Significance
The critical period of activity-dependent plasticity is considered to be an essential step for the refinement of synaptic connections in the mammalian visual cortex. Gene expression profiling of this period in the mouse indicates that the expression of most genes changes concomitantly with the association areas and that only a small number are region-specific, opening the way to further study of their possible role during normal and pathological development of vision.
Keywords: cerebral cortex, postnatal development, gene expression
Abstract
In contrast to the prenatal development of the cerebral cortex, when cell production, migration, and layer formation dominate, development after birth involves more subtle processes, such as activity-dependent plasticity that includes refinement of synaptic connectivity by its stabilization and elimination. In the present study, we use RNA-seq with high spatial resolution to examine differential gene expression across layers 2/3, 4, 5, and 6 of the mouse visual cortex before the onset of the critical period of plasticity [postnatal day 5 (P5)], at its peak (P26), and at the mature stage (P180) and compare it with the prefrontal association area. We find that, although genes involved in early developmental events such as cell division, neuronal migration, and axon guidance are still prominent at P5, their expression largely terminates by P26, when synaptic plasticity and associated signaling pathways become enriched. Unexpectedly, the gene expression profile was similar in both areas at this age, suggesting that activity-dependent plasticity between visual and association cortices are subject to the same genetic constraints. Although gene expression changes follow similar paths until P26, we have identified 30 regionally enriched genes that are prominent during the critical period. At P180, we identified several hundred differentially expressed gene isoforms despite subsiding levels of gene expression differences. This result indicates that, once genetic developmental programs cease, the remaining morphogenetic processes may depend on posttranslational events.
The cerebral cortex in most mammals develops region-specific cytoarchitecture, including establishment of basic neuronal connections, before birth. Neurons in all neocortical areas are generated and migrate into appropriate layers before or shortly after birth but then refine their synaptic connections by the process of activity-dependent plasticity (1–3). The visual cortex in mice has several well-defined periods under which various developmental processes take place. The period from birth to postnatal day 14 (P14) is characterized by the development of retinotopy and the creation of binocular responses (4, 5) whereas the period of P21–P35 is known as the “critical period” for ocular dominance plasticity where binocularity and binocular matching develop, with its peak at P26 (4, 6). Although great progress has been made in elucidating changes in connectivity and function during the postnatal maturation of the mouse visual cortex, the transcriptional programs enabling these developmental events are still unknown.
We previously established a pipeline, using Next-Gen sequencing, to determine transcriptional programs at high spatial resolution in embryonic tissue (7). More recently, work by Fertuzinhos et al. (8) has compared gene expression differences both temporally (P4–P180) and spatially (cortical layers) in the somatosensory cortex to provide a comprehensive view of the maturation process between and within layers in this region. However, very little is known about the transcriptional profiles that govern the development of the occipital and frontal cortices.
In the present study, we mapped the transcripts involved in the various maturation processes of these neuronal populations and identified the unique set of genetic markers that defines them in the adult brain. Toward this end, we dissected layers 2/3, 4, 5, and 6 of the cerebral cortex in both the occipital visual and frontal association cortices at P5, P26, and P180. These ages were chosen because they span the critical period of the visual cortex. Indeed, we found many interesting gene candidates that may be involved in critical developmental processes, including several genes enriched during the critical period that have layer- or area-specific expression.
Results
RNA Collection, Sequencing, and Differential Gene Expression.
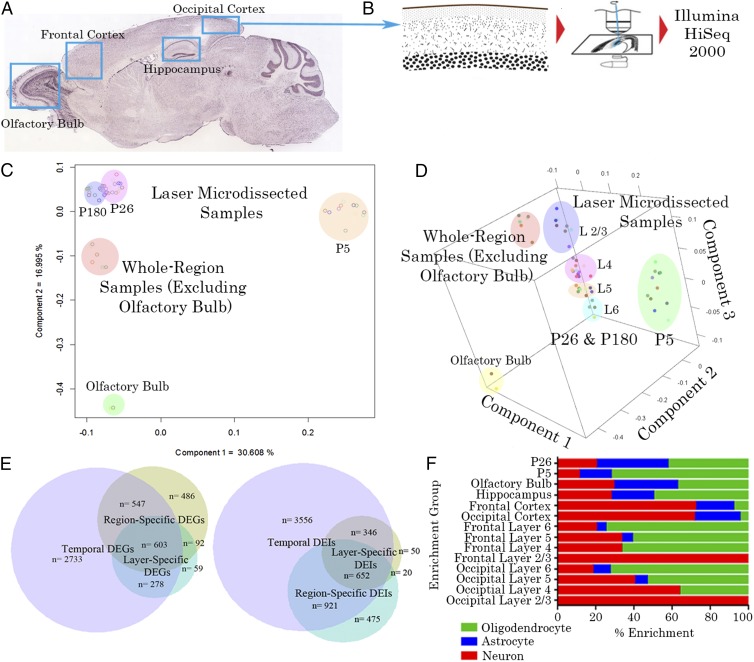
Tissue from layers 2/3, 4, 5, and 6 of the frontal cortex (FC) (prospective association) and occipital cortex (OC) (prospective visual) were obtained at P5, P26, and P180 by laser capture microdissection from six mouse brains (Fig. 1 A and B). As evident below, biological replicates were highly correlated (R2 > ∼0.95) so that additional animals for this particular experiment were deemed unnecessary. To augment these results, we sequenced gross-dissected tissue from these same cortical regions as well as the hippocampus (HC) and olfactory bulb (OB) from two additional animals. Pairwise comparisons were made between layers and regions and across time points to identify significantly differentially expressed genes (DEGs). Genes with a Benjamini–Hochberg-corrected P value (bhp) of <0.05 and fold changes greater than ±2× were considered to be differentially expressed. Because many comparisons were made between biological replicates, we created seven clusters of DEGs depending on whether they were differentially expressed across layers, regions, or time (Table S1). In total, 1,032 genes were differentially expressed between layers, 1,768 across regions, and 4,161 over time. The number of genes that were differentially expressed over a combination of layers, regions, and time can be seen in Fig. 1E. It is evident that there is a larger difference in gene expression across time than between regions and cortical layers. A covariance principal component analysis (Fig. 1 C and D) was performed on all samples to determine clustering of biological replicates. The analysis revealed three components that corresponded well to the three variables that differed between replicates. Component 1 corresponds to age, component 2 to laser microdissection (LMD) versus whole-region dissections, and component 3 to cortical layer for laser-microdissected samples. These data show that the laser microdissection protocol can accurately isolate layers of the cortex from one another and that replicates from different animals and litters are well-correlated.
Fig. 1.
Region and layer-specific expression. (A) The brain regions sampled with bulk dissections (with coarse dots) and regions that were also microdissected (with fine dots). (B) In these areas, layers were isolated by laser microdissection. All sampled tissue was then converted to cDNA and sequenced on an Illumina HiSEq 2000 sequencer. (C) Two-dimensional plot of the two principal components of all samples, separating by age as well as layer- from region-dissected samples, where component 1 corresponds to age and component 2 corresponds to LMD versus whole-region dissections. (D) Three-dimensional principal component analysis (PCA) plot showing a third component (z axis) that divides LMD samples by cortical layer. (E) Venn diagram of genes differentially expressed across time, region, or cortical lamina. The majority of differentially expressed genes (DEGs) are differentially expressed across postnatal time points, with relatively few genes being uniquely differentially expressed across regions or layers. The majority of DEIs are differentially expressed across postnatal time points, but with a larger proportion of total isoforms differentially regulated either uniquely by age or in combination with layer and/or region. (F) Layer, region and age-specific enrichment for neuronal, astrocyte, and oligodendrocyte genes. Upper layers are particularly enriched in neuronal genes, with increasing contributions from astrocytes and oligodendrocytes through the deeper layers. Neuronal genes also dominate in the cortex whereas, in the hippocampus and olfactory bulb, there is a more equal distribution. Finally, the transcriptional profile at P5 is enriched in oligodendrocyte genes whereas, at P26, there is a greater proportion of astrocyte and neuronal genes. P180 is not shown because it was not particularly enriched in any cell-specific genes due to the overall similarity of gene expression to P26.
Novel Transcriptionally Active Sites.
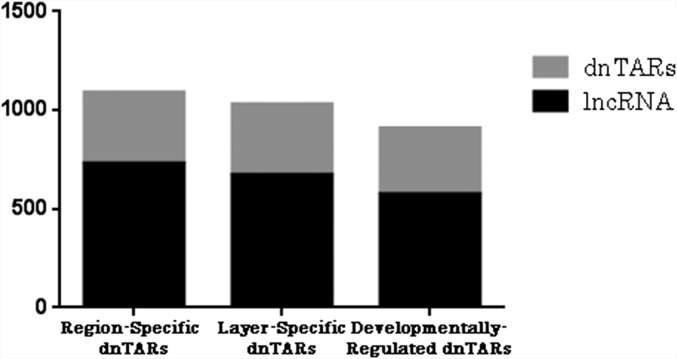
We chose to examine novel transcriptionally active regions (nTARs) in an attempt to classify the temporal and regional variation in RNA expression within areas of the genome outside of coding regions. Because such areas potentially contain gene enhancers, promoters, and long-noncoding and micro-RNAs, differential transcription of these regions could have a variety of effects on the cell. nTARs were identified based on lack of overlap with known genes and minimum 0.5-bp coverage with 100-bp minimum length. Comparisons were made across layers, regions, and time, and the resulting differentially expressed nTARs (dnTARs) were merged across these same groupings to reveal a total of 3,029 dnTARs. There were 1,090 region-specific dnTARs, 1,031 layer-specific dnTARs, and 908 dnTARs across time. A total of 798 nTARs were differentially regulated between P5 and P26, and 825 between P26 and P180, which indicates that, although there is not much difference in the transcriptional profile of protein-coding genes between P26 and P180, there is a substantial shift in expression of noncoding transcripts after P26. There were two region-specific, two layer-specific, and two dnTARs over time that overlapped with known enhancers and 65 region-specific, 114 layer-specific, and 62 temporal dnTAR regions that overlapped with known promoters. The majority of nTARs, including 731/1,090(67.1%) of region-specific dnTARs, 672/1,031 (65.2%) of layer-specific dnTARs, and 574/908 (63.2%) of temporal dnTARs were comprised of long noncoding RNA (lncRNA) (Fig. 2).
Fig. 2.
Breakdown of novel transcriptionally active regions (nTARs) by type. Three columns show nTARs that had differential expression across brain regions and cortical layers and over time, respectively. It can be seen that the majority of the nTARs in all columns are made up of long noncoding RNA (lncRNA).
Functional Analysis of DEGs by Layer.
All functional analyses were performed using the Bingo application within Cytoscape software for Genomic Ontogeny (GO) annotation or the Database for Annotation, Visualization and Integrated Discovery (DAVID) for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. All terms and pathways listed were significant at P < 0.05. Table S2 shows a sample of highly significant GO terms that are enriched for each layer, divided by FC and OC areas. Briefly, layer 2 was enriched for such terms as phosphorylation (OC), nitric oxide-mediated signal transduction (OC), and positive regulation of actin filament bundle assembly (FC); layer 4 for potassium ion transport (FC), negative regulation of appetite, and cerebral cortex radially oriented cell migration (OC); layer 5 for intermediate filament bundle assembly (FC), regulation of exocytosis (OC), and regulation of cyclic nucleotide biosynthetic process (frontal cortex); and layer 6 for cell adhesion (occipital cortex), integrin-mediated signaling pathway (occipital cortex), and neuron recognition (frontal cortex).
Briefly, layer 2 was enriched for calcium signaling pathway (both areas), MAPK signaling pathway (both areas), and long-term potentiation (FC), perhaps reflecting increased synaptic plasticity in this layer; layer 4 for neuroactive ligand–receptor interaction (OC); and layer 5 for tight junction pathway (occipital) and amyotrophic lateral sclerosis pathway (frontal). Layer 6 had no significantly enriched pathways (Table S3).
DEGs by Region.
Frontal cortex was enriched for such terms as gamma-aminobutyric acid secretion, sleep, and regulation of fatty acid beta-oxidation; olfactory bulb for generation of neurons, negative regulation of cell differentiation, and gliogenesis; occipital cortex for peptidyl-serine phosphorylation, synaptic vesicle transport, and intermediate filament bundle assembly; and hippocampus for neurogenesis, central nervous system neuron differentiation, and regulation of exocytosis. Few pathways were differentially enriched between regions, suggesting that the functional differences between these regions may be in the activity regulation of such pathways rather than in the expression levels of their constituents. No pathways were enriched for frontal cortex or hippocampus, but the olfactory bulb had several, including the ribosome, gap junction, and alanine, aspartate, and glutamate metabolism pathways; and occipital cortex was enriched for the ErbB signaling and glioma pathways (Table S4).
DEGs by Age.
Terms such as cell motility, RNA splicing, and chromatin modification were enriched at P5, and regulation of synaptic plasticity, glycolysis, and sodium ion export were enriched for P26. No terms were significantly enriched for P180, indicating that there is little further refinement of gene expression in development after P26. The ribosome, axon guidance, and terpenoid backbone synthesis pathways were all highly enriched at P5 whereas Alzheimer’s disease, oxidative phosphorylation, and phosphatidylinositol signaling system pathways were all highly enriched at P26 (Table 1). Once again, no pathways reached significance at P180. Overall, although the list of genes differentially expressed between P5 and P26 was largely similar between FC and OC (2,165/3,265 or 66.3%), only 3/30 critical-period genes were commonly differentially regulated between cortical areas. However, the general transcriptional profile of these regions at P26 was still highly similar (Table S5).
Table 1.
Critical period-regulated genes by region
| Gene | Enrichment region | ABA confirmed? |
| Fabp7 | O4 | N/A |
| Hba-a2 | F6 | N/A |
| Dcakd | O2 | N/A |
| AK217863 | O4 | N/A |
| Tubb2a | F6 | N/A |
| Serf1 | O4 | N/A |
| Scrt1 | F6 | N/A |
| DQ712046 | O6 | N/A |
| Cldn5 | F2,O4,O5,F6 | N |
| Tfrc | F2 | N/A |
| Apc | O4,O5 | N |
| AK205190 | F2 | N/A |
| Apcdd1 | F2,F4,F6 | N/A |
| AK215842 | F6 | N/A |
| Trp53i11 | F6 | Y |
| Gatm | F5 | N/A |
| Nnat | F5,F6 | Y |
| Peg5 | F5,F6 | Y |
| Prss12 | F6 | Y |
| Marcksl1 | O2,F2,F4,O5,O6,F6 | Y |
| Ptn | O2 | N/A |
| Cd9 | F6 | Y |
| Zfp580 | O2 | N/A |
| Hbb-b1 | F6 | N/A |
| Beta-s | F6 | N/A |
| Slc25a22 | F6 | N/A |
| Defb1 | O6 | N/A |
| Mir715 | O6,F6 | N/A |
| Id3 | F6 | N |
The final column indicates whether the pattern of expression was confirmed by the Allen Brain Atlas.
Cluster Analysis.
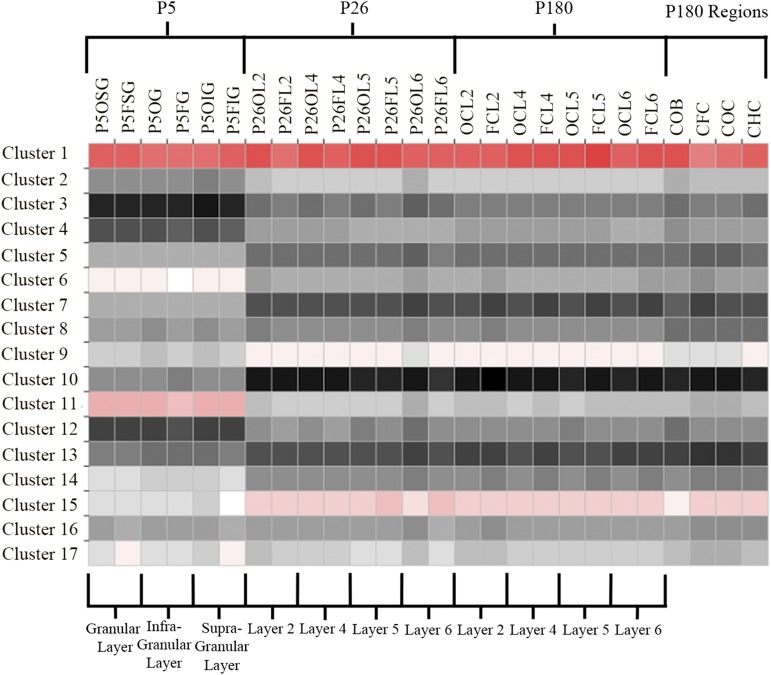
To determine genes that are expressed in functional, regional, and/or temporal groups, we performed a cluster analysis, which revealed a variety of primarily time-dependent clusters, highlighting the importance of such modules in developmental processes. Using the autosome cluster analysis platform (9), with 100 iterations and a P value of P = 0.05, we found 25 clusters, 17 of which had more than 50 genes represented. The average expression levels of each cluster across samples can be seen in Fig. 3. These clusters have very specific patterns of expression across samples, which can be summarized for the top 5 clusters as (i) a high degree of expression over all samples, with particular enrichment in genes involved in mitochondrial and ATP transport-related genes; (ii) expression increasing with time (P180 > P26 > P5), with enrichment in Ras protein signal transduction genes; (iii) expression higher in P180 and P26 versus P5, with gene enrichment in sodium ion transport and cell membrane organization; (iv) low expression at P5 and in olfactory bulb, with enrichment for genes involved in neurotransmitter reuptake and secretion; and (v) high expression at P5 and low expression at P26 and P180. Similarly specific patterns across time, layer, and region are seen throughout the gene clusters (Table S6).
Fig. 3.
Weighted gene coexpression networks. Expression level of each cluster is indicated over each layer/region/age combination. Note that most of these clusters have unique expression patterns over time, region, layer, or a combination of these variables. Codes in column headings are named by age (P5, P26, or nil for P180), area (F, O, HC, and OB), and layer (SG, G, IG, or a number). See Table S18 for definitions of sample names.
Cell-Specific Gene Expression.
We used the cell-specific gene expression data of Cahoy et al. (10) to determine enrichment of neuronal, astrocyte, and oligodendrocyte genes in our layer, region, and age-specific lists. It should be noted that the differences in cell-specific genes across areas (as well as layers and time), do not represent an increased importance of such genes or the presence of these cells in these areas, but rather that there are a larger suite of genes that are specific to the cell types in this particular region. For example, the fact that there are more oligodendrocyte-specific genes in the hippocampus versus the cortex may be interpreted as oligodendrocytes in the hippocampus expressing a larger number of genes that are specific to hippocampal oligodendrocytes and those in the cortex expressing fewer that are specific to cortical oligodendrocytes. In terms of temporal differences, at P5, the expression profile is dominated by oligodendrocyte genes, changing to a more balanced profile at P26 (Fig. 1F). P180 is not shown here because the paucity of P180-specific genes led to no overlap with the cell-specific genes identified in ref. 10. The cortex has a large proportion of neuronal-specific genes, with little unique contribution from oligodendrocytes, whereas the olfactory bulb and hippocampus have a roughly equal contribution from each. Layer 2/3-specific genes are entirely neuronal, with lesser neuronal contributions moving deeper, until oligodendrocyte genes become the majority in layers 5 and 6.
Isoform Analysis.
Because different isoforms of a gene can be independently regulated, and as such regulation may be important for cell- or developmental-specific processes, we reran all samples using cufflinks (11), which allowed us to compare different isoforms of the same gene and also determine differences in overall isoform expression compared with gene expression. Dividing the differentially expressed isoforms (DEIs) into seven groups based on pattern of expression as for DEGs, the majority of isoforms can be seen to be differentially expressed across time points, as for genes, but a larger proportion of total isoforms are differentially regulated by layer or region than in the gene data, indicating that differential splicing may be important in the definition of different cell types and populations (Fig. 1E and Table S7). We also ran GO and KEGG analyses with the gene data to determine whether there are functional differences in DEIs between regions and cortical layers and over time (Tables S8–S13). The most significant finding was that there were many differentially expressed isoforms between P26 and P180, in contrast to the whole-gene data (Tables S12 and S13). P180 cortical cells were enriched for positive regulation of translational termination, positive regulation of potassium ion transport, and G protein signaling, coupled to cAMP nucleotide second messenger and for pathways such as alanine, aspartate and glutamate metabolism, pyruvate metabolism, and prion diseases.
High Spatial Resolution RNA-seq and Quantitative Transcriptome Analysis.
RNA-seq is now accepted as the standard for high-throughput RNA quantification. We decided to leverage our spatial transcriptome analysis with the differential in situ hybridization data provided by the Allen Brain Atlas (ABA) for the same brain regions. We took the top 100 enriched genes from the ABA for the OC, FC, HC, and OB by fold enrichment and compared them with the level of fold enrichment as well as the cardinal rank among the top enriched genes for the brain regions in our dataset. Of the top 100 ABA genes from the OC, FC, HC, and OB, only 17/100, 2/100, 34/100, and 61/100, respectively, were also present in our dataset at significant levels of enrichment (Tables S14–S17). At least two factors may contribute to the low level of concordance: one being differences in defining boundaries of cortical areas, and the other being inherent to the qualitative nature of in situ hybridization. However, despite the somewhat low number of genes in common between the two datasets based on our enrichment list, we did find that most of the top genes in the ABA set were in fact enriched in our dataset (71/100, 32/100, 97/100, and 97/100 for OC, FC, HC and OB, respectively) at subsignificant levels. Again, upon careful inspection, we found that the difference in the FC was due to the areal boundary, which included the OB on several occasions. In this respect, we found that our RNA-seq data across regions and ages could better serve future quantitative analysis and could augment the data available from the ABA.
Discussion
The mammalian cerebral cortex undergoes important processes postnatally, such as the refinement of synaptic connections in the visual cortex, with synaptic pruning continuing until the fourth postnatal month in mice (12), fifth year in macaque (13), and the 30th year in humans (14). The overproduction of synapses, followed by their elimination, is a general property of neural development that has been seen in diverse structures, such as the neuromuscular junction in the chick (15, 16) and the prefrontal cortex in primates (13). To determine the genes that may be involved in these processes, we examined the transcriptional landscape spanning the critical period in the mouse OC visual area and compared it with the FC association area. We isolated genetic determinants in specific neuronal populations situated in the deep, middle, and superficial layers in these regions and identified molecular pathways that were differentially expressed not only across development, but also across cortical layers in these regions. In addition, we discovered several genes that have a specific layer/regional or temporal expression pattern, which may play roles in region-specific development.
Our data constitute a significant advance over available gene expression resources that use alternate methodologies (17). The advantages of RNA-seq over older methods, such as higher sensitivity and dynamic range, have been previously established by several authors (18, 19). A direct comparison between our data and data generated by qualitative or relative methods that depend on probe hybridization would not be fair. Still, we found good concordance across areas when we inspected the top 100 enriched genes as identified by the differential in situ hybridization data in the Allen Brain Atlas (ABA). Our dataset allowed quantitative assessments, and we were therefore able to produce a similar enrichment list for different brain regions. RNA-seq however, is not limited to known transcripts and allows for the discovery of nTARs, as well as splice isoform and complex transcriptome detection. Finally, our quantitative data also represent an excellent normative resource for future studies involving mouse models of neurological and psychiatric disorders.
Gene expression changes in the prefrontal association and primary visual areas in the postnatal mouse develop in parallel, at least until P26. This result may not seem intuitive, but there are precedents in primates where different sensory and association areas develop and eliminate overproduced synapses synchronously (20) and express their major neurotransmitter receptors on the same schedule (21). It is significant that this schedule is not related to time of birth or exposure to the visual environment because it retains the same tempo in prematurely delivered animals. Despite similar timing of expression changes in the two areas examined, there are some notable genes that are region-specific, particularly during the critical period for the visual cortex.
In addition, our data enabled us to characterize the various cortical layers, regions, and ages by their gene expression profile. Overall, we found that, although at P5 there are many highly expressed genes indicating that the cortex is still in an immature state, most of these developmental processes are complete by P26, which has minimal differences in gene level expression compared with the adult P180 cortex. Surprisingly, there were a large number of differentially expressed isoforms between P26 and P180, which suggests that further development after the critical period may depend on differential splicing events. In terms of layer-specific gene expression, we unexpectedly found layer 2/3 to be enriched in pathways and genes involved in cytoskeletal assembly, calcium signaling pathway, MAPK signaling pathway, and long-term potentiation, which suggest that these neurons may be capable of high levels of synaptic plasticity. This result stands in contrast to previous reports indicating that layer 4 is particularly privileged in terms of plasticity (22, 23). We found also that layer 5 is enriched for such terms as intermediate filament bundle assembly and regulation of exocytosis, which may be related to the fact that it contains projection neurons that are generally larger and have long-range axons projecting to subcortical targets, necessitating increased production of axonal proteins. In terms of regional differences, both the hippocampus and olfactory bulb, two regions known for continuing neuronal production into adulthood, are particularly enriched in genes related to neurogenesis and gliogenesis, confirming the sensitivity of our analysis to pick up such differences.
We found that, both in adulthood and much more so at P26, there were few differentially expressed genes between the FC and OC, indicating common expression patterns across these two functionally distinct areas. However, developmentally regulated area-specific genes were more enriched at P26. Although these genes were spread across a variety of functional categories, there were, unsurprisingly, several involved in synaptic plasticity and associated pathways (Tubb2a, Marcksl1, Ptn), which may be important for area-specific processes of synaptic pruning and activity-dependent plasticity during the critical period. There were other genes, including transcriptional inhibitors (Id3, Scrt1) and genes related to processes of neuronal differentiation (Nnat, Peg5), that could also be related to the final stages of maturation of specific cell types. More difficult to explain was the presence of hemoglobin genes (Hba-a2, Hbb-b1, and Beta-s), a tumor suppressor (Apc), a gene involved in the glycine metabolic pathway (Gatm), a micro-RNA (Mir715), and a protease (Prss12) although future research will determine the specific roles these genes may have in the critical period and associated plasticity. The frontal cortex was also significantly enriched overall for GABAergic genes, which could be related to an increased importance of inhibition in this region. Across regions and layers at P5, cells were found to express genes involved in cell motility, RNA splicing, chromatin modification, and axon guidance, reflecting epigenetic changes, the final stages of cell migration, and axonal growth in the early postnatal animal. Both cortices at P26 were enriched for genes involved in synaptic plasticity, sodium ion transport, and the phosphatidylinositol signaling system, supporting previous work indicating that the dominant process at this age is synaptic/structural plasticity (24) and the refinement of cortical and subcortical connectivity.
No terms or pathways were significantly enriched in P180 animals in the gene expression data although several categories were differentially expressed in the splice isoform data, such as positive regulation of translational termination, positive regulation of potassium ion transport, and G protein signaling, indicating that posttranscriptional processes may be responsible for the further refinement of receptor complement and signaling pathways necessary for the development of the mature cortex. We found, in general, that there is a large difference between differentially expressed genes and differentially expressed isoforms between the various regions and cortical layers, suggesting that, although gene expression differences were moderate between cell populations, there were many more differential splicing events that delineate them. Fig. 1E also indicates region, layer, and developmental differences in cell-specific genes. At P5, oligodendrocyte genes dominate, which shows the importance of developmental myelination processes at this age. By P26, there was a more balanced profile, with equal contributions from all three cell types. There were also some significant laminar differences, with neuronal genes dominating layer 2/3-specific gene expression and then having a decreasing contribution in layers 4–6, when oligodendrocyte and astrocyte genes became more prominent, indicating that there may be more layer-specific oligodendrocyte and astrocyte markers at increasing cortical depth. Not only neurons, but also astrocytes and oligodendrocytes, may express different genes across regions as well because there were many region-specific markers in all three cell types across regions, although more oligodendrocyte genes were found in the hippocampus and olfactory bulb and more neuronal genes in the cortex.
The RNA-seq data obtained in the present analysis provide new insights and a resource for examining region- and cortical layer-specific development and function in mice. We expect that the functional significance of some of these genes may help us understand region-specific synaptic refinement. In addition, as similar datasets from other species become available, such transcriptional differences between cortical laminae and areas can provide insight into how species differences in brain structure and function have evolved and how they may become dysfunctional.
Methods
High-Resolution RNA-seq.
The flash-frozen brains of all animals were sectioned, and cortical lamina were dissected by laser microdissection (LMD). RNA purification, sequencing, and differential gene expression were performed as described in Ayoub et al. (7) (Fig. 1 A and B). All procedures were approved by the Institutional Animal Care and Use Committee of Yale University and conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (25).
Statistical Methods.
Pairwise comparisons were made between layers, between regions, and across time points to identify significantly differentially expressed genes (DEGs). A likelihood ratio test was used to calculate P values for genes between samples, using a conservative Benjamini–Hochberg-corrected P value (bhp) of 10−5 as the cutoff.
Full details on experimental methods can be found in SI Methods.
SI Methods
Animals.
We used C57BL/6 males, which were killed at P5, P26, or P180 (n = 2 per age group). Mice were weaned at P28 and singly housed until killing at P180. Mice were killed by cervical dislocation and decapitation, and the brain was quickly extracted and flash frozen in OCT gel on dry ice-isopentane slurry. Brains were stored at −80 °C until further processing. All procedures were approved by the Institutional Animal Care and Use Committee of Yale University and conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (25).
Laser Microdissection.
LMD was performed on 20-µm-thick cryostat sections mounted on PEN slides (Leica Microsystems) as previously detailed in Ayoub et al. (7). Samples were microdissected on a fully motorized LMD6000 microscope (Leica) under a 10× objective. Tissue cut by the laser fell into the tops of 650-µL microcentrifuge tubes containing 35 µL of 0.1% β-mercaptoethanol in RLT buffer (Qiagen). Layers were defined based on the morphology and density of cells. Layers 2/3 and 6 are easily seen with toluidine staining because their cells are much denser than neighboring layers. The upper border of layer 2/3 was defined as the edge of the dense band of cells, excluding the acellular layer 1. Layer 6 consisted of only the cellular layer and excluded the white matter and subventricular zone. Layer 4 has smaller cells than layer 2/3 and is denser than layer 5, making it easily recognizable. Because the laser cuts a wide track into the tissue that ablates the cells within it, the laser line was placed on the borders of the above-defined layers and provided a buffer zone of cells that were ablated rather than included in the samples of the layers on either side. In this way, the purity of the samples was ensured so that they were enriched only for cells in the middle of the corresponding layer. Sections from one animal were used for each biological replicate, and the two replicates in each area came from different litters of pups, which were killed at different times. After dissection, the tubes were briefly centrifuged to bring all of the tissue and buffer to the bottom of the tube, and the tissue pieces were disrupted by vortexing and stored on dry ice temporarily until being placed in a −80° freezer until RNA extraction. Extraction was done using an RNeasy Microkit (Qiagen) according to the manufacturer’s instructions provided for laser microdissected tissue. Up to four tubes were combined at this stage to obtain an RNA concentration sufficient for library preparation and sequencing (>20 ng of total RNA per sample).
RNA Sequencing.
The quality of the RNA samples was checked using a BioAnalyzer (Agilent). Libraries for mRNA-seq were made using the standard Illumina protocol. Four samples were multiplexed per sequencing lane and then sequenced at a 75-bp paired-end read length using an Illumina HiSeq 2000 sequencer at the Yale Center for Genomic Analysis. Sequencing data were further processed on the Yale High Performance Computing (HPC) cluster as previously reported (7). Reads were aligned to the mouse genome (mm9), and their normalized abundance was calculated using the RPKM (reads per kilobase of exon model per million mapped reads) measure, from which composite gene models were created to get a total value for each expressed gene.
Statistical Methods.
Pairwise comparisons were made between layers, between regions, and across time points to identify significantly differentially expressed genes (DEGs). A likelihood ratio test was used to calculate P values for genes between samples, using a conservative Benjamini–Hochberg-corrected P value (bhp) of 10−5 as the cutoff. Both biological replicates were pooled for each pairwise analysis. Genes with a bhp of <0.05 and fold changes greater than ±2× were considered to be differentially expressed. In all, 48 samples were included in the analysis, comprising 4 layers (2/3, 4, 5 and 6) × 2 cortical areas (frontal and occipital) × 3 ages (P5, P26, and P180) × 2 technical replicates each. Within each age and area, every layer was compared with every other to identify layer-specific genes (six comparisons each); at each age, each layer in the occipital cortex was compared with its cognate layer in the frontal cortex; and each layer in each area was compared across time points. This comparison yielded 36 DEG lists for the first group of comparisons, 12 for the second, and 24 for the third, for a total of 72 comparisons made. Using the conservative bhp P value and a fold-cutoff of >2×, the number of DEGs was relatively small for each comparison. Some comparisons yielded as few as nine DEGs (P5, infragranular versus supergranular layers) and others as many as 2,201 (frontal layer 2, P5 versus P180). Most comparisons between layers of the same age and region yielded ∼150 DEGs (range 19–397), most comparisons between regions of the same age ∼500 DEGs (range 12–1042), and most comparisons across ages ∼1,000 DEGs (range 12–2201) (Table S18). For the in vitro morphology analysis, a 3 × 2 ANOVA was performed with posttransfection day and conditions as factors. Sidak’s multiple comparisons test was used for post hoc comparisons between individual groups.
Functional Analysis.
GO annotation terms for DEGs were acquired using the Bingo application within the open source platform Cytoscape (www.cytoscape.org/), which is used for visualization and analysis of complex networks. For each comparison of interest, Bingo listed GO terms that were enriched in that gene set, giving the term itself, as well as a bhp-corrected P value for each. GO terms with a corrected P value of <0.05 were considered to be significantly enriched. In addition, the functional annotation tool of the online resource DAVID (david.abcc.ncifcrf.gov/) was used for pathway analysis. For each list of DEGs, significantly differentially expressed pathways were identified, and bhp-corrected P values were generated based on the KEGG pathway annotation. Pathways with a bhp of <0.05 were considered to be differentially expressed between the samples.
Cluster Analysis.
Heatmaps, unsupervised hierarchical clustering, and principal component analyses were generated in R for gene sets. jColorGrid (jcolorgrid.sourceforge.net) was used to create other heatmaps for DEGs of interest. In addition, the GeneMania application within Cytoscape was used to create gene networks for several genes of interest based on coexpression, colocalization, genetic interaction, pathway, physical interaction, predicted interaction, and shared protein domain data. To obtain clusters of genes based on coexpression, we used the autosome cluster analysis platform (9), with 100 iterations and a P value of P = 0.05. We then accepted all clusters with greater than 50 genes in each for display and downstream analysis.
Supplementary Material
Acknowledgments
We thank S. Reilly for help with the RNA-seq; M. Pappy and S. Rodriguez for technical assistance; C. Castaldi, D. Harrison, and K. Bilguvar for assistance with RNA-seq at the Yale Center for Genome Analysis (supported by NIH Grant RR1989s); and members of the P.R. laboratory for critical comments on the manuscript. This work was supported by Natural Sciences and Engineering Research Council of Canada Fellowship 403855-2011 and the Department of Psychology, Yale University (to J.B.); NIH Grants DA02399 (to P.R. and A.E.A.), EY002593 (to P.R.), and R01NS014841 (to P.R.); and the Kavli Institute for Neuroscience at Yale University (P.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509323112/-/DCSupplemental.
References
- 1.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10(9):647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 2.Kerschensteiner D. Spontaneous network activity and synaptic development. Neuroscientist. 2013;20(3):272–290. doi: 10.1177/1073858413510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274(5290):1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 4.Espinosa JS, Stryker MP. Development and plasticity of the primary visual cortex. Neuron. 2012;75(2):230–249. doi: 10.1016/j.neuron.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huberman AD, Speer CM, Chapman B. Spontaneous retinal activity mediates development of ocular dominance columns and binocular receptive fields in v1. Neuron. 2006;52(2):247–254. doi: 10.1016/j.neuron.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon JA, Stryker MP. Experience-dependent plasticity of binocular responses in the primary visual cortex of the mouse. J Neurosci. 1996;16(10):3274–3286. doi: 10.1523/JNEUROSCI.16-10-03274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayoub AE, et al. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108(36):14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fertuzinhos S, et al. Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Reports. 2014;6(5):938–950. doi: 10.1016/j.celrep.2014.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman AM, Cooper JB. AutoSOME: A clustering method for identifying gene expression modules without prior knowledge of cluster number. BMC Bioinformatics. 2010;11:117. doi: 10.1186/1471-2105-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo Y, Lin A, Chang P, Gan WB. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005;46(2):181–189. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Bourgeois JP, Rakic P. Changes of synaptic density in the primary visual cortex of the macaque monkey from fetal to adult stage. J Neurosci. 1993;13(7):2801–2820. doi: 10.1523/JNEUROSCI.13-07-02801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petanjek Z, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci USA. 2011;108(32):13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Changeux JP, Danchin A. Selective stabilisation of developing synapses as a mechanism for the specification of neuronal networks. Nature. 1976;264(5588):705–712. doi: 10.1038/264705a0. [DOI] [PubMed] [Google Scholar]
- 16.Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 17.Lyckman AW, et al. Gene expression patterns in visual cortex during the critical period: synaptic stabilization and reversal by visual deprivation. Proc Natl Acad Sci USA. 2008;105(27):9409–9414. doi: 10.1073/pnas.0710172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232(4747):232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- 21.Lidow MS, Rakic P. Neurotransmitter receptors in the proliferative zones of the developing primate occipital lobe. J Comp Neurol. 1995;360(3):393–402. doi: 10.1002/cne.903600303. [DOI] [PubMed] [Google Scholar]
- 22.Maffei A, Nelson SB, Turrigiano GG. Selective reconfiguration of layer 4 visual cortical circuitry by visual deprivation. Nat Neurosci. 2004;7(12):1353–1359. doi: 10.1038/nn1351. [DOI] [PubMed] [Google Scholar]
- 23.De Paola V, et al. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49(6):861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 24.Majewska A, Sur M. Motility of dendritic spines in visual cortex in vivo: Changes during the critical period and effects of visual deprivation. Proc Natl Acad Sci USA. 2003;100(26):16024–16029. doi: 10.1073/pnas.2636949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. National Research Council (2011) Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC), 8th Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.