Abstract
Background
Although metabolomic approaches have begun to document numerous changes that arise in end stage renal disease (ESRD), how these alterations relate to established metabolic phenotypes in uremia is unknown.
Methods
In 200 incident hemodialysis patients we used partial least squares discriminant analysis to identify which among 166 metabolites could best discriminate individuals with or without diabetes, and across tertiles of body mass index, serum albumin, total cholesterol, and systolic blood pressure.
Results
Our data do not recapitulate metabolomic signatures of diabetes and obesity identified among individuals with normal renal function (e.g. elevations in branched chain and aromatic amino acids) and highlight several potential markers of diabetes status specific to ESRD, including xanthosine-5-phosphate and vanillylmandelic acid. Further, our data identify significant associations between elevated tryptophan and long-chain acylcarnitine levels and both decreased total cholesterol and systolic blood pressure in ESRD. Higher tryptophan levels were also associated with higher serum albumin levels, but this may reflect tryptophan’s significant albumin binding. Finally, an examination of the uremic retention solutes captured by our platform in relation to 24 clinical phenotypes provides a framework for investigating mechanisms of uremic toxicity.
Conclusions
In sum, these studies leveraging metabolomic and metabolic phenotype data acquired in a well-characterized ESRD cohort demonstrate striking differences from metabolomics studies in the general population, and may provide clues to novel functional pathways in the ESRD population.
Electronic supplementary material
The online version of this article (doi:10.1186/s12882-015-0100-y) contains supplementary material, which is available to authorized users.
Keywords: Dialysis, Metabolism, Metabolomics, Uremic toxins
Background
End stage renal disease (ESRD) is characterized by various metabolic disturbances linked to adverse outcomes, but the nature of these associations is poorly understood or even counterintuitive [1]. For example, low serum albumin has consistently been linked to cardiovascular mortality in dialysis [2, 3], yet the association is not simply a result of inadequate nutrition, as intradialytic parenteral nutrition does not always improve survival in malnourished ESRD patients [4], and alternative pathways including inflammation are known to contribute [5]. Dyslipidemia is also common in ESRD, including atherogenic increases in carbamylated and oxidized-LDL cholesterol. Clinical trials, however, have failed to demonstrate a survival benefit with statin therapy in ESRD, despite substantial reductions in LDL cholesterol [6, 7]. Indeed, several studies have identified an inverse association between cholesterol levels and uremic cardiovascular risk [8, 9]. Similar paradoxical associations, sometimes described as examples of ‘reverse epidemiology’ in ESRD, have been noted with body mass index (BMI) and blood pressure [8, 10–12] and survival on hemodialysis.
Metabolomic approaches enable high resolution interrogation of metabolic phenotypes. For example, studies have begun to highlight specific metabolites associated with obesity and diabetes risk in the general population, particularly branched-chain and aromatic amino acids, but also short chain acylcarnitines, the glutamate/glutamine ratio, and bile acids [13–23]. Ultimately, these findings may lead to a more refined understanding of underlying disease mechanisms. Whether these and other associations identified in the burgeoning metabolomics literature extend to the ESRD population is unknown. While initial applications of metabolomics to nephrology research have generated a broad view of uremia [24] and the immediate effects of the hemodialysis procedure [25, 26], no studies have explicitly examined the cross-sectional relationship between uremic metabolite alterations and metabolic or other clinical phenotypes captured in a given ESRD study population.
We recently performed liquid chromatography-mass spectrometry (LC-MS) based metabolite profiling on plasma obtained from incident hemodialysis subjects in the Accelerated Mortality on Renal Replacement (ArMORR) cohort, and highlighted baseline levels of long-chain acylcarnitines as markers of future cardiovascular death in a longitudinal study [27]. Using this data set, we now leverage the rich clinical phenotyping available in ArMORR to examine cross-sectional associations between plasma metabolite levels and select metabolic phenotypes. Specifically, we sought to identify associations between metabolites and metabolic phenotype data in this well-characterized ESRD cohort, hypothesizing that these findings would differ from metabolomic associations derived from the general population. The aim of such work is to gain new insights into the functional metabolic pathways that underlie the poorly understood associations between clinical outcomes in ESRD and variables such as BMI, albumin, cholesterol, and blood pressure. Further, we examined the correlation between the subset of metabolites measured by our platform that are known uremic retention solutes and numerous clinical variables captured in ArMORR. Taken together, these results provide a new perspective on the metabolic disturbances that accompany and arise in ESRD.
Methods
Study population
The Accelerated Mortality on Renal Replacement (ArMORR) study is a prospective cohort study of 10,044 incident hemodialysis patients in any of 1,056 U.S. centers operated by Fresenius Medical Care North America between June 2004 and August 2005. Full details have been previously published [27–31]. All subjects underwent 1 year of follow-up except for those who died (15.2 %), voluntarily discontinued dialysis (5 %), underwent kidney transplantation (3 %), recovered renal function (4 %), or transferred to a dialysis unit outside the Fresenius system (12 %). Clinical data were prospectively collected by physicians at the point-of-care and included demographics, coexisting conditions, and routine laboratory studies. Plasma samples drawn at the beginning of a dialysis session within 14 days of initiation of outpatient hemodialysis, and that would otherwise have been discarded after routine clinical testing, were saved and stored in liquid nitrogen. We previously evaluated the associations between metabolite profile and mortality in the ArMORR cohort in a study of 100 randomly selected patients who died within the first year of dialysis from a cardiovascular cause, and 100 patients who were alive 1 year after starting dialysis, frequency matched for age, sex, and race [27]. We included this entire group of individuals in the current study. This study was approved by the Massachusetts General Hospital IRB, which waived the need for informed consent because all personal identifiers were removed from the blood samples and clinical data before transfer to the investigators.
Metabolite profiling
We applied two distinct LC-MS based methods to distinct plasma aliquots for each study subject. Amino acids, amino acid derivatives, urea cycle intermediates, nucleotides and other positively charged polar metabolites were profiled as previously described [14]. Briefly, 10 μL of plasma were extracted with 90 μL of 74.9:24.9:0.2 vol/vol/vol acetonitrile/methanol/formic acid containing valine-d8 (Sigma-Aldrich; St Louis, MO). After centrifugation, supernatants underwent hydrophobic interaction chromatography using a 150 × 2.1 mm Atlantis hydrophobic interaction chromatography column (Waters, Milford, MA), and MS data were acquired on a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX, Foster City, CA) using ESI and MRM in the positive ion mode. Organic acids, sugars, bile acids, and other negatively charged polar metabolites were profiled as previously described [32]. Briefly, 30 μL of plasma were extracted with the addition of four volumes of 80:20 vol/vol methanol/water containing isotope-labeled inosine-15 N4, supernatants underwent chromatography on a 150 × 2.0 mm Luna NH2 column (Phenomenex; Torrance, CA), and MS data were acquired using a 5500 QTRAP triple quadrupole mass spectrometer (AB SCIEX; Foster City, CA) using ESI and MRM in the negative ion mode.
Statistical analyses
We first examined the association between plasma metabolites and pre-selected metabolic phenotypes: diabetes status, BMI, serum albumin, total cholesterol, and systolic blood pressure (SBP). We focused on these phenotypes because of their strong links to metabolism and their established, but incompletely understood, associations with ESRD mortality [33–39]. To investigate differences in metabolite profiles across these phenotypes, continuous outcomes measures (BMI, albumin, total cholesterol, and SBP) were divided into tertiles. Diabetes status was classified as “yes” or “no”.
Because of the number of metabolites measured by our LC-MS platform (166 total, Additional file 1: Table S1), many with significant inter-correlations, we performed partial least squares discriminant analysis (PLS-DA) to visualize the linear components that discriminate individuals across the categories (tertiles or class) of metabolic phenotypes. PLS-DA was performed on log transformed and auto-scaled (mean-centered and divided by the standard deviation of each metabolite) data using MetaboAnalyst 2.0 [40, 41]. This program performs PLS regression using the plsr function provided by the pls package in R, and classification and cross validation using the corresponding wrapper function offered by the caret package. Variable Importance in Projection (VIP) scores generated by this program estimate the importance of each variable in the projection used within the PLS model. A variable with a VIP score greater than 1 can be considered important in a given model. The metabolites with the highest VIP scores were further analyzed by comparing their levels across classes using Mann-U-Whitney and Kruskal-Wallis tests as appropriate.
Finally, heatmaps were created to represent Pearson correlation (r) and P-value matrices between available clinical variables in the cohort and the subset of metabolites measured by our platform that have previously been described as uremic retention solutes (http://www.uremic-toxins.org) [27, 42–45].
To account for multiple testing, we used a Bonferroni corrected significance threshold of P < 0.0003 (0.05/166) for comparisons of individual metabolites across phenotype class. For the correlation matrices, we used an adjusted significance threshold of P < 0.00007 (0.05/[29 metabolites* 24 clinical variables]). A sensitivity analysis was performed to gauge the influence of case–control status (i.e. whether or not a given individual died of a cardiovascular cause within one year of starting dialysis) on the associations described herein. Stratified analysis by case status did not alter the statistical significance of the models described. Therefore, we did not stratify models by mortality status and analyzed the entire group of 200 individuals together as a cohort. All analyses were performed using SAS software version 9.1.3 (SAS Institute) and MetaboAnalyst 2.0 software (www.metaboanalyst.ca).
Results and discussion
Cohort characteristics
As shown in Table 1, the mean age of the study population was 69.5 years and 69 % of subjects were white. There was an equal representation of males and females and nearly half of the individuals had a history of diabetes or had diabetes listed as their cause of ESRD (49 %). The mean BMI was 26.5 kg/m2 (SD ±7.6 kg/m2) and the mean SBP was 144 mmHg (±27 mmHg). A minority of individuals reported a lipid disorder (12 %) and the median total cholesterol level was 162 mg/dL (quartile1-quartile3, 127–188 mg/dL). The median serum albumin level was 3.6 g/dl (3.2-3.8 g/dL).
Table 1.
Baseline characteristics of the study sample
| Variable | Total n = 200 |
|---|---|
| Age, years | 69.5 ± 13.6 |
| Male | 53 % (106) |
| Race | |
| White | 69 % (138) |
| Black | 24 % (48) |
| Other | 7 % (14) |
| Coexisting conditions | |
| Coronary artery disease | 18 % (36) |
| Lipid disorders | 12 % (23) |
| Congestive heart failure | 18 % (35) |
| Cause of end-stage renal disease | |
| Diabetesa | 49 % (97) |
| Hypertensive renal disease | 38 % (75) |
| Glomerulonephropathy | 4 % (7) |
| Vascular access | |
| Fistula | 24 % (47) |
| Graft | 13 % (25) |
| Catheter | 59 % (118) |
| Body Mass Index (kg/m2) | 26.5 ± 7.6 |
| Systolic blood pressure (mmHg) | 144.4 ± 27.3 |
| Diastolic blood pressure (mmHg) | 72.5 ± 18.6 |
| Urea reduction ratio | 68.6 ± 10.3 |
| Laboratory data | |
| N-terminal pro-BNP (ng/L) | 8029 (3,757 – 21,404) |
| Troponin T (μg/L) | 0.14 (0.04 – 0.13) |
| Total cholesterol (mg/dL) | 162 (127 – 188) |
| LDL (mg/dL) | 83 (63 – 103) |
| HDL (mg/dL) | 41 (32 – 47) |
| Triglycerides (mg/dL) | 157 (93 – 199) |
| Creatinine (mg/dL) | 5.7 (4.0 – 7.0) |
| White blood cell (109/L) | 8.3 (6.2 – 9.8) |
| Hemoglobin (g/dL) | 10.4 (9.5 – 11.2) |
| Albumin (g/dL) | 3.5 (3.2 – 3.8) |
| Ferritin (ng/mL) | 285.7 (81.5 – 346.0) |
| Transferrin saturation (%) | 20.2 (13.0 – 24.0) |
| Phosphorus (mg/dL) | 4.5 (3.4 – 5.4) |
| Parathyroid hormone (pg/mL) | 268.6 (124.0 – 235.0) |
Categorical data are percentages (counts). Counts may not equal total n due to missing data. Clinical measures are means ± SD. Laboratory values are median (quartile 1 – quartile 3)
BNP, brain natriuretic peptide
aIncludes all patients with diabetes
Examination of metabolite profiles and select metabolic phenotypes
The PLS-DA approach allowed us to visualize and extract the metabolites that best separated individuals (Figs. 1,2,3,4 and 5; left panels) across phenotype tertile or class (Table 2). Because four of the clinical phenotypes we studied (BMI, serum albumin, total cholesterol, and SBP) are continuous measures, we recognize that tertile cut-offs do not demarcate distinct physiologic or pathophysiologic processes. Thus, plot overlap across tertiles was expected. Variable importance in projection (VIP) provided a score for each metabolite, ranking the metabolites according to their predictive power in the PLS model; the fifteen metabolites with the highest VIP scores for each plot are shown in Figs. 1, 2, 3, 4 and 5 (right panels), and the levels of these metabolites across tertiles (or class) of the phenotypes with corresponding test statistics are shown in Tables 3-7.Values are median peak area for the metabolites (quartile 1, quartile 3)*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Fig. 1.
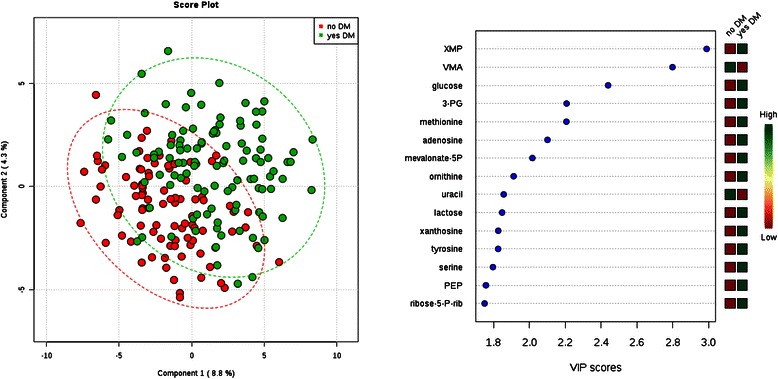
Comparison of metabolite profiles and diabetes status. Study subjects were grouped by diabetes status (yes/no). Left: Partial least squares discriminant analysis (PLS-DA) score plot for the study population separated by phenotype class. Oval outlines denote 95 % confidence intervals. Right: The variable importance in projection (VIP) scores for the 15 metabolites with the greatest score. The colored boxes on the right indicate the direction of metabolite alterations across phenotype class. Abbreviations: XMP, Xanthosine-5-phosphate; VMA, vanillylmandelic acid; 3-PG, 3-phosphoglycerate; mevalonate-5P, mevalonate-5-phosphate; PEP, phosphoenolpyruvate; ribose-5P, ribose-5-phosphate and ribulose-5-phosphate
Fig. 2.
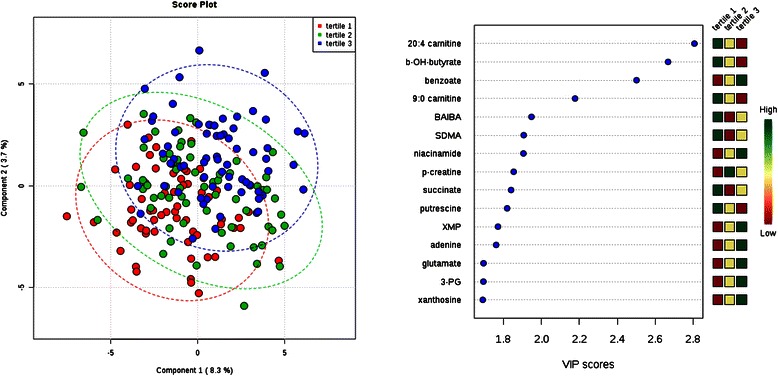
Comparison of metabolite profiles across tertile of body mass index (BMI). Study subjects were grouped by BMI tertile (tertile 1 = lowest BMI). Left: Partial least squares discriminant analysis (PLS-DA) score plot for the study population separated by phenotype class. Oval outlines denote 95 % confidence intervals. Right: The variable importance in projection (VIP) scores for the 15 metabolites with the greatest score. The colored boxes on the right indicate the direction of metabolite alterations across phenotype tertile. Abbreviations: BAIBA, β-aminoisobutyric acid; SDMA, symmetric dimethylarginine; 3-PG, 3-phosphoglycerate; b-OH-butyrate, β-hydroxybutryate; p-creatine, phosphocreatine; XMP, Xanthosine-5-phosphate
Fig. 3.
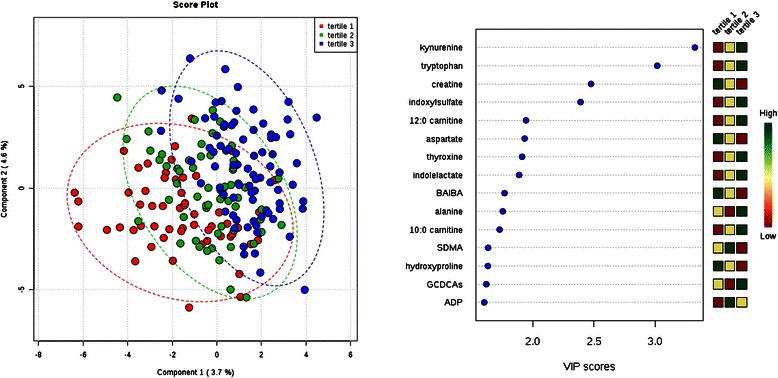
Comparison of metabolite profiles across tertile of serum albumin. Study subjects were grouped by albumin tertile (tertile 1 = lowest). Left: Partial least squares discriminant analysis (PLS-DA) score plot for the study population separated by phenotype class. Oval outlines denote 95 % confidence intervals. Right: The variable importance in projection (VIP) scores for the 15 metabolites with the greatest score. The colored boxes on the right indicate the direction of metabolite alterations across phenotype tertile. Abbreviations: BAIBA, β-aminoisobutyric acid; SDMA, symmetric dimethylarginine; GCDCAs, glycodeoxycholate and glycochenodeoxycholate; ADP, adenosine diphosphate
Fig. 4.
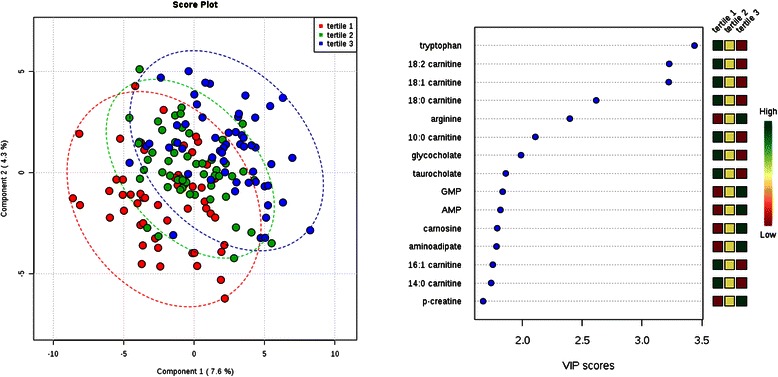
Comparison of metabolite profiles across tertile of total cholesterol. Study subjects were grouped by cholesterol tertile (tertile 1 = lowest). Left: Partial least squares discriminant analysis (PLS-DA) score plot for the study population separated by phenotype class. Oval outlines denote 95 % confidence intervals. Right: The variable importance in projection (VIP) scores for the 15 metabolites with the greatest score. The colored boxes on the right indicate the direction of metabolite alterations across phenotype tertile. Abbreviations: GMP, guanosine monophosphate; AMP, adenosine monophosphate; p-creatine, phosphocreatine
Fig. 5.
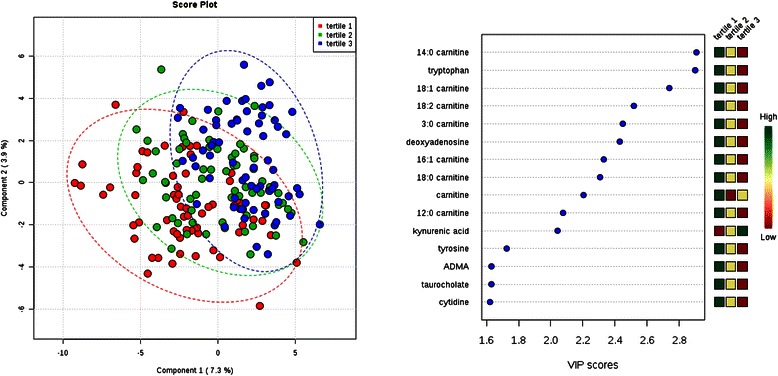
Comparison of metabolite profiles across tertile of systolic blood pressure. Study subjects were grouped by systolic blood pressure tertile (tertile 1 = lowest). Left: Partial least squares discriminant analysis (PLS-DA) score plot for the study population separated by phenotype class. Oval outlines denote 95 % confidence intervals. Right: The variable importance in projection (VIP) scores for the 15 metabolites with the greatest score. The colored boxes on the right indicate the direction of metabolite alterations across phenotype tertile. Abbreviations: ADMA, asymmetric dimethylarginine
Table 2.
Primary metabolic phenotypes by tertiles
| Tertile | Measure | |
|---|---|---|
| Body mass index (kg/m2) | 1 | 20.2 ± 3.9 |
| 2 | 25.6 ± 1.3 | |
| 3 | 34.2 ± 7.6 | |
| Albumin (g/dl) | 1 | 3.1 (2.8, 3.2) |
| 2 | 3.6 (3.4, 3.7) | |
| 3 | 4.0 (3.8, 4.2) | |
| Cholesterol (mg/dl) | 1 | 116.0 (106.0-126.0) |
| 2 | 154.0 (146.0-163.0) | |
| 3 | 209.5 (188.0-236.0) | |
| Systolic blood pressure (mmHg) | 1 | 116.1 ± 10.0 |
| 2 | 142.2 ± 7.0 | |
| 3 | 174.6 ± 20.2 |
Clinical measures are mean ± SD and laboratory values are median (quartile 1 to quartile 3)
Total n = 200
Table 3.
Cross sectional analysis of diabetes status and select metabolites
| no DM | DM | P | |
|---|---|---|---|
| XMP | 13 585 (8 386, 29 579) | 62 101 (14 261, 158 938) | 5.0E-7* |
| VMA | 173 153 (107 165, 279 690) | 104 186 (67 902, 164 683) | 1.5E-6* |
| Glucose | 2 935 (1 046, 5 294) | 6 245 (2 777, 9 807) | 3.1E-6* |
| 3-PG | 2 475 150 (1 882 036, 333 2264) | 3 767 372 (2 250 378, 5 598 373) | 4.9E-5* |
| Methionine | 195 026 (152 633, 239 841) | 243 230 (195 219, 283 910) | 4.5E-5* |
| Adenosine | 1 942 (1 463, 3 457) | 3 078 (1 706, 5 137) | 8.6E-4 |
| mevalonate-5P | 192 139 (136 927, 291 930) | 292 904 (165 128, 433 871) | 2.8E-4* |
| ornithine | 1 385 224 (1 083 311, 1 721 772) | 1 657 626 (1 351 270, 1 913 549) | 3.2E-4* |
| uracil | 6 888 567 (4 490 348, 9 622 913) | 4 784 283 (3 740 917, 6 592 407) | 5.7E-4 |
| lactose | 3 140 699 (2 178 153, 4 835 219) | 4 519 336 (2 900 447, 6 650 384) | 3.8E-4 |
| xanthosine | 10 839 (7 518, 22 311) | 22 007 (8 845, 36 474) | 1.0E-3 |
| tyrosine | 222 479 (174 354, 277 562) | 247 875 (210 621, 309 004) | 2.2E-3 |
| serine | 352 671 (280 146, 415 607) | 381 134 (319 491, 476 035) | 4.6E-3 |
| PEP | 312 003 (189 719, 522 172) | 462 016 (257 705, 925 860) | 1.3E-3 |
| ribose-5-P-rib | 144 443 (104 797, 208 719) | 180 078 (116 088, 322 799) | 8.7E-3 |
Values are median peak area for the metabolites (quartile 1, quartile 3)
*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Table 7.
Cross sectional analysis of systolic blood pressure tertiles and select metabolites
| 1 | 2 | 3 | P | |
|---|---|---|---|---|
| 14:0 carnitine | 10 549 (7 384, 17 510) | 7 855 (6 040, 11 626) | 6 824 (4 844, 9 486) | 9.0E-6* |
| tryptophan | 433 424 (297 696, 573 865) | 366 630 (261 744, 444 478) | 285 101 (229 380, 350 075) | 1.2E-6* |
| 18:1 carnitine | 86 406 (56 919, 127 989) | 70 273 (47 764, 105 108) | 52 275 (32 871, 76 097) | 8.7E-6* |
| 18:2 carnitine | 55 715 (35 972, 84 528) | 44 392 (31 334, 69 607) | 30 733 (21 812, 54 084) | 5.5E-5* |
| 3:0 carnitine | 770 205 (556 166, 976 463) | 604 794 (378 770, 724 282) | 549 619 (381 463, 708 095) | 5.4E-5* |
| deoxyadenosine | 882 (605, 1 330) | 831 (455, 1 011) | 571 (441, 787) | 7.5E-5* |
| 16:1 carnitine | 70 042 (50 052, 97 483) | 53 904 (39 997, 77 945) | 47 577 (36 257, 68 016) | 4.2E-4 |
| 18:0 carnitine | 12 434 (8 777, 17 403) | 10 456 (7 790, 15 356) | 8 806 (6 509, 12 276) | 4.2E-4 |
| carnitine | 4 940 542 (4 375 772, 5 974 275) | 4 311 636 (3 616 956, 4 936 736) | 4 279 513 (3 743 960, 4 756 471) | 2.1E-5* |
| 12:0 carnitine | 39 844 (22 621, 61 584) | 33 433 (22 258, 39 850) | 27 094 (17 488, 43 033) | 8.5E-3 |
| kynurenic acid | 63 185 (35 805, 129 532) | 86 131 (60 982, 133 796) | 116 334 (67 598, 169 586) | 9.4E-4 |
| tyrosine | 255 780 (216 337, 319 582) | 231 195 (200 964, 273 325) | 223 332 (177 881, 270 817) | 8.4E-3 |
| ADMA | 309 751 (281 198, 370 574) | 290 947 (261 258, 344 552) | 284 594 (248 154, 328 619) | 1.1E-2 |
| taurocholate | 103 888 (39 457, 284 746) | 65 463 (26 657, 196 110) | 59 392 (25 323, 157 090) | 1.8E-2 |
| cytidine | 60 033 (36 540, 80 779) | 51 105 (32 838, 73 636) | 44 821 (26 784, 61 200) | 2.1E-2 |
Values are median peak area for the metabolites (quartile 1, quartile 3)
*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
For individuals with or without diabetes, glucose was one of the strongest discriminating metabolites, with higher levels among the individuals with diabetes (Fig. 1). Xanthosine-5-phosphate (XMP) and vanillylmandelic acid (VMA), however, had even higher VIP scores than glucose, with XMP levels higher and VMA levels lower among the individuals with diabetes (P = 5.0 × 10−7 and 1.5 × 10−6 for XMP and VMA, respectively). Other significant differences included higher levels of 3-phosphoglyceric acid (P = 4.9 × 10−5), methionine (P = 4.5 × 10−5), mevalonate-5-phosphate (P = 2.8 × 10−4), and ornithine (P = 3.2 × 10−4) among individuals with diabetes. Tyrosine, an aromatic amino acid, had a nominal association with diabetes status (P = 1.0 × 10−3). There were no significant associations between tyrosine levels, or any aromatic or branched-chain amino acid levels, with BMI (Table 4). In fact, none of these metabolites had even a nominal association with BMI in univariate comparisons (data not shown). Overall, the ability of metabolite profiles to discriminate individuals across tertiles of BMI was poor (Fig. 2). Among the top metabolites by VIP score, only 20:4 carnitine (arachidonyl carnitine), β-hydroxybutyrate, and benzoate had nominal P-values in univariate analysis (Table 4).Values are median peak area for the metabolites (quartile 1, quartile 3)*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Table 5.
Cross sectional analysis of albumin tertiles and select metabolites
| 1 | 2 | 3 | P | |
|---|---|---|---|---|
| kynurenine | 425 998 (298 054, 535 460) | 478 927 (395 468, 596 735) | 552 251 (433 248, 679 481) | 1.8E-4* |
| tryptophan | 273 066 (223 467, 413 194) | 315 338 (255 914, 423 927) | 393 551 (301 885, 501 066) | 2.5E-4* |
| creatine | 2 157 292 (1 397 576, 3 355 087) | 1 907 461 (1 145 455, 2 791 140) | 1 570 693 (1 182 023, 2 037 120) | 4.9E-3 |
| indoxylsulfate | 42 510 713 (24 175 943, 58 375 318) | 55 564 881 (34 527 474, 67 400 369) | 57 398 073 (33 359 446, 80 148 935) | 1.3E-2 |
| 12:0 carnitine | 28 261 (18 195, 40 622) | 30 496 (18 540, 44 940) | 36 915 (24 245, 50 553) | 2.7E-2 |
| aspartate | 36 448 (25 083, 49 840) | 34 626 (24 685, 48 518) | 26 533 (20 120, 36 067) | 1.9E-3 |
| thyroxine | 2 332 (1 929, 2 738) | 2 523 (1 879, 2 916) | 2 712 (2 115, 3 244) | 1.8E-2 |
| indolelactate | 12 722 (8 573, 19 334) | 15 627 (11 171, 22 311) | 16 269 (11 922, 22 700) | 3.6E-2 |
| BAIBA | 46 979 (27 423, 106 593) | 42 313 (24 146, 80 713) | 34 051 (23 583, 57 272) | 1.0E-1 |
| alanine | 1 280 822 (985 715, 1 604 978) | 1 249 845 (1 028 428, 1 494 869) | 1 337 859 (1 117 636, 1 758 661) | 6.9E-2 |
| 10:0 carnitine | 33 566 (21 210, 61 857) | 48 413 (25 775, 68 324) | 47 884 (31 469, 72 628) | 5.8E-2 |
| SDMA | 896 522 (765 176, 1 090 288) | 925 219 (772 578, 1 063 241) | 824 248 (730 601, 974 498) | 5.2E-2 |
| hydroxyproline | 235 578 (162 155, 374 510) | 203 966 (149 720, 306 721) | 198 496 (150 564, 259 207) | 1.5E-1 |
| GCDCAs | 487 186 (203 751, 773 938) | 421 065 (248 114, 748 019) | 715 307 (356 712, 1 064 255) | 1.4E-2 |
| ADP | 280 998 (155 075, 610 214) | 539 330 (278 012, 877 605) | 455 070 (164 681, 1 239 020) | 1.2E-2 |
Values are median peak area for the metabolites (quartile 1, quartile 3)
*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Values are median peak area for the metabolites (quartile 1, quartile 3)*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Table 4.
Cross sectional analysis of BMI tertiles and select metabolites
| 1 | 2 | 3 | P | |
|---|---|---|---|---|
| 20:4 carnitine | 296 (246, 378) | 263 (216, 324) | 253 (207, 292) | 7.8E-3 |
| b-OH-butyrate | 508 950 (249 015, 1 818 943) | 395 440 (234 716, 895 354) | 343 616 (172 233, 717 989) | 2.9E-2 |
| benzoate | 15 216 (11 526, 24 891) | 18 382 (12 187, 28 788) | 21 271 (14 892, 34 600) | 4.1E-2 |
| 9:0 carnitine | 17 878 (11 801, 23 836) | 16 514 (9 560, 24 702) | 13 239 (9 472, 20 371) | 5.7E-2 |
| BAIBA | 49654 (26452, 107799) | 36205 (24704, 57849) | 38844 (23864, 74057) | 1.3E-1 |
| SDMA | 942527 (769351, 1098799) | 852551 (750082, 970184) | 826704 (729114, 1062038) | 7.4E-2 |
| niacinamide | 209151 (170065, 248736) | 214718 (175857, 281191) | 246744 (187545, 297341) | 6.8E-2 |
| p-creatine | 42402 (31941, 51249) | 47705 (39559, 61036) | 48786 (37472, 57166) | 4.0E-2 |
| succinate | 17 776 284 (15 961 017, 22 206 915) | 15 637 602 (12 262 309, 19 434 811) | 16 309 449 (12 852 577, 19 680 091) | 6.1E-3 |
| putrescine | 68559 (44891, 109979) | 54285 (18371, 87225) | 53 504 (12105, 93032) | 1.3E-1 |
| XMP | 21 819 (11 029, 53 381) | 20 694 (8 856, 143 900) | 37 771 (12 370, 121 441) | 1.7E-1 |
| adenine | 90 351 (61 426, 165 720) | 109 819 (79 107, 201 534) | 116 489 (76 249, 194 627) | 1.1E-1 |
| glutamate | 870 295 (691 881, 1 109 208) | 992 968 (715 437, 1 375 593) | 1 031 076 (714 204, 1 443 438) | 1.2E-1 |
| 3-PG | 2 824 102 (1 855 508, 4 336 931) | 2 788 537 (1 930 889, 5 190 633) | 3 294 065 (2 358 954, 5 383 792) | 9.2E-2 |
| xanthosine | 12 923 (7 425, 24 088) | 10 883 (7 604, 33 249) | 19 991 (8 413, 30 597) | 1.7E-1 |
Values are median peak area for the metabolites (quartile 1, quartile 3)
*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Thus, our data from an ESRD population do not recapitulate observations derived in the general population linking diabetes and obesity with elevations in branched chain and aromatic amino acids (except for the nominal association with tyrosine noted above), short chain acylcarnitines, the glutamate/glutamine ratio, or bile acids [2]. One potential explanation is that kidney function (and dysfunction) has a direct effect on select metabolites. The interaction between kidney function and amino acid metabolism, in particular, has been closely examined. For example, the kidney is known to make a substantial contribution to whole body tyrosine appearance via intra-organ phenylalanine hydroxylation [46]. Further, the metabolic acidosis that results from renal failure leads to increased leucine oxidation [47], and indeed, generalized muscle catabolism. The significant associations we did identify with diabetes status in ESRD, e.g. XMP (a purine breakdown product) and VMA (a catecholamine metabolite), have not been reported in the general population. To what extent these alterations reflect distinct pathophysiologic processes in uremia warrant further investigation.
For individuals in different tertiles of serum albumin, PLS-DA demonstrated moderate separation across groups (Fig. 3). Tryptophan and its downstream metabolite kynurenine were significantly higher in individuals with higher serum albumin levels (P = 1.8 × 10−4 and 2.5 × 10−4, respectively). Two additional tryptophan metabolites, indoxyl sulfate and indole lactate, had a trend for higher levels among individuals with higher serum albumin levels. Given recent observations that ascribe a functional role for tryptophan metabolism, specifically through indoleamine-2, 3-dioxygenase (IDO), in modulating the immune system and vascular tone [48, 49], it is an appealing link between metabolism, nutritional status, inflammation, and cardiovascular risk in ESRD. However, these data should be interpreted with caution. Because tryptophan and its catabolites are hydrophobic, their positive correlation with albumin may reflect their significant protein-binding [50] – our LC-MS method measures total, not free, plasma metabolite levels. A similar mechanism may underlie the trend for association between thyroxine and albumin levels. By contrast, creatine, the metabolite with the third highest VIP score, had a trend for higher levels among individuals with lower serum albumin levels. In a study that examined spent media from cultured muscle cells treated with mitochondrial respiratory chain inhibitors, as well as plasma obtained from individuals with respiratory chain diseases, creatine levels were reproducibly elevated [51]. In cell culture, extracellular creatine was inversely correlated with the intracellular phosphocreatine/creatine ratio, suggesting that elevated plasma creatine may signal a low energetic state in tissues using the creatine phosphate shuttle. Interestingly, elevated plasma creatine and decreased plasma phosphocreatine levels both have a nominal association with increased 1-year cardiovascular mortality among incident dialysis patients in ArMORR [27]. These observations raise the possibility that impaired mitochondrial respiration in muscle is linked to the pathogenesis of hypoalbuminemia in dialysis.
For individuals across tertiles of total cholesterol and SBP, PLS-DA again demonstrated moderate separation across tertiles (Figs. 4 and 5). For both analyses, long-chain acylcarnitines were among the metabolites with the highest VIP scores, with 18:2 carnitine (linoleylcarnitine, P = 9.0 × 10−7), 18:1 carnitine (oleoylcarnitine, P = 6.0 × 10−7), and 18:0 carnitine (stearoylcarnitine, P = 7.7 × 10−5) all significantly higher in individuals with lower total cholesterol levels (Table 6,) and linoleylcarnitine (P = 5.5 × 10−5) and oleoylcarnitine (P = 8.7 × 10−6) significantly higher in individuals with lower SBP (Table 7); select short-chain and medium-chain acylcarnitines were also significantly elevated among individuals with lower SBP. Like albumin, low cholesterol and blood pressure are established predictors of increased mortality in ESRD [8, 9, 11, 12]. These findings are consistent with our observation that long-chain acylcarnitines are associated with 1-year cardiovascular mortality in ESRD [27], and motivate further interest in long-chain acylcarnitines as markers, or even mediators, of altered lipid metabolism and cardiovascular function in uremia. Tryptophan, one of the strongest discriminators of albumin tertile, was also a top hit in regards to total cholesterol and SBP, with higher tryptophan significantly associated with lower levels for both. Finally, higher arginine levels were significantly associated with higher total cholesterol and higher deoxyadenosine levels were significantly associated with lower SBP.Values are median peak area for the metabolites (quartile 1, quartile 3)*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Table 6.
Cross sectional analysis of total cholesterol tertiles and select metabolites
| 1 | 2 | 3 | P | |
|---|---|---|---|---|
| tryptophan | 440 068 (345 130, 586637) | 323192 (257580, 429806) | 274624 (223120, 324798) | 1.7E-9* |
| 18:2 carnitine | 60 494 (38 250, 107 981) | 49 737 (27 163, 73 344) | 32 084 (19 167, 53 863) | 9.0E-7* |
| 18:1 carnitine | 88 418 (58 090, 144 102) | 69 442 (40 568, 103 682) | 47 134 (33 127, 74 763) | 6.0E-7* |
| 18:0 carnitine | 12 789 (10 118, 18 240) | 10 879 (7 872, 14 622) | 8 466 (6 103, 12 433) | 7.7E-5* |
| arginine | 175 576 (136 362, 259 524) | 187 566 (132 389, 324 585) | 321 420 (216 851, 491 848) | 9.6E-6* |
| 10:0 carnitine | 52 498 (28 731, 92 694) | 40 556 (26 190, 67 085) | 34 244 (20 094, 52 420) | 7.0E-3 |
| glycocholate | 193 580 (60 425, 672 001) | 105 938 (61 765, 208 013) | 76 985 (42 135, 222 324) | 5.6E-3 |
| taurocholate | 128 124 (30 180, 379 765) | 91 894 (35 067, 188 948) | 47 535 (24 706, 128 025) | 1.9E-2 |
| GMP | 640 409 (341 616, 884 723) | 544 860 (409 493, 969 998) | 831 864 (565 936, 1 246 583) | 8.9E-3 |
| AMP | 2 898 935 (2 275 762, 3 795 325) | 3 153 490 (2 679 559, 4 251 281) | 3 708 724 (3 046 005, 4 670 016) | 1.6E-3 |
| carnosine | 23 811 (15 639, 77 524) | 34 287 (14 160, 119 781) | 64 617 (29 846, 195 284) | 6.5E-3 |
| aminoadipate | 139 402 (119 264, 184 086) | 167 760 (130 029, 210 530) | 186 821 (144 705, 242 940) | 3.3E-3 |
| 16:1 carnitine | 68 049 (46 259, 99 787) | 65 354 (35 653, 77 665) | 49 527 (39 008, 71 026) | 2.1E-2 |
| 14:0 carnitine | 9 893 (6 819, 17 108) | 8 969 (5 659, 12 834) | 7 848 (5 373, 10 960) | 3.1E-2 |
| p-creatine | 42 102 (30 390, 51 406) | 46 713 (36 982, 55 510) | 50 600 (37 472, 70 045) | 1.8E-2 |
Values are median peak area for the metabolites (quartile 1, quartile 3)
*P-value significant at the Bonferroni adjusted level of 3.0 × 10−4
Relation of uremic solutes with twenty-four clinical phenotypes
To examine the specificity of the long-chain acylcarnitine-phenotype associations described above, we looked more broadly at the correlation between all of the metabolites captured on our platform that are uremic retention solutes and 24 phenotypes captured in ArMORR. Correlations between previously reported uremic retention solutes measured by our LC-MS platform, including long-chain acylcarnitines, and a broad array of clinical phenotypes measured in ArMORR are shown in Fig. 6 (full results are shown in Additional file 1: Tables S2 and S3). The correlations for glucose, as measured by the mass spectrometer, provide an anchor for interpreting this data – LC-MS glucose had a strong positive correlation with glucose measured by the clinical laboratory (r = 0.80, P <10−15), and a modest negative correlation with serum sodium (r = −0.21, P = 7.0 × 10−3) that did not reach statistical significance, but is well recognized in the clinic (i.e. dilutional hyponatremia due to hyperglycemia). Aside from the correlation between the two glucose measures, the strongest correlations identified by this broader examination of uremic metabolites were indeed between long-chain acylcarnitines and lipid traits (total and LDL cholesterol) and blood pressure. Among lipid phenotypes, long-chain acylcarnitines such as oleoylcarnitine were more strongly correlated with total (r = −0.45, P = 2.6 × 10−9) and LDL cholesterol (r = −0.43., P = 1.7 × 10−8) than with HDL cholesterol (r = −0.15, P = 5.7 × 10−2) or triglycerides (r = −0.27, P = 5.0 × 10−4). Similarly, oleoylcarnitine was more strongly correlated with SBP (r = −0.38, P = 3.4 × 10−8) than diastolic blood pressure (r = −0.25, P = 3.0 × 10−3). Long-chain acylcarnitines were not significantly correlated with any non-lipid or non-blood pressure phenotype.
Fig. 6.
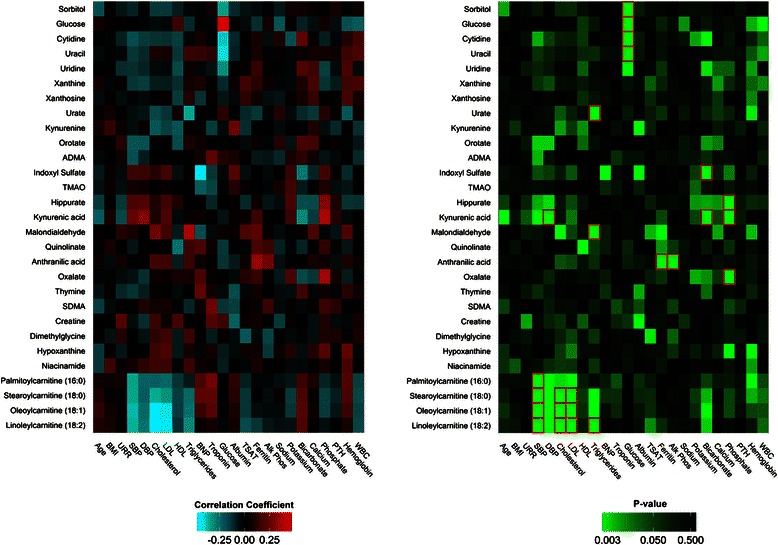
Relation of uremic solutes with 24 clinical phenotypes in the ArMORR cohort. Correlation coefficient (r, left) and P values (right) generated from Pearson correlation analyses of metabolites and available clinical variables in the ArMORR cohort. Squares outlined in red on the P value matrix represent values significant at the Bonferroni adjusted level of P < 7.4 × 10−5. Abbreviations: ADMA, asymmetric dimethylarginine; TMAO, trimethylamine-N-oxide; SDMA, symmetric dimethylarginine; BMI, body mass index; URR, urea reduction ratio; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL, low density lipoprotein; HDL, high density lipoprotein; BNP, b-type natriuretic peptide, TSAT, transferrin saturation; Alk Phos, alkaline phosphatase; PTH, parathyroid hormone; WBC, white blood cell
Among the other uremic retention solutes included in our analysis, only kynurenic and anthranilic acid (both tryptophan metabolites downstream of IDO) had more than one significant correlation with a clinical variable (Fig. 6). Kynurenic acid was significantly correlated with diastolic blood pressure (r = 0.32, P = 5.0 × 10−6), bicarbonate (r = −0.31, P = 1.1 × 10−5), and phosphorous (r = 0.38, P = 5.7 × 10−8), whereas anthranilic acid was significantly correlated with alkaline phosphatase (r = 0.29, P = 6.3 × 10−5) and ferritin (r = 0.35, P = 9.7 × 10−7), a frequently used marker of inflammation in hemodialysis. Notably, the other metabolites that had a nominal association with ferritin included two other tryptophan metabolites, kynurenic acid (r = 0.16, P = 2.6 × 10−2) and quinolinate (r = 0.20, P = 6.3 × 10−3), and malondialydehyde (r = 0.24, P = 9.0 × 10−4), an oxidative stress marker. These findings reinforce a potential link between the IDO pathway and uremic inflammation. Sorbitol (r = −0.34, P = 3.4 × 10−5), cytidine (r = −0.40, P = 1.1 × 10−6), and uracil (r = −0.42, P = 1.7 × 10−7) were significantly correlated with plasma glucose; urate (r = −0.35, P = 5.2 × 10−6) and malondialdehyde (r = 0.38, P = 5.7 × 10−7) were significantly correlated with triglycerides; hippurate (r = 0.31, P = 7.0 × 10−6) and oxalate (r = 0.36, P = 1.6 × 10−7) were significantly correlated with phosphorous; and indoxyl sulfate (r = −0.28, P = 6.8 × 10−5) was significantly correlated with bicarbonate. Indoxyl sulfate, which has been found to have several proinflammatory effects in model systems [52], had no correlation with ferritin (nor transferrin saturation, nor white blood cell count), but had a trend for a negative correlation with BNP (r = −0.44, P = 4.0 × 10−4).
Although significant work has been devoted to understanding the in vitro effects of uremic retention solutes, less is known about how many of these metabolites relate to clinical traits in humans. Thus, in addition to demonstrating the specificity of select long-chain acylcarnitine associations, our examination of uremic metabolites against clinical phenotypes provides a resource for future uremic toxin research.
Limitations
Our study has several limitations. First, by examining cross-sectional relationships between metabolite levels and various clinical phenotypes, our study is unable to address causation. Instead, by applying parsimonious selection procedures and conservative adjusted significance thresholds, we seek to highlight notable associations for future investigations. Second, study subjects were selected in the context of a case–control study of 1-year cardiovascular mortality, raising the possibility of confounding by case status. However, as noted in the Methods, the statistical significance of the findings are not changed when analyzed stratified by case status. Nevertheless, we acknowledge that our sample is not a random selection of incident dialysis patients, potentially limiting the generalizability of our findings. Third, residual confounding from other sources is likely to influence the associations described, as multivariate adjustments were not pursued. However, because the relationship between various clinical phenotypes and metabolite levels in uremia is unknown, we did not want to obscure novel biological associations by statistical adjustment. Thus, we present these raw associations as a framework for interpreting and appropriately adjusting select findings in future metabolomics studies in ESRD. Finally, our results require replication in an independent sample, ideally selected randomly and including both incident and prevalent dialysis patients.
Conclusions
In this report, we describe the small molecule alterations that accompany various metabolic phenotypes in ESRD. For phenotypes like diabetes and obesity that have been examined using metabolomics in the general population, our data reinforce the notion that unique pathophysiologic processes arise at ESRD onset. Our finding that long-chain acylcarnitines are strongly and inversely correlated with cholesterol and blood pressure corroborate their potential value as markers of cardiovascular risk in ESRD, whereas the association of tryptophan levels across several phenotypes is of interest given recent studies that assign a functional role for tryptophan metabolism in inflammation and vascular biology. Ultimately, we hope that the breadth of the data presented herein serves as a springboard for investigating mechanisms of uremic toxicity and identifying therapeutic targets to improve the unacceptably high morbidity and mortality attributable to ESRD.
Acknowledgements
SK received support from the National Kidney Foundation Young Investigator award and NIH award KL2TR001100; EPR was supported by the Extramural Grant Program by Satellite Healthcare, a not-for-profit renal care provider, and NIH grant K08-DK-090142. REG received support from NIH 2R01DK081572 and NIH 1R01 HL098280; RT received support from NIH award K24 DK094872.
Abbreviations
- ESRD
End stage renal disease
- LDL
Low density lipoprotein
- BMI
Body mass index
- LC-MS
Liquid chromatography-mass spectrometry
- ArMORR cohort
Accelerated Mortality on Renal Replacement cohort
- SBP
Systolic blood pressure
- PLS-DA
Partial least squares discriminant analysis
- VIP
Variable importance in projection
- XMP
Xanthosine-5-phosphate
- VMA
Vanillylmandelic acid
- IDO
Indoleamine-2,3-dioxygenase
- BNP
b-type natriuretic peptide
Additional file
Metabolites measured within the sample set. Table S2. Pearson correlation coefficients for select metabolites and available clinical variables in the ArMORR cohort. Table S3. P-values from Pearson correlations using select metabolites and available clinical variables in the ArMORR cohort.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SK and EPR designed the study, performed the statistical analysis, and wrote the manuscript. EPR and CBC performed the MS experiments. JJD, GO, ASM helped with sample processing, data analysis, and generating figures. REG aided with study design, data analysis, and manuscript drafting. RT established the cohort from which plasma samples were derived. All authors read and approved the final manuscript.
Contributor Information
Sahir Kalim, Phone: (617) 726-5050, Email: skalim@mgh.harvard.edu.
Clary B. Clish, Email: clary@broadinstitute.org
Joseph J. Deferio, Email: joseph.deferio@downstate.edu
Guillermo Ortiz, Email: gortiz@partners.org.
Alexander S. Moffet, Email: amoffet2@illinois.edu
Robert E. Gerszten, Email: rgerszten@partners.org
Ravi Thadhani, Email: rthadhani@partners.org.
Eugene P. Rhee, Email: eprhee@partners.org
References
- 1.Kopple JD. How to reconcile conventional and altered risk factor patterns in dialysis patients. Semin Dial. 2007;20(6):602–5. doi: 10.1111/j.1525-139X.2007.00362.x. [DOI] [PubMed] [Google Scholar]
- 2.Kalantar-Zadeh K, Kilpatrick RD, Kuwae N, McAllister CJ, Alcorn H, Jr, Kopple JD, et al. Revisiting mortality predictability of serum albumin in the dialysis population: time dependency, longitudinal changes and population-attributable fraction. Nephrol Dial Transplant. 2005;20(9):1880–8. doi: 10.1093/ndt/gfh941. [DOI] [PubMed] [Google Scholar]
- 3.Kovesdy CP, Kalantar-Zadeh K. Why is protein-energy wasting associated with mortality in chronic kidney disease? Semin Nephrol. 2009;29(1):3–14. doi: 10.1016/j.semnephrol.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cano NJ, Fouque D, Roth H, Aparicio M, Azar R, Canaud B, et al. Intradialytic parenteral nutrition does not improve survival in malnourished hemodialysis patients: a 2-year multicenter, prospective, randomized study. J Am Soc Nephrol. 2007;18(9):2583–91. doi: 10.1681/ASN.2007020184. [DOI] [PubMed] [Google Scholar]
- 5.Dukkipati R, Kopple JD. Causes and prevention of protein-energy wasting in chronic kidney failure. Semin Nephrol. 2009;29(1):39–49. doi: 10.1016/j.semnephrol.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Fellstrom BC, Jardine AG, Schmieder RE, Holdaas H, Bannister K, Beutler J, et al. Rosuvastatin and cardiovascular events in patients undergoing hemodialysis. N Engl J Med. 2009;360(14):1395–407. doi: 10.1056/NEJMoa0810177. [DOI] [PubMed] [Google Scholar]
- 7.Wanner C, Krane V, Marz W, Olschewski M, Mann JF, Ruf G, et al. Atorvastatin in patients with type 2 diabetes mellitus undergoing hemodialysis. N Engl J Med. 2005;353(3):238–48. doi: 10.1056/NEJMoa043545. [DOI] [PubMed] [Google Scholar]
- 8.Kalantar-Zadeh K, Block G, Humphreys MH, Kopple JD. Reverse epidemiology of cardiovascular risk factors in maintenance dialysis patients. Kidney Int. 2003;63(3):793–808. doi: 10.1046/j.1523-1755.2003.00803.x. [DOI] [PubMed] [Google Scholar]
- 9.Lowrie EG, Lew NL. Death risk in hemodialysis patients: the predictive value of commonly measured variables and an evaluation of death rate differences between facilities. Am J Kidney Dis. 1990;15(5):458–82. doi: 10.1016/S0272-6386(12)70364-5. [DOI] [PubMed] [Google Scholar]
- 10.Kopple JD, Zhu X, Lew NL, Lowrie EG. Body weight-for-height relationships predict mortality in maintenance hemodialysis patients. Kidney Int. 1999;56(3):1136–48. doi: 10.1046/j.1523-1755.1999.00615.x. [DOI] [PubMed] [Google Scholar]
- 11.Iseki K, Miyasato F, Tokuyama K, Nishime K, Uehara H, Shiohira Y, et al. Low diastolic blood pressure, hypoalbuminemia, and risk of death in a cohort of chronic hemodialysis patients. Kidney Int. 1997;51(4):1212–7. doi: 10.1038/ki.1997.165. [DOI] [PubMed] [Google Scholar]
- 12.Port FK, Hulbert-Shearon TE, Wolfe RA, Bloembergen WE, Golper TA, Agodoa LY, et al. Predialysis blood pressure and mortality risk in a national sample of maintenance hemodialysis patients. Am J Kidney Dis. 1999;33(3):507–17. doi: 10.1016/S0272-6386(99)70188-5. [DOI] [PubMed] [Google Scholar]
- 13.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–26. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menni C, Fauman E, Erte I, Perry JR, Kastenmuller G, Shin SY, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62(12):4270–6. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floegel A, Stefan N, Yu Z, Muhlenbruch K, Drogan D, Joost HG, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–48. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–31. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wewalka M, Patti ME, Barbato C, Houten SM, Goldfine AB. Fasting Serum Taurine-conjugated Bile Acids are Elevated in Type 2 Diabetes: and Do Not Change with Intensification of Insulin. J Clin Endocrinol Metab. 2014;99(4):1442–51. doi: 10.1210/jc.2013-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerhard GS, Styer AM, Wood GC, Roesch SL, Petrick AT, Gabrielsen J, et al. A role for fibroblast growth factor 19 and bile acids in diabetes remission after Roux-en-Y gastric bypass. Diabetes Care. 2013;36(7):1859–64. doi: 10.2337/dc12-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes. 2013;8(1):52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore SC, Matthews CE, Sampson JN, Stolzenberg-Solomon RZ, Zheng W, Cai Q, et al. Human metabolic correlates of body mass index. Metabolomics. 2014;10(2):259–69. doi: 10.1007/s11306-013-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atzler D, Schwedhelm E, Zeller T. Integrated genomics and metabolomics in nephrology. Nephrol Dial Transplant. 2014;29(8):1467–74. doi: 10.1093/ndt/gft492. [DOI] [PubMed] [Google Scholar]
- 25.Rhee EP, Souza A, Farrell L, Pollak MR, Lewis GD, Steele DJ, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21(6):1041–51. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato E, Kohno M, Yamamoto M, Fujisawa T, Fujiwara K, Tanaka N. Metabolomic analysis of human plasma from haemodialysis patients. Eur J Clin Invest. 2011;41(3):241–55. doi: 10.1111/j.1365-2362.2010.02398.x. [DOI] [PubMed] [Google Scholar]
- 27.Kalim S, Clish CB, Wenger J, Elmariah S, Yeh RW, Deferio JJ, et al. A plasma long-chain acylcarnitine predicts cardiovascular mortality in incident dialysis patients. J Am Heart Assoc. 2013;2(6) doi: 10.1161/JAHA.113.000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutierrez OM, Mannstadt M, Isakova T, Rauh-Hain JA, Tamez H, Shah A, et al. Fibroblast growth factor 23 and mortality among patients undergoing hemodialysis. N Engl J Med. 2008;359(6):584–92. doi: 10.1056/NEJMoa0706130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng M, Wolf M, Lowrie E, Ofsthun N, Lazarus JM, Thadhani R. Survival of patients undergoing hemodialysis with paricalcitol or calcitriol therapy. N Engl J Med. 2003;349(5):446–56. doi: 10.1056/NEJMoa022536. [DOI] [PubMed] [Google Scholar]
- 30.Teng M, Wolf M, Ofsthun MN, Lazarus JM, Hernan MA, Camargo CA, Jr, et al. Activated injectable vitamin D and hemodialysis survival: a historical cohort study. J Am Soc Nephrol. 2005;16(4):1115–25. doi: 10.1681/ASN.2004070573. [DOI] [PubMed] [Google Scholar]
- 31.Thadhani R, Tonelli M. Cohort studies: marching forward. Clin J Am Soc Nephrol. 2006;1(5):1117–23. doi: 10.2215/CJN.00080106. [DOI] [PubMed] [Google Scholar]
- 32.Rhee EP, Clish CB, Ghorbani A, Larson MG, Elmariah S, McCabe E, et al. Wang TJ. Gerszten RE. A combined epidemiologic and physiologic metabolomics approach improves chronic kidney disease prediction. J AM Soc Nephrol: Fox CS; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilpatrick RD, McAllister CJ, Kovesdy CP, Derose SF, Kopple JD, Kalantar-Zadeh K. Association between serum lipids and survival in hemodialysis patients and impact of race. J Am Soc Nephrol. 2007;18(1):293–303. doi: 10.1681/ASN.2006070795. [DOI] [PubMed] [Google Scholar]
- 34.Kalantar-Zadeh K, Kilpatrick RD, McAllister CJ, Greenland S, Kopple JD. Reverse epidemiology of hypertension and cardiovascular death in the hemodialysis population: the 58th annual fall conference and scientific sessions. Hypertension. 2005;45(4):811–7. doi: 10.1161/01.HYP.0000154895.18269.67. [DOI] [PubMed] [Google Scholar]
- 35.Kalantar-Zadeh K, Kopple JD, Kilpatrick RD, McAllister CJ, Shinaberger CS, Gjertson DW, et al. Association of morbid obesity and weight change over time with cardiovascular survival in hemodialysis population. Am J Kidney Dis. 2005;46(3):489–500. doi: 10.1053/j.ajkd.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Bradbury BD, Fissell RB, Albert JM, Anthony MS, Critchlow CW, Pisoni RL, et al. Predictors of early mortality among incident US hemodialysis patients in the Dialysis Outcomes and Practice Patterns Study (DOPPS) Clin J Am Soc Nephrol. 2007;2(1):89–99. doi: 10.2215/CJN.01170905. [DOI] [PubMed] [Google Scholar]
- 37.Trivedi H, Xiang Q, Klein JP. Risk factors for non-fatal myocardial infarction and cardiac death in incident dialysis patients. Nephrol Dial Transplant. 2009;24(1):258–66. doi: 10.1093/ndt/gfn426. [DOI] [PubMed] [Google Scholar]
- 38.Kovesdy CP, Kalantar-Zadeh K. Introduction: the reverse epidemiology controversy. Semin Dial. 2007;20(6):485. doi: 10.1111/j.1525-139X.2007.00363.x. [DOI] [PubMed] [Google Scholar]
- 39.Rhee CM, Leung AM, Kovesdy CP, Lynch KE, Brent GA, Kalantar-Zadeh K. Updates on the management of diabetes in dialysis patients. Semin Dial. 2014;27(2):135–45. doi: 10.1111/sdi.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40(Web Server issue):W127–33. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server issue):W652–60. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanholder R, Abou-Deif O, Argiles A, Baurmeister U, Beige J, Brouckaert P, et al. The role of EUTox in uremic toxin research. Semin Dial. 2009;22(4):323–8. doi: 10.1111/j.1525-139X.2009.00574.x. [DOI] [PubMed] [Google Scholar]
- 43.Vanholder R, Baurmeister U, Brunet P, Cohen G, Glorieux G, Jankowski J. A bench to bedside view of uremic toxins. J Am Soc Nephrol. 2008;19(5):863–70. doi: 10.1681/ASN.2007121377. [DOI] [PubMed] [Google Scholar]
- 44.Vanholder R, De Smet R, Glorieux G, Argiles A, Baurmeister U, Brunet P, et al. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–43. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 45.Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, et al. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012;23(7):1258–70. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tessari P, Deferrari G, Robaudo C, Vettore M, Pastorino N, De Biasi L, et al. Phenylalanine hydroxylation across the kidney in humans rapid communication. Kidney Int. 1999;56(6):2168–72. doi: 10.1038/sj.ki.4491156. [DOI] [PubMed] [Google Scholar]
- 47.Reaich D, Channon SM, Scrimgeour CM, Daley SE, Wilkinson R, Goodship TH. Correction of acidosis in humans with CRF decreases protein degradation and amino acid oxidation. Am J Physiol. 1993;265(2 Pt 1):E230–5. doi: 10.1152/ajpendo.1993.265.2.E230. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Liu H, McKenzie G, Witting PK, Stasch JP, Hahn M, et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16(3):279–85. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34(3):137–43. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jourde-Chiche N, Dou L, Cerini C, Dignat-George F, Vanholder R, Brunet P. Protein-bound toxins--update 2009. Semin Dial. 2009;22(4):334–9. doi: 10.1111/j.1525-139X.2009.00576.x. [DOI] [PubMed] [Google Scholar]
- 51.Shaham O, Slate NG, Goldberger O, Xu Q, Ramanathan A, Souza AL, et al. A plasma signature of human mitochondrial disease revealed through metabolic profiling of spent media from cultured muscle cells. Proc Natl Acad Sci U S A. 2010;107(4):1571–5. doi: 10.1073/pnas.0906039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pletinck A, Glorieux G, Schepers E, Cohen G, Gondouin B, Van Landschoot M, et al. Protein-bound uremic toxins stimulate crosstalk between leukocytes and vessel wall. J Am Soc Nephrol. 2013;24(12):1981–94. doi: 10.1681/ASN.2012030281. [DOI] [PMC free article] [PubMed] [Google Scholar]


