Significance
The cyanobacterium Prochlorococcus is the most abundant photosynthetic organism in the oceans, driving marine food webs and biogeochemistry. Estimates of Prochlorococcus cell mortality and cell production are critical to determine how organic matter is transferred into the food web. Using novel high-resolution sampling methods on two winter-time cruises, we show that the daily production of Prochlorococcus cells in surface waters of the subtropical Pacific gyre is consistently balanced by their nightly consumption by other organisms. These synchronized loss processes suggest that Prochlorococcus-derived organic matter stabilizes multiple species interactions, from viruses to grazers. The observed resilience of this synchronized food web dynamic as temperature increased within the gyre suggests that ecosystem stability may persist in future warmer oceans.
Keywords: cyanobacteria, cell division, mortality, flow cytometry, SeaFlow
Abstract
Theoretical studies predict that competition for limited resources reduces biodiversity to the point of ecological instability, whereas strong predator/prey interactions enhance the number of coexisting species and limit fluctuations in abundances. In open ocean ecosystems, competition for low availability of essential nutrients results in relatively few abundant microbial species. The remarkable stability in overall cell abundance of the dominant photosynthetic cyanobacterium Prochlorococcus is assumed to reflect a simple food web structure strongly controlled by grazers and/or viruses. This hypothesized link between stability and ecological interactions, however, has been difficult to test with open ocean microbes because sampling methods commonly have poor temporal and spatial resolution. Here we use continuous techniques on two different winter-time cruises to show that Prochlorococcus cell production and mortality rates are tightly synchronized to the day/night cycle across the subtropical Pacific Ocean. In warmer waters, we observed harmonic oscillations in cell production and mortality rates, with a peak in mortality rate consistently occurring ∼6 h after the peak in cell production. Essentially no cell mortality was observed during daylight. Our results are best explained as a synchronized two-component trophic interaction with the per-capita rates of Prochlorococcus consumption driven either directly by the day/night cycle or indirectly by Prochlorococcus cell production. Light-driven synchrony of food web dynamics in which most of the newly produced Prochlorococcus cells are consumed each night likely enforces ecosystem stability across vast expanses of the open ocean.
Potential interdependencies between species diversity and ecosystem stability have gained increased focus due to global changes in species distributions and abundances (1). Strong predator–prey interactions are predicted to enhance the number of coexisting species and limit fluctuations in abundances (2, 3), whereas competition for limited resources is predicted to reduce biodiversity, in some instances, to the point of ecological instability (3, 4). Mechanisms underlying ecosystem stability remain challenging to characterize on relevant temporal and spatial scales, in part because few empirical data are available to test these theories.
Our focus is on the microbial communities within surface waters of the vast oligotrophic gyre of the north Pacific Subtropical Ocean. Here, competition for low concentrations of essential nutrients is hypothesized to result in relatively few abundant microbial species, typified by their extremely small cell sizes and streamlined genomes (5). The cyanobacterium Prochlorococcus numerically dominates the photosynthetic community in these regions, with a relatively constant cell abundance close to half a billion cells per liter despite a population doubling time of approximately one day (6). Such constant cell numbers are predicted when both prey and predators are abundant and broadly dispersed and when the predator–prey interactions are tightly coupled (7). Understanding the stability of this coupling is important as loss processes determine how Prochlorococcus cell production is transferred into the marine food web. Estimates of cell production and cell mortality rates for Prochlorococcus remain sparse, however, as they are commonly derived from incubations in bottles (8) or measures of cell cycle progression over the day/night cycle (9), both of which are labor intensive and thus limited in their broad-scale applicability (10). Here we estimated Prochlorococcus cell production and cell mortality rates continuously across a 2,900-km transect in the northeast Pacific Ocean in winter (October–November 2011) using a shipboard-based flow cytometer, SeaFlow (11) and show that, throughout the gyre, rates of Prochlorococcus cell production and cell mortality are both tightly synchronized to the day/night cycle, which likely helps stabilize open ocean ecosystems.
Results and Discussion
Temperature Regulates the Rates of Prochlorococcus Cell Division.
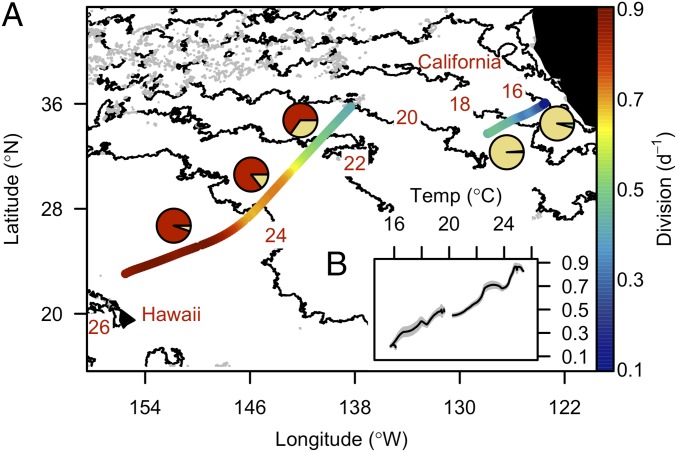
Prochlorococcus division rates were estimated using a size-structured population model (12, 13) with the continuously measured size distribution of Prochlorococcus as input (SI Appendix, Figs. S1–S4). Continuous, daily averaged division rates of natural Prochlorococcus populations along the transect increased from 0.2 to 0.9 d−1 (Fig. 1A) and were linearly correlated with temperature (R2 = 0.97, P < 0.001; Fig. 1B). These computed division rates were similar to our independent estimates based on cell cycle analyses (SI Appendix, Figs. S2 and S3). Lower division rates were encountered in cooler waters (16–19 °C) at the edge of the north Pacific gyre and further inshore where the high-light-adapted Prochlorococcus ecotype I (eHLI) dominated (>95% of all Prochlorococcus 16S rDNA barcoded sequences), and were similar to those previously observed in that region (<0.4 d−1) (14) (Fig. 1A and SI Appendix, Fig. S5). Higher division rates were observed in the warmer subtropical gyre waters, which were dominated by the high-light-adapted Prochlorococcus ecotype II (eHLII) (65–94% of all Prochlorococcus 16S rDNA barcoded sequences) and were similar to, or higher than, the few individual measurements available from the Sargasso Sea (>0.7 d−1) (14), another warm water ecosystem.
Fig. 1.
Temperature determines the rate of Prochlorococcus cell division in surface waters of the Northeast Pacific Ocean. (A) Daily averaged division rate of Prochlorococcus (division, d−1, color scale), monthly averaged satellite-based sea surface temperature for October (°C, contour line), and Prochlorococcus ecotype composition (pie charts based on percent of each Prochlorococcus ecotype relative to total Prochlorococcus ecotypes represented in 16S rRNA gene amplicons from TAG sequencing; yellow, red, and green for eHLI, eHLII, and eLLI, respectively) during the northward cruise to California (4–9 October 2011) and southward cruise to Hawaii (29 October–2 November 2011). Daily division rates were calculated as the sum of hourly division rates over a 24-h period using a 1-h rolling window. (B) Relationship between daily division rates and surface temperature during the survey. Vertical gray bars represent SDs (n = 24).
The highest in situ division rate (0.9 d−1) occurred at 25 °C in low-latitude waters (25°N; Fig. 1B) and is remarkably similar to the highest division rate reported for a cultured representative of eHLII (MIT9312) grown under nutrient-replete conditions (SI Appendix, Fig. S6). Thus, the eHLII-dominated field population in the subtropical gyre was likely dividing at near-maximal rates, despite low ambient nutrient availability commonly found in this region (15). Similar to other ecotypes (16, 17), Prochlorococcus eHLII must possess efficient nutrient acquisition mechanisms and reduced physiological nutrient demands that allow it to maintain such high division rates in the face of nutrient scarcity. Under laboratory conditions, the growth rate of Prochlorococcus is positively correlated with temperature across the range of conditions tested in this study (18). In the field, Prochlorococcus is most abundant at warmer low latitudes (6). Both observations suggest a central role of temperature in regulating the rate of Prochlorococcus cell division in surface waters.
Synchronized Oscillations in the Rates of Prochlorococcus Cell Production and Cell Mortality.
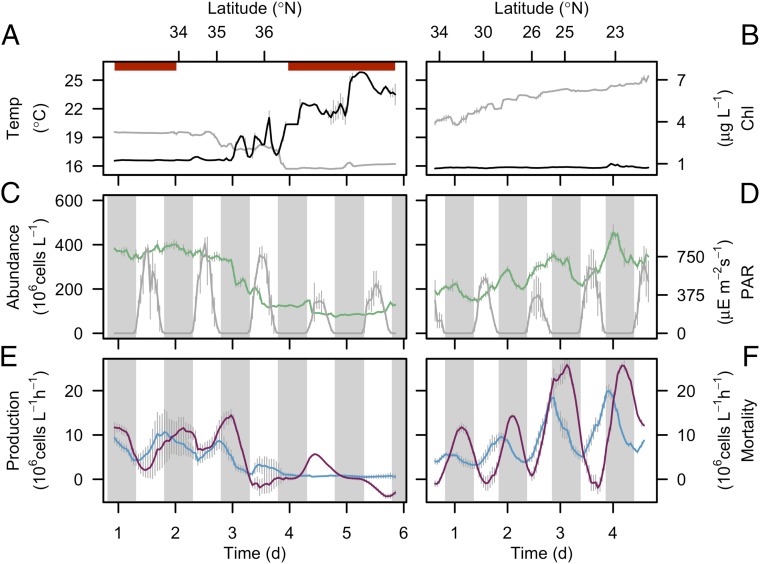
Despite the significant differences in daily division rates along the transect, maximum cell abundance (∼400 × 106 cells/L) was comparable in both cooler and warmer waters (Fig. 2 A–D). In warm waters (>19 °C), cell numbers were tightly synchronized to the day/night cycle, with a net increase in abundance during the day and a net decrease at night (Fig. 2 C, day 1, and D). No such synchrony was observed in the colder waters (<19 °C; Fig. 2C, day 3–6). The day/night pattern observed in warm waters differs from those observed in other warm water regions such as the equatorial Pacific, where cell abundances increase during the first half of the night and decrease or remain stable during the day (19). These different patterns of cell abundance over the day/night cycle indicated an underlying difference in the timing of cell division and/or cell loss processes among these regions.
Fig. 2.
Synchrony of Prochlorococcus mortality rates at high cell production rates. (A and B) Hourly averaged temperature (°C, gray line) and bulk chlorophyll a concentrations (µg/L, black line), (C and D) hourly averaged cell abundances (106 cells/L, green line) and hourly averaged PAR at 5 m depth (µE/m2 per second, gray line), and (E and F) hourly rates of cell production (106 cells/L per hour, blue line) and cell mortality (106 cells/L per hour, purple line) during the northward cruise to California (4–9 October 2011) (Left) and southward cruise to Hawaii (29 October–2 November 2011) (Right). Vertical gray bars represent SDs (n = 20 for temperature, chlorophyll a concentrations, PAR, and cell abundances, n = 24 for cell production and mortality rates). The gray regions indicate night. Horizontal red bars in A represent periods on station. Note that the left panels are shown according to chronology of sampling, as opposed to ascending sea surface temperature in Fig. 1B.
Our use of continuous measurements of cell size allowed us to examine hourly cell division over the daily cycle. The peak in cell division consistently occurred near dusk, regardless of ecotype composition (Fig. 1A), and was reflected in the declining mean cell size of the Prochlorococcus population (SI Appendix, Fig. S7). Although most previous studies report essentially stable or decreasing cell abundance in Prochlorococcus with essentially no cell division during the day (6, 19), our data revealed that a significant proportion of the population underwent cell division (SI Appendix, Fig. S7 C and D), similar to populations in the Indian Ocean (20). Our estimates of cell production (Fig. 2 E and F), computed by multiplying our independent measures of cell abundance by our estimates of division rates, accounted for most of the observed changes in Prochlorococcus cell abundances during the day (Fig. 2D) and indicated that loss processes were negligible during this period.
The net loss rate of Prochlorococcus cells from a given location is a combination of biological factors (i.e., consumption by grazers and/or lysis by viruses) and physical factors (i.e., the removal or addition of cells through physical transport). In our study, the greatest source of physical transport was due to sampling from a moving ship, with other sources of physical transport considered as negligible for our calculations. Physically driven rates of changes in Prochlorococcus abundance was estimated based on the spatial gradient of Prochlorococcus abundance along the cruise track (SI Appendix, SI Materials and Methods). Biologically driven rates of cell loss (i.e., cell mortality) were based on the difference between the cell production and the rates of change of cell abundance over time, after correcting for physical transport (SI Appendix, Fig. S8). Hourly mortality rates were near zero during the day and peaked at night in subtropical gyre waters (<34°N; Fig. 2 E, day 1, and F), with a more stochastic pattern observed further inshore (>34°N; Fig. 2E, day 2–6). In the subtropical gyre, the peak in mortality consistently occurred about 6 h after dusk (Fig. 2F) such that the bulk of the daily Prochlorococcus cell production (ranging from 88 × 106 to 289 × 106 cells/L per day; SI Appendix, Fig. S9) was consumed over the course of the night. This 90°-phase difference (i.e., 6 h in a 24-h cycle) between the rates of cell production and cell mortality was consistent across 2,240 km of the gyre, regardless of water temperature or Prochlorococcus ecotype composition (Fig. 1A). The consistency of the phase and period of the oscillations in the rates of cell mortality was particularly surprising across such a large spatial scale given the diversity of potential sources of Prochlorococcus mortality (e.g., viruses and grazers) (21, 22). A similar diel pattern of cell production and cell mortality was observed in the same region the previous year (November 2010; SI Appendix, Fig. S10), suggesting that these synchronized oscillations may be relatively stable, at least over an annual cycle.
Light-Driven Prochlorococcus Cell Mortality.
In dynamical systems theory, a synchrony between two coupled oscillators indicates that the periodicity of one signal entrains the phase and period of the other. The periodic biological signal in the subtropical gyre is the light-driven photosynthesis and division rate of Prochlorococcus that produces a daily pulse of new carbon. This pulse entrains a synchronous, phase-coupled transfer of the carbon into the food web. Synchronized predator–prey cycles such as those observed here are predicted to promote food web stability (7). No such synchrony was observed in the colder waters where the abundance of Prochlorococcus was significantly lower (<150 × 106 cells/L), and the phytoplankton assemblage was dominated by larger sized phytoplankton (SI Appendix, Fig. S11). As predicted by systems theory, changes in Prochlorococcus cell abundance over time were more stochastic in this region (Fig. 2E, day 3–6).
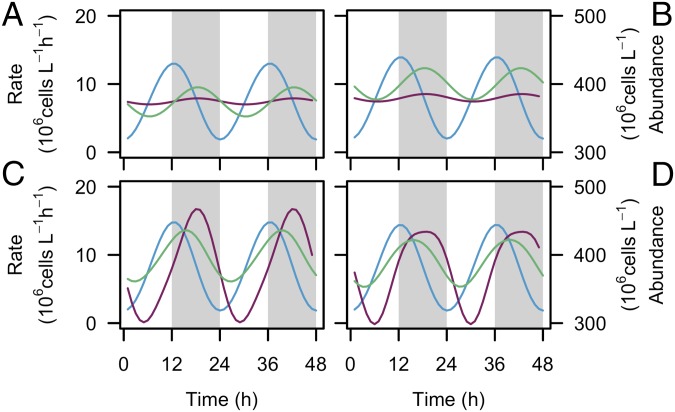
The synchronized oscillations in the rates of cell production and cell mortality observed in the subtropical gyre mirrors two-component models of virus/host or predator/prey systems, although compressed over the course of a daily cycle. We explored different models to determine which best emulates the observed oscillations in Prochlorococcus mortality rates characterized by limited cell loss during daylight. Classical models commonly assume that mortality rate depends exclusively on prey abundance, with a constant per-capita rate of host infection or prey consumption (23, 24). These models produce a diel oscillation in mortality rates but do not explain the negligible cell loss during the day (Fig. 3 A and B). There is growing evidence that viral infection rates in the cyanobacteria Synechococcus and other marine phytoplankton depend on light either directly or indirectly via the host’s cell cycle (25, 26). Mixotrophic protists, such as crysophytes, can graze at night and photosynthesize during the day (27), some of which consume Prochlorococcus in oligotrophic regions (22). Modeled cell mortality emulates our observations if we assume that the per-capita rate of viral infection varies over the day/night cycle or that grazing only occurs at night (Fig. 3 C and D). The relative contribution of grazers and viruses to overall Prochlorococcus mortality in natural communities has not been quantified. However, Prochlorococcus abundances in the Sargasso Sea were observed to decrease dramatically during the latter half of the night, similar to the timing observed here; this nearly identical period of cell loss was experimentally linked to grazing activity rather than viruses (14). Regardless of whether the mortality we observed in the subtropical gyre is caused by viruses or by grazers, our results indicate that the day/night cycle, either directly or indirectly, restricts mortality to the night, allowing Prochlorococcus abundance to recover each day and maintain a stable population over vast regions of the ocean.
Fig. 3.
Light dependency of Prochlorococcus mortality forces. Modeled Prochlorococcus mortality rate (purple line), production rate (blue line), and cell abundance (green line). (A and C) Stable solution for a virus/host model (23) where the per-capita rates of virus infection (A) remain constant over the day/night cycle or (C) vary according to Prochlorococcus cell division rates. (B and D) Stable solution for a predator–prey model (24) where the per-capita rates of predation (B) remain constant over the day/night cycle or (D) vary according to light intensity. The gray regions indicate night.
Concluding Remarks.
The observed synchrony between cell production and cell mortality means that each day a pulse of Prochlorococcus-derived organic matter enters the ecosystem, either as dissolved organic matter derived from viral lysis, or as particulate organic matter grazed by higher trophic level organisms, or a combination of both. We estimate that Prochlorococcus pulses of carbon could account for more than 75% of the daily photosynthetic organic carbon production in the surface layer of the subtropical gyre during this time of year (SI Appendix, Fig. S9). A recent study by Ottesen et al. (28) documented synchronous gene expression patterns across bacterial communities within the subtropical gyre and hypothesized that these patterns could result from light-driven pulses of dissolved organic matter released by Prochlorococcus. Our results indicate that a mortality-driven pulsed delivery of Prochlorococcus-derived organic compounds through the food web may serve as a stabilizing factor of additional trophic levels across vast expanses of the subtropical ecosystem. The next step is to determine whether similar patterns are observed in the subtropical gyre throughout the year and whether they are conserved across different oceanic regions.
Materials and Methods
Phytoplankton Abundance and Composition.
Flow cytometry samples were collected underway from the continuous seawater flow-through system (5 m depth) during the northward cruise to California (4–9 October 2011) and during the southward cruise to Hawaii (16–18 November 2010 and 29 October–2 November 2011). Continuous underway measurements of Prochlorococcus abundances and cell size were made using SeaFlow (11), and data were analyzed using the R package flowPhyto (29) version 2.4.1, which uses statistical clustering methods to discriminate phytoplankton populations. Forward light scatter measured by the instrument was converted to cell volume using an empirical relationship between light scatter measured by SeaFlow and cell volume measured by a Coulter counter for different exponentially growing phytoplankton cultures (SI Appendix, SI Materials and Methods).
Estimated Cell Division Rates of Prochlorococcus.
Hourly division rates of Prochlorococcus were estimated using the R package ssPopModel version 0.1.1, a modified version of the size-structured matrix population model developed by Sosik et al. (12). The model is based on the assumptions that (i) cell growth is determined by light exposure, (ii) the probability of a cell dividing depends on size, (iii) all cells within a discrete size class have the same probability to change to another size class, and (iv) a cell divides into two daughter cells, each half the size of the mother cell. The model predicts the cell size distribution over the course of the day using the cell size/cell division relationships and the light dependence of cell division (SI Appendix, SI Materials and Methods). We assumed that there was no differential mortality of Prochlorococcus based on cell size. Size distribution-based division rate estimates of Synechococcus have been shown to be similar between undiluted incubations (higher grazing pressure) and diluted incubations (lower grazing pressure) (30), supporting the assumption that size-selective grazing is not important. To establish the accuracy of our estimates of size distribution-based division rate estimates, we compared size-based estimates of Prochlorococcus division rates (h−1) with rates of changes in cell number for laboratory cultures and with cell cycle-based estimates of division rates for natural Prochlorococcus populations sampled near the beginning and end of the transect (SI Appendix, SI Materials and Methods). Estimates of Prochlorococcus cell production was obtained by multiplying hourly averaged cell abundances by the hourly estimates of cell division rates.
Estimated Cell Mortality Rates of Prochlorococcus.
The rate of change in the abundance of Prochlorococcus, C, measured by the instrument from a moving ship reflects changes in C due to local time-dependent biological processes (i.e., division and mortality) and physical processes (i.e., advection and diffusion) occurring at a particular location and an advective component due to the ship moving through a spatial gradient of C. Given that the greatest source of physical transport was due to sampling from a moving ship, with other sources of physical transport considered as negligible for our calculations, mortality rate is given by
where g is the mortality rate due to biological processes such as grazing and viral infection, µ is the division rate, C is the cell abundance, vship is the velocity of the ship and ∂C/∂x is the spatial gradient of C along the cruise track, x. To approximate the spatial gradient of C, we assumed that Prochlorococcus abundances remain constant from day to day at a given location, as observed from previous studies (31, 32) and during the first day on station (Fig. 2C, from day 1 to day 2). Daily averaged C was calculated using a 24-h running mean of hourly averaged C. The daily distance was calculated as the distance between the location at t0 and the location at t + 24 h. A 1-h rolling window was then used to determine the start of each 24-h period in the time series.
Idealized Ecological Models.
We used two different mathematical models that describe two-trophic-level microbial systems (i.e., virus/host and predator/prey systems) in continuous culture, based on our estimates of Prochlorococcus division rates (SI Appendix, SI Materials and Methods). We modeled the dynamics of host/virus interactions according to Levin’s model (23) where the per capita adsorption rate of the viruses was set either as a constant or defined to maximize cell lysis 8 h after the peak of Prochlorococcus cells in S phase (i.e., 2 h before dusk). The dynamics of the interactions between predator and prey was modeled using Lokta–Volterra equations (24), where the per-capita predation rate was set either as a constant or defined such that the predation rate was inversely correlated to irradiance. Abundances were simulated for 100 d to reach a system at equilibrium.
Estimated Daily Carbon Production of Prochlorococcus.
Cell volume was converted to carbon per cell using a conservative estimate of 325 fg C/µm3 (33). This yields median values of carbon content per cell of 25 and 50 fg C/cell in the subtropical gyre and near the California coast, respectively. These estimates are in agreement with direct measurements made on cultures (49 fg/cell) (34). The carbon per cell was multiplied by the estimated daily cell production at the end of the southward transect (near Hawaii) to obtain the mean daily carbon production (0.59 mmol C/m3 per day). Our estimate of daily carbon production represents more than 75% of the seasonal gross primary production at station ALOHA (0.75 mmol C/m3 per day) (35), which is based on 18O-labeled O2 in bottles over a 24-h incubation period.
Prochlorococcus Ecotype Composition.
DNA samples were collected underway from the continuous seawater flow-through system. Genomic DNA was extracted from 500-mL water samples filtered through a 0.2-µm Supor filter following the protocol described in Demir-Hilton et al. (36) for the northward cruise and through a 0.2-µm GTTP Isopore membrane filter using the QIAGEN DNeasy tissue kit according to the manufacturer’s protocol for gram-negative bacterial cells for the southward cruise. Filters were first thawed and vortexed vigorously with lysis buffer using a bead beater (Bio-Spec Products) for 30 s at 2,100 × g (Eppendorf 5418 with rotor FA-45-18-11), and the buffer with cells was then collected and processed according to the manufacturer’s protocol. Barcoded V1-V2 region 16S rRNA data were produced and analyzed using a previously published method (37) that incorporates pplacer (38) version 1.1.15. The number of Prochlorococcus amplicons produced were 1,382, 34,351, 40,046, 30,738, and 38,153 (ordered by ascending sea surface temperature).
Auxiliary Data.
Monthly averaged fields of satellite-based sea surface temperature were obtained from the NASA website (disc.sci.gsfc.nasa.gov/giovanni). Photosynthetic active radiation (PAR) was measured using an underwater Biospherical Instruments light sensor with log amplifier. Underway sea surface temperature was measured using a shipboard Sea-Bird Electronics SBE-21 thermosalinograph, and underway chlorophyll a fluorescence was measured using a shipboard WET Labs ECO FLRTD chlorophyll fluorometer.
Supplementary Material
Acknowledgments
We thank the officers, crew aboard the Thomas G. Thompson and Western Flyer, and J. Koester for assistance during the cruise onboard the T.G. Thompson; C. Olson for helping with DNA extractions; J. Saunders for providing Prochlorococcus cultures (MED4); and S. Coesel for comments on the manuscript. We also thank H. Sosik and K. Hunter-Cevera for sharing the original version of the matrix population model. This work was supported by National Science Foundation Grant OTIC-1154074 and Gordon and Betty Moore Foundation Grant GBMF3776 (to E.V.A.) and the David and Lucille Packard Foundation and Gordon and Betty Moore Foundation Grant GBMF3788 (to A.Z.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424279112/-/DCSupplemental.
References
- 1.McCann KS. The diversity-stability debate. Nature. 2000;405(6783):228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 2.Rooney N, McCann K, Gellner G, Moore JC. Structural asymmetry and the stability of diverse food webs. Nature. 2006;442(7100):265–269. doi: 10.1038/nature04887. [DOI] [PubMed] [Google Scholar]
- 3.Allesina S, Tang S. Stability criteria for complex ecosystems. Nature. 2012;483(7388):205–208. doi: 10.1038/nature10832. [DOI] [PubMed] [Google Scholar]
- 4.Loreau M, de Mazancourt C. Biodiversity and ecosystem stability: A synthesis of underlying mechanisms. Ecol Lett. 2013;16(Suppl 1):106–115. doi: 10.1111/ele.12073. [DOI] [PubMed] [Google Scholar]
- 5.Swan BK, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110(28):11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63(1):106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasseur DA, Fox JW. Phase-locking and environmental fluctuations generate synchrony in a predator-prey community. Nature. 2009;460(7258):1007–1010. doi: 10.1038/nature08208. [DOI] [PubMed] [Google Scholar]
- 8.Landry MR, Hassett RP. Estimating the grazing impact of marine micro-zooplankton. Mar Biol. 1982;67(3):283–288. [Google Scholar]
- 9.Vaulot D, Marie D, Olson RJ, Chisholm SW. Growth of prochlorococcus, a photosynthetic prokaryote, in the equatorial pacific ocean. Science. 1995;268(5216):1480–1482. doi: 10.1126/science.268.5216.1480. [DOI] [PubMed] [Google Scholar]
- 10.Laws EA. Evaluation of in situ phytoplankton growth rates: A synthesis of data from varied approaches. Annu Rev Mar Sci. 2013;5:247–268. doi: 10.1146/annurev-marine-121211-172258. [DOI] [PubMed] [Google Scholar]
- 11.Swalwell JE, Ribalet F, Armbrust EV. SeaFlow: A novel underway flow-cytometer for continuous observations of phytoplankton in the ocean. Limnol Oceanogr Methods. 2011;9:466–477. [Google Scholar]
- 12.Sosik HM, Olson RJ, Neubert MG, Shalapyonok A, Solow AR. Growth rates of coastal phytoplankton from time-series measurements with a submersible flow cytometer. Limnol Oceanogr. 2003;48(5):1756–1765. [Google Scholar]
- 13.Caswell H. Matrix Population Models: Construction, Analysis and Interpretation. Sinauer; Sunderland, MA: 1989. [Google Scholar]
- 14.Worden A, Binder B. Application of dilution experiments for measuring growth and mortality rates among Prochlorococcus and Synechococcus populations in oligotrophic environments. Aquat Microb Ecol. 2003;30(2):159–174. [Google Scholar]
- 15.Yasunaka S, et al. Mapping of sea surface nutrients in the North Pacific: Basin-wide distribution and seasonal to interannual variability. J Geophys Res Oceans. 2014;119(11):7756–7771. [Google Scholar]
- 16.Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103(33):12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Mooy BAS, et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature. 2009;458(7234):69–72. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- 18.Johnson ZI, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311(5768):1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 19.Binder BJ, DuRand MD. Diel cycles in surface waters of the equatorial Pacific. Deep Sea Res Part II Top Stud Oceanogr. 2002;49(13-14):2601–2617. [Google Scholar]
- 20.Zubkov MV, Quartly GD. Ultraplankton distribution in surface waters of the Mozambique Channel-flow cytometry and satellite imagery. Aquat Microb Ecol. 2003;33(2):155–161. [Google Scholar]
- 21.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424(6952):1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann M, Zubkov MV, Scanlan DJ, Lepère C. In situ interactions between photosynthetic picoeukaryotes and bacterioplankton in the Atlantic Ocean: Evidence for mixotrophy. Environ Microbiol Rep. 2013;5(6):835–840. doi: 10.1111/1758-2229.12084. [DOI] [PubMed] [Google Scholar]
- 23.Levin BR, Stewart FM, Chao L. Resource-limited growth, competition, and predation: A model and experimental studies with bacteria and bacteriophage. Am Nat. 1977;111(977):3–24. [Google Scholar]
- 24.Lotka AJ. Analytical note on certain rhythmic relations in organic systems. Proc Natl Acad Sci USA. 1920;6(7):410–415. doi: 10.1073/pnas.6.7.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao CC, Green S, Stein B, Golden SS. Diel infection of a cyanobacterium by a contractile bacteriophage. Appl Environ Microbiol. 2005;71(8):4276–4279. doi: 10.1128/AEM.71.8.4276-4279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thyrhaug R, Larsen A, Brussaard CPD, Bratbak G. Cell cycle dependent virus production in marine phytoplankton. J Phycol. 2002;38(2):338–343. [Google Scholar]
- 27.Graham JR, Myers J. The role of photosynthesis in the physiology of Ochromonas. J Cell Physiol. 1956;47(3):397–414. doi: 10.1002/jcp.1030470307. [DOI] [PubMed] [Google Scholar]
- 28.Ottesen EA, et al. Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345(6193):207–212. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 29.Ribalet F, Schruth DM, Armbrust EV. flowPhyto: Enabling automated analysis of microscopic algae from continuous flow cytometric data. Bioinformatics. 2011;27(5):732–733. doi: 10.1093/bioinformatics/btr003. [DOI] [PubMed] [Google Scholar]
- 30.Hunter-Cevera KR, et al. Diel size distributions reveal seasonal growth dynamics of a coastal phytoplankter. Proc Natl Acad Sci USA. 2014;111(27):9852–9857. doi: 10.1073/pnas.1321421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaulot D, Marie D. Diel variability of photosynthetic picoplankton in the equatorial Pacific. J Geophys Res C Oceans. 1999;104(C2):3297–3310. [Google Scholar]
- 32.Cavender-Bares K, Mann E, Chisholm S, Ondrusek M, Bidigare R. Differential response of equatorial Pacific phytoplankton to iron fertilization. Limnol Oceanogr. 1999;44(2):237–246. [Google Scholar]
- 33.DuRand MD, Olson RJ, Chisholm SW. Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep Sea Res Part II Top Stud Oceanogr. 2001;48(8-9):1983–2003. [Google Scholar]
- 34.Cailliau C, Claustre H, Vidussi F, Marie D, Vaulot D. Carbon biomass, and gross growth rates as estimated from 14C pigment labelling, during photoacclimation in Prochlorococcus CCMP 1378. Mar Ecol Prog Ser. 1996;145:209–221. [Google Scholar]
- 35.Quay P, Stutsman J, Feely R, Juranek L. Net community production rates across the subtropical and equatorial Pacific Ocean estimated from air-sea delta C-13 disequilibrium. Global Biogeochem Cycles. 2009;23(2):1–15. [Google Scholar]
- 36.Demir-Hilton E, et al. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 2011;5(7):1095–1107. doi: 10.1038/ismej.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergin KL, et al. High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J. 2013;7(7):1322–1332. doi: 10.1038/ismej.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsen FA, Kodner RB, Armbrust EV. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538–538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.