for the DGC assay of SesA (incubation: 0 min). NAD served as the internal control. (B, Left) Difference spectrum of the absorption spectra of native SesB Pb and Pt. (Middle) White light-induced difference spectrum of the chromophore (C15-E – C15-Z) for the urea/acid-denatured SesB. (Right) Control HPLC chromatograms for the PDE assays of SesB (incubation: 0 min). NAD served as the internal control. (C, Left) Difference spectrum of the absorption spectra of native SesC Pb and Pg. (Middle) White light-induced difference spectrum of the chromophore (C15-E – C15-Z) for the urea/acid-denatured SesC. (Right) Control HPLC chromatograms for the DGC/PDE assays of SesC (incubation: 0 min). NAD served as the internal control. (D) SesB is a PDE (c-di-GMP→pGpG) that is not affected by ATP. NAD served as the internal control. Reaction mixtures including 100 µM c-di-GMP with 100 µM ATP were incubated for 10 min under blue light (Left) or teal light (Right).
Spectral Properties.
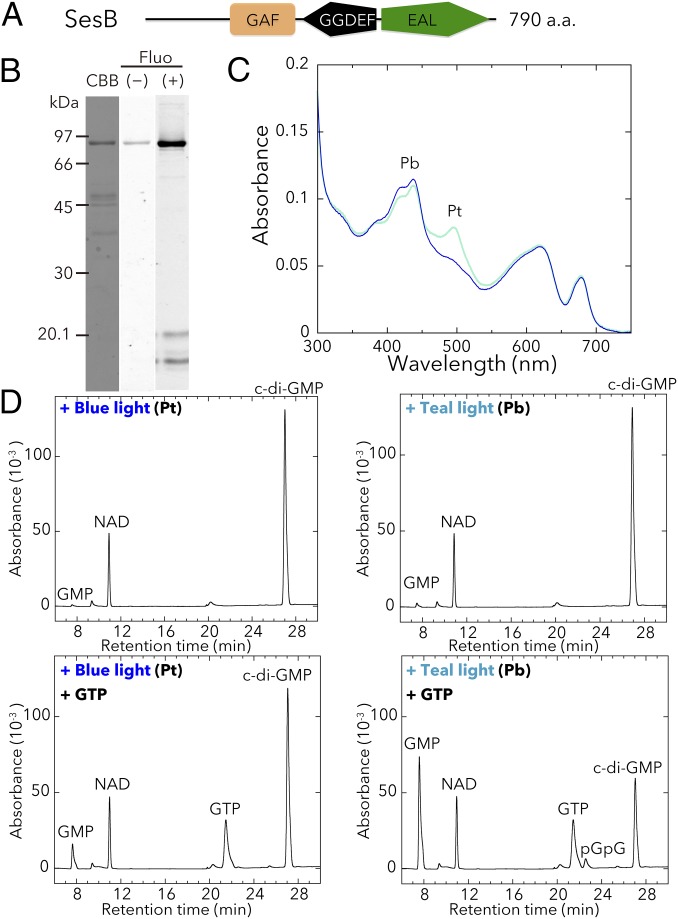
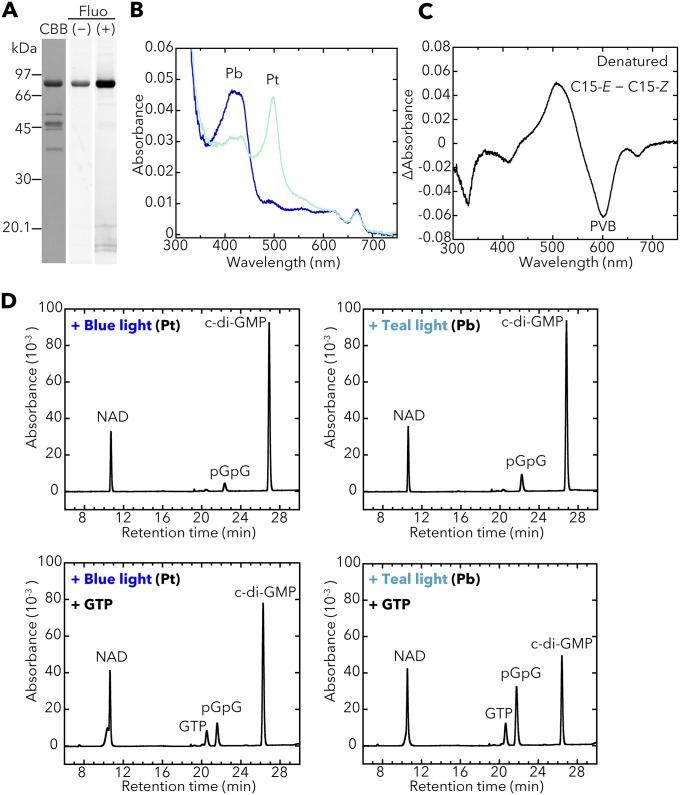
We isolated the full-length T. vulcanus SesB holoprotein (∼93.6 kDa; Fig. 2A) from a cyanobacterial expression system (Fig. 2B) and showed that it reversibly photoconverts between a blue light-absorbing form (Pb; λmax 417 nm) and a teal light-absorbing form (Pt; λmax 498 nm) (Fig. 2C and Fig. S1B). The light-induced difference spectrum of acid/urea-denatured SesB shows that its chromophore is phycoviolobilin (PVB). These results are consistent with our previous analyses on the CBCR-GAF domain of T. elongatus SesB (28).
Fig. 2.
Photobiochemical properties of T. vulcanus SesB. The full-length SesB holoprotein was prepared in and purified from the cyanobacterial expression system. (A) Domain composition of SesB (Tlr1999) deduced by SMART. (B) SDS/PAGE of SesB after CBB staining and the fluorescence in the gel before (–) and after (+) Zn2+ addition. (C) Absorption spectra of native SesB Pb (blue line) after irradiation with teal light, and SesB Pt (teal line) after irradiation with blue light. (D) HPLC chromatograms assessing SesB PDE activity (c-di-GMP→pGpG→GMP). NAD served as the internal control. The reaction mixtures including 100 μM c-di-GMP (substrate) were incubated for 10 min under blue light (Left) or teal light (Right). Addition of 100 μM GTP (Lower) under both light conditions enhanced PDE activity, compared with reactions performed without GTP (Upper).
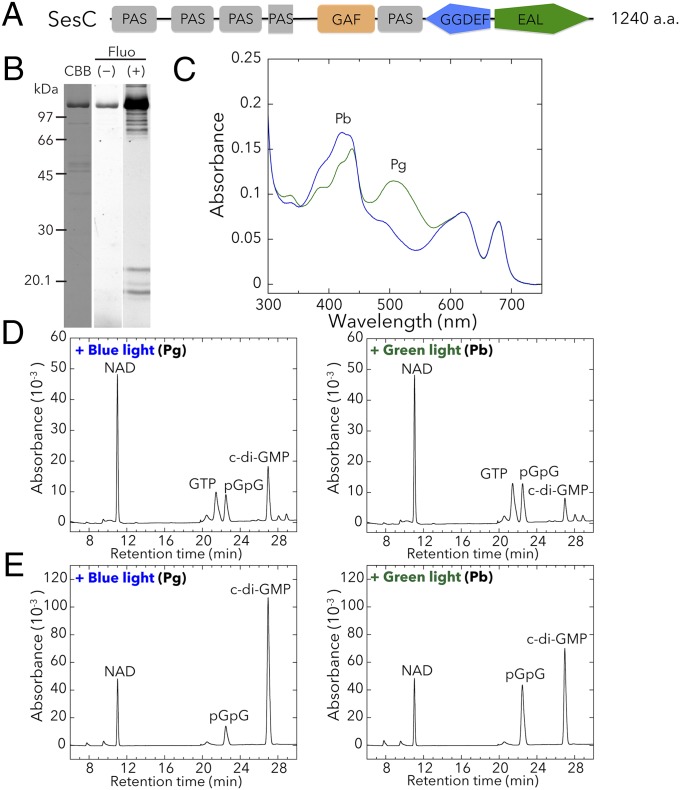
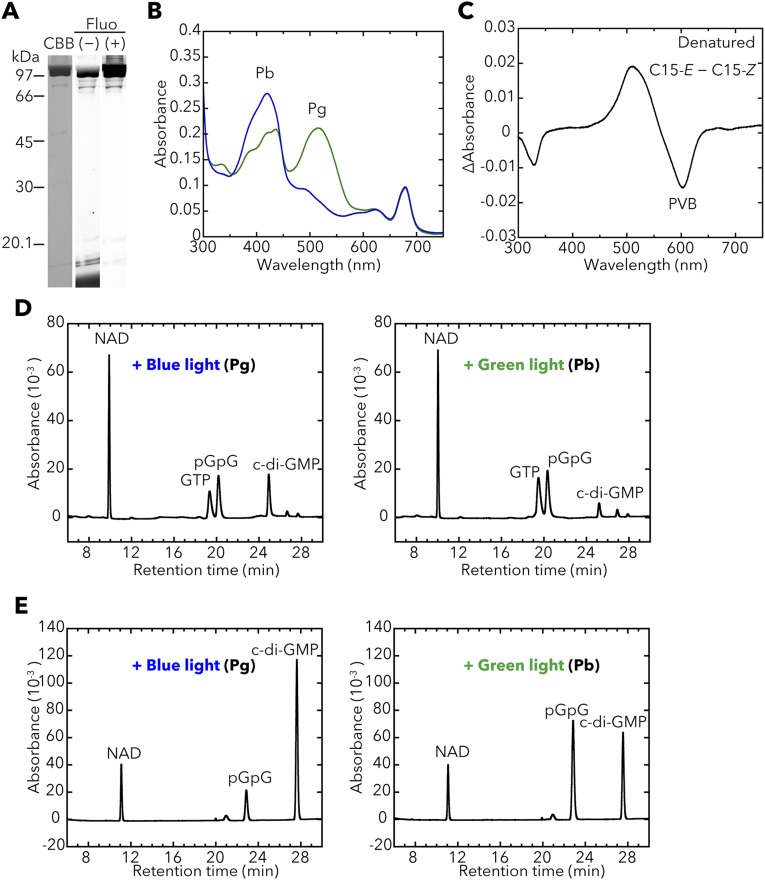
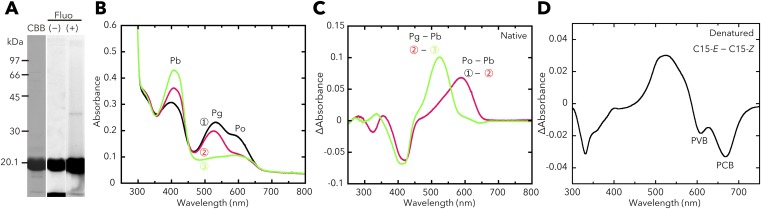
We also prepared the full-length T. vulcanus SesC holoprotein (∼145.7 kDa; Fig. 3A) from cyanobacterial cells (Fig. 3B) and showed that it reversibly photoconverts between a blue light-absorbing form (Pb; λmax 415 nm) and a green light-absorbing form (Pg; λmax 522 nm) (Fig. 3C and Fig. S1C). The bound chromophore is PVB.
Fig. 3.
Photobiochemical properties of T. vulcanus SesC. The full-length SesC holoprotein was prepared in and purified from the cyanobacterial expression system. (A) Domain composition of SesC (Tlr0911) deduced by SMART. (B) SDS/PAGE of SesC after CBB staining and the fluorescence in the gel before (–) and after (+) Zn2+ addition. (C) Absorption spectra of native SesC Pb (blue line) after irradiation with green light, and SesC Pg (green line) after irradiation with blue light. (D) HPLC chromatograms assessing SesC DGC activity (GTP→c-di-GMP). NAD served as the internal control. Reaction mixtures, including 100 μM GTP, were incubated for 10 min under blue light (Left) or green light (Right). pGpG was produced from c-di-GMP, the DGC product, via the accompanying SesC PDE activity. (E) HPLC chromatograms assessing SesC PDE activity (c-di-GMP→pGpG→GMP). NAD served as the internal control. The reaction mixtures, including 100 μM c-di-GMP, were incubated for 10 min under blue light (Left) or green light (Right).
Although there are one and three amino acid substitutions in the T. vulcanus and T. elongatus SesB and SesC homologs, respectively (Fig. S2), T. elongatus SesB and SesC showed spectral properties similar to the corresponding ones in T. vulcanus (Figs. S3 and S4). When prepared in E. coli, all three CBCR proteins (SesA, SesB, and SesC) contain both PVB and phycocyanobilin (PCB) (Fig. S5) (12, 28), and SesA and SesC show two independent photoconversions (Fig. S6) (29). Conversely, when these CBCRs are expressed in cyanobacteria their chromophore is PVB, and they have only a single photoconversion cycle that occurs in a similar spectral window, although their effective wavelengths are distinct.
Fig. S2.
(A) Amino acid sequence alignment of T. vulcanus and T. elongatus SesB. The degenerate motif in the GGDEF domain of SesB is underlined in magenta. The residue that differs in the two sequences is shaded red. (B) Amino acid sequence alignment of T. vulcanus and T. elongatus SesC. The residues that differ in the two sequences are shaded red.
Fig. S3.
Photobiochemical properties of the full-length T. elongatus SesB holoprotein expressed in and purified from the cyanobacterial expression system. (A) SDS/PAGE. CBB, Coomassie brilliant blue stain; Fluo, fluorescence before (–) and after (+) Zn2+ addition. (B) Photoconversion. The SesB Pb absorption spectrum induced by irradiation with teal light is shown in blue. The SesB Pt absorption spectrum induced by irradiation with blue light is shown in teal. (C) The white light-induced difference spectra of the chromophore (C15-E – C15-Z) for the urea/acid-denatured SesB. (D) SesB is a PDE (c-di-GMP→pGpG) activated by teal light and GTP. NAD served as the internal control. Reaction mixtures, including 100 µM c-di-GMP, were incubated for 10 min under blue light (Left) or teal light (Right). Addition of 100 µM GTP (Lower) under both light conditions enhanced PDE activity, compared with reactions performed without GTP (Upper).
Fig. S4.
Photobiochemical properties of the full-length T. elongatus SesC holoprotein expressed in and purified from the cyanobacterial expression system. (A) SDS/PAGE. (B) Photoconversion. The SesC Pb absorption spectrum induced by irradiation with green light is shown in blue. The SesC Pg absorption spectrum induced by blue-light irradiation is shown in green. (C) The white light-induced difference spectra of the chromophore (C15-E – C15-Z) for the urea/acid-denatured SesC. (D) SesC is a DGC (GTP→c-di-GMP) activated by blue light. NAD served as the internal control. Reaction mixtures, including 100 µM GTP, were incubated for 10 min under blue light (Left) or green light (Right). pGpG was produced from the DGC product, c-di-GMP, via SesC PDE activity. (E) SesC is a PDE (c-di-GMP→pGpG) activated by green light. NAD served as the internal control. The reaction mixtures, including 100 μM c-di-GMP, were incubated for 10 min under blue light (Left) or green light (Right).
Fig. S5.
Spectral properties of the full-length T. elongatus SesB and SesC holoproteins prepared in and purified from the E. coli expression system. (A) SesB. (Left) SDS/PAGE of SesB after CBB staining and fluorescence before (–) and after (+) Zn2+ addition. (Middle) Absorption spectra of native SesB Pb (blue line) after irradiation with teal light, and SesB Pt (teal line) after irradiation with blue light. (Right) Difference spectrum for SesB Pb and Pt. (B) SesC. (Left) SDS/PAGE of SesC after CBB staining and fluorescence before (–) and after (+) Zn2+ addition. (Middle) Absorption spectra of native SesC Pb (blue line) after irradiation with green and orange light, and SesC Pg and an orange light-absorbing form (Po) (green line) after irradiation with blue light. (Right) Difference spectrum for SesC Pb and Pg plus Po.
Fig. S6.
Two independent photoconversions of SecC PCB and PVB. The SesC-GAF protein was prepared in and purified from the E. coli expression system. (A) SDS/PAGE of SecC-GAF after CBB staining and fluorescence before (–) and after (+) Zn2+ addition. (B) Absorption spectra of native SesC-GAF after irradiation with blue light (black line; 1), then orange light (red line; 2), and finally green light (green line; 3). (C) Difference spectrum for the native SesC-GAF Pb and Po absorption spectra (red line), representing photoconversion of the PCB-bearing population, and the difference spectrum for the SesC-GAF Pb and Pg absorption spectra (green line), representing photoconversion of the PVB-bearing population. (D) The white light-induced difference spectra of the chromophore (C15-E – C15-Z) for the urea/acid-denatured SesC-GAF, representing the chromophore composition (PCB vs. PVB).
DGC and PDE Activities.
SesB and SesC have GGDEF-type DGC and EAL-type PDE domains that might be involved for signal (c-di-GMP) output. We measured the DGC and PDE activities of the full-length T. vulcanus SesB and SesC holoproteins from the cyanobacterial expression system and found that SesB has no DGC activity under blue- or teal-light conditions but has PDE activity that is enhanced under teal light rather than under blue light (Fig. 2D), indicating that SesB degrades c-di-GMP mainly when exposed to teal light. Therefore, SesB represents a previously unidentified type of CBCR, that is, one that can degrade c-di-GMP. Addition of GTP further substantially enhances this newly uncovered PDE activity of SesB under blue and teal light (Fig. 2D), whereas ATP has no effect (Fig. S1D). In SesB, the consensus GGDEF motif (16) is replaced with GSDEF (Fig. S2A). As mentioned above, SesB does not show DGC activity, suggesting that its degenerate GGDEF domain may act instead as a GTP-binding domain to regulate its PDE activity, which would be functionally similar to that of the degenerate GGDEF domain in CC3396 (PdeA) from Caulobacter crescentus, which has the noncanonical motif GEDEF (21). The teal-on/blue-off PDE activity of SesB is compatible with the blue-on/green-off DGC activity of SesA (8), in that c-di-GMP is increased under blue light and decreased under teal and green light.
SesC has DGC activity that is enhanced under blue light and decreased under green light (Fig. 3D). It also has PDE activity that is enhanced under green light and decreased under blue light (Fig. 3E), which indicates that SesC is a blue/green sensor/regulator of c-di-GMP levels. SesC is, therefore, the first protein, to our knowledge, whose photosensory CBCR-GAF domain appears to regulate two distinct output activities—c-di-GMP synthesis and degradation. Interestingly, the spectral dependency of SesC is also compatible with the aforementioned regulation of c-di-GMP levels by light color.
We confirmed that the SesB and SesC homologs from T. elongatus have photobiochemical properties similar to the corresponding ones in T. vulcanus. T. elongatus SesB is a teal light- and GTP-activated PDE (Fig. S3), and T. elongatus SesC is a blue light-induced DGC/green light-induced PDE bifunctional protein (Fig. S4).
Single-Gene Mutagenesis.
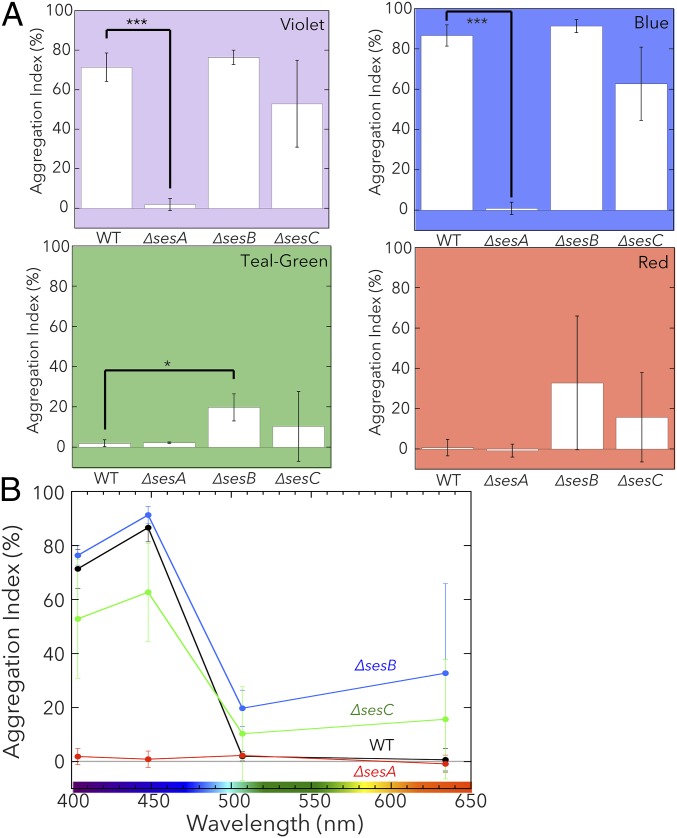
In contrast to the very closely related T. elongatus, T. vulcanus shows cellulose-dependent cell aggregation under blue light and at a low temperature (31 °C) that is not optimal for viability and replication (8, 24). To assess the roles of T. vulcanus SesA, SesB, and SesC in cell aggregation, we disrupted their genes to create strains that lack sesA and/or sesB and/or sesC (ΔsesA, ΔsesB, and ΔsesC, respectively). We cultured these strains at 31 °C for 48 h under violet (λmax 404 nm), blue (λmax 448 nm), teal-green (λmax 507 nm), or red light (λmax 634 nm) at a photon flux density of 5 μmol photon⋅m–2⋅s–1 (Fig. S7). In all cases, the strains were also irradiated with red light (30 μmol photon⋅m–2⋅s–1) to support phototrophic viability without affecting the activities of SesA, SesB, and SesC. To determine the effect of disrupted Ses genes, we calculated the relative number of aggregated to total cells (reported as the aggregation index; %) (24) (Fig. 4A). We also present the aggregation indexes for the mutants as a function of light wavelength (Fig. 4B).
Fig. S7.
Energy spectra of the LEDs used for the cell-aggregation assays. A.U., arbitrary units.
Fig. 4.
Light-induced cell aggregation of the T. vulcanus single gene-disrupted mutants. (A) Aggregation index values for wild type (WT) and its single gene-disrupted mutants (∆sesA, ∆sesB, and ∆sesC) (error bars report the SDs for three biological replicates). Cells were cultured at 31 °C for 48 h under light of a single wavelength (violet light, Upper Left; blue light, Upper Right; teal-green light, Lower Left). The cells were also cultured under only photosynthetic red light (Lower Right). Statistical significance was determined using Student’s t tests (*P < 0.05; ***P < 0.001). (B) Aggregation indexes for the single gene-disrupted mutants as a function of light wavelength using the data shown in A.
Aggregation of wild-type T. vulcanus is strictly dependent on violet- and blue-light irradiation, because no aggregation occurred under teal-green and red light. ΔsesA did not aggregate under any of the light conditions, which is a finding consistent with our previous report (8). ΔsesB showed apparently enhanced cell aggregation under all light conditions, which was significantly different from that of wild type, when cells were exposed to teal-green light. These results indicate that SesB is a negative regulator of cell aggregation and fine-tune the effective light range allowed for cell aggregation upon blue/violet-light irradiation. For ΔsesC, the dependency of cell aggregation on the specific color of light appeared to be weakened but not significantly different from that for wild type. However, the ΔsesC aggregation index fluctuated compared with that of wild type, indicating that SesC improves the specificity of the c-di-GMP signal in the cell-aggregation system.
Double- and Triple-Gene Mutagenesis.
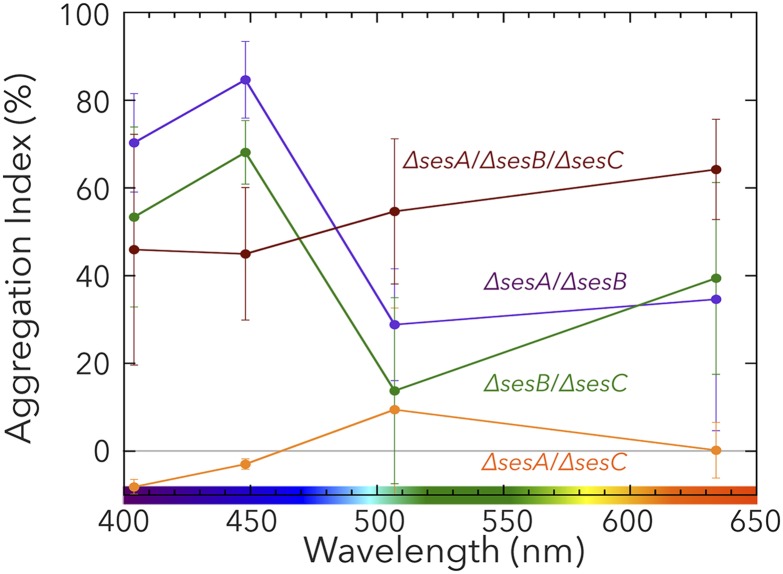
ΔsesA/ΔsesB restored cell aggregation (Fig. 5, violet line), even though ΔsesA could not aggregate under any light-color conditions (Fig. 4). The restored cell aggregation was up-regulated by blue light and down-regulated by teal-green light. Because the cell-aggregation results for ΔsesA/ΔsesB are consistent with the observation that SesC is a dual sensor/regulator of c-di-GMP levels, we hypothesized that SesC in ΔsesA/ΔsesB was responsible for cell aggregation. This hypothesis was strengthened by assessing the effect of ΔsesA/ΔsesB/ΔsesC on cell aggregation. Disruption of all three genes resulted in partial cell aggregation, which was accompanied by the loss of color dependency (Fig. 5, brown line). The uncoupling of the light-color dependency and cell aggregation for ΔsesA/ΔsesB/ΔsesC also implies that no other photoreceptor is involved in cell aggregation. Moreover, because ΔsesA/ΔsesB/ΔsesC can still aggregate, a light-independent, parallel c-di-GMP signaling pathway(s) operating on an unidentified target(s) appears to be present. Thus, SesC improves the specificity of the c-di-GMP signal that induces cell aggregation, possibly by sequestering c-di-GMP from a parallel c-di-GMP pathway(s). In addition to sesA, sesB, and sesC, six other genes exist in the T. vulcanus genome that encode a GGDEF domain, and some may be involved in a parallel pathway(s).
Fig. 5.
Aggregation indexes for the T. vulcanus double (∆sesA/∆sesB, ∆sesA/∆sesC, and ∆sesB/∆sesC) and triple (∆sesA/∆sesB/∆sesC) gene-disrupted mutants as a function of light wavelength. Experiments were performed and the data are plotted as described in the legend for Fig. 4.
Because neither ΔsesA/ΔsesC (Fig. 5, orange line) nor ΔsesA (Fig. 4B, red line) aggregated, it appears that in the absence of SesC, only SesA is capable of triggering cell aggregation and that SesB can only negatively regulate cell aggregation to counteract the effects of SesA, although we cannot rule out that the expression of sesC might be impaired in ΔsesA, as described for RcaE/IflA (30). Cell aggregation of ΔsesB/ΔsesC is increased under blue light and decreased under teal-green light (Fig. 5, dark green line), which is consistent with the presence of SesA providing DGC activity. Under teal-green or red light, however, SesA appears to work as a negative regulator of cell aggregation (compare the results for ΔsesB/ΔsecC and ΔsesA/ΔsesB/ΔsesC in Fig. 5), which implies that SesA might bind and sequester c-di-GMP in the allosteric product-inhibition site of its GGDEF domain (31) that is produced by a parallel pathway(s). In summary, none of the mutants displayed the cell aggregation-related wild-type color sensitivity, which, therefore, underscores our proposal that all three photoreceptors are needed for color-sensitive cell aggregation.
Heterologous Expression of DGC and PDE.
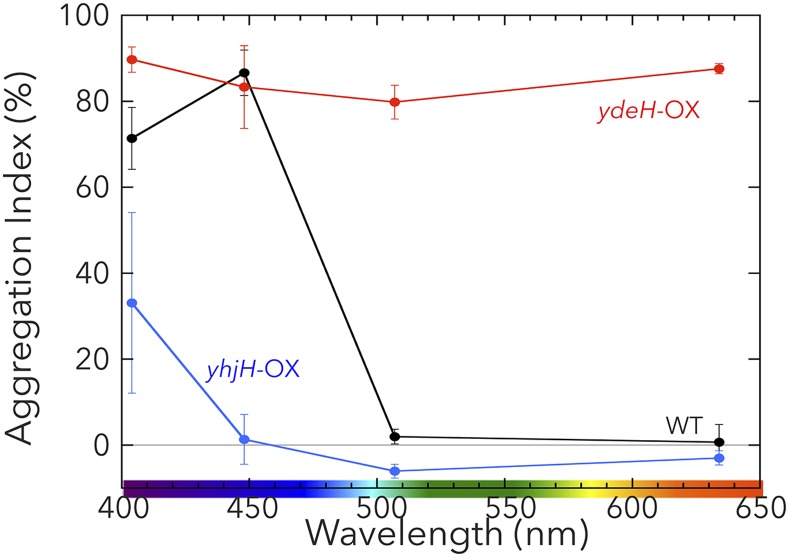
To confirm that c-di-GMP is an activating factor for cell aggregation, we created two heterologous expression mutants, one that expressed DGC-encoding ydeH (32) and one that expressed PDE-encoding yhjH (33), both from E. coli in wild-type T. vulcanus. The mutant that expressed ydeH showed strong cell aggregation under all light colors (Fig. 6). Conversely, the mutant that expressed yhjH showed greatly decreased cell aggregation compared with that of wild type under all light colors (Fig. 6). These results demonstrate that c-di-GMP is, indeed, a critical factor that can trigger cell aggregation and that the dependency of light color on cell aggregation at low temperature(s) is regulated by the c-di-GMP levels produced/degraded by SesA, SesB, and SesC.
Fig. 6.
Aggregation indexes for the heterologous DGC/PDE expression mutants (ydeH-OX and yhjH-OX) as a function of light wavelength. ydeH-OX, the T. vulcanus mutant expressing the E. coli DGC, ydeH. yhjH-OX, the T. vulcanus mutant expressing the E. coli PDE, yhjH. The data for wild type are shown as a black line for comparison. Experiments were performed and the data are plotted as described in the legend for Fig. 4.
Discussion
For this report, we identified and characterized a color-sensitive, cyanobacterial c-di-GMP signaling system composed of three CBCRs: (i) SesA, a blue light-activated DGC; (ii) SesB, a teal light- and GTP-activated PDE; and (iii) SesC, a bifunctional CBCR with DGC activity induced by blue light and PDE activity induced by green light (Fig. 7A). A large number of photoreceptors have been found in cyanobacteria, and the results herein demonstrate that some may work in concert. Multiple blue-light receptors, such as PixA (6, 7), PixJ (4), PixD (34), and Cph2 (5), are known to regulate Synechocystis phototaxis, but their combined effects have not been evaluated. In moss and ferns, chloroplast relocation is regulated by a phytochrome or neochrome photoreceptor with activity regulated by a red/far-red photocycle and a blue light-receptor phototropin (35). However, the effects of these light colors likely cannot be discriminated by moss and ferns, because the activities of the two photoreceptors have been assumed to work under very low light intensity (36).
Fig. 7.
Coordination of the CBCRs SesA, SesB, and SesC that form a color-sensitive, highly specific c-di-GMP signaling system. (A) A signaling model for cell aggregation of Thermosynechococcus modulated SesA, SesB, and SesC. Under blue light, the DGC activities of SesA and SesC increase and the PDE activities of SesB and SesC decrease, leading to an increase in c-di-GMP levels. C-di-GMP binds to the PilZ domain of cellulose synthase Tll0007 and activates it, resulting in cell aggregation. Under teal-green light, the DGC activities of SesA and SesC are decreased and the PDE activities of SesB and SesC are increased, leading to a decrease in c-di-GMP levels. In the absence of c-di-GMP, the cellulose-synthesizing activity of Tll0007 is silenced and cell aggregation is not triggered. Thermosynechococcus probably contains a light-independent c-di-GMP signaling pathway(s), which has not been characterized. (B) The difference spectra for the photoforms of SesA, SesB, and SesC. SesA produces c-di-GMP under blue light but SesB degrades it under teal light, thereby confining the wavelength range that induces an increase in c-di-GMP levels to shorter wavelengths. SesC produces c-di-GMP at shorter wavelengths than does SesA, and degrades it at longer wavelengths than does SesB. SesC can, therefore, broaden the effective wavelength range without impacting the color specificity of c-di-GMP net synthesis.
SesA is the main trigger of cell aggregation in T. vulcanus, because ΔsesA did not aggregate under any of the tested light conditions (Fig. 4). SesB serves to counteract the activity of SesA, because the presence of SesB appears to be necessary for cell-aggregation suppression under all light-color conditions in ΔsesA (Fig. 4) and ΔsesA/ΔsesC (Fig. 5). More specifically, SesB prevents cell aggregation of the teal-green light–irradiated wild-type strain by degrading c-di-GMP, as shown in ΔsesB (Fig. 4). Thus, we concluded that SesB is a light color-specificity enhancer. It is of note that SesB Pt has the narrowest absorption peak, owing to the trapped-twist form of its chromophore (Fig. 2C, teal line) (29, 37). Even though teal light-absorbing CBCRs are commonly found, their physiological roles have not been delineated. It is very likely that, because the SesB Pt peak is sharp and blue-shifted compared with the Pg peaks, SesB can confine the cell-aggregation response to irradiation by shorter wavelengths of light, resulting in color specificity.
We concluded that SesC is a signaling-specificity enhancer because ΔsesC is less sensitive to cell-aggregation responses under all tested light colors (Fig. 4). SesC seems to have an auxiliary backup-type role in comparison with the SesA/SesB pair, because the contribution of SesC to cell aggregation is most noticeable only when sesA and sesB are disrupted (Fig. 5). SesC may help sequester c-di-GMP generated in the cell aggregation-signaling pathway from another parallel c-di-GMP signaling pathway(s). SesC responds to a wider range of light wavelengths than do SesA and SesB (Fig. 7B). SesC produces c-di-GMP under shorter-wavelength conditions than does SesA. SesC also degrades c-di-GMP under longer-wavelength conditions than does SesB. Because of these properties, SesC should broaden the effective range of light wavelengths without deteriorating color specificity. The coordination of the three CBCRs is, therefore, crucial for light wavelength-sensitive cell aggregation. This study provides the first clue, to our knowledge, as to why many CBCRs and c-di-GMP synthesis/degradation proteins are needed in a simple bacterial cell.
Physiologically, photosynthesis is driven by visible light, but blue light also damages the manganese cluster of the oxygen-evolving complex of photosystem II in plants and cyanobacteria (38). Cell aggregation is effective in protection against blue light-induced damage by self-shading especially at relatively low temperature (suboptimal conditions for damage repair), whereas cell aggregation should be avoided to perform photosynthesis effectively under other light conditions. Thus, survival and photosynthetic production could be optimized by the complex light color-sensitive regulatory system.
SesB and SesC as Targets of Intramolecular Signaling.
The SesB GGDEF domain seems to respond to the presence of GTP, and its CBCR-GAF domain responds to blue and teal light. Although further work is needed to confirm the binding of GTP to the GGDEF domain of SesB, both domains seem to regulate its single signal-output domain, which is the EAL domain (Fig. 2D). In SesC, a single CBCR-GAF domain regulates the DGC activity of the GGDEF domain and the PDE activity of the EAL domain in an opposing manner (Fig. 3 D and E). Many GGDEF and EAL domains are present in tandem in a single polypeptide chain (approximately one-third of all GGDEF domains and approximately two-thirds of all EAL domains) (16). In certain of these hybrid proteins, either a GGDEF or EAL domain is the enzymatically active domain; although the other domain is inactive, it may modulate the activity of the neighboring domain. For example, in C. crescentus CC3396 (PdeA), the inactive GGDEF domain still binds GTP and, thereby, enhances the activity of the EAL domain (21). SesB is a composite protein, with an inactive GGDEF domain that can still bind GTP and an N-terminal photosensory CBCR-GAF domain that, under blue and teal light, regulates the PDE activity of its C-terminal EAL domain.
There are a few other GGDEF/EAL hybrid proteins known to have DGC and PDE activities (16). Shewanella woodyi DGC and, probably, Vibrio parahaemolyticus ScrC, regulate both their GGDEF and EAL domain activities independently but with the aid of interacting partner proteins (39, 40). Notably, SesC is the first example, to our knowledge, of a protein containing GGDEF and EAL domains that regulates the activities of these domains via an intramolecular photosensory domain. Generally, proteins containing active GGDEF and EAL domains are dimeric and their dimerization interfaces often modulate their DGC and PDE activities (17, 31, 32). GAF domains including those of CBCRs often transduce the input signal via a rotary movement of a connecting α-helix toward a neighboring output domain (41, 42). Further characterization of SesB and SesC should clarify their intramolecular signaling and any possible intermolecular signaling with interacting proteins, which then might be used to design chimeric sensor proteins.
C-Di-GMP Signaling Specificity.
A major feature of c-di-GMP signaling is redundancy; many c-di-GMP synthesis/degradation domains (GGDEF/EAL/HD-GYP) are found in bacterial genomes, including those of cyanobacteria. A second feature of c-di-GMP signaling is its specificity; c-di-GMP produced or degraded by an individual DGC or PDE regulates a subset of all possible c-di-GMP–regulated responses, even though canonical second messengers, such as c-di-GMP, are thought to be a diffusible intracellular pool of molecules (16). The great specificity of c-di-GMP signaling may be accomplished by intracellular compartmentalization of the various DGC/PDE and c-di-GMP receptor proteins, so that these proteins would have available only a local supply of c-di-GMP (43). Thermosynechococcus spp. may be a suitable system for elucidating the basis for c-di-GMP signaling specificity, because the number of Thermosynechococcus spp. c-di-GMP synthesis/degradation genes is only 10; the cell has a large, elongated rod-like shape, which is suitable for localization studies, and it is thermally stable, which is suitable for biochemical studies.
Materials and Methods
Experimental details are described in SI Materials and Methods, including information on used primers (Table S1) and expression and purification of the CBCRs and their characterization by absorption spectroscopy and in vitro DGC/PDE assays. Information on how the cyanobacterial mutant strains were created and how they were characterized using the cell-aggregation assay is also presented therein.
Table S1.
Primers used in this study
| Purpose | Sequence, 5′–3′ |
| SesB (FL) | CACATATGATTGAGAAACTCCTTGT |
| taggATCCGGTTTGATTGGCAGCACT | |
| SesC (GAF) | GCCATATGGATCTTGAAACCGTTCTC |
| CTGGATCCTAATGTAGCGCACTCACTAAT | |
| SesC (FL) | CACATATGATCATTGACGAATTTG |
| ATGGATCCAGCAAATCCTAAGGCAGG | |
| Introduction of the TEV recognition site into pTCH2031V | TTGAAAATACAAATTTTCCCGACCCATTTGCTGTCCACCA |
| AATTTGTATTTTCAATCCATGATTGAGAAACTCCTTGTTT | |
| Introduction of the TEV recognition site into pET28a | TGAAAATACAGATTTTCGCCGCTGCTGTGATGATGATGA |
| AAATCTGTATTTTCAGAGCCATATGATTGAGAAACTCC | |
| rrnB terminator | CGCAGAAGCGGTCTGATAAA |
| GCGTTCACCGACAAACAACA | |
| ∆sesB | TCTCTATCGCGATTTCGC |
| TGTTGAGGGTGATGATGC | |
| ∆sesC | TCGTCAATGATCATTCCTG |
| TGCCTATGAATTTGCGGG | |
| GTGTTAACGGAACTGCAACAATTGGG | |
| ATCTACGGCCAAAGTGAG | |
| ydeH-OX | AGGAGAGACCATATGATCAAGAAGACAACGG |
| GTGTTCAAACGCAAGTTAAACTCGGTTAATCACAT | |
| yhjH-OX | GAGGAATAAACCATGATAAGGCAGGTTATCC |
| GAGGTTAACAGATCTTTATAGCGCCAGAACCGC |
FL, full length; GAF, GAF domain; OX, overexpression.
SI Materials and Methods
Plasmid Constructions.
Primers used are listed in Table S1. Plasmids were constructed using the In-Fusion System (TaKaRa). To express Thermosynechococcus elongatus or Thermosynechococcus vulcanus SesA, SesB, and SesC individually in E. coli C41 (DE3) or in the cyanobacterium Synechocystis sp. PCC 6803, protein-encoding DNA was cloned into pET28a (Novagen) or pTCH2031V, respectively (28). The protease recognition site in each original plasmid for removal of the N-terminal His tag was replaced with one for tobacco etch virus (TEV) protease, which could be used for future purification. The E. coli rrnB terminator sequence was inserted after the C terminus of the protein-encoding region incorporated into pTCH2031V.
For disruption of sesB (tlr1999) in the thermophilic cyanobacterium T. vulcanus strain RKN (equivalent to National Institute for Environmental Studies 2134) that shows positive phototaxis, a kanamycin-resistance cassette was inserted at the EcoRV site inside the protein-coding region. For disruption of sesC (tlr0911) in T. vulcanus, most of the protein-encoding region was deleted and a spectinomycin/streptomycin-resistance cassette was inserted in its place. The protocol for disruption of sesA (tlr0924) in T. vulcanus has been reported (8). For expression of E. coli ydeH encoding DGC in T. vulcanus, the DNA construct included the psaA promoter (–4 to –622 of psaA of T. vulcanus), ydeH protein-encoding region, and E. coli rrnB terminator sequence and was introduced downstream of tlr2443 as a neutral site. For expression of E. coli PDE yhjH in T. vulcanus, a similar construct was used except that the synthetic promoter trc replaced the psaA promoter.
Cyanobacterial Strains and Cultures.
The thermophilic cyanobacterium T. vulcanus strain RKN that shows positive phototaxis was cultured at 45 °C or 31 °C in BG11 medium as described (8). Culture density was monitored at 730 nm. Transformations of T. vulcanus were performed according to ref. 8. The antibiotic concentration in BG11 medium for selection of transformants was 5 μg⋅mL–1 chloramphenicol, 80 μg⋅mL–1 kanamycin, or 10 μg⋅mL–1 spectinomycin plus 5 μg⋅mL–1 streptomycin.
Protein Purification.
E. coli and Synechocystis protein-expressing cells were harvested by centrifugation at 4,450 × g for 10 min, suspended in 50 mM Hepes⋅NaOH (pH 7.5), 300 mM NaCl, 10% (wt/vol) glycerol, 0.5 mM Tris(2-carboxylethyl)phosphine, and then frozen at −80 °C. After thawing, cells were subjected to three rounds of disruption using a French press (5501-M; Ohtake) at 1,500 kg⋅cm–2. The homogenate from the cyanobacterial cells was centrifuged at 12,000 × g for 10 min and then at 194,100 × g for 30 min. The E. coli homogenate was centrifuged at 194,100 × g for 30 min. Each supernatant was filtered and then loaded onto a nickel-affinity His-Trap chelating column (GE Healthcare). Proteins were eluted with a linear gradient of 30–430 mM imidazole in 20 mM Hepes⋅NaOH (pH 7.5), 300 mM NaCl, 10% (wt/vol) glycerol, 0.5 mM Tris(2-carboxylethyl)phosphine. EDTA (1 mM) was added to the pooled peak fractions, which were then dialyzed against 20 mM Hepes⋅NaOH (pH 7.5), 300 mM NaCl, 10% (wt/vol) glycerol, 1 mM DTT.
SDS/PAGE and Spectroscopy.
SDS/PAGE, followed by Zn2+-enhanced fluorescence detection and Coomassie brilliant blue staining, of recombinant SesB and SesC was performed as described (28). The zinc-induced fluorescence from a protein band in SDS/PAGE gels indicates that a linear tetrapyrrole (bilin) is covalently bound to the protein. Absorption spectra were recorded using a UV-2600PC spectrophotometer (Shimadzu). For the photoconversion of SesB, a blue-light light-emitting diode (LED) (λmax 392 nm; half-bandwidth, 12 nm) and a teal-light LED (λmax 512 nm; half-bandwidth, 34 nm) were used. For the photoconversion of SesC, a blue-light LED and a green-light LED (λmax 539 nm; half-bandwidth, 36 nm) were used. To determine the covalently bound chromophore compositions, the Pt form of SesB and Pg form of SesC proteins (C15-E forms) were first denatured in 8 M urea (pH 2.0) at room temperature in the dark and then irradiated with white light for 3 min. The difference spectra allow distinguishing PVB (negative peak at ∼600 nm) from PCB (negative peak at ∼660 nm) (28).
DGC and PDE Activity Assays.
The reaction mixture for T. elongatus SesB contained 50 mM Tris⋅HCl (pH 8.0), 10 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, 100 μM GTP (Sigma-Aldrich) for DGC or 100 μM c-di-GMP (Biolog) for PDE. For some of the PDE assays, 100 μM GTP or ATP was added. Each reaction was initiated by addition of preirradiated protein (final concentration ∼3 μM) to a prewarmed reaction mixture and incubated at 45 °C under blue or teal light. Each reaction was stopped by addition of EDTA (final concentration 20 mM) and immediately heated at 95 °C for 5 min, followed by centrifugation at 20,400 × g for 5 min. Each supernatant was subjected to HPLC. Reaction mixtures for T. elongatus SesC (0.2 μM), T. vulcanus SesA (0.1 μM), T. vulcanus SesB (0.3 μM), and T. vulcanus SesC (0.1 μM) contained 50 mM Tris⋅HCl (pH 8.0), 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 100 μM GTP for DGC or 100 μM c-di-GMP for PDE. DGC activities of T. elongatus and T. vulcanus SesC were measured at pH 6.8.
Nucleotides were separated by reversed-phase HPLC through a C18 column (150 mm × 6 mm i.d.; DAISOPAK SP-120-5-ODS-AP; Daiso). Samples (20 μL) were injected and eluted at 1.4 mL⋅min–1 using buffer A (100 mM potassium phosphate, 4 mM tetrabutylammonium hydrogen sulfate, pH 6.0) and buffer B (75% buffer A, 25% methanol) (20) and the gradient protocol: 0.0, 0; 2.5, 0; 5.0, 30; 14.0, 40; 25.0, 100; 32.0, 100; 33.0, 50; and 34.0, 0 [the first of each set of values is the time (min) and the second in each set is the percentage of buffer B]. Nucleotides were detected at 254 nm. The internal standard NAD (final concentration 50 μM) was added to each sample just before an injection.
Cell-Aggregation Assays.
Cultures of wild-type T. vulcanus and its disrupted mutants were incubated at 45 °C (OD730 0.5–2) and then diluted to give a culture OD730 of 0.2. These samples were then incubated at 31 °C for 48 h under photosynthetic red light (λmax 634 nm; 30 μmol photon⋅m–2⋅s–1; Valore Corp.; Fig. S7) and, at the same time, some samples were irradiated with violet, blue, or teal-green light (λmax 404, 448, or 507 nm, respectively; 5 μmol photon⋅m–2⋅s–1; Valore; Fig. S7). The results of the cellulose-dependent cell-aggregation assay, described as follows, are reported as an aggregation index (%) (24). Briefly, after the irradiation period, cell suspensions were thoroughly mixed and aliquots were transferred to cuvettes. The samples were held at room temperature for 30 min, during which time most of the aggregated cells had precipitated to the bottom of the cuvettes. Then the OD730 of each sample was measured (denoted ODNA; i.e., OD730 of nonaggregated cells remaining in the culture medium). Next, cellulase (12.5 U⋅mL–1; Worthington Biochemical) was added to each cuvette sample, which were then incubated for 30 min at 37 °C to completely disperse the aggregated cells. The OD730 of each sample (denoted ODtotal) was then measured. The aggregation index (%) is defined as ((ODtotal – ODNA)/ODtotal) × 100.
Supplementary Material
Acknowledgments
We thank Drs. Yu Kanesaki and Hirofumi Yoshikawa (Genome Research Center, NODAI Research Institute, Tokyo University of Agriculture) for providing the nucleic acid sequences of T. vulcanus sesB and sesC. This work was supported by Grants-in-Aid for Japan Society for the Promotion of Science Fellows (to G.E.) and for Scientific Research (to R.N. and M.I.); by Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency (R.N.); and by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency (M.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504228112/-/DCSupplemental.
References
- 1.Gan F, et al. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science. 2014;345(6202):1312–1317. doi: 10.1126/science.1256963. [DOI] [PubMed] [Google Scholar]
- 2.Kehoe DM, Gutu A. Responding to color: The regulation of complementary chromatic adaptation. Annu Rev Plant Biol. 2006;57:127–150. doi: 10.1146/annurev.arplant.57.032905.105215. [DOI] [PubMed] [Google Scholar]
- 3.Hirose Y, et al. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc Natl Acad Sci USA. 2013;110(13):4974–4979. doi: 10.1073/pnas.1302909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshihara S, Ikeuchi M. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol Sci. 2004;3(6):512–518. doi: 10.1039/b402320j. [DOI] [PubMed] [Google Scholar]
- 5.Savakis P, et al. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol. 2012;85(2):239–251. doi: 10.1111/j.1365-2958.2012.08106.x. [DOI] [PubMed] [Google Scholar]
- 6.Song JY, et al. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA. 2011;108(26):10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narikawa R, et al. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011;52(12):2214–2224. doi: 10.1093/pcp/pcr155. [DOI] [PubMed] [Google Scholar]
- 8.Enomoto G, et al. Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J Biol Chem. 2014;289(36):24801–24809. doi: 10.1074/jbc.M114.583674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeuchi M, Ishizuka T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci. 2008;7(10):1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 10.Rockwell NC, Lagarias JC. A brief history of phytochromes. ChemPhysChem. 2010;11(6):1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA. 2011;108(29):11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012;51(7):1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 13.Narikawa R, Enomoto G, Win N-N, Fushimi K, Ikeuchi M. A new type of dual-Cys cyanobacteriochrome GAF domain found in cyanobacterium Acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry. 2014;53(31):5051–5059. doi: 10.1021/bi500376b. [DOI] [PubMed] [Google Scholar]
- 14.Narikawa R, et al. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Sci Rep. 2015;5:7950. doi: 10.1038/srep07950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci USA. 2008;105(28):9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barends TR, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459(7249):1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 18.Gomelsky M, Hoff WD. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 2011;19(9):441–448. doi: 10.1016/j.tim.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Agostoni M, Koestler BJ, Waters CM, Williams BL, Montgomery BL. Occurrence of cyclic di-GMP-modulating output domains in cyanobacteria: An illuminating perspective. MBio. 2013;4(4):e00451-13. doi: 10.1128/mBio.00451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187(5):1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280(35):30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 22.Ryan RP, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA. 2006;103(17):6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Mandalari C, Losi A, Gärtner W. Distance-tree analysis, distribution and co-presence of bilin- and flavin-binding prokaryotic photoreceptors for visible light. Photochem Photobiol Sci. 2013;12(7):1144–1157. doi: 10.1039/c3pp25404f. [DOI] [PubMed] [Google Scholar]
- 24.Kawano Y, et al. Cellulose accumulation and a cellulose synthase gene are responsible for cell aggregation in the cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol. 2011;52(6):957–966. doi: 10.1093/pcp/pcr047. [DOI] [PubMed] [Google Scholar]
- 25.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22(1):3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 26.Morgan JL, McNamara JT, Zimmer J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol. 2014;21(5):489–496. doi: 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narikawa R, Kohchi T, Ikeuchi M. Characterization of the photoactive GAF domain of the CikA homolog (SyCikA, Slr1969) of the cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol Sci. 2008;7(10):1253–1259. doi: 10.1039/b811214b. [DOI] [PubMed] [Google Scholar]
- 28.Enomoto G, Hirose Y, Narikawa R, Ikeuchi M. Thiol-based photocycle of the blue and teal light-sensing cyanobacteriochrome Tlr1999. Biochemistry. 2012;51(14):3050–3058. doi: 10.1021/bi300020u. [DOI] [PubMed] [Google Scholar]
- 29.Rockwell NC, Martin SS, Lagarias JC. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry. 2012;51(17):3576–3585. doi: 10.1021/bi300171s. [DOI] [PubMed] [Google Scholar]
- 30.Bussell AN, Kehoe DM. Control of a four-color sensing photoreceptor by a two-color sensing photoreceptor reveals complex light regulation in cyanobacteria. Proc Natl Acad Sci USA. 2013;110(31):12834–12839. doi: 10.1073/pnas.1303371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan C, et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101(49):17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zähringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure. 2013;21(7):1149–1157. doi: 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 33.Pesavento C, et al. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22(17):2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okajima K, et al. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J Biochem. 2005;137(6):741–750. doi: 10.1093/jb/mvi089. [DOI] [PubMed] [Google Scholar]
- 35.Hughes J. Phytochrome cytoplasmic signaling. Annu Rev Plant Biol. 2013;64:377–402. doi: 10.1146/annurev-arplant-050312-120045. [DOI] [PubMed] [Google Scholar]
- 36.Kawai H, et al. Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature. 2003;421(6920):287–290. doi: 10.1038/nature01310. [DOI] [PubMed] [Google Scholar]
- 37.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry. 2014;53(19):3118–3130. doi: 10.1021/bi500037a. [DOI] [PubMed] [Google Scholar]
- 38.Nishiyama Y, Allakhverdiev SI, Murata N. Protein synthesis is the primary target of reactive oxygen species in the photoinhibition of photosystem II. Physiol Plant. 2011;142(1):35–46. doi: 10.1111/j.1399-3054.2011.01457.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu N, et al. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry. 2012;51(10):2087–2099. doi: 10.1021/bi201753f. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol. 2008;190(3):851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narikawa R, et al. Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc Natl Acad Sci USA. 2013;110(3):918–923. doi: 10.1073/pnas.1212098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasser C, et al. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci USA. 2014;111(24):8803–8808. doi: 10.1073/pnas.1321600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol. 2011;407(5):633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]