Abstract
With the increasing appreciation for the human microbiome coupled with the global rise of antibiotic resistant organisms, it is imperative that new methods be developed to specifically target pathogens. To that end, a novel computational approach was devised to identify compounds that reduce the activity of urease, a medically important enzyme of Helicobacter pylori, Proteus mirabilis, and many other microorganisms. Urease contains a flexible loop that covers its active site; Glide was used to identify small molecules predicted to lock this loop in an open conformation. These compounds were screened against the model urease from Klebsiella aerogenes and the natural products epigallocatechin and quercetin were shown to inhibit at low and high micromolar concentrations, respectively. These molecules exhibit a strong time-dependent inactivation of urease that was not due to their oxygen sensitivity. Rather, these compounds appear to inactivate urease by reacting with a specific Cys residue located on the flexible loop. Substitution of this cysteine by alanine in the C319A variant increased the urease resistance to both epigallocatechin and quercetin, as predicted by the computational studies. Protein dynamics are integral to the function of many enzymes; thus, identification of compounds that lock an enzyme into a single conformation presents a useful approach to define potential inhibitors.
Keywords: catechol, epigallocatechin, Glide, quercetin
1. INTRODUCTION
Urease is a key enzyme in medically and agriculturally significant organisms. This enzyme catalyzes the hydrolysis of urea to ammonia and carbamic acid, which then decomposes to bicarbonate and another molecule of ammonia.1–5 In agriculture, urea is both a plant metabolite and a common component of fertilizer.6 Excessive urease activity present in soil bacteria can rapidly metabolize exogenously applied urea, leading to unproductive volatilization of ammonia and harmful soil alkalinization.6 For many pathogenic bacteria, urease is a virulence factor that is associated with ammonia encephalopathy, hepatic coma, pyelonephritis, and urinary stone formation.3, 7, 8 Most notably, Helicobacter pylori cells are able to colonize the stomach lining by taking advantage of the pH increase from urea hydrolysis, thus locally neutralizing the acidic environment. H. pylori infection can lead to duodenal or peptic ulcers and gastric cancer, and, surprisingly, this microorganism is found in gastric samples for up to 50% of the world’s population.9–11 The primary method of treatment against H. pylori utilizes a proton pump inhibitor and two antibiotics, amoxicillin and clarithromycin.12 With our increasing appreciation for the human microbiome13 and the rise of antibiotic resistance around the world14 it is becoming increasingly important to develop new, specific methods for inhibiting pathogens. Urease provides an excellent target, as it is required for H. pylori survival within the stomach. Given the environmental and medical implications of urease, identifying compounds that inhibit urease’s enzymatic function offers an exciting approach to develop both agriculturally useful fertilizer amendments and potential therapeutic drugs.
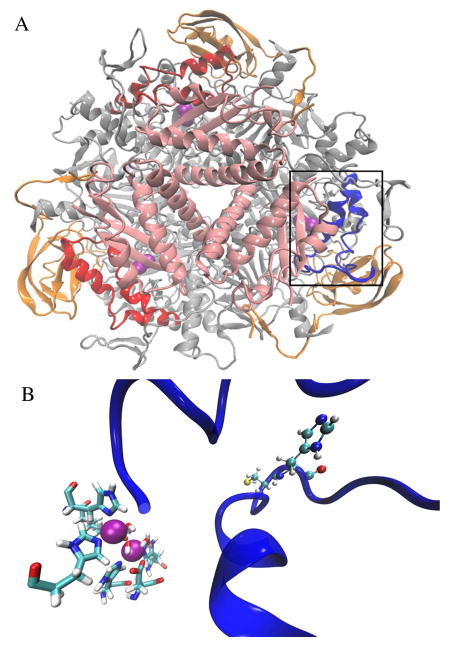
Irrespective of the urease source, the overall enzyme structures exhibit widespread similarities.1–5 Generally, bacterial ureases have three subunits in a trimer-of-trimers configuration (UreABC)3, as epitomized by the proteins from Klebsiella aerogenes (Fig. 1A) and Sporosarcina (formerly Bacillus) pasteurii.15–17 In some organisms, such as H. pylori, a fusion of two genes (corresponding to K. aerogenes ureA and ureB) results in only two subunits, in a [(UreAB)3]4 structure.18 In fungi and plants, all urease domains are encoded by a single gene, such as for jack bean (Canavalia ensiformis), with the trimeric protein forming back-to-back dimers [(α)3]2.19 The structures of the urease active sites are identical for all of these sources: two Ni2+ ions are bridged by a carbamylated Lys and water; one metal is further coordinated by two His and a terminal water molecule; and the second Ni2+ coordinates another two His, an Asp, and a water (Fig. 1B).1, 5, 15–19 The widespread structural similarities between the myriad of ureases ensure that an inhibitor for one isozyme will likely inhibit other urease isozymes. We utilized a novel computational approach to identify small molecules that reduce urease activity by preferentially binding to an open configuration of a flexible loop (residues Thr308-Arg336 of K. aerogenes urease)20 that covers the active site and contains a residue that is suggested to function in the catalytic mechanism.15
Figure 1.
(A) The overall structure of K. aerogenes urease is depicted in cartoon format. The three unique subunits are indicated by color; the trimer of alpha subunits (UreC) is depicted as gray, the beta subunits (UreB) as orange, and the gamma subunits (UreA) as pink. Ni pairs are shown as magenta Van der Waals spheres within the enzyme. The active site flaps, contained within the alpha subunits, are colored red, with exception of the wide-open active site flap (boxed), which is colored in blue. (B) Expanded view of the wide-open active site flap and the nickel metallocenter (magenta spheres) with its coordinating ligands. The coordination sphere consists of two His per nickel, a terminal water molecule per nickel, a hydroxide bridging the nickels, a carbamylated Lys bridging the nickels, and a single Asp residue, all depicted in licorice representation and colored by atom type. The Cys319 and His320 residues of the active site flap are shown in CPK and colored by atom type. The active site flap is again represented in blue cartoon representation.
To test our predictions, we utilized the model urease from K. aerogenes. The wealth of mechanistic and structural studies on K. aerogenes urease makes this isozyme an ideal initial test platform to verify computationally identified potential inhibitors and compare with actual experimental results.
2. MATERIALS AND METHODS
2.1. Docking
We ran a docking study on the wide-open flap state of K. aerogenes urease. This wide-open flap state was observed following a 100 nanosecond Molecular Dynamics (MD) simulation on the K. aerogenes urease structure 1EJX.20 The FF99SB force field was used in the AMBER suite of programs. The MD simulation was run in the isothermic, isobaric (NPT) ensemble in TIP3P water. We used two ligand libraries from the ZINC database: the ZINC natural products (ZNP) library with 180,313 ligands and the ZINC drug database (ZDD) with 2,924 ligands.21 Docking calculations and ligand preparation were conducted using the Schrödinger suite of programs, version 9.3.5 (Schrödinger, LLC). A 46 Å x 46 Å x 46 Å grid with 1 Å spacing was generated in Glide22 centered on the active site residues, the two nickels and the active site covering flap in order to dock the ligands. Default parameters were used in Glide with the exception of a 40 Å ligand length, 5 poses per ligand, 50,000 poses retained per run and no post-docking minimization. Of the 5 poses per ligand, only the best scoring pose was retained for further analysis. Docking calculations were performed using the Glide SP scoring function and the ligands receiving docking scores < −7.7 were extracted. A docking score cutoff of −6.0 – determined by the quercetin control – was initially employed, but this resulted in an extraction of 85,038 ligand poses. To address this overabundance, the top 100 unique ligands of each sublibrary were observed, and the highest-scoring ligand, kanamycin, was selected as the cutoff. A ligand length selection criterion was enforced, restricting the length of the ligand to less than 40 Å. The 40 Å length was calculated using a 3D structure with an active site cavity of approximately 12 Å. Note that this value was chosen to encompass the entire ligand library; as such, no ligands were lost due to the 40 Å cutoff. The control ligands were built using GaussView version 5.023 and optimized at the B3LYP/6-31+G* level of theory using the Gaussian0924 suite of programs. A frequency calculation was performed to ensure an energy minimum was located on the potential energy surface. The XYZ coordinates of quercetin and epigallocatechin were converted to SDF formatted files in order to dock the ligands to urease with Glide. Glide produced five possible conformational isomers and docked each to the active site.
2.2 Molecular similarity
To calculate molecular similarity, we used the OpenBabel software suite to generate a Molecular Access System (MACCS) key fingerprint for each of the six compounds, then used these fingerprints to calculate the Tanimoto coefficient between each pair.
2.3. Site directed mutagenesis
The C319A variant urease was obtained by using Quick Change site-directed mutagenesis (Stratagene) with pKK17 as the template, 5′-GCTGATGGTCGCCCACCATCT-3′ as the forward primer, and 5′-CGACTACCAGCGGGT GGTAGA-3′ as the reverse primer.25 The C319A mutation was confirmed both by sequencing and by the reduced specific activity of the purified protein.25
2.4. Urease assay
The activity of urease was determined by quantification of ammonia released by urea hydrolysis.26 Enzyme was incubated in 50 mM urea, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and 0.5 mM ethylenediaminetetraacetic acid (EDTA), pH 7.8, at 37°C. Ammonia was measured by the formation of indophenol and monitored at 625 nm.26 One unit of urease activity is the amount of enzyme needed to hydrolyze 1 μmol of urea per min at 37°C.
2.5. Protein Purification
K. aerogenes urease and its C319A variant were purified from E. coli BL21(DE3) cells containing pKK17 or pC319A by standard procedures (Table 1).27 Cells were grown aerobically in lysogeny broth (LB) at 37°C to an OD600 of 0.4. Nickel ions (1 mM) and 0.5 mM isopropyl β-D-1-thiogalactopyranoside were added to the culture and growth was continued another ~16 h aerobically at 37°C. Cells were harvested by centrifugation (11,000 g, 4°C, 10 min), resuspended in 20 mM tris(hydroxymethyl)aminomethane (Tris)/1 mM EDTA/1 mM β-mercaptoethanol (BME)/500 mM NaCl/pH 7.5, lysed by sonification, and the membranes were removed by centrifugation (100,000 g, 4°C, 1 h). Membrane- and cell-free extracts were loaded onto to a DEAE-Sepharose column (2.5 cm diameter, 60 ml bed volume) and eluted by a 0-1.5 M KCl gradient in 20 mM sodium phosphate/1 mM EDTA/1 mM BME/pH 7.4. The fractions with the highest activity were pooled and adjusted to a KCl concentration of 1.5 M. Pooled fractions were loaded onto a phenyl-Sepharose column (2.5 cm diameter, 50 ml bed volume) and step eluted with 20 mM sodium phosphate/1 mM EDTA/1 mM BME/pH 7.4. The fractions with the highest activity were pooled and dialyzed against 20 mM Tris/1 mM EDTA/1 mM BME/pH 7.4 to exchange the buffer. Protein concentrations were quantified by using Coomassie protein reagent (Bio-Rad) with bovine serum albumin as the standard.
Table 1.
The E. coli strain and plasmids used in this study.
| Strain | Genotype | Source |
|---|---|---|
| BL21 (DE3) | dcm ompT hsdS(rB− mB−) galλ(DE3) | Stratagene |
| Plasmids | ||
| pKK17 | K. aerogenes urease cluster (ureDABCEFG) inserted into pKK22-3 | 39 |
| pLEM12 | ureCC319A variant of pKK17 | This study |
2.6. Urease inactivation assays
The K. aerogenes urease inactivation studies were performed in 100 mM HEPES, pH 7.8, at 37°C, containing the indicated concentrations of inhibitors. Anaerobic experiments were conducted in a Coy anaerobic chamber (81% N2/7% H2/12% CO2).
2.7. Dialysis
The K. aerogenes urease protein was dialyzed in 10,000 molecular weight cut off cellulose tubing against 20 mM Tris/1 mM EDTA/25 mM NaCl/pH 7.8 (6 times); or sequentially, 20 mM Tris/1 mM EDTA/25 mM NaCl/pH 7.8 (2 times), 20 mM Tris/1 mM EDTA/25 mM NaCl/1 mM dithiothreitol (DTT)/pH 7.8 (2 times), followed by 20 mM Tris/1 mM EDTA/25 mM NaCl/pH 7.8 (2 times). All dialyses were conducted at 4°C for ~8 h with 500-fold volume equivalents of buffer.
3. RESULTS
3.1. Identification of potential urease inhibitors
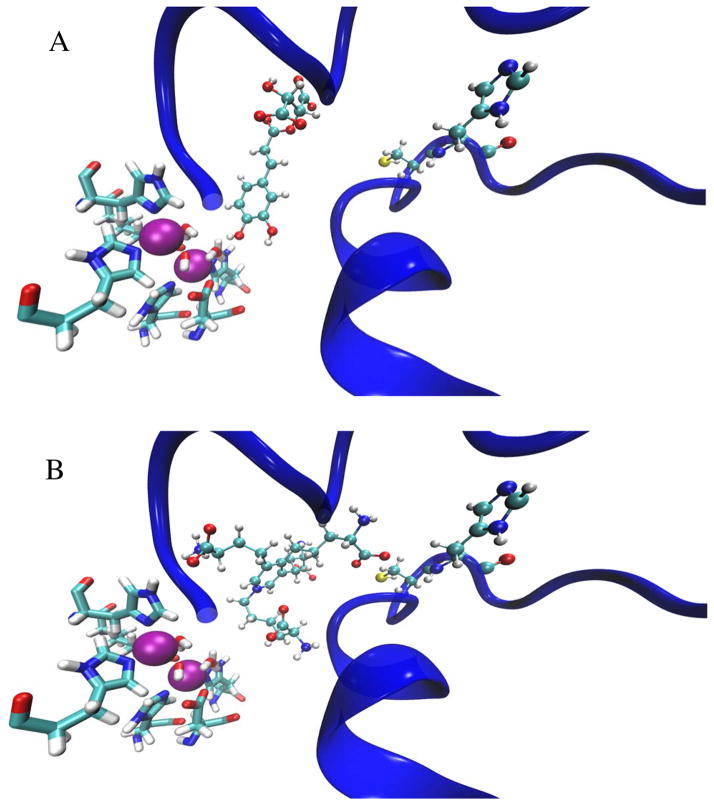
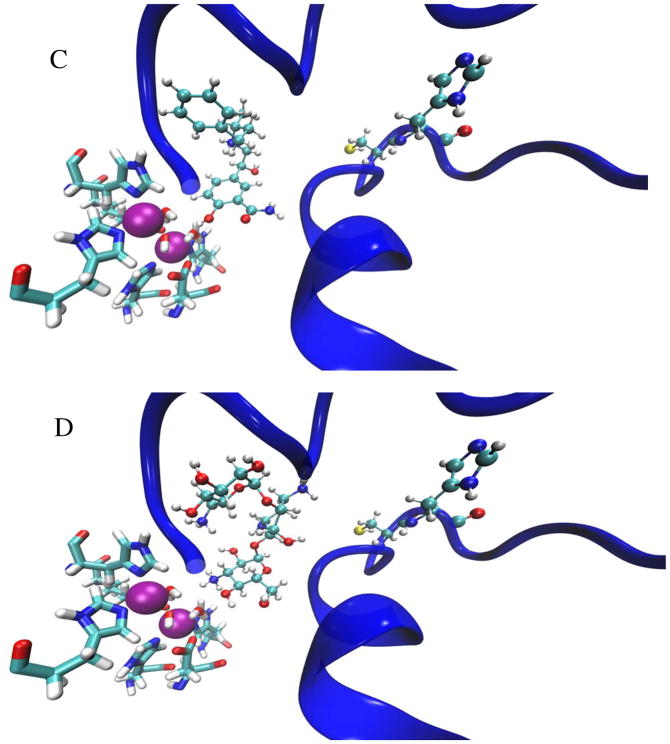
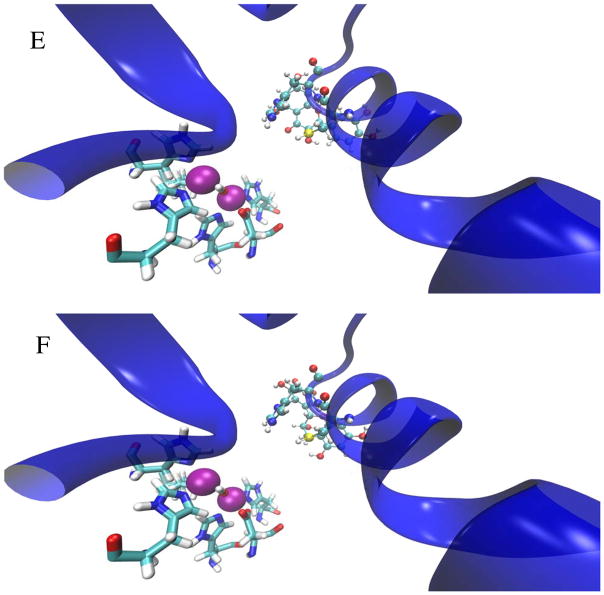
Molecular docking experiments identified several compounds that potentially lock urease in an open state by preventing the flap from closing (Table 2). The urease active-site region without an inhibitor (Fig. 1B) can be visually compared to depictions containing each of the potential docking ligands (Fig. 2A–2D) as well as to images of the computationally-bound epigallocatechin and quercetin (Fig. 2E–2F). Whereas the Nε nitrogen of His320 is located 5.1 Å from Ni-1 and 6.0 Å from Ni-2 in wild-type enzyme, these distances increase to 8.1 Å and 9.1 Å in the epigallocatechin-bound protein and 8.1 Å and 8.7 Å in the quercetin-bound enzyme. Similar interactions were predicted for each of compounds in Table 2.
Table 2.
Docking scores, masses, and structures of four potential urease ligands obtained from the ZINC natural products and ZINC drug databases along with two control ligands.
| Entry (ZN Number) [Sublibrary] | Docking score | Molecular weight | Structure |
|---|---|---|---|
| Caftaric Acid 02389524 Drug Database |
−10.2 | 310.21 |
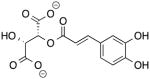
|
| Desmosine 35024527 Drug Database |
−10.1 | 523.59 |
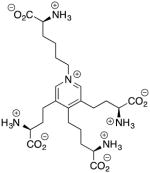
|
| Labetalol 00000416 Natural Products |
−8.1 | 329.42 |
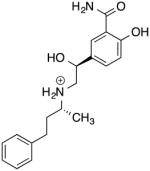
|
| Kanamycin 08214590 Natural Products |
−7.7 | 488.54 |
|
| Quercetin 03869685 Control |
−6.4 | 302.24 |
|
| Epigallocatechin 03870339 Control |
−6.0 | 306.27 |
|
Figure 2.
Depictions of the best-scoring docking poses for each of 4 potential urease ligands (panels A–D) and the 2 control ligands (panels E–F). In each figure, the active site flap is shown in a blue cartoon representation, the Ni pair is shown as magenta-colored Van der Waals spheres, the coordination sphere is shown in licorice representation colored by atom type, and the docked ligand – as well as the Cys319 and His320 residues – are shown as CPK representations colored by atom type. Shown in order are docked images of caftaric acid (A), desmosine (B), labetalol (C), kanamycin (D), quercetin (E), and eipgallocatechin (F).
3.2 Molecular similarity
To evaluate the molecular similarity between the four compounds of interest and the two control compounds in Table 2, we determined the Tanimoto coefficient scores between each possible pair (Table 3). We chose a Tanimoto similarity cutoff of 0.85, as molecule pairs with scores below this typically do not exhibit chemical similarity. With this cutoff only the control molecules quercetin and epigallocatechin (T = 0.87) demonstrated a possibility for MACCS similarity. Along with similarity scoring between compound pairs, we also performed a similarity search of the ZNP and ZDD library for each of the compounds, determining the number of ligands from each library that feature Tanimoto coefficients greater than 0.85 (Table 4). Taking into account the sizes of the ZNP and ZDD libraries, few similar ligands were found for any of the six molecules. In relative terms, desmosine and labetalol received especially few hits, with desmosine scoring no hits when compared to the ZDD. Caftaric acid produced a comparatively higher number of ZNP hits, though no hits were found in the ZDD. Kanamycin produced a sizable quantity of overall hits, scoring the most hits of the four new compounds from ZNP and the most hits of all six compounds from ZDD. The two control compounds, quercetin and epigallocatechin, scored the most overall hits.
Table 3.
Tanimoto similarity scores for each pair of molecules.
| Tanimoto Scores (MACCS) | Caftaric Acid | Desmosine | Labetalol | Kanamycin | Quercetin | Epigallocatechin |
|---|---|---|---|---|---|---|
| Caftaric Acid | 1.00 | 0.29 | 0.36 | 0.37 | 0.61 | 0.53 |
| Desmosine | 0.29 | 1.00 | 0.49 | 0.43 | 0.25 | 0.27 |
| Labetalol | 0.36 | 0.49 | 1.00 | 0.53 | 0.36 | 0.40 |
| Kanamycin | 0.37 | 0.43 | 0.53 | 1.00 | 0.40 | 0.47 |
| Quercetin | 0.61 | 0.25 | 0.36 | 0.40 | 1.00 | 0.87 |
| Epigallocatechin | 0.53 | 0.27 | 0.40 | 0.47 | 0.87 | 1.00 |
Table 4.
Number of compounds with Tanimoto similarity scores > 0.85 in ZINC Natural Products (ZNP) and ZINC Drug Database (ZDD) libraries.
| Entry | ZNP | ZDD |
|---|---|---|
| Caftaric Acid | 16 | 0 |
| Desmosine | 5 | 0 |
| Labetalol | 4 | 4 |
| Kanamycin | 69 | 7 |
| Querctin | 285 | 4 |
| Epigallocatechin | 83 | 4 |
For the four new compounds of interest, we took the ligands determined to be chemically similar from the ZNP and ZDD libraries and determined the mean, median, and standard deviation of their docking scores (Table 5). Note that the docking score for each of the four compounds exceeds that of the mean docking scores for the similar compounds; in particular, caftaric acid and labetalol exceed the means by greater than a standard deviation.
Table 5.
Mean, median and standard deviation of docking score for all compounds with Tanimoto similarity scores greater than 0.85.
| Entry | Mean | Median | St Dev | Docking Score |
|---|---|---|---|---|
| Caftaric Acid | −5.7 | −5.3 | 1.8 | −10.2 |
| Desmosine | −9.3 | −9.9 | 1.2 | −10.1 |
| Labetalol | −7.4 | −7.7 | 0.5 | −8.1 |
| Kanamycin | −7.5 | −7.5 | 0.7 | −7.7 |
3.3. Assessment of urease inhibition by target compounds
The compounds depicted in Table 2 were screened as inhibitors against K. aerogenes urease, a particularly well-characterized representative of this enzyme family. Caftaric acid, desmosine, kanamycin, and labetalol, each examined at concentrations up to 600 μM, showed little to no inhibitory activity against K. aerogenes urease (data not shown). In contrast, however, the catechols epigallocatechin and quercetin cause urease to lose activity in a time dependent manner at micromolar concentrations (Fig. 3A and 4A). The extent of inhibition continued to increase even up to 4 h of exposure, indicating that loss of activity is due to a slowly accumulating degradation product, that the compounds of interest are slow-binding inhibitors, or that a secondary reaction is required for inactivation. Each of these possibilities was examined.
Figure 3.
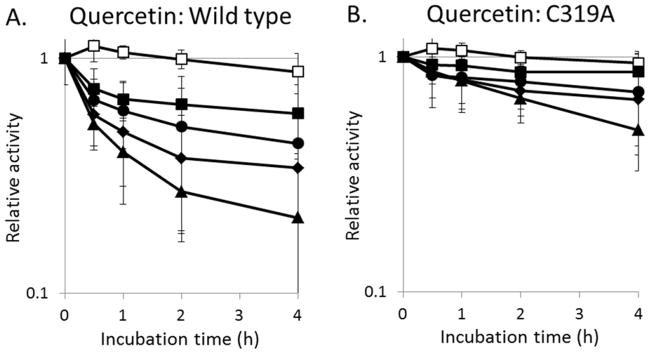
Comparison of the rates of urease inactivation by selected concentrations of quercetin. The (A) wild-type enzyme and (B) C319A variant urease were incubated with 0 (□), 22 (■), 66 (●), 200 (◆), or 600 μM (▲) quercetin aerobically in 100 mM HEPES, pH 7.8, at 37°C and the remaining activities measured at the times indicated. Error bars represent the standard deviation of three independent experiments.
Figure 4.
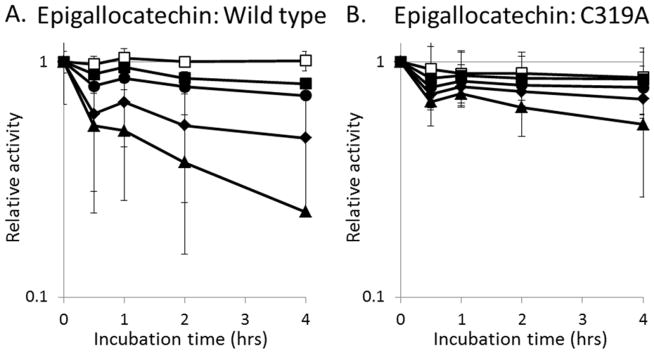
Comparison of the rates of urease inactivation by selected concentrations of epigallocatechin. The (A) wild-type enzyme and (B) C319A variant urease were incubated with 0 (□), 3 (■), 9 (●), 27 (◆), or 81 μM (▲) epigallocatechin aerobically in 100 mM HEPES, pH 7.8, at 37°C and the remaining activities measured at the times indicated. Error bars represent the standard deviation of three independent experiments.
3.4. Epigallocatechin and quercetin are oxygen labile, but oxidation products do not account for their inhibition of urease
Epigallocatechin-gallate is reported to have a half-life in solution of less than 30 min, with the compound autooxidizing and forming dimers.28–30 Indeed, epigallocatechin undergoes a rapid reaction in buffer containing oxygen as revealed by absorption changes in its spectrum, whereas these spectral changes do not take place in the absence of oxygen (Fig. 5). Even more dramatic spectral changes have been reported for quercetin, which forms a variety of degradation products including 3,4-dihydroxybenzoate, 2,4,6-trihydroxybenzoate, and 1,3,5-trihydroxybenzene in aerobic solutions.31, 32 However, quercetin and epigallocatechin retain the ability to inhibit urease even under anaerobic conditions (Fig. 6).
Figure 5.
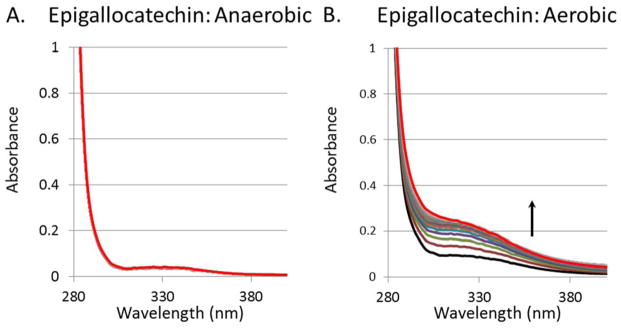
Epigallocatechin spontaneously oxidizes in aerobic solutions. The UV-Vis spectra of (A) anaerobic epigallocatechin (1.6 mM) and (B) anaerobic epigallocatechin (1.6 mM) after a 5 min sparge with air. All spectra were obtained over 2 h at room temperature (25°C) using buffer containing 100 mM HEPES, pH 7.8.
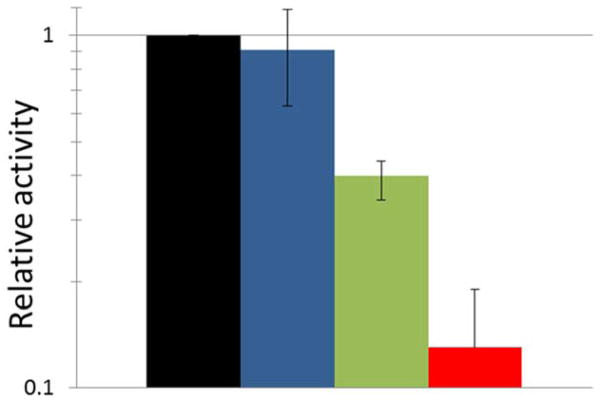
Figure 6.
K. aerogenes urease is inhibited by quercetin and epigallocatechin when using anaerobic conditions. Urease was incubated anaerobically with no inhibitor (blue), 600 μM quercetin (green), or 81 μM epigallocatechin (red) for 4 h in 100 mM HEPES, pH 7.8, at 37°C and assayed for enzyme activity. The black bar represents the relative activity at time zero. Error bars represent the standard deviation of three independent experiments. The Student T-test for two independent means with a significance level of 0.05 provides p values of 0.003 and 0.00001 for quercetin and epigallocatechin, respectively, versus no inhibitor.
3.5. Effects of inhibitors on a variant urease lacking the active site cysteine residue
As shown in Figure 2E and 2F, epigallocatechin and quercetin are predicted to prop open the flap covering the urease active site and feature at least one atom that is within 4 Å of Cys319, one of ten cysteine residues in the enzyme. To test whether this residue participates in the inhibition by these compounds, we used a C319A variant that was previously shown to retain ~60% of the activity of the wild-type enzyme.25 Consistent with the hypothesis that these compounds would be less effective when using enzyme lacking Cys319, the C319A urease was less sensitive to loss of activity by epigallocatechin and quercetin (Fig. 3B, 4B). These results suggest that quercetin and epigallocatechin may bind to the flap region and then form a covalent attachment to Cys319. Confounding this interpretation, these compounds were still able to inhibit the enzyme weakly using C319A urease, raising the possibility that other sites of interaction play a role in activity loss. Additional evidence consistent with covalent adduct formation by these compounds is that quercetin- and epigallocatechin-dependent activity loss of the enzyme was not reversed upon extensive dialysis or by the addition of thiols (e.g., extended dialysis against buffer containing 1 mM DTT; data not shown). These results suggest that after binding to the enzyme the catechols react with a side chain to covalently modify the protein in a manner that is not simply reversed by reductant; however, additional studies are required to confirm such covalent modification of the protein by these compounds.
4. DISCUSSION
We have provided evidence suggesting a new mechanism for urease activity loss in which compounds first bind to the enzyme so as to prop open the active site flap and then covalently modify the Cys residue in the flap polypeptide. Two compounds appearing to inactivate K. aerogenes urease by this mechanism are epigallocatechin and quercetin.
Epigallocatechin-gallate, a component of green tea, was previously reported to inhibit urease from H. pylori and Staphylococcus saprophyticus;33, 34 however, the time dependence of inhibition and covalent adduct formation, as demonstrated here, were not investigated in those studies. Similarly, quercetin and its glycosides were previously identified as H. pylori urease inhibitors,35, 36 but the potential for time dependent activity loss and covalent attachment were not examined. By contrast, inactivation of jack bean urease by other catechols, such as baicalin and scutellarin, was shown to be time dependent, with thiols protecting against inhibition;37, 38 notably, these compounds also were proposed to bind near the flap Cys residue. In contrast to the thiol-dependent reversibility of catechol inhibitors using the jack bean enzyme, however, thiols alone were unable to reverse epigallocatechin- or quercetin-dependent inhibition of K. aerogenes urease, consistent with inhibitor-dependent modification of the bacterial protein.
In conclusion, this work has demonstrated the feasibility of reducing urease activity by identifying compounds that bind to the open-loop conformation of the protein and then covalently react with a residue on this loop. Because protein dynamics are integral to the function of many enzymes, the use of compounds that lock an enzyme into a single conformation presents a useful approach to identify potential new inhibitors and inactivators.
Acknowledgments
We would like to thank Gemma Reguera (Michigan State University) for the generous use of anaerobic equipment. This work was generously funded by National Institutes of Health Grant (DK045686 to R.P.H. and GM066859 to K.M.M.). M.S.M. thanks the National Science Foundation (NSF) for support in the form of a graduate research fellowship, the UF Disability Resource Center, Juan Contreras and Sean Jones.
ABBREVIATIONS
- BME
β-mercaptoethanol
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- LB
lysogeny broth
- Tris
tris(hydroxymethyl)aminomethane
- ZDD
ZINC drug database
- ZNP
ZINC natural products
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript
References
- 1.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1(3):207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krajewska B, Ureases I. Functional catalytic and kinetic properties: A review. Journal of Molecular Catalysis B: Enzymatic. 2009;59:9–21. [Google Scholar]
- 3.Boer JL, Mulrooney SB, Hausinger RP. Nickel-dependent metalloenzymes. Arch Biochem Biophys. 2014;544:142–152. doi: 10.1016/j.abb.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witte C-P. Urea metabolism in plants. Plant Sci. 2011;180(3):431–438. doi: 10.1016/j.plantsci.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Zambelli B, Musiani F, Benini S, Ciurli S. Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Acc Chem Res. 2011;44(7):520–530. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]
- 6.Mobley HL, Hausinger RP. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989;53(1):85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins CM, D’Orazio SE. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9(5):907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 8.Burne RA, Chen YY. Bacterial ureases in infectious diseases. Microbes Infect. 2000;2(5):533–542. doi: 10.1016/s1286-4579(00)00312-9. [DOI] [PubMed] [Google Scholar]
- 9.Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 10.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stingl K, Altendorf K, Bakker EP. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 2002;10(2):70–74. doi: 10.1016/s0966-842x(01)02287-9. [DOI] [PubMed] [Google Scholar]
- 12.Sharma VK, Bailey DM, Raufman JP, Elraie K, Metz DC, Go MF, Schoenfeld P, Smoot DT, Howden CW. A survey of internal medicine residents’ knowledge about Helicobacter pylori infection. Am J Gastroenterol. 2000;95(8):1914–1919. doi: 10.1111/j.1572-0241.2000.02247.x. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 14.Neu HC. The crisis in antibiotic resistance. Science. 1992;257(5073):1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 15.Jabri E, Carr MB, Hausinger RP, Karplus PA. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268(5213):998–1004. [PubMed] [Google Scholar]
- 16.Pearson MA, Michel LO, Hausinger RP, Karplus PA. Structures of Cys319 variants and acetohydroxamate-inhibited Klebsiella aerogenes urease. Biochemistry. 1997;36(26):8164–8172. doi: 10.1021/bi970514j. [DOI] [PubMed] [Google Scholar]
- 17.Benini S, Rypniewski WR, Wilson KS, Miletti S, Ciurli S, Mangani S. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure. 1999;7(2):205–216. doi: 10.1016/S0969-2126(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 18.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8(6):505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 19.Balasubramanian A, Ponnuraj K. Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J Mol Biol. 2010;400(3):274–283. doi: 10.1016/j.jmb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Roberts BP, Miller BR, 3rd, Roitberg AE, Merz KM., Jr Wide-open flaps are key to urease activity. J Am Chem Soc. 2012;134(24):9934–9937. doi: 10.1021/ja3043239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52(7):1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 23.Dennington R, Keith T, Millam J. GaussView, Version 5. Semichem, Inc; Shawnee Mission, KS, USA: 2009. [Google Scholar]
- 24.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro Fo, Bearpark MJ, Heyd J, Brothers EN, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ãdn, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09. Gaussian, Inc; Wallingford, CT, USA: 2009. [Google Scholar]
- 25.Martin PR, Hausinger RP. Site-directed mutagenesis of the active site cysteine in Klebsiella aerogenes urease. J Biol Chem. 1992;267(28):20024–20027. [PubMed] [Google Scholar]
- 26.Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Analytical Chemistry. 1967;39(8):971–974. [Google Scholar]
- 27.Carter EL, Hausinger RP. Characterization of the Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J Bacteriol. 2010;192(9):2294–2304. doi: 10.1128/JB.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J, Lu H, Meng X, Ryu JH, Hara Y, Yang CS. Stability, cellular uptake, biotransformation, and efflux of tea polyphenol (-)-epigallocatechin-3-gallate in HT-29 human colon adenocarcinoma cells. Cancer Res. 2002;62(24):7241–7246. [PubMed] [Google Scholar]
- 29.Sang S, Lee MJ, Hou Z, Ho CT, Yang CS. Stability of tea polyphenol (-)-epigallocatechin-3-gallate and formation of dimers and epimers under common experimental conditions. J Agric Food Chem. 2005;53(24):9478–9484. doi: 10.1021/jf0519055. [DOI] [PubMed] [Google Scholar]
- 30.Krook MA, Hagerman AE. Stability of Polyphenols Epigallocatechin Gallate and Pentagalloyl Glucose in a Simulated Digestive System. Food Res Int. 2012;49(1):112–116. doi: 10.1016/j.foodres.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenkevich IG, Eshchenko AY, Makarova SV, Vitenberg AG, Dobryakov YG, Utsal VA. Identification of the products of oxidation of quercetin by air oxygen at ambient temperature. Molecules. 2007;12(3):654–672. doi: 10.3390/12030654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramesova S, Sokolova R, Degano I, Bulickova J, Zabka J, Gal M. On the stability of the bioactive flavonoids quercetin and luteolin under oxygen-free conditions. Anal Bioanal Chem. 402(2):975–982. doi: 10.1007/s00216-011-5504-3. [DOI] [PubMed] [Google Scholar]
- 33.Matsubara S, Shibata H, Ishikawa F, Yokokura T, Takahashi M, Sugimura T, Wakabayashi K. Suppression of Helicobacter pylori-induced gastritis by green tea extract in Mongolian gerbils. Biochem Biophys Res Commun. 2003;310(3):715–719. doi: 10.1016/j.bbrc.2003.09.066. [DOI] [PubMed] [Google Scholar]
- 34.Loes AN, Ruyle L, Arvizu M, Gresko KE, Wilson AL, Deutch CE. Inhibition of urease activity in the urinary tract pathogen Staphylococcus saprophyticus. Lett Appl Microbiol. 2014;58(1):31–41. doi: 10.1111/lam.12153. [DOI] [PubMed] [Google Scholar]
- 35.Shabana S, Kawai A, Kai K, Akiyama K, Hayashi H. Inhibitory activity against urease of quercetin glycosides isolated from Allium cepa and Psidium guajava. Biosci Biotechnol Biochem. 2010;74(4):878–880. doi: 10.1271/bbb.90895. [DOI] [PubMed] [Google Scholar]
- 36.Xiao ZP, Wang XD, Peng ZY, Huang S, Yang P, Li QS, Zhou LH, Hu XJ, Wu LJ, Zhou Y, Zhu HL. Molecular docking, kinetics study, and structure-activity relationship analysis of quercetin and its analogous as Helicobacter pylori urease inhibitors. J Agric Food Chem. 2012;60(42):10572–10577. doi: 10.1021/jf303393n. [DOI] [PubMed] [Google Scholar]
- 37.Tan L, Su J, Wu D, Yu X, Su Z, He J, Wu X, Kong S, Lai X, Lin J, Su Z. Kinetics and mechanism study of competitive inhibition of jack-bean urease by baicalin. Sci World J. 2013;2013:879501. doi: 10.1155/2013/879501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu DW, Yu XD, Xie JH, Su ZQ, Su JY, Tan LR, Huang XQ, Chen JN, Su ZR. Inactivation of jack bean urease by scutellarin: elucidation of inhibitory efficacy, kinetics and mechanism. Fitoterapia. 2013;91:60–67. doi: 10.1016/j.fitote.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Colpas GJ, Brayman TG, Ming LJ, Hausinger RP. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry. 1999;38(13):4078–4088. doi: 10.1021/bi982435t. [DOI] [PubMed] [Google Scholar]