Significance
TRAIL (TNF-related apoptosis-inducing ligand) is a promising antitumor agent effective in a very small subset of lung cancer patients with low toxicity. However, the majority of lung tumors are TRAIL-resistant and very little is known about how tumor cells acquire resistance to TRAIL. Here, we show that continuous exposure to subtoxic concentrations of TRAIL induces NF-κB–dependent up-regulation of miR-21, miR-30c, and miR-100, which by silencing caspase-8, caspase-3, TRAF7, and FoxO3a further strengthens the NF-κB signaling, inducing acquired TRAIL resistance. Our findings imply that combinatory therapies of NF-κB inhibitors and TRAIL might be a useful therapy to improve the response of lung cancer to TRAIL.
Keywords: microRNAs, acquired TRAIL-resistance, lung cancer
Abstract
TRAIL (TNF-related apoptosis-inducing ligand) is a promising anticancer agent that can be potentially used as an alternative or complementary therapy because of its specific antitumor activity. However, TRAIL can also stimulate the proliferation of cancer cells through the activation of NF-κB, but the exact mechanism is still poorly understood. In this study, we show that chronic exposure to subtoxic concentrations of TRAIL results in acquired resistance. This resistance is associated with the increase in miR-21, miR-30c, and miR-100 expression, which target tumor-suppressor genes fundamental in the response to TRAIL. Importantly, down-regulation of caspase-8 by miR-21 blocks receptor interacting protein-1 cleavage and induces the activation of NF-κB, which regulates these miRNAs. Thus, TRAIL activates a positive feedback loop that sustains the acquired resistance and causes an aggressive phenotype. Finally, we prove that combinatory treatment of NF-κB inhibitors and TRAIL is able to revert resistance and reduce tumor growth, with important consequences for the clinical practice.
TRAIL (TNF-related apoptosis-inducing ligand) is a member of the TNF superfamily and is considered a potential anticancer agent, as it shows selective high cytotoxicity toward tumor cells and little or no toxicity against normal cells (1). The anticancer activity of TRAIL is ascribable to its ability to induce apoptosis through the binding of DR4 and DR5 and the recruitment of intracellular apoptosis-initiating caspase-8 through Fas-associated death domain for the assembly of the death-inducing signaling complex (DISC). The cleavage of caspase-8 in the DISC is a critical upstream event in TNF family ligand-induced apoptosis. The other two receptors, DcR1 and DcR2, are “decoy receptors” and lack the ability to initiate the apoptotic cascade. Cross-linking of TRAIL to its death receptors leads to the recruitment of Fas-associated death domain, procaspase-8, and procaspase-10 to form DISC (2, 3). Recombinant TRAIL and agonistic antibodies directed at DRs in phase-II clinical trials have shown monotherapy activity with isolated responses reported in follicular lymphoma (4), adenocarcinoma of the lung (5), and synovial sarcoma (6).
However, recent studies have demonstrated that many types of cancer cells possess primary or acquired resistance to TRAIL. Moreover, TRAIL has been found to activate a prosurvival transcription factor, NF-κB, and enhance metastasis in apoptosis-resistant cancer cells (7–9). Receptor interacting protein (RIP) kinases constitute a family of seven members, all of which contain a kinase domain. These kinases are crucial regulators of cell survival and death (10, 11). Particularly, RIP1 serine/threonine kinase plays an important role in the TRAIL dualism. In apoptosis-sensitive cells, caspase-8 cleaves RIP1 in response to TNF, resulting in rapid RIP1 depletion and induction of apoptosis. However, in apoptosis-resistant cells, RIP1 is constitutively active and activates the NF-κB pathway (10, 12). In line with this finding is the observation that the activation of NF-κB is blocked by RIP dominant-negative mutants (13, 14), whereas cultured cells lacking RIP1 are highly sensitive to TNF-induced cell death, probably because they are unable to activate the prosurvival transcription factor NF-κB (15). However, little is known about the involvement of these proteins in acquired TRAIL resistance.
Lung cancer is the leading cause of cancer-related mortality, not only in the United States but also around the world. Most stage-III nonsmall cell lung cancer (NSCLC) express either or both TRAIL receptors, raising the possibility of using TRAIL as an anticancer drug, but understanding the molecular mechanisms of primary and acquired TRAIL resistance is necessary for a successful TRAIL therapy (7, 8). The most well-known small noncoding RNAs (ncRNAs) are microRNAs (miRNAs), single-stranded RNAs of 19–25 nt in length that negatively regulate gene expression by translational inhibition or degradation of the mRNA target. A large body of evidence suggests that miRNAs are involved in development of chemosensitivity or resistance in various tumors (16, 17).
Here, we show that acquired TRAIL resistance is mediated by a positive feedback loop involving the NF-κB transcription factor, miR-21, miR-30c, and miR-100 and their respective targets caspase-8, caspase-3, Forkhead Box O3a (FoxO3a), and TNF receptor-associated factor 7 (TRAF-7). Importantly, the combination of TRAIL and NF-κB inhibitors drastically triggers apoptosis in TRAIL-resistant cells in vitro and in vivo, suggesting that combinatory treatment could be effective in overcoming TRAIL resistance in lung cancer.
Results
MicroRNA Dysregulation in Cells with Acquired TRAIL Resistance.
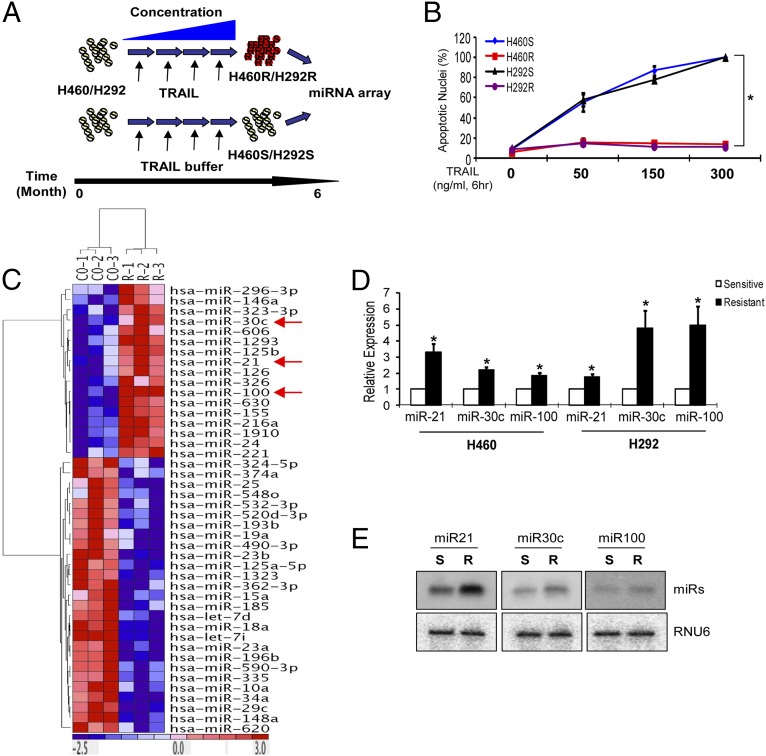
Although the many molecules that regulate the TRAIL signaling cascade are known, the mechanisms by which tumor cells become more resistant to TRAIL is still not clear. To this end, we generated TRAIL-resistant cells (H460R and H292R) by exposing H460 and H292-sensitive cells (H460S and H292S) to stepwise increases in TRAIL concentrations (1–500 ng/mL) over a period of 6 mo to select cells capable of growing at high concentrations of TRAIL (Fig. 1A). The establishment of the resistant cells was verified by counting the apoptotic nuclei after TRAIL treatment (Fig. 1B). TRAIL receptor isoforms analyzed by Western blot (SI Appendix, Fig. S1A) or quantitative RT-PCR (qRT-PCR) (SI Appendix, Fig. S1B) revealed comparable levels of expression in TRAIL-sensitive and -resistant cells. Moreover, sequencing did not show any mutations, possible because of chronic exposure to the drug, in the DR4 and DR5 cDNAs of both H460R and H292R compared with the sensitive parental cells. We have previously reported the involvement of miRNAs in the innate resistance of NSCLC cell lines to TRAIL (16). Therefore, we analyzed the miRNA expression profile in H460R versus H460S using the nanoString technology. Several miRNAs were found dysregulated (Fig. 1C) and we focused on three miRNAs with the highest fold change in expression: miR-21, miR-30c, and miR-100. qRT-PCR and Northern blot analyses confirmed the overexpression of the three miRNAs in H460R compared with H460S cells (Fig. 1 D and E).
Fig. 1.
Identification of dysregulated miRNAs in cells with acquired TRAIL resistance. (A) A schematic diagram showing generation of acquired TRAIL-resistant cells. (B) Cell death measurement of H460R and H292R cells after TRAIL treatment. H406S/R and H292S/R cells were exposed to TRAIL at the indicated time and concentration. Cell death rates were determined by counting apoptotic nuclei after staining with Hoechst dye. Bars indicate mean ± SD (n = 3). P values were obtained by two tailed Student t test (*P < 0.01). (C) MiRNA heatmap showing dysregulated miRNAs in H460S versus H460R cells. P values were obtained by ANOVA test (<0.001). (D) qRT-PCR analysis of miR-21, miR-30c, and miR-100 in H460S/R and H292S/R cells. Bars indicate mean ± SD (n = 3). P values were obtained by two tailed Student t test (*P < 0.01). (E) Northern blot analysis showing miR-21, miR-30c, and miR-100 up-regulation in H460R compared with H460S cells.
MiR-21, miR-30, and miR-100 Modulate the Response to TRAIL by Targeting Fundamental Tumor-Suppressor Genes.
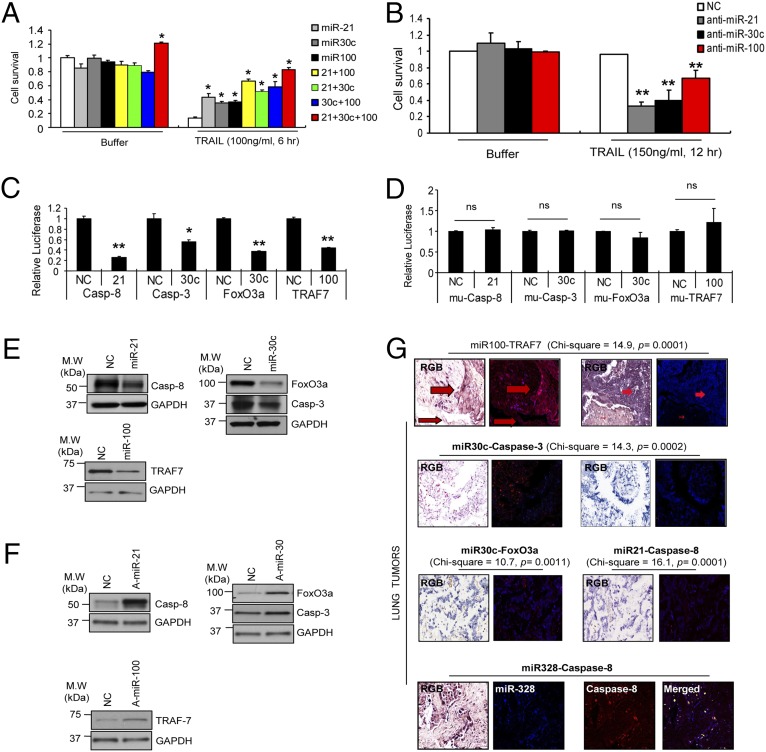
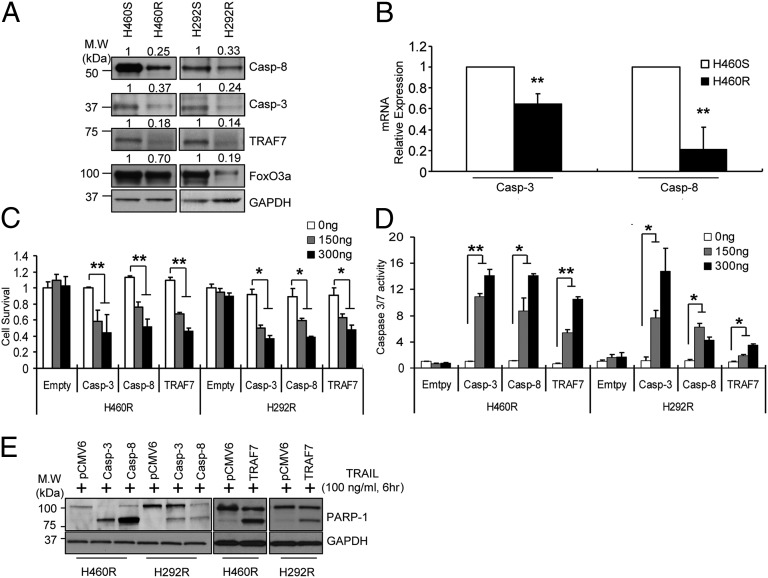
To investigate the role of miR-21, miR-30c, and miR-100 in acquired TRAIL resistance, we overexpressed these miRNAs in H460S cells. Enforced expression of each individual miRNA drastically reduced the sensitivity to TRAIL-induced apoptosis, as assessed by MTS and caspase-3/7 assays. Moreover, combined ectopic expression of all three miRNAs further suppressed TRAIL-induced cell death (Fig. 2A and SI Appendix, Fig. S1C). The antiapoptotic effect of the three microRNAs was confirmed by Western blot analysis for poly(ADP-ribose) polymerase (PARP)-1 cleavage (SI Appendix, Fig. S1D). Conversely, knockdown of miR-21, miR-30c, and miR-100 by anti-miR oligonucleotides in H460R cells induced a very low proliferation rate (Fig. 2B) and increased caspase-3/7 activity (SI Appendix, Fig. S1E). Identical results were obtained in H292S and H292R cells (SI Appendix, Fig. S1 F–I), suggesting that miR-21, miR-30c, and miR-100 are involved in the resistance to TRAIL-induced cell death. A gene-expression profile was analyzed to highlight the global changes in gene expression in H460R compared with H460S cells. Down-regulation of important tumor suppressor genes, such as caspase-8 and caspase-9, and up-regulation of oncogenes, such as PI3K and Cyclins, were observed (SI Appendix, Fig. S2). Our search for mRNA targets of miR-21, miR-30c, and miR-100 using Targetscan and Pictar computational tools revealed that 3′ UTRs of human caspase-8, caspase-3, TRAF-7, and FoxO3a genes contained evolutionarily conserved binding sites specific for these miRNAs (SI Appendix, Fig. S3A). We focused on these genes because they are tumor suppressors with defined roles in apoptosis and in the TRAIL pathway. TRAF-7, the unique, noncanonical member of the TRAF family, physically associates with RelA/p65 (p65) member of the NF-κB transcription factor family inducing its polyubiquitination, which results in lysosomal degradation of the protein, repressing NF-κB activation (18). The forkhead transcription factors (FoxOs) play pivotal roles in tumorigenesis and in mediating chemotherapy sensitivity (19). To verify that caspase-8, caspase-3, FoxO3a, and TRAF-7 are direct targets of miR-21, miR-30c, and miR-100, 3′UTRs were cloned downstream of the luciferase ORF. These reporter constructs were cotransfected along with the respective miRNA in HEK293 cells. Increased expression of these miRNAs significantly reduced luciferase expression, measured as relative luciferase activity (Fig. 2C). Target gene repression was rescued by deletions of the complementary seed sites (Fig. 2D), confirming that caspase-8, caspase-3, FoxO3a, and TRAF-7 are direct targets of miR-21, miR-30c, and miR-100. To determine whether these miRNAs affect the target gene expression in the H460S cellular environment, we analyzed the consequences of their ectopic expression in H460S cells. MiR-21, miR-30c, or miR-100 enforced expression significantly reduced the endogenous levels of caspase-8, caspase-3, FoxO3a, and TRAF-7 (Fig. 2E). Intriguingly, by qRT-PCR, we found that only caspase-3 and caspase-8 (SI Appendix, Fig. S3B) were down-regulated in H460S cells after miRNA overexpression, indicating that miR-21 and miR-30c induce the degradation of caspase-8 and caspase-3 mRNAs, whereas FoxO3a and TRAF-7 are regulated at the translational level. Conversely, stable knockdown of miR-21, miR-30c, and miR-100 by lentivector-based anti-miRNAs in H460R cells or knockdown by anti-miR oligonucleotides, confirmed by qRT-PCR, increased mRNA (SI Appendix, Fig. S3C) and protein levels of the target genes (Fig. 2F). To verify these findings in vivo, in situ hybridization analysis was performed using 5′-dig-labeled LNA probes on lung cancer tissues, followed by immunohistochemistry for the target proteins. These analyses were done initially on serial sections so as to compare the results on the same groups of cells, followed by coexpression analysis. Coexpression analysis indicated that when both the miRNA and protein were found in the same core, they were being expressed in different groups of cancer cells (Fig. 2G and SI Appendix, Fig. S3D). Next, to evaluate the contribution of the target genes in acquired TRAIL resistance, we compared their endogenous levels in H460R and H292R versus H460S and H292S cells. Western blot analysis clearly indicated a down-regulation of the target genes in the resistant cell lines (Fig. 3A). As expected, caspase-3 and caspase-8 (Fig. 3B) but not FoxO3a and TRAF-7 mRNAs (SI Appendix, Fig. S4A) were also down-regulated in H460R and H292R cells. Furthermore, enforced expression of caspase-8, caspase-3, TRAF-7, and FoxO3a in H460R and H292R cells, decreased cell viability (Fig. 3C and SI Appendix, Fig. S4B), increased caspase-3/7 activity (Fig. 3D and SI Appendix, Fig. S4C), and enhanced PARP-1 cleavage (Fig. 3E) compared with the cells transfected with an empty vector. These data suggest that these genes play a fundamental role in TRAIL-induced cell death.
Fig. 2.
MiR-21, miR-30c, and miR-100 confer acquired TRAIL-resistance by silencing caspase-8, caspase-3, FoxO3a, and TRAF-7. (A) Increased cell survival in TRAIL-sensitive H460 cells in response to TRAIL after miR-21, miR-30c, and miR-100 overexpression or combinatorial expression of the miRNAs. The error bars indicate mean ± SD (n = 5) and the P values were calculated by two-tailed Student t test (*P < 0.001). (B) Decreased cell survival in TRAIL-resistant cells after miR-21, miR-30c, and miR-100 knockdown by anti-miR oligonucelotides in response to TRAIL. The error bars indicate mean ± SD (n = 3) and the P values were calculated by two-tailed Student t test (**P < 0.02). (C and D) caspase-8, caspase-3, FoxO3a, and TRAF-7 3′UTRs are direct targets of miR-21, miR-30c, and miR-100. pGL3-caspase-3, pGL3-caspase-8, pGL3-FoxO3a, and pGL3-TRAF-7 luciferase constructs, containing wild-type (C) or mutated (D) 3′UTRs, were transfected into HEK293 cells. Bars indicate mean ± SD (n = 3) and the P values were addressed by two-tailed Student t test (*P < 0.01, **P < 0.005); ns, not significant. (E) Down-regulation of caspase-8, caspase-3 FoxO3a, and TRAF-7 by ectopic expression of miR-21, miR-30c, and miR-100. (F) Increased expression of the target genes in lenti–anti-miR-21, anti–miR-30c, and anti–miR-100 H460R stable infected cells. (G) Coexpression analysis of miRNAs and target genes. The top row shows coexpression analysis of miR-100 and TRAF-7. The first and third images are the regular light microscopy views after colocalization analysis of the miRNA and protein (RGB). The small arrows point to the stroma and large arrows to the cancer cells. The other panels show the Nuance fluorescent-based images where the miRNA is fluorescent blue and the protein fluorescent red. Note that the expression of TRAF-7 and miR-100 are mutually exclusive; strong TRAF-7- expression is seen in the cancer cells in the first set of images, whereas strong miR-100 expression is seen in the stroma of those images and only in the cancer cells in the last set of images. The second row shows miRNA-30c (blue/fluorescent blue) and caspase-3 coanalysis (red/fluorescent red). In the first set of images, only caspase-3 expression is evident, whereas in an adjacent core only miR-30c expression is evident (second set of images). The third row shows the Nuance converted individual protein expression (fluorescent red), the individual miR-30c (fluorescent blue) and the merged images of miR-30c/FoxO3a and miR-21/caspase-8. Note the lack of coexpression in the merged images (no fluorescent yellow). The bottom images show coexpression between miR-328 (made by most of the cancer cores) and caspase-8, as a positive control for coexpressions. The Pearson’s χ2 test was done using the InStat Software for Windows (v3.36) testing the null hypothesis that the probability of the expression of a given protein (caspase-3, -8, TRAF7, or Fox03a) in cancer cells would be independent of the expression of the specific miRNA of interest. The 2 × 2 table analysis used the continuity correction and the null hypothesis was rejected if the significance level at 1 df was below 1%. In each case listed the P value for rejection of the null hypothesis was strongly significant. (Magnification, 400×.)
Fig. 3.
Caspase-8, caspase-3, and TRAF-7 overexpression increases sensitivity to TRAIL in cells with acquired resistance. (A) Down-regulation of caspase-8, caspase-3, FoxO3a, and TRAF-7 in TRAIL-resistant cells. The bands were quantified by ImageJ software and the relative values normalized by the value of each GAPDH. (B) qRT-PCR showing caspase-3 and caspase-8 mRNAs down-regulation in H460R compared with H460S cells. (C–E) Proliferation (C), caspase-3/7 assay (D), and Western blot showing PARP-1 cleavage (E) after caspase-3, caspase-8, and TRAF-7 overexpression in TRAIL-resistant cells. Error bars indicate mean ± SD (n = 3) and the P values were calculated by two-tailed Student t test (*P < 0.02, **P < 0.01).
NF-κB Transcriptionally Activates miR-21, miR-30c, and miR-100.
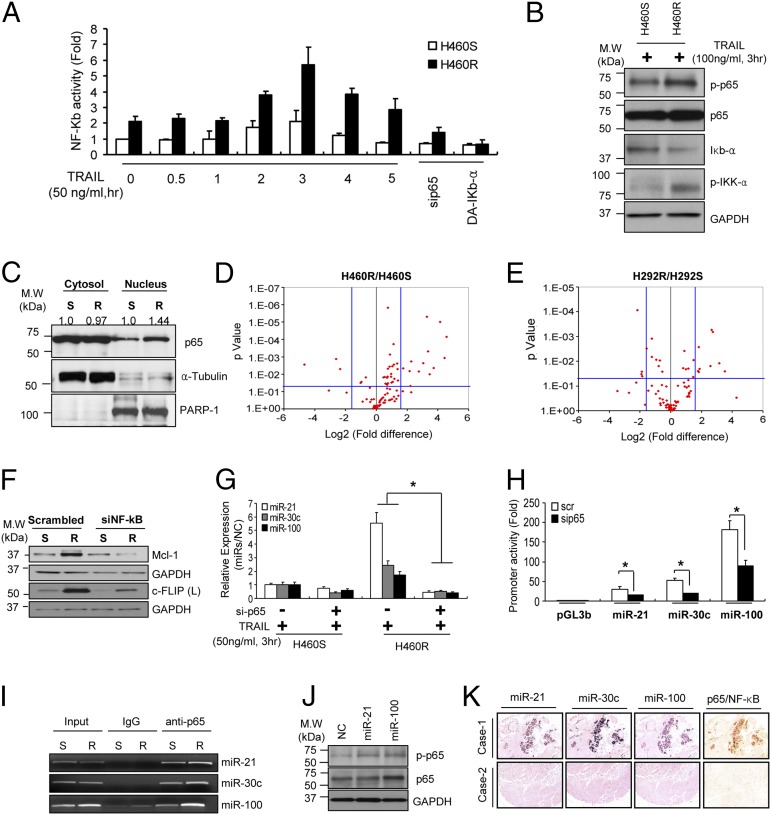
The transcription factor NF-κB is broadly associated with oncogenesis, including control of metastasis and angiogenesis (20, 21). It has been shown that primary TRAIL-resistant cells usually have higher NF-κB/p65 activity (7, 22–25); therefore, we examined NF-κB activation in acquired TRAIL-resistant cells. A pGL4.3-luc2p/NF-κB construct containing five copies of an NF-κB response element that drives transcription of the luciferase reporter gene was transfected into H460S and H460R cells stimulated with TRAIL at different times. Basal NF-κB activity was higher in H460R compared with H460S cells, with the highest peak after 3 h of TRAIL treatment. A drop in NF-κB activity was observed after NF-κB silencing or the overexpression of dominant active IκB-α (Fig. 4A). Moreover, Western blot confirmed a higher phosporylation of NF-κB and IKK-α in H460R cells, whereas IκB-α was down-modulated in H460R cells after TRAIL stimulation compared with control cells (Fig. 4B). Similar results were observed in H292R cells (SI Appendix, Fig. S5A). Next, we analyzed NF-κB nuclear translocation in both H460S and H460R cells. Cells were incubated with TRAIL for 3 h and then nucleo-cytosol separation was performed. Western blot showed higher NF-κB expression in the nuclei of H460R compared with the H460S cells (Fig. 4C). Hundreds of NF-κB target genes have been identified by ChIP, as well as bioinformatic analyses based on predicted binding sites. We used a qRT-PCR array (SABiosciences) to profile the expression of 84 key genes responsive to NF-κB in H460S and H460R (Fig. 4 D and E). We found that antiapoptotic genes, such as BCL2A1, BIRC2, XIAP, and genes involved in inflammation and immune responses were up-modulated in H460R and H292R cells (SI Appendix, Table S1). In addition, Western blot analysis showed increased expression of NF-κB transcriptionally activated TRAIL antagonists, such as Mcl-1 and c-FLIP, which were suppressed after NF-κB down-regulations (9, 26–29) in H460R compared with H460S cells (Fig. 4F). Then, we addressed whether NF-κB could be involved in miR-21, miR-30c, and miR-100 transcriptional activation. First, endogenous levels of miR-21, miR-30c, and miR-100 were higher in TRAIL-resistant cells following TRAIL-stimulation and consistently decreased after NF-κB knockdown (Fig. 4G). By bioinformatics search (PROMO database: alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3; TESS database: www.cbil.upenn.edu/tess), we found that miR-21, miR-30c, and miR-100 had NF-κB binding sites located ∼−1,280 bp (miR-21), ∼3,730 bp, ∼4,180 bp (miR-30c), and ∼−3,600 bp (miR-100) upstream of the premiRNAs 5′ ends, respectively. To determine whether the identified NF-κB sites were located in promoter regions, we cloned the NF-κB binding sites into the pGL3basic vector, which harbors the promoterless luciferase gene. These constructs were cotransfected along with NF-κB siRNA in H460R cells. Subsequent luciferase assays showed that NF-κB silencing gave rise to a 40–50% reduction in luciferase activity, whereas inactivation of the NF-κB binding site by site-direct mutagenesis reduced luciferase activity compared with cells transfected with NF-κB wild type (Fig. 4H and SI Appendix, Fig. S5B). Because we noticed that the promoter regions were responsive to NF-κB modulation, to verify a direct binding of NF-κB on miR-21, miR-30c, and miR-100 promoters we carried out ChIP assays. As expected, ChIP assays of H460R cells showed considerable NF-κB binding at the predicted analyzed regions for miR-21, miR-30c, and miR-100 (Fig. 4I). Taken together, these data led us to conclude that NF-κB is the transcription factor responsible for miR-21, miR-30c, and miR-100 transcriptional activation in NSCLC.
Fig. 4.
NF-κB directly activates miR-21, miR-30c and miR-100. (A) Increased NF-κB promoter activity in H460R cells in response to TRAIL stimulation. NF-κB activity was assessed by measuring firefly luciferase activity. (B) Western blot analysis showing NF-κB activation in H460R after TRAIL treatment. (C) Cytosol-nucleus fractionation of H460S and H460R cells to determine NF-κB localization. H460S and H460R cells were treated with 50 ng/mL of TRAIL. After 3 h a cytosol-nucleus extraction kit (Cell Biolabs) was used to separate nuclear and cytosolic proteins. Anti-tubulin and anti–PARP-1 antibodies were used for cytosolic and nuclear marker, respectively. (D and E) Volcano plots of NF-κB target genes. Total RNAs of H460S/R (D) and H292S/R (E) were subjected to qRT-PCR in plates equipped with primer sets for 84 NF-κB target genes. P values were obtained by three independent experiments. Fold-difference is reported as H460R/H460S and H292R/H292S. (F) Western blot analysis of NF-κB antiapoptotic targets in control and NF-κB knockdown H460S and H460R cells. (G) NF-κB silencing reduces miR-21, miR-30c, and miR-100 expression levels. The values were normalized with a negative control. Error bars represent mean ± SD (n = 4) and P values were calculated by two-tailed Student t test or ANOVA test (*P < 0.001). (H) NF-κB activity on miR-21, miR-30c, and miR-100 promoters. The promoter regions of miR-21, miR-30c, or miR-100 containing NF-κB binding sites were subcloned into pGL3-luciferase vectors. The constructs were transfected into H460R cells along with a scrambled or NF-κB p65 siRNA. After 48 h, promoter activity was assessed by measuring firefly luciferase activity. Error bars represent mean ± SD (n = 3) and P values were obtained by two-tailed Student t test (*P < 0.01). (I) ChIP showing a direct interaction between NF-κB/p65 and miR-21, miR-30c, and miR-100 promoter regions. (J) Increased phosphorylation of NF-κB/p65 in H460S cells overexpressing miR-21 and miR-100. (K) Coexpression analysis of miRNAs and NF-κB/p65. The images are serial sections from core 1 (Upper) and core 2 (Lower). Note that the same cancer cells in the top row express miR-21, miR-30c, miR-100, and NF-κB/p65. The cancer cells in the core in the bottom row do not express both miRNA and the protein. (Magnification, 100×.)
Finally, because caspase-8 and TRAF-7 inhibit NF-κB activation and they are directly down-regulated by miR-21 and miR-100, we questioned if miR-21 and miR-100 overexpression could induce NF-κB activation. In this regard, the pGL4.3-luc2p/NF-κB construct was cotransfected along with miR-21 and miR-100 in H292S cells. Luciferase assays showed an increased luciferase activity compared with the cells transfected with a scrambled miRNA (SI Appendix, Fig. S5C). Moreover, immunoblot analysis displayed an increase in NF-κB phosphorylation after miR-21 and miR-100 enforced expression (Fig. 4J). To test whether this was a general mechanism and not specific to these cells we analyzed NF-κB phosphorylation levels in cells with primary TRAIL resistance. Intriguingly, NF-κB phosphorylation was higher in A549 and Calu-1 compared with the H460S and H292S cell lines in response to TRAIL stimulation (SI Appendix, Fig. S5D). Moreover, NF-κB silencing induced a down-regulation of miR-21, miR-30c, and miR-100 also in the cells with de novo TRAIL resistance (SI Appendix, Fig. S5 E and F). Importantly, caspase-8, caspase-3, and TRAF-7 overexpression sensitized also primary resistant NSCLC cells to TRAIL treatment (SI Appendix, Fig. S5 G–J). To verify the correlation between miRNAs and NF-κB in lung cancer tissues, in situ hybridization analysis was performed using 5′-dig–labeled LNA probes for miR-21 and miR-100 followed by immunohistochemistry analysis for NF-κB/p65. As shown in Fig. 4K, miR-21, miR-30c, and miR-100 showed strong correlation with NF-κB protein in lung cancer tissues (Fig. 4K and SI Appendix, Fig. S5K). These data suggest the activation of NF-κB by miR-21 and miR-100 and the presence of a positive feedback loop.
NF-κB and TRAIL Signaling.
It is known that the TNF receptor-1 associated death-domain protein (TRADD)-TNF-receptor associated factor (TRAF)-NF-κB signaling cascade acts downstream of TRAIL receptors. TRADD has a survival role in TRAIL signaling and deficiency of TRADD sensitizes cells to TRAIL-induced apoptosis in innate TRAIL resistance (30). TRAF-2 has recently been shown to have a positive role in the canonical pathway that activates NF-κB (23). AKT is one of the main effectors of NF-κB activation (31, 32). We found an increased phosphorylation of AKT and ERKs and increased expression of TRADD and TRAF-2 in H460R and H292R cells in response to TRAIL stimulation compared with H460S (SI Appendix, Fig. S6 A–C). Knockdown of TRAF-2, TRADD, or NF-κB in H460R cells decreased cell survival rate, increased caspase-3/7 activity and induced PARP-1 cleavage (SI Appendix, Fig. S6 D–F), suggesting that NF-κB–dependent cell survival signal is conserved in acquired TRAIL-resistant cells. Interestingly, AKT and not ERKs inhibition sensitized the resistant cells to TRAIL-induced cell death (SI Appendix, Fig. S6 G and H).
Caspase 8-Dependent RIP1 Cleavage Plays an Important Role in Acquired TRAIL Resistance.
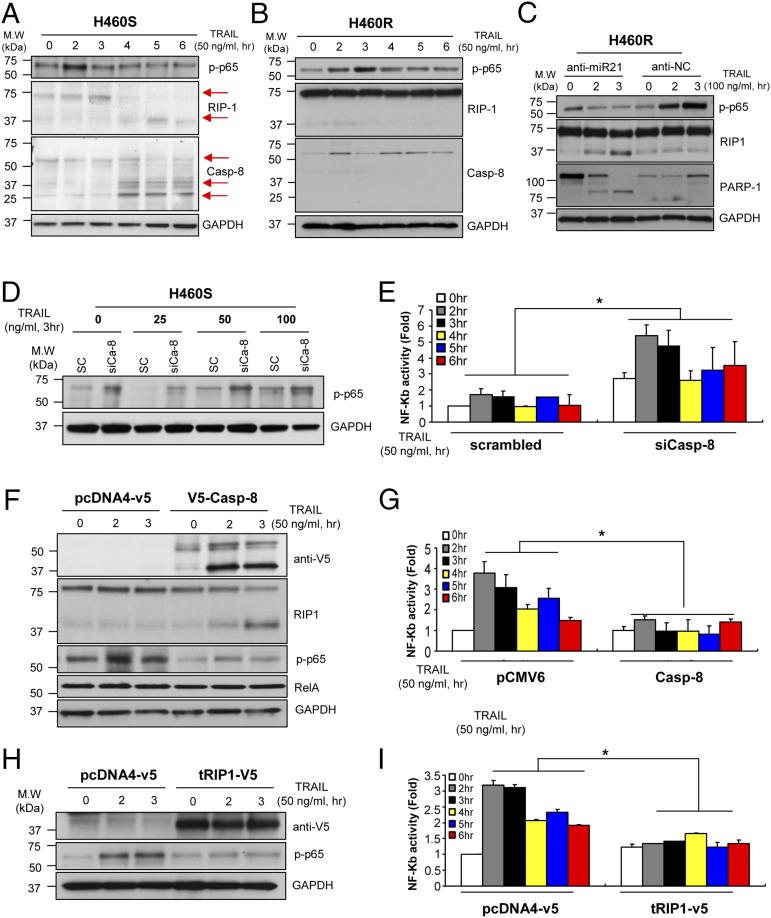
The death domain kinase RIP1, a key component of the TNF signaling complex, is cleaved by caspase-8 in TNF-induced apoptosis, resulting in the blockage of TNF-induced NF-κB activation (10, 13–15, 33). Therefore, we investigated the role of caspase-8–dependent RIP1 cleavage in the acquired TRAIL-resistant system. First, H460S cells were exposed to TRAIL at different times for up to 6 h and, as expected, a decrease in NF-κB phosphorylation and an increase in cleaved RIP1 and caspase-8 activation starting 4 h after TRAIL-treatment were observed (Fig. 5A). Conversely, RIP1 cleavage and caspase-8 activation were not detected in H460R cells in response to TRAIL stimulation (Fig. 5B). Remarkably, we observed a decrease in NF-κB phosphorylation and enhanced RIP1 cleavage in H460R cells transfected with anti–miR-21 in response to TRAIL treatment (Fig. 5C). In addition, phosphorylation levels of NF-κB were higher in caspase-8 knockdown H460S cells (Fig. 5D). Conversely, ectopic expression of pCMV-caspase-8 in H460R cells suppressed NF-κB phosphorylation and increased RIP1 cleavage (Fig. 5F). In addition, NF-κB promoter activity was enhanced in H292S cells transfected with caspase-8 siRNA and reduced in H292R cells overexpressing caspase-8 (Fig. 5 E and G). Finally, we cloned the RIP1 death domain into a pcDNA4-V5 vector (tRIP1-V5). Overexpression of this construct in H460R cells significantly decreased NF-κB phosphorylation (Fig. 5H). Moreover, NF-κB promoter activity was suppressed in H292R cells after enforced expression of cleaved RIP1 compared with the cells transfected with an empty vector (Fig. 5I). MTS and caspase-3/7 assays showed that overexpression of RIP1 death domain induced apoptosis in both H460R and H292R cells (SI Appendix, Fig. S7). Taken together, these data strongly suggest that caspase-8–dependent RIP1 cleavage driven by miR-21 blocks NF-κB activation in NSCLC cells with acquired TRAIL resistance and the use of an anti–miR-21 oligonucleotide can sensitize acquired resistant cells to TRAIL-induced apoptosis.
Fig. 5.
Caspase-8–dependent RIP1 cleavage blocks NF-κB activation in TRAIL-resistant cells. (A and B) Increased NF-κB phosphorylation in H460R cells (B) and not in H460S (A) cells after TRAIL treatment. The sensitive and resistant cells were exposed to 50 ng/mL of TRAIL at the indicated time points. Cells were harvested and analyzed by Western blot for p-p65, RIP1 cleavage, and caspase-8 activation. (C) MiR-21 knockdown by anti-miR oligonucleotides in H460R cells reduces p-p65 phosphorylation and increases RIP1 and PARP-1 cleavage. (D and E) Increased NF-κB phosphorylation in H460S (D) and activity in H292S (E) after caspase-8 knockdown. (F and G) Increased RIP1 cleavage and reduced NF-κB phosphorylation in H460R (F) and activity in H292R (G) cells overexpressing caspase-8. (H and I) Enforced expression of the C-terminal fragment of RIP1 (tRIP1) inhibit NF-κB activation. pcDNA4-tRIP1-v5 vector were transfected into H460R cells. After 24 h, cells were stimulated with 50 ng of TRAIL for 2 and 3 h and analyzed by Western blot (H) and luciferase assay to assess NF-κB activity (I). Error bars indicate mean ± SD (n = 4) and P values were calculated by ANOVA test (*P < 0.001).
Acquired TRAIL Resistance Induces Epithelial-Mesenchymal Transition and Aggressive Cancer Phenotype.
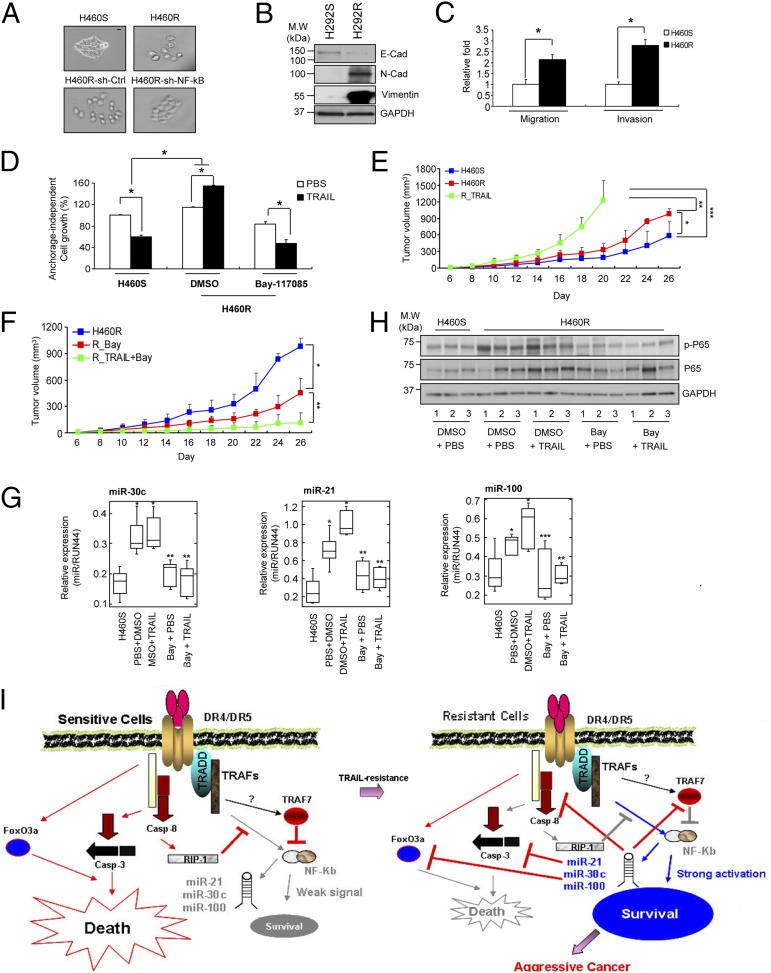
Epithelial-mesenchymal transition (EMT) is associated with reduced sensitivity to many chemotherapeutic drugs (34). We observed an impressive change in cellular shape of H460R cells from an epithelial phenotype to a spindle-fibroblastoid morphology, which was rescued by the knockdown of NF-κB (Fig. 6A). Therefore, we further investigated whether this morphological change could be because of an EMT. We assessed the expression of key EMT-associated markers and observed increased expression of mesenchymal markers and decreased E-cadherin expression (Fig. 6B and SI Appendix, Fig. S8A). In addition, H460R and H292R showed higher migratory and invasive capabilities compared with the parental cells (Fig. 6C and SI Appendix, Fig. S8B). Anchorage-independent growth is one of the hallmarks of cell transformation. Thus, H460R and H292R were plated in soft agar to verify their colony formation ability. H460R and H292R cells presented higher growth rates than their control cells. Interestingly, they displayed increased colony growth in response to TRAIL but the colony growth was dramatically reduced when they were cotreated with TRAIL and the irreversible NF-κB inhibitor Bay-117085 (Fig. 6D and SI Appendix, Fig. S8C). Finally, to analyze tumor growth and the response to TRAIL-induced apoptosis in vivo, we injected H460S and H460R in the right side of nude mice. Mice injected with H460R cells presented tumors slightly but significantly enlarged compared with those injected with the H460S cells. Furthermore, tumor size dramatically increased when H460R-injected mice were treated with TRAIL (Fig. 6E and SI Appendix, Fig. S8D). Of note, combinatory treatment of TRAIL and Bay-117085 showed a highly antitumor effect compared with the mice treated only with the inhibitor (Fig. 6F and SI Appendix, Fig. S8E). The same results were obtained in mice injected with H292R cells (SI Appendix, Fig. S8 F and G). MiR-21, miR-30c, and miR-100 expression in the tumor samples was analyzed by qRT-PCR. MiRNA expression levels were higher in H460R-derived tumors and reduced in Bay-117085 or Bay-117085 + TRAIL-treated groups (Fig. 6G). Consistently, NF-κB phosphorylation was reduced in the Bay-117085 or Bay-117085 + TRAIL-treated tumors (Fig. 6H). These results indicate that acquired TRAIL resistance induces a more aggressive cancer phenotype in vitro and in vivo and the use of NF-κB inhibitors can improve the response of cancer cells to TRAIL-induced apoptosis.
Fig. 6.
Acquired TRAIL resistance induces EMT. (A) Morphological changes in H460R compared with H460S. The phenotype is rescued after NF-κB silencing. (Scale bar, 20 μm.) (B) Western blot showing EMT markers in H292S and H292R cells. (C) Enhanced migration and invasion capacity of acquired TRAIL-resistant cells. Bars show mean ± SD (n = 4) and P values were calculated by paired Student t test (*P < 0.02). (D) Anchorage independent growth assay of TRAIL-resistant cells in response to TRAIL and the NF-κB inhibitor, Bay 117085 treatments. Bars show mean ± SD (n = 3) and P values were calculated by paired Student t test (*P < 0.01). (E) Growth curve of engrafted tumors injected with H460S and H460R. 1 × 106 of H460S or H460R cells were subcutaneously injected into three groups of nude mice. One group of the H460R injected mice was treated intraperitoneally with 10 mg/kg of TRAIL for 2 wk. Error bars indicate mean ± SD (n = 4) and P values were calculated by paired Student t test (*P = 0.022, **P = 0.005, and ***P = 0.003). (F) Growth curve of engrafted tumors in nude mice injected with H460R cells; 1 × 106 of H460R cells were subcutaneously injected into three groups of nude mice. Next, 5 mg/kg of Bay-117085 were administered intraperitoneally twice per week for 2 wk and 10 mg/kg of TRAIL were injected intraperitoneally daily for 2 wk. Combination of TRAIL and the NF-κB inhibitor Bay-117085 reduced tumor volumes compared with a control group treated with PBS and DMSO only. Error bars show mean ± SD (n = 4) and P values were calculated by paired Student t test (*P = 0.027, **P = 0.006). (G) qRT-PCR showing miR-21, miR-30c, and miR-100 expression in the tumors. Three tumors were isolated from each group and prepared for qRT-PCR analysis. P values were calculated by ANOVA test or paired Student t test. (*P < 0.001, **P < 0.005, and (***P = 0.011). (H) Significant phospho-p65 (p-p65) down-regulation in the xenograft tumors treated with TRAIL plus Bay-117085. Tumors from three mice of each group were minced and analyzed by Western blot with the indicated antibodies. (I) Working model: NF-κB induces acquired TRAIL resistance and EMT through miR-21, miR-30c, and miR-100 activation. In TRAIL-sensitive cells, the binding of TRAIL to its receptors induces cell death through the canonical caspase-cascade. In cells with acquired TRAIL-resistance subtoxic concentrations of TRAIL are not enough to induce apoptosis and lead to NF-κB activation, which in turn, transcriptionally regulates miR-21, miR-30c and miR-100. Down-regulation of caspase-8, casapse-3, FoxO3a and TRAF-7 by these microRNAs causes acquired resistance to TRAIL and EMT.
Our working model summarizes the results of this study. In TRAIL-sensitive cells, caspase-8 cleaves RIP1; therefore, NF-κB activation is weak and apoptosis is triggered after TRAIL treatment. Continuous TRAIL treatment induces constitutive NF-κB activation, which leads to the accumulation of miR-21, miR-30c, and miR-100. Caspase-8 and TRAF-7 down-modulation by miR-21 and miR-100 further strengthens the NF-κB signaling, establishing a positive feedback loop that leads to TRAIL resistance and EMT (Fig. 6I).
Discussion
Drug resistance is a major obstacle in cancer therapy, and it can be either intrinsic or acquired during therapy. So far, no strategy has been found to overcome resistance, which is based on highly complex and individually variable biological mechanisms. Lung cancer still represents a very deadly disease in strong need of new, effective therapeutic approaches. The long-term survival for patients with advanced high-grade lung cancer has been limited by the occurrence of resistance to chemotherapeutic drugs. In this context, TRAIL may represent an alternative therapeutic molecule. Several TRAIL agonists have been used in clinical trials showing efficacy in a small fraction of lung cancer patients (5, 6). However, the majority of NSCLC patients are resistant or, as with other molecularly targeted agents, resistance is likely to develop later during therapy. Despite massive studies for intrinsic TRAIL resistance, the molecular mechanisms underlying the acquired TRAIL resistance is still unclear and little is known on how lung cancer can acquire resistance to TRAIL (7, 35). Therefore, it is fundamental to identify biomarkers to predict the response to the drug and to improve its therapeutic efficacy, using drug combinations that not only synergize with TRAIL but that might also overcome resistance as it arises (36). To understand the mechanisms involved in acquired TRAIL resistance, we established TRAIL-resistant human isogenic cell lines (H460R and H292R) from the parental TRAIL-sensitive (H460S and H292S) cells. Alterations in the expression levels of TRAIL-R1 and TRAIL-R2 or decoy receptors have been associated with the modulation of TRAIL-induced cell death in different systems (37–39). Both cell lines H460R and H292R had similar TRAIL receptor expression levels compared with the corresponding sensitive cells, and did not show any mutation in their sequences, suggesting that the resistance may be linked to the alteration of downstream pathways.
miRNAs, single-stranded RNAs of 19–25 nt in length, are key players in cancer onset and progression. Recent data demonstrate that selective modulation of miRNA activity can improve the response to chemotherapy (17, 40). Therefore, we compared the miRNA expression profile of H460R and H460S cells. Several miRNAs, including miR-21, miR-30c, and miR-100, were found drastically up-regulated in TRAIL-resistant compared with TRAIL-sensitive cells. miR-21 is a very well-known oncomiR that inhibits multiple tumor-suppressor genes in different tumors including NSCLC (41, 42). MiR-30 family is regulated by EGF and MET receptors, and induces resistance to tyrosine kinase inhibitors (40). High expression of miR-100 is associated with poor survival in NSCLC (43). We demonstrated that enforced expression of miR-21, miR-30c, and miR-100 in TRAIL-sensitive cells is sufficient to induce resistance to the drug by directly targeting fundamental tumor-suppressor genes and effectors of the TRAIL pathway, such as caspase-8, caspase-3, TRAF-7, and FoxO3a. Noteworthy is that up-regulation of miR-21, miR-30c, and miR-100 was also observed in NSCLC cell lines with de novo TRAIL-resistance, suggesting that these miRNAs may play a role in both the primary and acquired TRAIL-resistance. Furthermore, we demonstrated that enforced expression of miR-21, miR-30c, and miR-100 in TRAIL-sensitive cells is sufficient to induce resistance to the drug by directly targeting fundamental tumor-suppressor genes and effectors of the TRAIL pathway, such as caspase-8, caspase-3, TRAF-7, and FoxO3a.
Besides inducing apoptosis through the activation of the caspase signaling pathway, TRAIL is also known to activate NF-κB in innate TRAIL-resistant cells (44, 45). However, the role of caspase-8 in NF-κB activation is controversial. Caspase-8 is required for NF-κB activation in HeLa and HEK293 cells in response to TNF stimulation (46). Several other studies have shown that active caspase-8 cleaves RIP1 and cleaved RIP1 prevents NF-κB activation and activates downstream caspases (13, 14, 33, 47). In lung TRAIL-sensitive cells, RIP1 is cleaved in a caspase-8–dependent process and is unable to activate NF-κB signaling. We proved that in acquired TRAIL-resistant cells down-modulation of caspase-8 by miR-21 blocks RIP1 cleavage, leading to NF-κB activation and acquired TRAIL-resistance. Of note, miR-21 knockdown in H460R cells enhanced RIP1 cleavage and increased cells sensitivity, demonstrating that modulation of miRNAs is able to revert the sensitivity of cancer cells to TRAIL treatment. TRAFs were initially discovered as adaptor proteins that couple the TNF receptor family to signaling pathways. A recent study reported that TRAF-7 induces cell death by repressing NF-κB activation (18). We proved that miR-100, by down-regulating TRAF-7, induces NF-κB activation in cells with acquired TRAIL-resistance. Therefore, in the sensitive cells TRAIL triggers apoptosis through caspase activation and the NF-κB signaling is weak because of the cleavage of RIP1 by caspase-8. In the resistant cells, down-modulation of caspase-8 and TRAF-7 by miR-21 and miR-100 activates NF-κB, which in turn, in a positive feedback loop, induces the transcriptional activation of miR-21and miR-100. In a previous study we reported that miR-221/222 induced the activation of the AKT pathway through phosphatase and tensin homolog deleted on chromosome 10 silencing, increasing the resistance to TRAIL-induced apoptosis. Intriguingly, we found an up-regulation of miR-221 in the H460R cells (Fig. 1C), suggesting that it could play a role also in acquired TRAIL resistance. As expected, AKT activation was observed in cells with acquired TRAIL resistance and inhibition of this pathway sensitized the cells to the drug. Of note, recent literature reported that AKT is a key regulator of NF-κB activation and, although initially believed to belong to distinct signaling pathways, the NF-κB and AKT signaling pathways can converge (31, 32). It is known that there is a strict correlation between acquired drug resistance and EMT. Recently, it has been reported that NF-κB constitutive activation is involved in EMT of NSCLC cells through the up-regulation of master-switch transcription factors (48). We found a morphological change and a more aggressive phenotype in H460R and H292R cells (Fig. 6). Analysis of epithelial and mesenchymal markers confirmed that acquired TRAIL resistance induced epithelial to mesenchymal transition in both cell lines. Genetic inhibition of the NF-κB pathway affects both the initiation and the maintenance of lung cancer, identifying this pathway as a promising therapeutic target (49). Remarkably, TRAIL treatment made acquired TRAIL-resistant cells more aggressive but combinatorial treatment of resistant cells with TRAIL and NF-κB inhibitors increased apoptosis and reduced cell proliferation in vitro and in vivo, compared with cells treated with TRAIL or NF-κB inhibitor alone. Taken together, these data identify NF-κB as a potential companion drug target, together with TRAIL, in lung cancer and provide insight into the mechanisms by which tumor cells become resistant to TRAIL-induced apoptosis. In addition, the results not only suggest that, miR-21, miR-30c, and miR-100 expression levels could be used as prognostic tool to predict TRAIL sensitivity or resistance, but also that in the near future, the delivery and modulation of specific miRNAs could improve the response of lung cancer patients to TRAIL.
Experimental Procedures
Lung Cancer Samples and Cell Lines.
Sixty-nine cancer lung tissues were purchased from US Biomax. Human H460S/R, H292S/R, HEK293, Calu-1, and A549 cell lines were grown in RPMI 1640 containing 10% (vol/vol) heat-inactivated FBS and with 2 mM l-glutamine and 100 U/mL−1 penicillin–streptomycin.
Western Blot Analysis.
Detailed procedures are described in SI Appendix.
MiRNA Locked Nucleic Acid in Situ Hybridization of Formalin-Fixed, Paraffin-Embedded Tissue Section.
In situ hybridization was carried out on deparaffinized human lung tissues using previously a published protocol (50) and described in SI Appendix.
Generation of Stable Clones with miR-21, miR-30c, and miR-100 Down-Regulation.
The detailed experimental procedures are described in SI Appendix.
Soft Agar Colony Formation Assay.
The overall procedure of soft agar colony formation assay was performed according to the manufacturer’s protocol and detailed procedures are described in SI Appendix.
In Vivo Studies.
Animal studies were performed according to institutional guidelines. The detailed protocols are described in SI Appendix. Animal experiments were conducted after approval of the Institutional Animal Care and Use Committee, Ohio State University.
Statistical Analysis.
Student’s t test and one-way ANOVA were used to determine significance. All error bars represent the SEM. Statistical significance for all of the tests, assessed by calculating P value, was < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. D. Guttridge for supplying the DA IKB-α plasmid; Arianna Bottoni for quantitative RT-PCR and Nanostring assistance; and Sunhee Park (Curry School of Education at the University of Virginia) for statistical advice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE55860).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504630112/-/DCSupplemental.
References
- 1.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281(5381):1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5(11):876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8(10):782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- 4.Wakelee HA, et al. Phase I and pharmacokinetic study of lexatumumab (HGS-ETR2) given every 2 weeks in patients with advanced solid tumors. Ann Oncol. 2010;21(2):376–381. doi: 10.1093/annonc/mdp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbst RS, et al. A first-in-human study of conatumumab in adult patients with advanced solid tumors. Clin Cancer Res. 2010;16(23):5883–5891. doi: 10.1158/1078-0432.CCR-10-0631. [DOI] [PubMed] [Google Scholar]
- 6.Herbst RS, et al. Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J Clin Oncol. 2010;28(17):2839–2846. doi: 10.1200/JCO.2009.25.1991. [DOI] [PubMed] [Google Scholar]
- 7.Plantivaux A, Szegezdi E, Samali A, Egan L. Is there a role for nuclear factor kappaB in tumor necrosis factor-related apoptosis-inducing ligand resistance? Ann N Y Acad Sci. 2009;1171:38–49. doi: 10.1111/j.1749-6632.2009.04725.x. [DOI] [PubMed] [Google Scholar]
- 8.Spierings DC, et al. Expression of TRAIL and TRAIL death receptors in stage III non-small cell lung cancer tumors. Clin Cancer Res. 2003;9(9):3397–3405. [PubMed] [Google Scholar]
- 9.Wang X, et al. Akt-mediated eminent expression of c-FLIP and Mcl-1 confers acquired resistance to TRAIL-induced cytotoxicity to lung cancer cells. Mol Cancer Ther. 2008;7(5):1156–1163. doi: 10.1158/1535-7163.MCT-07-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meylan E, Tschopp J. The RIP kinases: Crucial integrators of cellular stress. Trends Biochem Sci. 2005;30(3):151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol. 2014;76:129–150. doi: 10.1146/annurev-physiol-021113-170259. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary PM, et al. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity. 1997;7(6):821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13(19):2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, et al. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol Cell Biol. 2000;20(18):6638–6645. doi: 10.1128/mcb.20.18.6638-6645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelliher MA, et al. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8(3):297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 16.Garofalo M, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13(3):57–66. doi: 10.1016/j.drup.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zotti T, et al. TRAF7 protein promotes Lys-29-linked polyubiquitination of IkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J Biol Chem. 2011;286(26):22924–22933. doi: 10.1074/jbc.M110.215426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010;9(6):1091–1096. doi: 10.4161/cc.9.6.11035. [DOI] [PubMed] [Google Scholar]
- 20.Bassères DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 21.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 22.Hall MA, Cleveland JL. Clearing the TRAIL for cancer therapy. Cancer Cell. 2007;12(1):4–6. doi: 10.1016/j.ccr.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: Induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281(5383):1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Kandasamy K, Srivastava RK. Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res. 2003;63(5):1059–1066. [PubMed] [Google Scholar]
- 25.Sheridan JP, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277(5327):818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 26.Ricci MS, et al. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell. 2007;12(1):66–80. doi: 10.1016/j.ccr.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, et al. Regulation of Mcl-1 by constitutive activation of NF-κB contributes to cell viability in human esophageal squamous cell carcinoma cells. BMC Cancer. 2014;14:98. doi: 10.1186/1471-2407-14-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21(16):5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panner A, James CD, Berger MS, Pieper RO. mTOR controls FLIPS translation and TRAIL sensitivity in glioblastoma multiforme cells. Mol Cell Biol. 2005;25(20):8809–8823. doi: 10.1128/MCB.25.20.8809-8823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JY, et al. TRADD is critical for resistance to TRAIL-induced cell death through NF-κB activation. FEBS Lett. 2011;585(14):2144–2150. doi: 10.1016/j.febslet.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Dan HC, et al. Akt-dependent regulation of NF-kappaB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22(11):1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng F, Liu L, Chin PC, D’Mello SR. Akt is a downstream target of NF-kappa B. J Biol Chem. 2002;277(33):29674–29680. doi: 10.1074/jbc.M112464200. [DOI] [PubMed] [Google Scholar]
- 33.Varfolomeev E, et al. Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem. 2005;280(49):40599–40608. doi: 10.1074/jbc.M509560200. [DOI] [PubMed] [Google Scholar]
- 34.McConkey DJ, et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 2009;28(3-4):335–344. doi: 10.1007/s10555-009-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 36.Dimberg LY, et al. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene. 2013;32(11):1341–1350. doi: 10.1038/onc.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol. 1998;161(6):2833–2840. [PubMed] [Google Scholar]
- 38.Jin Z, McDonald ER, 3rd, Dicker DT, El-Deiry WS. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem. 2004;279(34):35829–35839. doi: 10.1074/jbc.M405538200. [DOI] [PubMed] [Google Scholar]
- 39.Mitsiades N, Poulaki V, Mitsiades C, Tsokos M. Ewing’s sarcoma family tumors are sensitive to tumor necrosis factor-related apoptosis-inducing ligand and express death receptor 4 and death receptor 5. Cancer Res. 2001;61(6):2704–2712. [PubMed] [Google Scholar]
- 40.Garofalo M, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18(1):74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 42.Hatley ME, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang T, et al. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS ONE. 2012;7(8):e43268. doi: 10.1371/journal.pone.0043268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grunert M, et al. The adaptor protein FADD and the initiator caspase-8 mediate activation of NF-κB by TRAIL. Cell Death Dis. 2012;3:e414. doi: 10.1038/cddis.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider P, et al. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity. 1997;7(6):831–836. doi: 10.1016/s1074-7613(00)80401-x. [DOI] [PubMed] [Google Scholar]
- 46.Jun JI, et al. Role of FLASH in caspase-8-mediated activation of NF-kappaB: dominant-negative function of FLASH mutant in NF-kappaB signaling pathway. Oncogene. 2005;24(4):688–696. doi: 10.1038/sj.onc.1208186. [DOI] [PubMed] [Google Scholar]
- 47.Rébé C, et al. Caspase-8 prevents sustained activation of NF-kappaB in monocytes undergoing macrophagic differentiation. Blood. 2007;109(4):1442–1450. doi: 10.1182/blood-2006-03-011585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar M, et al. NF-κB regulates mesenchymal transition for the induction of non-small cell lung cancer initiating cells. PLoS ONE. 2013;8(7):e68597. doi: 10.1371/journal.pone.0068597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue W, et al. Response and resistance to NF-kappaB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1(3):236–247. doi: 10.1158/2159-8290.CD-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nuovo GJ, et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4(1):107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.