Abstract
AIM: To investigate whether ezetimibe ameliorates hepatic steatosis and induces autophagy in a rat model of obesity and type 2 diabetes.
METHODS: Male age-matched lean control LETO and obese and diabetic OLETF rats were administered either PBS or ezetimibe (10 mg/kg per day) via stomach gavage for 20 wk. Changes in weight gain and energy intake were regularly monitored. Blood and liver tissue were harvested after overnight fasting at the end of study. Histological assessment was performed in liver tissue. The concentrations of glucose, insulin, triglycerides (TG), free fatty acids (FFA), and total cholesterol (TC) in blood and TG, FFA, and TG in liver tissue were measured. mRNA and protein abundance involved in autophagy was analyzed in the liver. To investigate the effect of ezetimibe on autophagy and reduction in hepatic fat accumulation, human Huh7 hepatocytes were incubated with ezetimibe (10 μmol/L) together with or without palmitic acid (PA, 0.5 mmol/L, 24 h). Transmission electron microscopy (TEM) was employed to demonstrate effect of ezetimibe on autophagy formation. Autophagic flux was measured with bafilomycin A1, an inhibitor of autophagy and following immunoblotting for autophagy-related protein expression.
RESULTS: In the OLETF rats that received ezetimibe (10 mg/kg per day), liver weight were significantly decreased by 20% compared to OLETF control rats without changes in food intake and body weight (P < 0.05). Lipid parameters including TG, FFA, and TC in liver tissue of ezetimibe-administrated OLETF rats were dramatically decreased at least by 30% compared to OLETF controls (P < 0.01). The serum glucose, insulin, HOMA-IR, and lipid profiles were also improved by ezetimibe (P < 0.05). In addition, autophagy-related mRNA expression including ATG5, ATG6, and ATG7 and the protein level of microtubule-associated protein light chain 3 (LC3) were significantly increased in the liver in rats that received ezetimibe (P < 0.05). Likewise, for hepatocytes cultured in vitro, ezetimibe treatment significantly decreased PA-induced fat accumulation and increased PA-reduced mRNA and protein expression involved in autophagy (P < 0.05). Ezetimibe-increased autophagosomes was observed in TEM analysis. Immunoblotting analysis of autophagy formation with an inhibitor of autophagy demonstrated that ezetimibe-increased autophagy resulted from increased autophagic flux.
CONCLUSION: The present study demonstrates that ezetimibe-mediated improvement in hepatic steatosis might involve the induction of autophagy.
Keywords: Autophagy, Ezetimibe, Hepatic steatosis, Nonalcoholic fatty liver disease
Core tip: As an anti-hypercholesterolemia drug, ezetimibe is reported to improve metabolic disorders. Moreover, the hepatic expression of Niemann-Pick C1-like 1 protein, the target of ezetimibe, has led to increased interest in the effects, which have not been fully delineated, of ezetimibe on the liver. In the current study, ezetimibe treatment improved hepatic fat accumulation, which was accompanied by the induction of hepatic autophagy in obese and diabetic rats. In addition, in vitro hepatocytes treated with an inhibitor of autophagy showed that ezetimibe-induced autophagy resulted from an increase in autophagic flux. Therefore, ezetimibe-increased autophagy flux may play an important role in the improvement of hepatic steatosis.
INTRODUCTION
The increasing prevalence of nonalcoholic fatty liver disease (NAFLD) by up to 30% in Western countries has resulted in NAFLD becoming the most common feature of chronic liver disease[1-3]. Numerous studies have demonstrated a positive correlation between metabolic disorders such as insulin resistance and obesity and the progression of liver fibrosis, cirrhosis, and hepatocellular carcinoma[1,4]. Given the rapidly increasing prevalence of NAFLD and its positive relationship with metabolic syndrome, the prevention and attenuation of NAFLD is critical.
Ezetimibe decreases intestinal cholesterol incorporation by blocking Niemann-Pick C1-like 1 (NPC1L1) protein[5,6]. Given the association between dysregulated cholesterol metabolism and metabolic disorders, numerous human and animal studies have shown that the NPC1L1 inhibitor ezetimibe affects metabolic disorders including insulin resistance, type 2 diabetes, and atherosclerosis[7-11]. In addition to intestinal NPC1L1 expression, NPC1L1 is also highly abundant in the liver[5,12]. Consequently, fatty liver is improved by ezetimibe in obese subjects undergoing weight loss intervention[13] as well as in subjects with NAFLD[14] and nonalcoholic steatohepatitis (NASH) with dyslipidemia[15]. Ezetimibe administration ameliorates hepatic steatosis in diet-induced fatty liver animal models[16,17], fatty liver Shionogi mice[18,19], db/db mice[20], and Zucker obese fatty rats[7,8]. Moreover, NPC1L1-ablated mice are protected from high fat-induced fatty liver[21]. Together, these data support the possibility that NPC1L1 inhibition might be an effective method for treating NAFLD. However, more studies investigating the molecular and intracellular mechanisms by which ezetimibe regulates hepatic lipid metabolism and improves NAFLD are necessary.
Autophagy is the process of intracellular degradation via the formation of double-membrane structures known as autophagosomes, which is followed by their transport to lysosomes, fusion with the lysosomes to form autolysosomes, and finally, degradation of the contents enclosed within the inner autophagosomal membrane[22]. The role of autophagy is to maintain cellular homeostasis by routine turnover of cytoplasmic components under various stress conditions such as starvation, virus infection, and endoplasmic reticulum (ER) stress[23-26]. However, recent studies have suggested a new function and role for autophagy in hepatic lipid storage via a process termed lipophagy. Hepatic lipid accumulation by lipid challenge inhibits hepatic autophagy[27-29], and inhibition of autophagy by genetic knockdown or pharmacological methods increases hepatocyte triglyceride (TG) and cholesterol levels[27,29]. Importantly, these results may suggest a new mechanism for the treatment of hepatic steatosis.
In the present study, we investigated the effects of ezetimibe on glycemic control, hepatic fat accumulation, and the induction of liver autophagy. In both a rat model of obesity and diabetes and palmitic acid (PA)-treated hepatocytes, ezetimibe attenuated hepatic fat accumulation concomitant with increased hepatic autophagy.
MATERIALS AND METHODS
Animals
All animal experiments were conducted following the recommendations in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Ethics Committee for Animal Experiments of Kangbuk Samsung Hospital, Sungkyunkwan University. Male OLETF (n = 11) and age-matched LETO rats (n = 3) were purchased from Otsuka Pharmaceuticals (Tokushima, Japan), and experiments were conducted in a specific pathogen-free facility with a 12 h light/dark cycle at Kangbuk Samsung Hospital, Sungkyunkwan University. The OLETF rat is a model that represents late-onset hyperglycemia and exhibits a chronic disease course, mild obesity and clinical onset of diabetes mellitus[30,31]. Animals had unrestricted access to water and food (PicoLab Rodent Diet 20 5053, Purina Mills, Richmond, IN, United States). At 12 wk of age, rats were randomized and treated with either PBS or ezetimibe (10 mg/kg per day) via a stomach gavage for 20 wk. At the end of the study, the rats were fasted overnight and anesthetized with intraperitoneal Zoletil/Rompun. Blood was collected from the abdominal aorta, and liver tissues were dissected, immediately frozen in liquid nitrogen, and stored at -80 °C until further analysis.
Cell culture
Huh7 human hepatocytes (Korean Cell Line Bank, Seoul, South Korea) were cultured in high glucose DMEM (Gibco, Grand Island, NY, United States) containing 10% FBS (Gibco), 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco) at 37 °C in a 95% air/5% CO2 atmosphere. Ezetimibe was provided by Merck Sharp and Dohme Corp. (Rahway, NJ, United States). Hepatocytes were treated with or without ezetimibe (10 μmol/L, 1 h) and incubated with palmitic acid (PA, 0.5 mmol/L, 24 h; Sigma-Aldrich, St. Louis, MO, United States). Palmitic acid was prepared as previously described[32].
Measurement of metabolic parameters
Serum glucose and insulin were analyzed by enzymatic assay (Sigma-Aldrich and Crystal Chem, Downers Grove, IL, United States). The blood and liver TG levels were also measured by enzymatic assay (Sigma-Aldrich). Commercial kits were employed for the measurement of free fatty acid (FFA; Wako Pure Chemical Industries, Osaka, Japan) and total cholesterol (TC; Cayman Chemical Com., Ann Arbor, MI, United States). The liver metabolic parameters were normalized to their respective protein concentrations. The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated using the following formula: fasting glucose (mmol/L) × fasting insulin (mU/L)/22.5.
Histological Analysis and NAFLD activity score
Dissected liver tissues were fixed in 10% formalin buffer overnight. The tissues were then embedded in paraffin, sliced into 5-μm-thick sections, and stained with hematoxylin and eosin (HE). Digital images were captured with an Olympus BX51 light microscope (magnification, 200 ×, Tokyo, Japan). A pathologist blinded to the experimental conditions evaluated the NAFLD activity score (NAS). Three features of NAFLD, namely steatosis, lobular inflammation, and ballooning, were scored as described previously[33]. NAFLD activity score (NAS) was calculated as follows; steatosis (0-3), ballooning (0-2), and inflammation (0-3) were summed.
RNA analysis
The total RNA was extracted using an RNeasy Mini Kit (Invitrogen, Carlsbad, CA, United States). A high-capacity cDNA Kit (Applied Biosystems, Foster City, CA, United States) was used for cDNA synthesis. Real-time quantitative PCR (RT-PCR) was performed with a Roche Lightcycler 480 (Roche, Mannheim, Germany) using Roche real-time PCR master mix and UPL. The primer sequences used are listed in Table 1. The PCR parameters were as follows: pre-denaturation at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 10 s, and annealing/extension at 60 °C for 20 s. Expression of each target gene was normalized to housekeeping gene (GAPDH or β-actin) and expressed as the fold change relative to the control treatment. CT values of GAPDH or β-actin were not statistically different among groups.
Table 1.
Primers used for real-time quantitative-polymerase chain reaction
| Organism | Gene | Forward primer | Reverse primer |
| Human | ATG5 | CAACTTGTTTCACGCTATATCAGG | CACTTTGTCAGTTACCAACGTCA |
| ATG6 | GGATGGTGTCTCTCGCAGAT | TTGGCACTTTCTGTGGACAT | |
| ATG7 | CCGTGGAATTGATGGTATCTG | TCATCCGATCGTCACTGCT | |
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC | |
| Rat | β-actin | CCTGTATGCCTCTGGTCGTA | CCATCTCTTGCTCGAAGTCT |
| ATG5 | CTGTTCGATCTTCTTGCATCA | TCCTTTTCTGGAAAACTCTTGAA | |
| ATG6 | CAGGCGAAACCAGGAGAG | CGAGTTTCAATAAATGGCTCCT | |
| ATG7 | TTCTTAGAAGATTTGACTGGTCTTACA | TCACTCATGTCCCAGATCTCA |
ATG: Autophagy-related gene; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
Immunoblot analysis
Equal amounts of protein were loaded into each lane of a 4%-20% gradient SDS polyacrylamide gel, separated by SDS-polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (Millipore, Marlborough, MA, United States). The membranes were probed with the following primary antibodies: α-tubulin, ATG5, ATG6, LC3B, and GAPDH (Cell Signaling Technology, Danvers, MA, United States) followed by the appropriate secondary antibody. The immunoreactive bands were developed with the Amersham ECL plus system (Amersham-Pharmacia Biotech, Arlington Heights, IL, United States). Densitometry analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD, United States).
Electron microscopy
After washing with PBS, samples were fixed with 2.5% glutaraldehyde plus paraformaldehyde in 0.1 mol/L PBS (pH 7.4) for 2 h and washed three times for 30 min in 0.1 mol/L PBS. Next, glutaraldehyde-fixed specimens were treated with 1% OsO4 in 0.1 mol/L PBS for 2 h, dehydrated in increasing concentrations of ethanol (50%-100%), infiltrated with propylene oxide, and embedded in an EPON mixture. Polymerized sections were then cut, stained, and examined using transmission electron microscopy (JEM-1101, JEOL, Japan).
Statistical analysis
The data are expressed as the mean ± SE. Student’s t test were employed for comparisons of two matched groups using PASW Statistics 18 (SPSS Inc., Chicago, IL, United States). Statistical significance was defined as P < 0.05.
RESULTS
Ezetimibe affects hepatic steatosis without changing body weight and food intake in OLETF rats
As shown in Table 2, the body weight and daily food intake were not significantly different between control and ezetimibe-treated OLETF rats. Despite the lack of difference in body weight, the liver tissue weight was significantly decreased (20%) by ezetimibe in OLETF rats (Table 2). Likewise, blood and liver lipid levels including TG, FFA, and TC were significantly decreased in ezetimibe-treated OLETF rats (Table 2). Moreover, OLETF rats showed higher serum levels of glucose, insulin, HOMA-IR, TG, FFA, and TC than LETF animals, which were significantly reduced by ezetimibe (Table 2). In addition, histological analysis indicated that OLETF control rats showed larger lipid droplets in hepatocytes than age-matched LETO controls, which were attenuated by administration of ezetimibe (Figure 1A). Similar to these liver features, the NAFLD activity score (NAS) was also reduced by ezetimibe treatment (Figure 1B).
Table 2.
Metabolic characteristics of the LETO control and control or ezetimibe-treated OLETF
| LETO control (n = 3) | OLETF control (n = 5) | OLETF Ezetimibe (n = 6) | |
| Body weight (g) | 524.00 ± 1.15b | 617.67 ± 23.29 | 642.29 ± 20.47 |
| Daily food intake (g) | 3.61 ± 0.12b | 5.06 ± 0.30 | 4.62 ± 0.13 |
| Liver tissue weight (%BW) | 2.52 ± 0.05a | 3.60 ± 0.29 | 2.90 ± 0.10a |
| Serum concentration | |||
| Glucose (mmol/L) | 5.24 ± 0.01b | 10.23 ± 0.13 | 7.25 ± 0.08b |
| Insulin (ng/mL) | 0.10 ± 0.002b | 0.82 ± 0.08 | 0.29 ± 0.05b |
| HOMA-IR | 0.57 ± 0.01b | 3.84 ± 0.26 | 1.75 ± 0.04b |
| TG (mmol/L) | 4.56 ± 0.08b | 12.94 ± 0.94 | 9.83 ± 0.94a |
| FFA (mmol/L) | 0.37 ± 0.01b | 0.69 ± 0.02 | 0.51 ± 0.02b |
| TC (μmol/L) | 124.02 ± 4.34b | 466.22 ± 12.32 | 255.08 ± 5.10b |
| Liver concentration | |||
| TG (mmol/mg protein) | 14.75 ± 0.72b | 22.72 ± 1.21 | 6.66 ± 1.01b |
| FFA (nmol/mg protein) | 9.93 ± 0.64b | 18.90 ± 1.35 | 13.10 ± 0.60b |
| TC (nmol/mg protein) | 4.74 ± 0.41b | 17.81 ± 0.98 | 12.66 ± 0.50b |
Data are expressed as the mean ± SE.
P < 0.05,
P < 0.01 vs OLETF control. BW: Body weight; FFA: Free fatty acids; HOMA-IR: Homeostasis model assessment of insulin resistance; TC: Total cholesterol; TG: Triglycerides.
Figure 1.
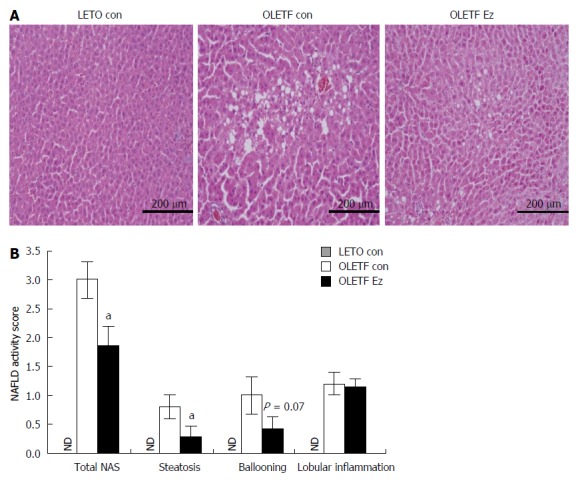
Ezetimibe improves hepatic steatosis in OLETF rats. A: Representative HE liver sections (scale bar, 200 μm; magnification × 200); B: NAFLD activity score. Data are expressed as the mean ± SE. aP < 0.05 vs OLETF control. ND: Not detected in LETO control (LETO con); white bar: OLETF control (OLETF con); black bar: OLETF ezetimibe group (OLETF Ez); NAFLD: Nonalcoholic fatty liver disease.
Ezetimibe induces autophagy in OLETF liver tissue
To address whether ezetimibe administration alters the catabolic autophagy process, we first examined the mRNA and protein expression of autophagy-related genes (ATG). Among identified 30 ATG genes[34], ATG5, ATG6, and ATG7 have been fully demonstrated using method of targeted deletion in animals and cells. In the process of autophagosome formation, ATG5 is conjugated and forms a complex with ATG12 and ATG16[35]. ATG6 and ATG7 are required for autophagy as a part of a lipid kinase complex or by specifically involvement in autophagosome formation[36,37]. In the present study, the hepatic mRNA expression of ATG5, ATG6, and ATG7 was significantly upregulated in ezetimibe-treated OLETF rats by at least 50% (Figure 2A), but no significant difference in protein levels was observed (Figure 2B and C). Microtubule-associated protein light chain 3 (LC3) is related to the extent of autophagosome formation[38]. Specifically, the level of LC3-II is closely associated with the number of autophagosomes present; thus, the ratio between LC3-I and LC3-II (LC3 conversion) can be used to measure the extent of autophagy[39]. In the current study, liver LC3 conversion (LC3-II/LC3-I) was decreased in OLETF controls compared to LETO controls, but ezetimibe administration significantly increased the relative ratio of LC3-II to LC3-I by 15% (Figure 2B and C).
Figure 2.
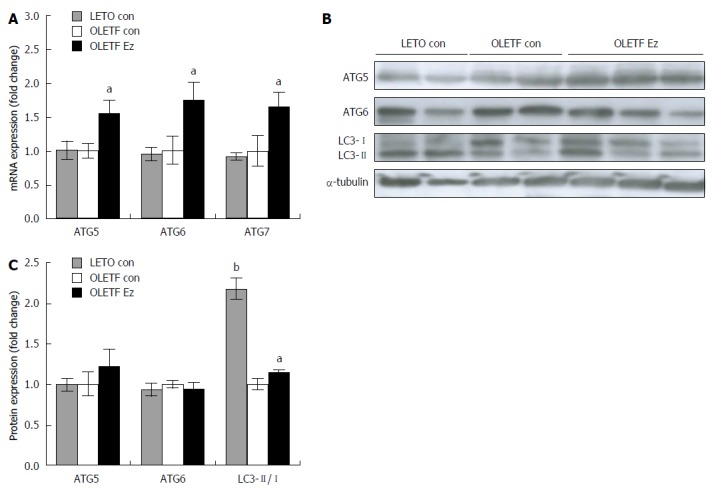
Ezetimibe increases autophagy makers in OLETF liver tissue. mRNA level (A) and protein expression (B and C) was measured and expressed as the fold change compared to OLETF control. ATG 5 and 6 protein expression was normalized by α-tubulin and LC3 protein abundance was expressed as the ratio between LC3-II (14 kDa) and LC3-I (16 kDa). Results are presented as mean ± SE. aP < 0.05, bP < 0.01 vs OLETF control. ATG: Autophagy-related gene; LETO con: LETO control; OLETF con: OLETF control; OLETF Ez: OLETF ezetimibe.
In vitro effects of ezetimibe on TG levels and autophagy in Huh7 hepatocytes
Human huh7 hepatocytes were pretreated with ezetimibe (10 μmol/L, 1 h) and incubated with PA (0.5 mmol/L, 24 h) to induce hepatic steatosis. As shown in Figure 3A, ezetimibe treatment significantly attenuated PA-increased TG levels, which was consistent with our animal study. PA treatment resulted in an approximately 20% decrease in mRNA expression of ATG5, ATG6, and ATG7, which had been increased by ezetimibe treatment (Figure 3B). In addition, ezetimibe treatment significantly increased the PA-induced reduction in LC3 protein abundance (Figure 3C and D). However, p62, specific substrates for autophagy turnover and degradation[40] was significantly elevated by PA which was attenuated by ezetimibe in PA-treated hepatocytes (Figure 3C and D). Transmission electron microscopy (TEM) was employed to investigate the effects of ezetimibe on hepatocyte autophagosome formation. As shown in Figure 4A, PA plus ezetimibe-treated hepatocytes exhibited increased formation of autophagosomes compared to the PA-treated cells. To determine if ezetimibe increases autophagic flux, cells were co-treated with bafilomycin A1 (BAF), an inhibitor of autophagy that inhibits the vacuolar type H+-ATPase, and immunoblotting for LC3 was performed. The results shown in Figure 4B and C indicate that the combination of PA and ezetimibe in the presence of BAF increased the ratio of LC3-II to LC-I, suggesting that ezetimibe affects autophagic flux in PA-treated hepatocytes.
Figure 3.
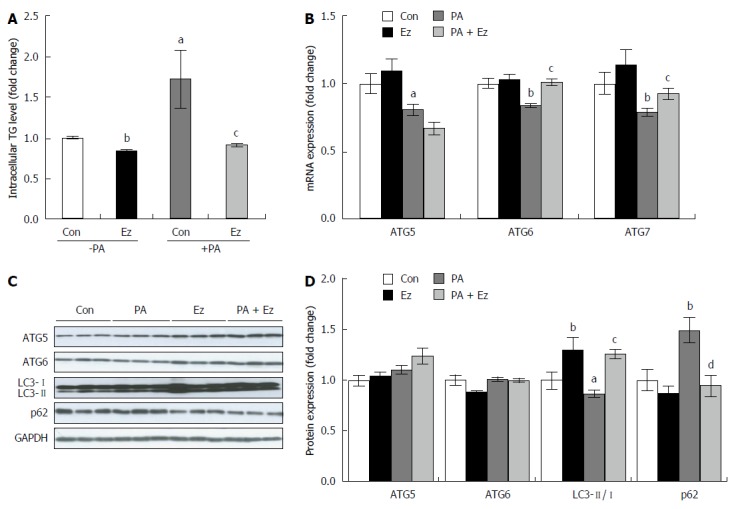
Ezetimibe treatment attenuates triglycerides accumulation and induces autophagy in hepatocytes. Hepatic triglyceride concentration (A) and mRNA (B) and protein expression (C and D) involved in autophagy were expressed as the mean ± SE. Experiments represent at least three independent experiments (n ≥ 9). aP < 0.05, bP < 0.01, vs control; cP < 0.05, dP < 0.01, PA vs PA + Ez. ATG: Autophagy-related gene; Con: Control; Ez: Ezetimibe; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; PA: Palmitic acid; TG: Triglycerides.
Figure 4.
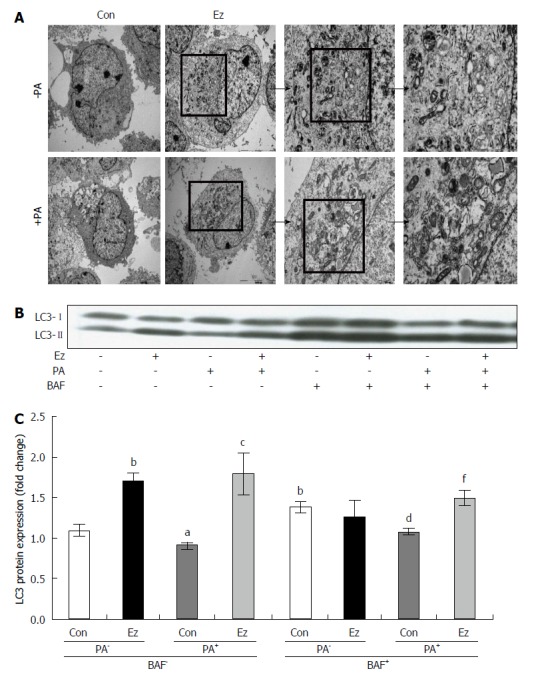
Ezetimibe increases autophagosome formation and autophagic flux in hepatocytes. Electron microscopy of hepatocytes demonstrated ezetimibe-induced autophagy (A). Representative blot (B) and quantitative analysis (C) expressed as LC3-II (14 kDa)/LC3-I (16 kDa) followed by sample/control (mean ± SE). aP < 0.05, bP < 0.01, vs control in the absence of PA and BAF; cP < 0.05, PA vs PA + Ez in the absence of BAF; dP < 0.01, vs control in the absence of PA but in the presence of BAF. fP < 0.01, PA vs PA + Ez in the presence of BAF. BAF: Bafilomycin A1; Con: Control; Ez: Ezetimibe; PA: Palmitic acid.
DISCUSSION
Ezetimibe is a drug used to lower blood cholesterol levels by targeting NPC1L1[5,6]. As described in previous studies showing ezetimibe-mediated improvement in metabolic syndrome[7-11], we found that chronic ezetimibe treatment improves glycemic control and leads to an increase in bioactive glucagon-like peptide-1 (GLP-1) and pancreatic β-cell mass in OLETF rats[41]. In addition, recent findings concerning hepatic NPC1L1 expression and its ability to mediate improvement in hepatic fat accumulation have attracted new research interest[42]. Previous studies have suggested that ezetimibe attenuates hepatic steatosis by decreasing hepatic oxidative stress and improving hepatic insulin sensitivity and lipid metabolism[7,8,16-20]. However, most studies used a mouse model in which hepatic NPC1L1 expression is undetectable. Therefore, these findings might be associated with the ability of ezetimibe to inhibit intestinal cholesterol uptake and the subsequent reduction in hepatic lipid trafficking, rather than a direct effect on liver and hepatic fat metabolism. In the current study, we investigated the role of ezetimibe in hepatic fat accumulation using both in vivo and in vitro models. Age-matched LETO rats were used to compare with OLETF rats. OLETF control rats showed bigger liver size, heavier body weight, and higher glucose and insulin levels than those of LETO controls. Ezetimibe administration in OLETF rats (n = 5) significantly decreased liver weight and serum and liver lipid parameters including TG, FFA, and TC levels, compared to OLETF control animals (n = 6). In addition to the improvement of hepatic fat accumulation, we also found that ezetimibe significantly decreased serum concentrations of glucose and insulin and HOMA-IR, an index of insulin resistance in OLETF rats, consistent with our previous study[41] and numerous other studies[7-9,13,17,20]. Moreover, in vitro hepatocytes, ezetimibe treatment significantly reduced PA-induced TG accumulation (P < 0.05). According to the most accepted hypothesis for hepatic steatosis, the multi-hits model, the accumulation of FFAs and TG in hepatocytes is the first hit which led up to subsequent hits to more advanced stages of liver injury[43]. Thus, ezetimibe-decreased concentrations of lipid parameters in the serum and liver might show the preventive effects of ezetimibe on the initial stage of NAFLD.
As a bulk degradation system, autophagy breaks down the plasma membrane and extracellular proteins in the lysosomal pathway via the formation of double-membrane structures known as autophagosomes[22]. This catabolic pathway has been described recently as a regulator of lipid storage and metabolism, suggesting that autophagy might be a potential therapeutic target for excessive fat accumulation[44]. Genetic and dietary animal models for obesity and hepatic steatosis decrease mRNA expression of autophagy-related genes and conditional ATG7-knockout mice improves hepatic lipid accumulation[27,45]. These animal studies reveal that autophagy play a major role in hepatic lipid metabolism. Moreover, deleted autophagy-related gene (ATG) leads to impaired insulin signaling and increased endoplasmic reticulum (ER) stress. Restoration of ATG in liver of this ATG deficient animals, decreases ER stress and improves hepatic insulin action[46]. It demonstrates that autophagy is involved in intracellular lipid storage, release of fatty acids from triglyceride within hepatocyte lipid droplets, lipotoxicity-induced ER stress, and hepatic insulin sensitivity. In addition, liver specific autophagy deficient mice show hepatocyte cell death and liver injury[47]. In autophagy deficient cells, hepatocyte survival is impaired and production of TNFα is increased, all of which leads to hepatocellular carcinoma[48]. Moreover, autophagy is associated with cell death by interacting with Fas-associated protein with death domain (FADD)[49]. It suggests that autophagy plays a critical role of tumor-suppression mechanism.
In the present study, ezetimibe administration decreased liver weight and lipid levels and improved serum metabolic parameters and histological features, in addition to increasing the expression of LC3-II, in a rat model of type 2 diabetes. The ezetimibe-induced autophagy that we observed was consistent with a recent study showing that ezetimibe plays a role in cholesterol homeostasis and liver degeneration in α1-antitrypsin deficiency[50]. To induce hepatic steatosis, we incubated Huh7 human hepatocytes with 0.5 mmol/L PA for 24 h. As shown in a previous study, PA treatment in hepatocytes significantly decreases autophagy, suggesting that hepatic autophagy is associated with hepatic fat accumulation[29]. In the present study, ezetimibe treatment increased PA-decreased autophagosome formation in Huh7 hepatocytes as determined by TEM and Western blotting of LC3-II and p62 expression. In an autophagic flux assay designed to measure changes in LC3-II in the presence of BAF, an inhibitor of late-phase autophagy that prevents fusion between autophagosomes and lysosomes, ezetimibe treatment increased the level of LC3-II. Taken together, these data indicate that ezetimibe induces autophagy in PA-treated hepatocytes. Despite important findings illustrating the involvement of ezetimibe in autophagy formation and the improvement of hepatic fat accumulation, our study has limitations; gender bias, small sample size, and not to demonstrate direct ezetimibe action to autophagy. Like most previous other studies, the present study used only male animals to demonstrate the influence of ezetimibe on liver to prevent confounding factors such as reproductive cycles and hormone fluctuations. To know exactly how ezetimibe improves hepatic steatosis in women, further studies with female animal model are necessary to be executed. In addition, further investigation is also needed to determine whether ezetimibe as autophagy inducer is directly involved in the improvement of hepatic steatosis by using liver-specific targeted ATG modification in animals with large sample number and precise methods such as electron microscopy, fluorescence microscopy, molecular assays, and use of chemical modulators.
In conclusion, ezetimibe treatment attenuates hepatic fat accumulation and improves hyperglycemia, which is accompanied by an increase in autophagy flux. To the best of our knowledge, this is the first study to show that ezetimibe induces autophagy in the liver of diabetic rats and PA-treated hepatocytes. Our findings suggest a possible target of ezetimibe action and the potential for the use of ezetimibe in the treatment of hepatic steatosis.
COMMENTS
Background
Nonalcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease representing fat accumulation with hepatocytes. NAFLD is strongly associated with other components of metabolic syndrome. Given the high prevalence of NAFLD and its positive correlation with metabolic syndrome, it is important to prevent fat accumulation in the liver. However, there is no satisfying therapeutic strategy for NAFLD.
Research frontiers
Ezetimibe, a Niemann-Pick C1-Like 1 inhibitor has been used as an agent for hypercholesterolemia. In addition to the favorable effects of ezetimibe on lipid metabolism, numerous studies demonstrate that ezetimibe improves other metabolic disorders such as hepatic steatosis and diabetes. However, the mechanisms by which ezetimibe influences hepatic fat accumulation are still unclear.
Innovations and breakthroughs
In this current study, ezetimibe administration improved glucose homeostasis and serum and hepatic lipid parameters, which is accompanied by increased autophagy-related gene and protein expression in liver of obese and diabetic rats. Authors also found that ezetimibe might affect autophagic flux in fatty acid-treated hepatocytes.
Applications
The results in the present study suggest a possible target of ezetimibe action and potential use of ezetimibe in the treatment of NAFLD.
Terminology
Autophagy is a cellular catabolic process by lysosome-dependent machinery, which role is to maintain cellular energy homeostasis. Ezetimibe is a lipid-lowering compound that selectively inhibits the intestinal cholesterol.
Peer-review
The manuscript describes the effect of ezetimibe on hepatic steatosis in a rat model of obesity and type II diabetes. Major conclusion of the manuscript is the induction of autophagy in the liver by application of ezetimibe and, therefore, a reduction of hepatic steatosis. The idea of ezetimibe as inducer of autophagy in the liver/hepatocytes is in line with a previous report by a different group earlier this year. There are several points that need to be addressed.
Footnotes
Supported by Samsung Biomedical Research Institute, Grant No. SBRI C-B1-111-3; National Research Foundation of Korea, No. 2012R1A1A2009143/2013027171; and Korean Diabetes Association (to Park CY, 2014S-1)
Institutional animal care and use committee: All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Kangbuk Samsung Hospital, Sungkyunkwan University (IACUC protocol number: 201010014).
Conflict-of-interest statement: The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 1, 2014
First decision: September 27, 2014
Article in press: January 8, 2015
P- Reviewer: Chen LZ, Cynis H S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
References
- 1.Clark JM. The epidemiology of nonalcoholic fatty liver disease in adults. J Clin Gastroenterol. 2006;40 Suppl 1:S5–10. doi: 10.1097/01.mcg.0000168638.84840.ff. [DOI] [PubMed] [Google Scholar]
- 2.Lazo M, Clark JM. The epidemiology of nonalcoholic fatty liver disease: a global perspective. Semin Liver Dis. 2008;28:339–350. doi: 10.1055/s-0028-1091978. [DOI] [PubMed] [Google Scholar]
- 3.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 4.Byrne CD. Dorothy Hodgkin Lecture 2012: non-alcoholic fatty liver disease, insulin resistance and ectopic fat: a new problem in diabetes management. Diabet Med. 2012;29:1098–1107. doi: 10.1111/j.1464-5491.2012.03732.x. [DOI] [PubMed] [Google Scholar]
- 5.Altmann SW, Davis HR, Zhu LJ, Yao X, Hoos LM, Tetzloff G, Iyer SP, Maguire M, Golovko A, Zeng M, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Calvo M, Lisnock J, Bull HG, Hawes BE, Burnett DA, Braun MP, Crona JH, Davis HR, Dean DC, Detmers PA, et al. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1) Proc Natl Acad Sci USA. 2005;102:8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deushi M, Nomura M, Kawakami A, Haraguchi M, Ito M, Okazaki M, Ishii H, Yoshida M. Ezetimibe improves liver steatosis and insulin resistance in obese rat model of metabolic syndrome. FEBS Lett. 2007;581:5664–5670. doi: 10.1016/j.febslet.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Nomura M, Ishii H, Kawakami A, Yoshida M. Inhibition of hepatic Niemann-Pick C1-like 1 improves hepatic insulin resistance. Am J Physiol Endocrinol Metab. 2009;297:E1030–E1038. doi: 10.1152/ajpendo.00343.2009. [DOI] [PubMed] [Google Scholar]
- 9.Takase H, Dohi Y, Okado T, Hashimoto T, Goto Y, Kimura G. Effects of ezetimibe on visceral fat in the metabolic syndrome: a randomised controlled study. Eur J Clin Invest. 2012;42:1287–1294. doi: 10.1111/eci.12000. [DOI] [PubMed] [Google Scholar]
- 10.Davis HR, Compton DS, Hoos L, Tetzloff G. Ezetimibe, a potent cholesterol absorption inhibitor, inhibits the development of atherosclerosis in ApoE knockout mice. Arterioscler Thromb Vasc Biol. 2001;21:2032–2038. doi: 10.1161/hq1201.100260. [DOI] [PubMed] [Google Scholar]
- 11.Mangili OC, Moron Gagliardi AC, Mangili LC, Mesquita CH, Machado Cesar LA, Tanaka A, Schaefer EJ, Maranhão RC, Santos RD. Favorable effects of ezetimibe alone or in association with simvastatin on the removal from plasma of chylomicrons in coronary heart disease subjects. Atherosclerosis. 2014;233:319–325. doi: 10.1016/j.atherosclerosis.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Davies JP, Scott C, Oishi K, Liapis A, Ioannou YA. Inactivation of NPC1L1 causes multiple lipid transport defects and protects against diet-induced hypercholesterolemia. J Biol Chem. 2005;280:12710–12720. doi: 10.1074/jbc.M409110200. [DOI] [PubMed] [Google Scholar]
- 13.Chan DC, Watts GF, Gan SK, Ooi EM, Barrett PH. Effect of ezetimibe on hepatic fat, inflammatory markers, and apolipoprotein B-100 kinetics in insulin-resistant obese subjects on a weight loss diet. Diabetes Care. 2010;33:1134–1139. doi: 10.2337/dc09-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H, Shima T, Yamaguchi K, Mitsuyoshi H, Minami M, Yasui K, Itoh Y, Yoshikawa T, Fukui M, Hasegawa G, et al. Efficacy of long-term ezetimibe therapy in patients with nonalcoholic fatty liver disease. J Gastroenterol. 2011;46:101–107. doi: 10.1007/s00535-010-0291-8. [DOI] [PubMed] [Google Scholar]
- 15.Yoneda M, Fujita K, Nozaki Y, Endo H, Takahashi H, Hosono K, Suzuki K, Mawatari H, Kirikoshi H, Inamori M, et al. Efficacy of ezetimibe for the treatment of non-alcoholic steatohepatitis: An open-label, pilot study. Hepatol Res. 2010;40:566–573. doi: 10.1111/j.1872-034X.2010.00644.x. [DOI] [PubMed] [Google Scholar]
- 16.Zheng S, Hoos L, Cook J, Tetzloff G, Davis H, van Heek M, Hwa JJ. Ezetimibe improves high fat and cholesterol diet-induced non-alcoholic fatty liver disease in mice. Eur J Pharmacol. 2008;584:118–124. doi: 10.1016/j.ejphar.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 17.Muraoka T, Aoki K, Iwasaki T, Shinoda K, Nakamura A, Aburatani H, Mori S, Tokuyama K, Kubota N, Kadowaki T, et al. Ezetimibe decreases SREBP-1c expression in liver and reverses hepatic insulin resistance in mice fed a high-fat diet. Metabolism. 2011;60:617–628. doi: 10.1016/j.metabol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Matono T, Koda M, Tokunaga S, Kato J, Sugihara T, Ueki M, Murawaki Y. Therapeutic effects of ezetimibe for non-alcoholic steatohepatitis in fatty liver shionogi-ob/ob mice. Hepatol Res. 2011;41:1240–1248. doi: 10.1111/j.1872-034X.2011.00888.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Sugimoto K, Fujisawa T, Shindo N, Minato S, Kamada Y, Hamano M, Ohishi M, Ikegami H, Rakugi H. Novel effect of ezetimibe to inhibit the development of non-alcoholic fatty liver disease in Fatty Liver Shionogi mouse. Hepatol Res. 2014;44:102–113. doi: 10.1111/hepr.12092. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda M, Nakamura T, Kataoka K, Nako H, Tokutomi Y, Dong YF, Yasuda O, Ogawa H, Kim-Mitsuyama S. Ezetimibe ameliorates cardiovascular complications and hepatic steatosis in obese and type 2 diabetic db/db mice. J Pharmacol Exp Ther. 2010;335:70–75. doi: 10.1124/jpet.110.170373. [DOI] [PubMed] [Google Scholar]
- 21.Jia L, Ma Y, Rong S, Betters JL, Xie P, Chung S, Wang N, Tang W, Yu L. Niemann-Pick C1-Like 1 deletion in mice prevents high-fat diet-induced fatty liver by reducing lipogenesis. J Lipid Res. 2010;51:3135–3144. doi: 10.1194/jlr.M006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tallóczy Z, Jiang W, Virgin HW, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding WX. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawano K, Hirashima T, Mori S, Saitoh Y, Kurosumi M, Natori T. Spontaneous long-term hyperglycemic rat with diabetic complications. Otsuka Long-Evans Tokushima Fatty (OLETF) strain. Diabetes. 1992;41:1422–1428. doi: 10.2337/diab.41.11.1422. [DOI] [PubMed] [Google Scholar]
- 31.Kawano K, Hirashima T, Mori S, Natori T. OLETF (Otsuka Long-Evans Tokushima Fatty) rat: a new NIDDM rat strain. Diabetes Res Clin Pract. 1994;24 Suppl:S317–S320. doi: 10.1016/0168-8227(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 32.Song Z, Song M, Lee DY, Liu Y, Deaciuc IV, McClain CJ. Silymarin prevents palmitate-induced lipotoxicity in HepG2 cells: involvement of maintenance of Akt kinase activation. Basic Clin Pharmacol Toxicol. 2007;101:262–268. doi: 10.1111/j.1742-7843.2007.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang SJ, Choi JM, Chae SW, Kim WJ, Park SE, Rhee EJ, Lee WY, Oh KW, Park SW, Kim SW, et al. Activation of peroxisome proliferator-activated receptor gamma by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PLoS One. 2011;6:e17057. doi: 10.1371/journal.pone.0017057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 37.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 40.Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin Immunopathol. 2010;32:431–436. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- 41.Yang SJ, Choi JM, Kim L, Kim BJ, Sohn JH, Kim WJ, Park SE, Rhee EJ, Lee WY, Oh KW, et al. Chronic administration of ezetimibe increases active glucagon-like peptide-1 and improves glycemic control and pancreatic beta cell mass in a rat model of type 2 diabetes. Biochem Biophys Res Commun. 2011;407:153–157. doi: 10.1016/j.bbrc.2011.02.129. [DOI] [PubMed] [Google Scholar]
- 42.Park SW. Intestinal and hepatic niemann-pick c1-like 1. Diabetes Metab J. 2013;37:240–248. doi: 10.4093/dmj.2013.37.4.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 44.Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 50.Yamamura T, Ohsaki Y, Suzuki M, Shinohara Y, Tatematsu T, Cheng J, Okada M, Ohmiya N, Hirooka Y, Goto H, et al. Inhibition of Niemann-Pick-type C1-like1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant α1-antitrypsin Z deposition. Hepatology. 2014;59:1591–1599. doi: 10.1002/hep.26930. [DOI] [PubMed] [Google Scholar]


