Abstract
Mutations of CSB account for the majority of Cockayne syndrome (CS), a devastating hereditary disorder characterized by physical impairment, neurological degeneration and segmental premature aging. Here we report the generation of a human CSB-knockout cell line. We find that CSB facilitates HR and represses NHEJ. Loss of CSB or a CS-associated CSB mutation abrogating its ATPase activity impairs the recruitment of BRCA1, RPA and Rad51 proteins to damaged chromatin but promotes the formation of 53BP1-Rif1 damage foci in S and G2 cells. Depletion of 53BP1 rescues the formation of BRCA1 damage foci in CSB-knockout cells. In addition, knockout of CSB impairs the ATM- and Chk2-mediated DNA damage responses, promoting a premature entry into mitosis. Furthermore, we show that CSB accumulates at sites of DNA double-strand breaks (DSBs) in a transcription-dependent manner. The kinetics of DSB-induced chromatin association of CSB is distinct from that of its UV-induced chromatin association. These results reveal novel, important functions of CSB in regulating the DNA DSB repair pathway choice as well as G2/M checkpoint activation.
Keywords: CSB, DNA damage checkpoint, DNA double-strand break repair
Introduction
DNA double-strand breaks (DSBs) are one of the most lethal forms of DNA damage and can promote tumorigenesis if not repaired properly. Eukaryotic cells have evolved a complex network to sense and repair DSBs. Ataxia telangiectasia mutated (ATM), a kinase, is responsible for transducing the DNA damage signal through phosphorylation of many proteins essential for the activation of the DNA damage checkpoint, cell cycle arrest, DNA repair or apoptosis (Shiloh, 2003; Lukas et al, 2011). Specifically, upon DSB induction, ATM phosphorylates the histone variant H2AX at serine 139, giving rise to γH2AX (Rogakou et al, 1998, 1999). γH2AX plays a key role in marking damaged chromatin and in directing the recruitment of DNA damage signaling and DNA repair proteins into repair centers, also known as ‘foci’ (Lukas et al, 2011; Chapman et al, 2012).
There exist two major pathways responsible for repairing DSBs: nonhomologous end joining (NHEJ) and homologous recombination (HR) (Chapman et al, 2012; McKerlie et al, 2013). NHEJ can ligate two broken ends in the absence of sequence homology, whereas HR, largely error-free, requires sequence homology and is often restricted to the S and G2 phases of the cell cycle during which sister chromatids are present. The choice of which DNA DSB repair pathway is utilized is highly regulated, and two tumor suppressor proteins BRCA1 and 53BP1 play pivotal roles in influencing the fate of the repair of DSBs by either HR or NHEJ (Chapman et al, 2012). BRCA1 promotes HR (Xie et al, 2007; Cao et al, 2009; Bouwman et al, 2010; Bunting et al, 2010), perhaps by facilitating DNA end resection (Bunting et al, 2010), an early step of HR marked by the generation of RPA-coated single-stranded DNA (RPA-ssDNA), whereas 53BP1 and its effector Rif1 are found to antagonize BRCA1 at DSBs to promote NHEJ (Xie et al, 2007; Cao et al, 2009; Bouwman et al, 2010; Bunting et al, 2010; Chapman et al, 2013; Di Virgilio et al, 2013; Escribano-Diaz et al, 2013). A perturbation in the recruitment of BRCA1 or 53BP1 to damaged chromatin can lead to an error in the choice of the DNA DSB repair pathway, which can promote genomic instability, an underlying hallmark of cancer and aging.
Cockayne syndrome (CS) is a devastating hereditary disorder characterized by physical impairment, neurological degeneration and segmental premature aging. The majority of CS cases are caused by mutations in the ERCC6 gene, which encodes Cockayne syndrome group B protein (CSB). CSB is required for transcription-coupled nucleotide excision repair (Troelstra et al, 1992; van der Horst et al, 1997) and has also been implicated in chromatin remodeling (Newman et al, 2006), oxidative damage repair (Stevnsner et al, 2008), interstrand crosslink repair (Iyama et al, 2015), mitochondrial function (Aamann et al, 2010; Scheibye-Knudsen et al, 2012), telomere maintenance (Batenburg et al, 2012) and transcription-associated DNA recombination (Gottipati & Helleday, 2009; Savolainen et al, 2010). CSB is also known to play key roles in transcription and modulation of the stress response (Velez-Cruz & Egly, 2013). CSB contains a central SWI/SNF-like ATPase domain and possesses an DNA-dependent ATPase activity that is important for its chromatin remodeling and UV repair functions (Citterio et al, 1998; Selzer et al, 2002; Cho et al, 2013).
CSB-deficient cells derived from CS patients are best known for their hypersensitivity to UV light because of their defect in transcription-coupled nucleotide excision repair (Troelstra et al, 1992; van der Horst et al, 1997). However, they are also sensitive to several other types of DNA damaging agents including ionizing radiation (IR) (Leadon & Cooper, 1993; Tuo et al, 2002, 2003), camptothecin (CPT) (Squires et al, 1993) and etoposide (Etop) (Elli et al, 1996), all of which are known to induce DNA DSBs. It has been suggested that a defect in base excision repair in CSB-deficient CS cells may contribute to their sensitivity to IR (Tuo et al, 2002, 2003). However, CSB has recently been implicated in the processing of CPT-induced R-loops into DNA DSBs (Sollier et al, 2014), suggesting that it may play a role in DNA DSB repair.
Most CSB-deficient cell lines derived from CS patients carry compound heterozygous CSB mutations, making them less than ideal for mutational analysis of CSB function and speak for a need for human CSB-knockout cells. Here we report the generation of a human CSB-knockout cell line, which we used to demonstrate that CSB has novel, important roles in regulating the choice of DNA DSB repair pathways. We show that CSB accumulates at sites of DNA DSBs in a transcription-dependent manner. Moreover, the loss of CSB promoted NHEJ-mediated repair of DNA DSBs but impaired HR-mediated repair of DNA DSBs. The absence of CSB promoted the recruitment of 53BP1 and Rif1 in S/G2 cells at the expense of blocking BRCA1 association with damaged chromatin. Introduction of wild-type CSB fully suppressed the increase in 53BP1 and Rif1 damage foci formation in CSB-knockout cells, whereas the introduction of CSB carrying a CS-associated W851R mutation in its conserved ATPase domain failed to do so. We propose that CSB represses NHEJ in S/G2 cells to facilitate the HR repair of DNA DSBs and that CSB’s ATPase activity is important for its role in regulating this choice of DNA DSB repair. In addition, we find that CSB is needed for the activation of the ATM- and Chk2-dependent DNA damage responses. Furthermore, we find that the ATPase activity of CSB, which is essential for its UV-induced chromatin association, is dispensable for its DSB-induced chromatin association, suggesting that CSB association with DSBs is distinct from its association with UV-induced damaged chromatin. Our work suggests that dysregulation of DNA DSB repair resulting from defects in CSB could play a role in CS pathology.
Results
Generation of a human CSB-knockout cell line
Most CSB-deficient cell lines derived from CS patients carry compound heterozygous CSB mutations, making them less than ideal for mutational analysis of CSB function. To address this problem, we decided to create a human CSB-knockout cell line by targeting exon 5, the largest exon of CSB, through recombinant adeno-associated virus (rAAV)-mediated gene targeting technology (Fig1A). For these studies, we selected the telomerized human retinal pigment epithelial (hTERT-RPE) cell line since these cells are diploid, wild-type for all known DNA repair genes and have successfully been utilized for gene targeting experiments (Kohli et al, 2004; Burkard et al, 2007; Di Nicolantonio et al, 2008). After the first round, we screened 280 clones and obtained two clones (L3A2 and M1D3) that were correctly targeted (Fig1B). The clone L3A2 was used in the second round of gene targeting. After screening 1,158 clones, 46 correctly targeted clones were obtained; however, only one of them (28-C4) corresponded to the desired genotype in which both copies of exon 5 had been disrupted (Fig1B). The other 45 clones comprised cells in which retargeting to the already (first round) targeted allele had occurred. The absence of CSB expression in the clone 28-C4 was subsequently confirmed by Northern (Fig1C) and Western analyses with two independent anti-CSB antibodies (Fig1D and E).
Figure 1.
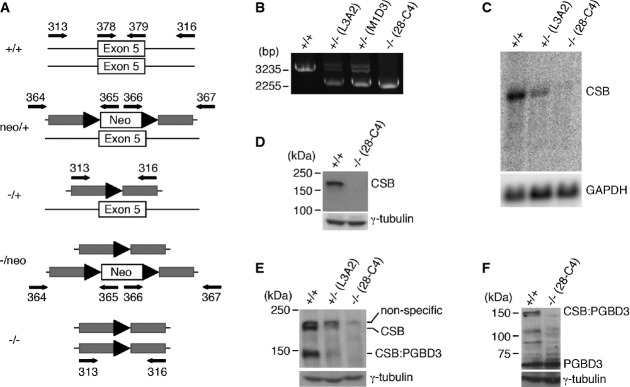
- Schematic diagram of rAAV-mediated gene targeting of exon 5 of CSB. Gray horizonal bars represent homology arms flanking exon 5 of CSB. Black arrowheads represent loxP sites.
- Analysis of PCR products amplified with the primer set 313 and 316 and genomic DNA isolated from parental hTERT-RPE cells, the heterozygote cell clones (L3A2 and M1D3) and the CSB homozygous cell clone (28-C4). Molecular weight markers corresponding to 3,225 bp and 2,255 bp are indicated on the left.
- Northern blot analysis of RNA isolated from parental hTERT-RPE cells, the heterozygous cell clone L3A2 and the homozygous cell clone 28-C4. GAPDH was used as a loading control.
- Western analysis with an antibody against the C-terminus of CSB (Abcam). γ-tubulin was used as a loading control in this and subsequent figures.
- Western analysis with an antibody against the N-terminus of CSB (Bethyl).
- Western analysis with an antibody against PGBD3.
Source data are available online for this figure.
Alternative splicing of exon 5 of CSB with the gene PGBD3, which is located within intron 5 of CSB, instead of exon 6 of CSB, gives rise to a CSB:PGBD3 fusion protein (Newman et al, 2008). Therefore, we also examined the expression of CSB:PGBD3 in 28-C4 cells. Western analysis with an antibody raised against either the N-terminus of CSB, which is present in CSB:PGBD3, or PGBD3 revealed no expression of the CSB:PGBD3 fusion protein in 28-C4 cells (Fig1E and F). On the other hand, expression of PGBD3 was not disrupted (Fig1F). These results demonstrated that CSB:PGBD3 is also knocked out in clone 28-C4, and hereafter, we refer to clone 28-C4 as CSB-knockout (CSB-KO) cells.
Loss of CSB promotes NHEJ but impairs HR, rendering cells sensitive to DNA DSB-inducing agents
We observed that the respective frequency of the random integration of two independent targeting vectors (CSB-rAAV and CCR5-rAAV) in the CSB-KO cells was a 2.65-fold and a 1.74-fold higher than in the parental hTERT-RPE cells (Fig2A), indicating that the loss of CSB may promote random integration. It has been suggested that the random integration of gene targeting vectors is mediated by NHEJ (Kotin et al, 1992; Kan et al, 2014), and therefore, we examined whether inhibition of DNA-PKcs, a kinase directly engaged in NHEJ, might affect the frequency of the random integration in CSB-KO cells. Treatment with NU7026, a specific inhibitor of DNA-PKcs (Veuger et al, 2003), severely impaired the frequency of random integration of either CSB-rAAV or CCR5-rAAV targeting vector in both parental and CSB-KO cells (Supplementary Fig S1A and B), although it did not abolish the increased frequency of random integration in CSB-KO cells (Supplementary Fig S1A and B). These results suggest that random integration is mediated at least in part by NHEJ. Although we cannot rule out the possibility that the increased frequency of random integration in CSB-KO cells is not epistatic to NHEJ deficiency, it is possible that the residual NHEJ activity in cells treated with the DNA-PKcs inhibitor might be sufficient to support the increased frequency of random integration in the CSB-KO cells.
Figure 2.
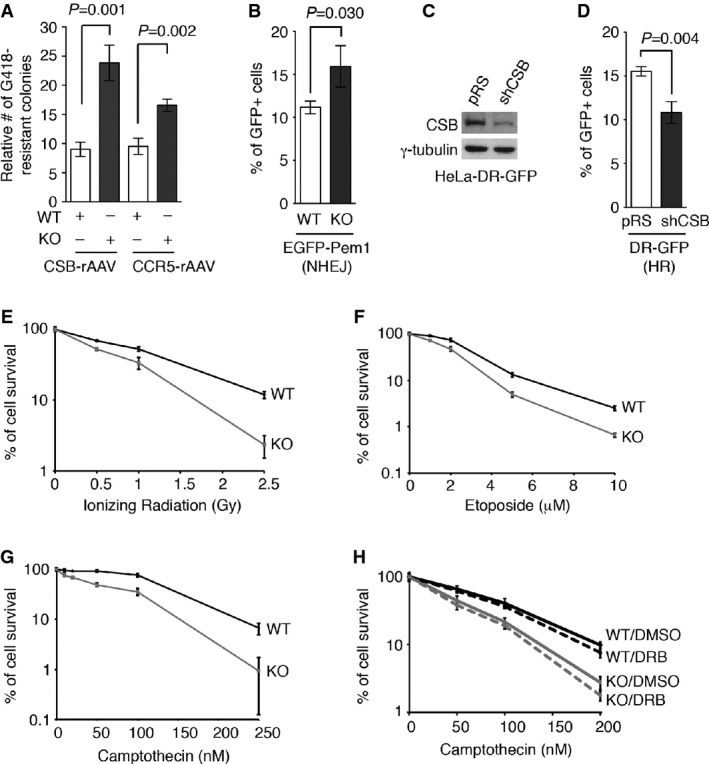
- A Quantification of the frequency of random integration of two independent rAAV targeting vectors (CSB-rAAV and CCR-rAAV). Standard deviations from three independent experiments are indicated.
- B NHEJ-mediated repair of I-SceI-induced DNA DSBs. The parental (WT) and the CSB-KO (KO) cells were cotransfected with pEGFP-Pem1-Ad2, pCherry and I-SceI expression constructs. The number of cells positive for both GFP and pCherry was normalized to the total number of pCherry-positive cells, giving rise to the percentage of normalized GFP-positive cells. Standard deviations from three independent experiments are indicated.
- C Western analysis of CSB in HeLa-DR-GFP transiently transfected with the vector alone or shCSB.
- D HR-mediated repair of I-SceI-induced DNA DSBs. HeLa-DR-GFP transiently expressing the vector alone or shCSB were cotransfected with pCherry and I-SceI expression constructs. The percentage of normalized GFP-positive cells was calculated as described in (B). Standard deviations from three independent experiments are indicated.
- E–G Clonogenic survival assays of the parental (WT) and the CSB-KO (KO) cells following various doses of ionizing radiation (IR) (E), etoposide (Etop) (F), or camptothecin (CPT) (G). Standard deviations from at least three experiments are indicated.
- H Clonogenic survival assays. Both WT and CSB-KO cells were treated with 50 μM DRB, prior to the addition of various doses of CPT. Standard deviations from at least three experiments are indicated.
Data information: P-values were determined using two-tailed unpaired Student’s t-test.
Source data are available online for this figure.
To further investigate the role of CSB in regulating NHEJ-mediated DNA DSB repair, we employed a well-established NHEJ reporter plasmid pEGFP-Pem1-Ad2 (Seluanov et al, 2004), which contains the GFP gene disrupted by the insertion of an I-SceI restriction enzyme site. Repair of I-SceI-induced DNA DSBs by NHEJ restores GFP expression in pEGFP-Pem1-Ad2. NHEJ-mediated repair of I-SceI-induced DSBs was significantly upregulated in CSB-KO cells when compared with parental cells (Fig2B), further supporting the notion that CSB negatively regulates NHEJ-mediated DSB repair.
An upregulation in NHEJ-mediated DSB repair can have a consequence on the repair of DSBs by HR, and therefore, we also asked whether loss of CSB might affect HR-mediated DSB repair. To address this question, we employed a HeLa cell line stably expressing a well-established HR reporter DR-GFP (Escribano-Diaz et al, 2013), which also contains the GFP gene disrupted by the insertion of an I-SceI restriction enzyme site. In this instance, however, repair of I-SceI-induced DNA DSBs by HR restores GFP expression in HeLa cells. The knockdown of CSB led to a significant reduction in HR-mediated repair of I-SceI-induced DSBs (Fig2C and D), suggesting that CSB facilitates HR-mediated DSB repair.
In support of the notion that CSB regulates DNA DSB repair pathway choice, CSB-KO cells were sensitive to a range of DSB-inducing agents including IR, Etop and CPT (Fig2E–G), in agreement with previous findings (Squires et al, 1993; Elli et al, 1996; Tuo et al, 2002, 2003). CPT is known to induce DSBs specifically in S phase (Ryan et al, 1991); however, we did not observe any significant difference in the cell cycle profile between parental and CSB-KO cells (Supplementary Fig S2), suggesting that the increased sensitivity of CSB-KO cells to CPT is not likely due to a change in their S phase profile.
It has been suggested that CPT can also induce DNA DSBs in a transcription-dependent manner (Sakasai et al, 2010), and therefore, we asked whether the hypersensitivity of CSB-KO cells to CPT might be dependent upon transcription. To address this question, we treated both parental and CSB-KO cells with the transcription inhibitor DRB for 1 h, followed by the incubation with CPT in the presence of DRB for another 1 h. Treatment with DRB resulted in a slight increase in the sensitivity of both parental and CSB-KO cells to CPT (Fig2H), which was not significantly different, suggesting that the increased sensitivity of CSB-KO cells to CPT is unlikely to be mediated solely by transcription-dependent damage.
Loss of CSB impairs the recruitment of BRCA1 and HR repair factors to sites of DNA damage
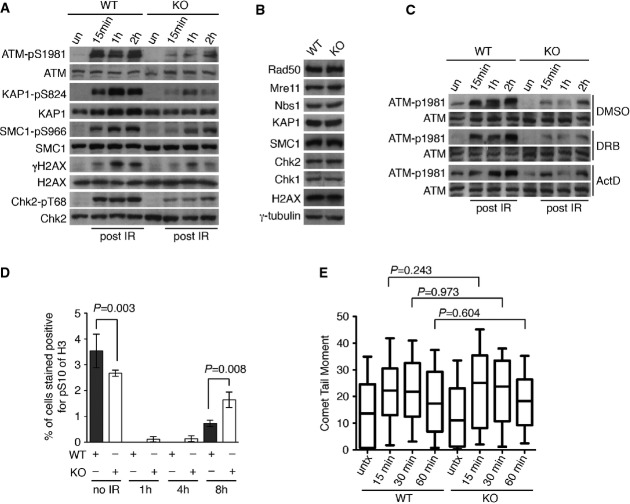
Analysis of indirect immunofluorescence with anti-γH2AX revealed that CSB-KO cells were able to form IR-induced γH2AX foci indistinguishable from the parental cells 1 h post-IR (2 Gy) exposure (Supplementary Fig S3A and B), suggesting that the absence of CSB does not prevent the recruitment of γ-H2AX to sites of DNA DSBs. However, we observed that the CSB-KO cells exhibited a small—but significant—accumulation of IR-induced γH2AX foci 4 and 8 h post-IR (Supplementary Fig S3B). Further analysis revealed that this increase in the formation of IR-induced γH2AX foci was restricted to cyclin A (a marker for cells in the S/G2 phases of the cell cycle)-positive CSB-KO cells (Fig3A; Supplementary Fig S3C), suggesting that CSB-KO cells may be compromised in HR-mediated repair of DSBs in S/G2, in agreement with our earlier finding that depletion of CSB impairs HR-mediated repair of I-SceI-induced DSBs (Fig2D).
Figure 3.

- Quantification of percentage of cyclin A-positive and cyclin A-negative cells exhibiting 10 or more IR-induced γH2AX foci. Both parental (WT) and CSB-KO (KO) cells were treated with 2 Gy IR and fixed 1 h, 4 h and 8 h post-IR. A total of at least 1,500 cells from three independent experiments were scored in blind.
- Quantification of the percentage of cells with 10 or more IR-induced BRCA1 foci. Cells (WT and KO) were treated with 2 Gy IR and fixed 1 h and 4 h post-IR. A total of at least 1,500 cells from three independent experiments were scored in blind.
- Quantification of percentage of cyclin A-positive and cyclin A-negative cells displaying 10 or more IR-induced BRCA1 foci. Cells were treated with 2 Gy IR and scored as described in (B).
- Quantification of the percentage of cyclin A-positive cells with 10 or more IR-induced RPA32-pS4/pS8 foci. Cells (WT and KO) were treated with 10 Gy IR and fixed 4 h and 8 h post-IR. A total of 750 cells from three independent experiments were scored in blind.
- Quantification of percentage of cells with 10 or more IR-induced Rad51 foci. Cells (WT and KO) were treated with 10 Gy IR and fixed 4 h and 8 h post-IR. A total of 750 cells from three independent experiments were scored in blind.
- Clonogenic survival assays of olaparib-treated parental (WT) and CSB-KO (KO) cells as indicated.
- Quantification of percentage of cells with 10 or more IR-induced Rad51 foci. Cells (WT and KO) were treated with actinomycin D (1 μg/ml) or DRB (50 μM) prior to 10 Gy IR and then fixed 8 h post-IR. A total of at least 1,500 cells from three independent experiments were scored in blind.
BRCA1, a tumor suppressor protein, plays a key role in directing DNA DSBs to HR repair (Xie et al, 2007; Cao et al, 2009; Bouwman et al, 2010; Bunting et al, 2010), and therefore, we examined the recruitment of BRCA1 to sites of DNA damage in CSB-KO cells. We observed a significant reduction in the formation of IR-induced BRCA1 foci in CSB-KO cells (Fig3B; Supplementary Fig S3D). Further analysis of dual indirect immunofluorescence with an anti-BRCA1 antibody in conjunction with an anti-cyclin A antibody revealed that the reduction in the formation of IR-induced BRCA1 foci in the knockout cells was largely confined to cyclin A-positive cells (Fig3C; Supplementary Fig S3E). On the other hand, the loss of CSB did not lead to any detectable change in the level of BRCA1 expression (Supplementary Fig S3F). Taken together, these results suggest that CSB is important for the recruitment of BRCA1 to sites of DNA damage in S/G2 cells.
The effect of the loss of CSB on the recruitment of proteins directly involved in DNA DSB repair was also examined. CSB-KO cells were compromised in forming not only IR-induced foci of RPA (Fig3D), a readout commonly used for DNA end resection (Huertas & Jackson, 2009; McKerlie et al, 2013), but also IR-induced foci of Rad51 (Fig3E), a HR recombinase. In addition, CSB-KO cells were sensitive to olaparib (Fig3F), a PARP1 inhibitor known to be toxic to cells deficient in HR (Chapman et al, 2013; Escribano-Diaz et al, 2013). Collectively, these results demonstrate that CSB plays an important role in facilitating HR repair in S/G2 cells.
As CSB is known to be involved in transcription, we also investigated whether the observed impairment of IR-induced Rad51 foci in CSB-KO cells might be transcription dependent. To address this question, we treated parental and CSB-KO cells with a transcription inhibitor (actinomycin D or DRB) prior to IR treatment. Pretreatment with actinomycin D or DRB severely impaired the formation of IR-induced Rad51 foci formation in both parental and CSB-KO cells that stained positive for cyclin A (Fig3G), in agreement with previous finding that Rad51 recruitment to sites of DNA DSBs is transcription dependent (Aymard et al, 2014). On the other hand, pretreatment with actinomycin D or DRB did not abolish the decrease in IR-induced Rad51 foci formation observed in CSB-KO cells (Fig3G), suggesting that transcription-dependent damage is not likely to be the main cause for the impaired Rad51 foci formation in the CSB-KO cells.
Loss of CSB leads to an accumulation of 53BP1 and Rif1 at sites of DNA damage in S/G2 cells
To investigate whether the observed defect in recruiting HR factors in CSB-KO cells might be associated with a concomitant increase in recruiting NHEJ-promoting factors to the sites of DSBs, the formation of IR-induced foci of 53BP1 and Rif1, both of which are known to inhibit BRCA1 and to promote NHEJ (Chapman et al, 2013; Di Virgilio et al, 2013; Escribano-Diaz et al, 2013; Feng et al, 2013; Zimmermann et al, 2013), was examined. Analysis of indirect immunofluorescence with anti-53BP1 revealed that CSB-KO cells were not only competent in forming IR-induced 53BP1 foci (Fig4A; Supplementary Fig S4A) but also displayed a significant supernumerary accumulation of IR-induced 53BP1 foci 4 and 8 h post-IR (Fig4A). Similarly, an excess accumulation of IR-induced Rif1 foci in CSB-KO cells (Fig4B; Supplementary Fig S4B) was observed. Again, the accumulation of IR-induced Rif1 foci was predominantly confined to CSB-KO cells staining positive for cyclin A (Fig4C; Supplementary Fig S4B), suggesting that the loss of CSB promotes NHEJ activity in S/G2 cells, which is in agreement with our earlier findings that the loss of CSB promoted NHEJ-mediated repair of I-SceI-induced DSBs (Fig2B).
Figure 4.
- Quantification of percentage of cells with 10 or more IR-induced 53BP1 foci. Cells (WT and KO) were treated with 2 Gy IR and fixed 1 h, 4 h and 8 h post-IR. A total of 1,500 cells from three independent experiments were scored in blind.
- Quantification of percentage of cells with 10 or more IR-induced Rif1 foci. Cells (WT and KO) were treated with 2 Gy IR and fixed 1 h and 8 h post-IR. A total of 750 cells from three independent experiments were scored in blind.
- Quantification of the percentage of cyclin A-positive and cyclin A-negative cells with 10 or more IR-induced Rif1 foci. Cells were treated and scored as described in (B).
- Clonogenic survival assays of olaparib-treated parental (WT) and CSB-KO (KO) cells stably expressing the vector alone, sh53BP1-A or sh53BP1-B as indicated.
- Clonogenic survival assays of UV-treated parental (WT) and CSB-KO (KO) cells stably expressing the vector alone, sh53BP1-A or sh53BP1-B as indicated.
- Quantification of percentage of cells with 10 or more IR-induced BRCA1 foci. Parental (WT) and knockout (KO) stably expressing the vector (pRS) alone or sh53BP1-A were treated with 2 Gy IR and fixed 1 and 8 h post-IR. A total of at least 1,500 cells from three independent experiments were scored in blind.
CSB-KO cells are sensitive to olaparib (Fig3F). To investigate whether the observed increase in NHEJ activity in S/G2 cells might contribute to the sensitivity of the CSB-KO cells to olaparib, 53BP1 was knocked down with two independent shRNA constructs (Supplementary Fig S4C). The knockdown of 53BP1 fully suppressed the sensitivity of the knockout cells to olaparib (Fig4D), and this suppression was specific to olaparib since the 53BP1 knockdown did not suppress the UV sensitivity of the CSB-KO cells (Fig4E). In addition, the knockdown of 53BP1 rescued the formation of IR-induced BRCA1 foci in the CSB-KO cells (Fig4F). Taken together, these results demonstrate that CSB is important for suppressing NHEJ in S/G2, which, in turn, supports the HR-mediated repair of DSBs.
Loss of CSB impairs the ATM-mediated DNA damage response and promotes a premature exit from the G2/M checkpoint
Upon the induction of DNA DSBs, ATM, a master regulator of the DNA damage response, is activated through its autophosphorylation at S1981 (Bakkenist & Kastan, 2003). To investigate whether loss of CSB might affect ATM activation, both parental and CSB-KO cells were exposed to 5 Gy of IR. The parental cells displayed a robust ATM phosphorylation at S1981 as early as 15 min post-IR (Fig5A), consistent with previous findings (Bakkenist & Kastan, 2003; McKerlie et al, 2013). In contrast, the level of ATM phosphorylation at S1981 was severely impaired in the CSB-KO cells after IR (Fig5A) although the level of ATM expression in the knockout cells was indistinguishable from that in the parental cells (Fig5A). A loss in the level of ATM phosphorylation at S1981 was also observed in the knockout cells following treatment with Etop (Supplementary Fig S5A). Furthermore, the IR-induced phosphorylation of KAP1, SMC1, H2AX and Chk2, downstream targets of ATM, was also impaired in the CSB-KO cells (Fig5A). Little change in Chk1 phosphorylation was detected in the CSB-KO cells (Supplementary Fig S5B). Loss of CSB also had little effect on the expression level of KAP1, SMC1, H2AX, Chk1 and Chk2 (Fig5B). Taken together, these results suggest that loss of CSB impairs ATM-mediated DNA damage response.
Figure 5.
- Western analysis of the parental (WT) and the CSB-KO (KO) cells that were either mock-treated or treated with 5 Gy IR. Immunoblotting was performed with anti-ATM-pS1981, anti-ATM, anti-KAP1-pS824, anti-KAP1, anti-SMC1-pS966, anti-SMC1, anti-γH2AX, anti-H2AX, anti-Chk2-pT68 and anti-Chk2 antibodies.
- Western analysis of WT and CSB-KO cells. Immunoblotting was performed with anti-Rad50, anti-Mre11, anti-Nbs1, anti-KAP1, anti-SMC1, anti-Chk2, anti-Chk1, anti-H2AX and anti-γ-tubulin antibodies. The anti-γ-tubulin blot was used as a loading control.
- Western analysis. WT and CSB-KO cells were treated with DMSO, DRB or actinomycin D (ActD) prior to 5 Gy IR. Immunoblotting was performed with anti-ATM-pS1981 and anti-ATM antibodies.
- Quantification of the percentage of cells staining positive for H3-pS10. For each cell line, at least 3,000 cells from three independent experiments were scored in blind. Standard deviations from three independent experiments are indicated. P-values were determined using two-tailed unpaired Student’s t-test.
- Quantification of comet tail moment. Both WT and CSB-KO cells were treated with 10 Gy IR and harvested for comet assays 15 min, 30 min and 1 h post-IR. At least 200 cells were scored for each sample. In the box and whisker plot, boxes extend from the 25th to 75th percentiles whereas whiskers are from the 10th to 90th percentiles. P-values were determined using non-parametric Mann-Whitney rank-sum t-test.
Source data are available online for this figure.
CSB has been suggested to play a role in transcription, and therefore, formally it was possible that the loss of CSB might affect the expression of DNA damage response factors important for the regulation of ATM activation. Following the induction of DNA DSBs, ATM activation requires the Mre11/Rad50/Nbs1 complex (Lee & Paull, 2004, 2005). Western analysis revealed that the levels of Mre11, Rad50 and Nbs1 expression in CSB-KO cells were indistinguishable from that in parental cells (Fig5B), suggesting that the compromised ATM activation observed in the CSB-KO cells is unlikely due to a loss in the level of the Mre11/Rad50/Nbs1 complex. Furthermore, we found that pretreatment with the transcription inhibitor DRB or actinomycin D did not abrogate the reduction in the level of ATM phosphorylation at S1981 in the CSB-KO cells (Fig5C), suggesting that the compromised ATM activation in the CSB-KO cells is not likely to be mediated by active transcription.
Although CSB-KO cells were able to enter an G2/M arrest immediately following the treatment with IR (Fig5D), they exhibited premature exit from the G2/M checkpoint (Fig5D). Earlier we have shown that CSB-KO cells promote NHEJ (Fig2B). To investigate whether an increase in NHEJ-mediated fast repair of DNA DSBs might contribute to the observed premature entry of CSB-KO cells into mitosis, we performed neutral comet assays with both parental (WT) and CSB-KO cells that were either mock or IR treated. The comet tail moment in the CSB-KO cells was indistinguishable from that in the parental (WT) cells 15 min, 30 min or 1 h after 10 Gy IR (Fig5E), suggesting that the premature exit of CSB-KO cells from the G2/M checkpoint is not likely to be due to a difference in the efficiency of fast DSB repair.
Inhibition of ATM abrogates IR-induced Rif1 foci formation in CSB-KO cells
The formation of IR-induced Rif1 foci is dependent upon ATM activation (Chapman et al, 2013; Escribano-Diaz et al, 2013). Moreover, CSB-KO cells exhibit an impairment in ATM activation (Fig5A) but are competent in forming IR-induced Rif1 foci (Fig4C; Supplementary Fig S4B). To investigate whether the ATM activity might mediate the accumulation of IR-induced Rif1 foci in the CSB-KO cells, we treated both parental and CSB-KO cells with KU55933, a specific inhibitor for ATM, prior to 2 Gy IR treatment. The preincubation with KU55933 abrogated the IR-induced Rif1 foci formation in both parental and CSB-KO cells (Supplementary Fig S4D), in agreement with previous findings (Chapman et al, 2013; Escribano-Diaz et al, 2013). Taken together, these results suggest that Rif1 recruitment to sites of DNA DSBs may not require a full level of ATM activation.
CSB, but not the CSB:PGBD3 fusion protein, is the main factor responsible for facilitating HR repair of DNA DSBs
The deletion of exon 5 of CSB leads to loss of expression of both CSB and the CSB:PGBD3 fusion protein from the CSB locus (Fig1E). To investigate whether CSB or CSB-PGBD3 was responsible for the observed defect in recruiting HR factors to sites of DSBs in the CSB-KO cells, we generated derivative CSB-KO cell lines stably expressing CSB, CSB:PGBD3 or an empty vector (Supplementary Fig S6A). The introduction of CSB, but not the CSB-PGBD3 fusion protein, was able to suppress the sensitivity of the knockout cells to olaparib (Supplementary Fig S6B). In addition, the introduction of CSB, but not the CSB:PGBD3 fusion, was able to rescue Etop-induced foci of RPA and Rad51 (Supplementary Fig S6C and D). From these results, we conclude that CSB is the main factor from the CSB locus responsible for promoting HR-mediated repair of DSBs.
Recruitment of DSB repair factors to sites of DNA damage is misregulated in cells derived from CS patients
To investigate whether the defect in HR-mediated repair of DSBs in the CSB-KO cells might be cell type specific, we examined the recruitment of DSB repair factors to sites of DSBs in two cell lines derived from CS patients lacking functional CSB (hTERT-GM10905 and GM16095). hTERT-GM10905 is a telomerase-immortalized CS cell line carrying a homozygous nonsense mutation at position 735 (R735X) of CSB, whereas GM16095 is a SV40-transformed CS cell line with heterozygous compound mutations of K377X and R857X (Batenburg et al, 2012). Through retroviral infection, two pairs of isogenic cell lines stably expressing either wild-type CSB or the vector alone were generated. The introduction of wild-type CSB into these two CS cell lines led to a significant decrease in the percentage of cells with IR-induced 53BP1 foci (Supplementary Fig S7A) and simultaneously resulted in a significant increase in the number of cyclin A-positive cells with IR-induced foci of BRCA1, RPA and Rad51 (Supplementary Fig S7B–D), suggesting that CS cells lacking functional CSB are also defective in HR-mediated DSB repair. In support of this notion, the introduction of CSB into GM16095 cells also enhanced cell survival in response to the treatment with olaparib (Supplementary Fig S7E), consistent with a previous report that CS cells are hypersensitive to PARP inhibition (Thorslund et al, 2005). Furthermore, the introduction of CSB into GM16905 cells suppressed their sensitivity to CPT and Etop (Supplementary Fig S7F and G). Taken together, these results demonstrate that DNA DSB repair is misregulated in CS cells lacking functional CSB.
CSB is found to accumulate at sites of DSBs in a transcription-dependent manner
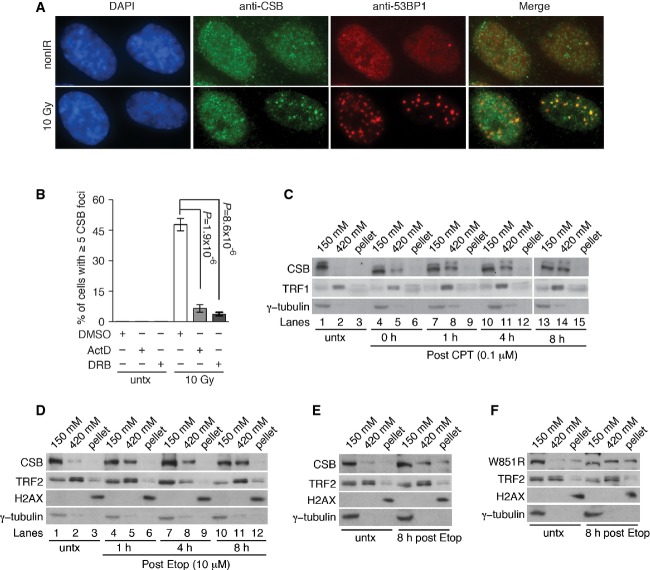
To investigate whether CSB may be associated with sites of DSBs, dual indirect immunofluorescence with an anti-CSB antibody in conjunction with an anti-53BP1 antibody in cells treated with no IR or 10 Gy IR was performed. About 40 to 50% of cells exhibited IR-induced damage foci of CSB 8 h post-IR, and these CSB damage foci all contained 53BP1 (Fig6A), a marker for DSBs (Daley & Sung, 2014; Panier & Boulton, 2014), suggesting that CSB accumulates at sites of DSBs.
Figure 6.
- A Analysis of indirect immunofluorescence with an anti-CSB antibody in conjunction with an anti-53BP1 antibody. hTERT-RPE cells were treated with 10 Gy IR and fixed 8 h post-IR. Cell nuclei were stained with DAPI (blue).
- B CSB accumulation at sites of DNA DSBs is severely impaired by actinomycin D or DRB treatment. Quantification of hTERT-RPE cells with five or more IR-induced CSB damage foci in cells. Cells were treated with actinomycin D (1 μg/ml, ActD) for 1 h or DRB (50 μM) for 2 h prior to 10 Gy IR and maintained in the drugs for 8 h post-IR. A total of at least 1,500 cells from three independent experiments were scored in blind. Standard deviations from three independent experiments are indicated. P-values were determined using two-tailed unpaired Student’s t-test
- C, D Analysis of differential salt extraction of chromatin. hTERT-RPE cells were treated with 0.1 μM CPT (C) or 10 μM Etop (D) for 1 h and then released from drugs for various time points as indicated. Immunoblotting was performed with anti-CSB antibody. The anti-TRF1, anti-TRF2, anti-H2AX and anti-γ-tubulin blots were used as controls for differential salt extraction.
- E, F Analysis of differential salt extraction of chromatin. The CSB-KO cells stably expressing either wild-type CSB (E) or mutant CSB-W851R (F) were treated with 10 μM etoposide (Etop) for 1 h and then released from Etop for 8 h. Immunoblotting was performed with anti-CSB, anti-TRF2, anti-H2AX and anti-γ-tubulin antibodies.
Source data are available online for this figure.
CSB is engaged in transcription, and we therefore asked whether transcription might regulate CSB accumulation at sites of DSBs. To address this question, cells were treated with the transcription inhibitor actinomycin D or DRB prior to 10 Gy IR treatment. Treatment with actinomycin D or DRB severely impaired the formation of IR-induced CSB damage foci (Fig6B), indicating that CSB accumulation at sites of DSBs is dependent upon active transcription.
The ATPase activity of CSB is dispensable for its DSB-induced chromatin association
To gain further insights into the CSB association with DSB-induced damaged chromatin, differential salt extraction of chromatin from hTERT-RPE cells that were mock-treated or treated with either CPT or Etop was performed. Treatment with CPT or Etop led to a significant increase in the association of CSB with chromatin (lane 2 versus lanes 5, 8, 11 and 14 in Fig6C and lane 2 versus lanes 5, 8 and 11 in Fig6D). At 8 h after release from treatment with either Etop or CPT, approximately 50% of the CSB was found associated with chromatin (Fig6C, lane 2 versus lane 14 and Fig6D, lane 2 versus lane 11), supporting the notion that CSB is recruited to damaged chromatin following the induction of DNA DSBs. On the other hand, proportionally, UV-induced CSB association with chromatin peaked 2 h post-UV treatment and was largely lost 4 h post-UV (Supplementary Fig S8A, lane 8 versus lane 11), in agreement with previous findings (Lake et al, 2010). Taken together, these results suggest that the kinetics of CPT- and Etop-induced CSB association with chromatin is distinct from that of UV-induced CSB association with chromatin.
CSB contains a conserved SWI/SNF-like ATPase domain and exhibits a DNA-dependent ATPase activity (Citterio et al, 1998) that is required for its UV-induced chromatin association (Lake et al, 2010). Amino acid substitutions in the conserved ATPase domain are found in CS patients, and the W851R mutation abrogates the ATPase activity of CSB and its UV-induced chromatin association (Lake et al, 2010). To investigate whether the ATPase activity of CSB might be important for its DSB-induced chromatin association, we generated derivative CSB-KO cells stably expressing either wild-type CSB, CSB carrying the W851R mutation or the vector alone (Supplementary Fig S8B). In undamaged (untreated) cells, we reproducibly observed chromatin association of mutant CSB-W851R at a level higher than that of wild-type CSB (Fig6E and F; Supplementary Fig S8C and D). Upon Etop treatment, mutant CSB-W851R was able to exhibit DSB-induced chromatin association (Fig6F). On the other hand, we failed to detect any increase in the proportion of CSB associated with chromatin following UV treatment (Supplementary Fig S8D) although wild-type CSB exhibited UV-induced chromatin association (Supplementary Fig S8C), in agreement with previous findings (Lake et al, 2010). Analysis of multiple protein markers, either cytoplasmic or chromatin bound (γ-tubulin, TRF2 or H2AX), revealed that the chromatin salt fractionation procedure was done consistently between the CSB-KO cells expressing CSB-W851R and the CSB-KO cells expressing wild-type CSB (Fig6E and F; Supplementary Fig S8C and D). Taken together, these results suggest that DSB-induced chromatin association of CSB is distinct from its UV-induced chromatin association and that the ATPase activity of CSB is dispensable for its DSB-induced chromatin association.
The ATPase activity of CSB is essential for suppressing NHEJ to facilitate HR-mediated repair of DSBs in S/G2 cells
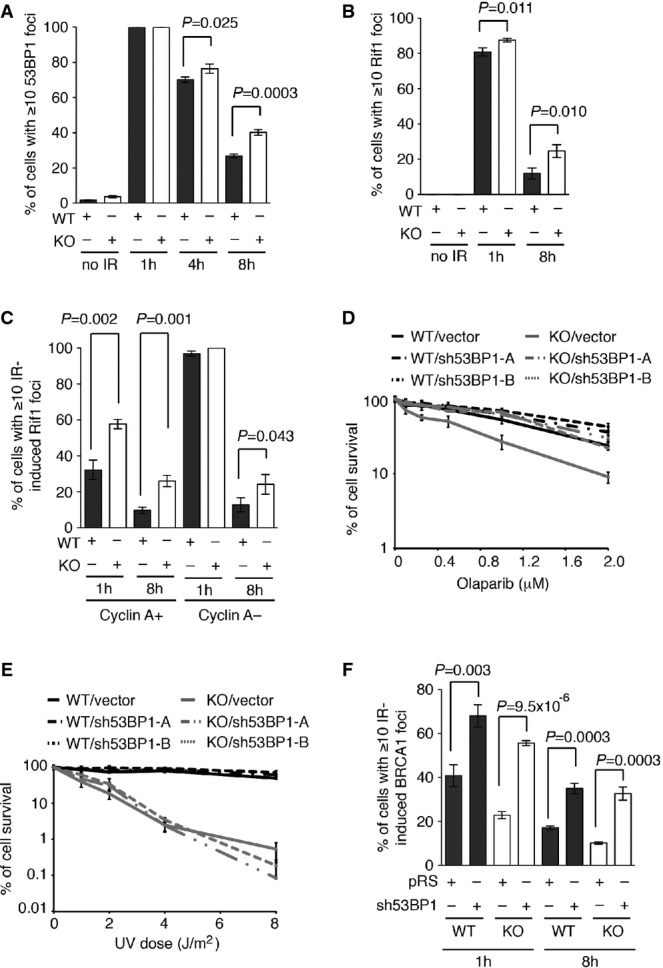
To investigate whether the ATPase activity of CSB might be important for regulating the choice of DNA DSB repair pathways, we examined the recruitment of 53BP1/Rif1 and BRCA1 to sites of DSBs in CSB-KO cells stably expressing vector alone, wild-type CSB or CSB harboring the W851R mutation. Introduction of wild-type CSB into CSB-knockout cells suppressed the number of cells with IR-induced foci of 53BP1 and Rif1 (Fig7A and B). This suppression was not detectable in CSB-KO cells expressing the mutant CSB-W851R (Fig7A and B). The reduction in IR-induced Rif1 foci, resulting from introduction of wild-type CSB, was only observed in the knockout cells staining positive for cyclin A (Fig7C), suggesting that the ATPase activity of CSB is important for suppressing NHEJ-mediated repair of DNA DSBs in S/G2 cells.
Figure 7.
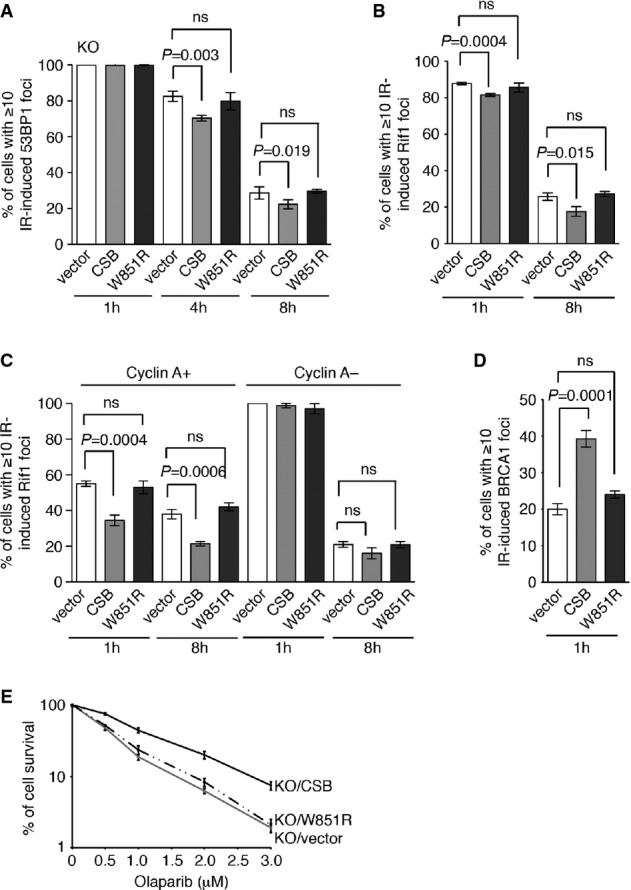
- Quantification of percentage of cells with 10 or more IR-induced 53BP1 foci. CSB-KO cells stably expressing wild-type CSB, CSB-W851R or the vector alone were treated with 2 Gy IR and fixed 1 h, 4 h and 8 h post-IR. A total of 1,500 cells from three independent experiments were scored in blind for each cell line.
- Quantification of percentage of cells with 10 or more IR-induced Rif1 foci. CSB-KO cells stably expressing wild-type CSB, CSB-W851R or the vector alone were treated with 2 Gy IR and fixed 1 h and 8 h post-IR. A total of 1,500 cells from three independent experiments were scored in blind for each cell line.
- Quantification of percentage of cyclin A-positive and cyclin A-negative cells with 10 or more IR-induced Rif1 foci. Cells were treated and fixed as described in (B). A total of 750 cells from three independent experiments were scored in blind for each cell line. Standard deviations from three independent experiments are indicated.
- Quantification of percentage of cells with 10 or more IR-induced BRCA1 foci. Cells were treated with 2 Gy IR and fixed 1 h post-IR. A total of 1,500 cells from three independent experiments were scored in blind for each cell line.
- Clonogenic survival assays of olaparib-treated CSB-KO cells stably expressing wild-type CSB, CSB-W851R or the vector alone as indicated.
Additionally, analysis of indirect immunofluorescence with anti-BRCA1 antibody revealed that introduction of wild-type CSB rescued the formation of IR-induced BRCA1 foci in CSB-KO cells, whereas CSB carrying the W851R mutation failed to do so (Fig7D). Furthermore, while introduction of wild-type CSB into CSB-KO cells promoted cell survival after treatment with olaparib (Fig7E), CSB carrying the W851R mutation was unable to suppress the sensitivity of the knockout cells to olaparib (Fig7E). Taken together, these results suggest that while the ATPase activity of CSB is not important for chromatin recruitment, it is important for its ability to facilitate the HR-mediated repair of DSBs.
The ATPase activity of CSB is important for the maintenance of the G2/M checkpoint
When introduced into the CSB-KO cells, wild-type CSB was able to rescue the level of IR-induced ATM phosphorylation at S1981, most noticeable at 15 min post-IR treatment (Fig8A). On the other hand, no rescue was detected in the CSB-KO cells complemented with CSB carrying the W851R mutation (Fig8A). In addition, we reproducibly observed a rescue in the level of Chk2 phosphorylation 1 h post-IR treatment in CSB-KO cells complemented with wild-type CSB and such a rescue was not seen in CSB-KO cells complemented with CSB carrying the W851R mutation (Fig8A). Furthermore, the introduction of wild-type CSB into CSB-KO cells was able to suppress their premature exit from the G2/M checkpoint, whereas introduction of CSB carrying a W851R mutation failed to do so (Fig8B). Introduction of CSB carrying a W851R mutation also failed to suppress the sensitivity of CSB-KO cells to IR exposure (Fig8C). Collectively, these results suggest that the ATPase activity of CSB is important for facilitating the maintenance of the G2/M checkpoint and cell survival in response to the induction of DNA DSBs.
Figure 8.
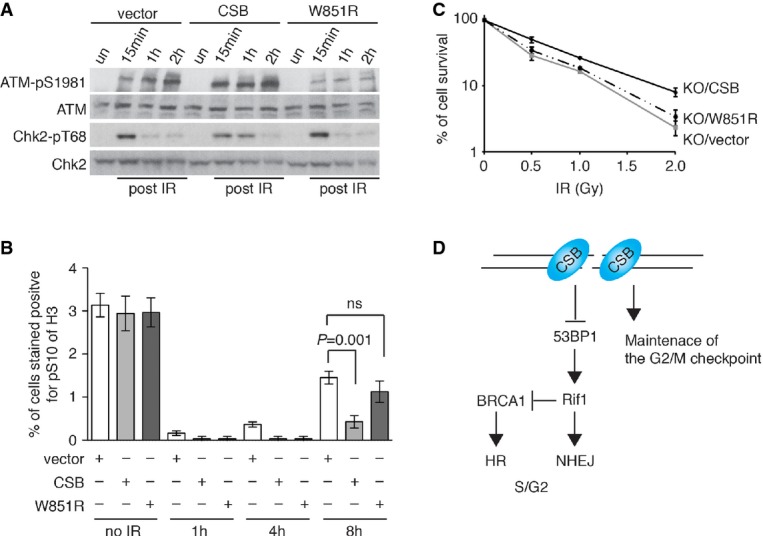
- Western analysis of CSB-KO cells stably expressing wild-type CSB, CSB-W851R or the vector alone as indicated. Cells were either mock-treated or treated with 5 Gy IR. Immunoblotting was performed with anti-ATM, anti-ATM-pS1981, anti-Chk2 and anti-Chk2-pT68 antibodies.
- Quantification of the percentage of cells stained positive for H3-pS10. Cells were either mock-treated or treated with 2 Gy IR. For each cell line, at least 3,000 cells from three independent experiments were scored in blind. P-values were determined using two-tailed unpaired Student’s t-test.
- Clonogenic survival assays of IR-treated CSB-KO (KO) cell stably expressing wild-type CSB, CSB-W851R or the vector alone as indicated.
- Model for the role of CSB in repressing NHEJ and maintaining the G2/M checkpoint. See text for more information.
Data information: (B, C) Standard deviations from three independent experiments are indicated.
Source data are available online for this figure.
Discussion
In this report, we uncover a novel but important function of CSB in regulating the choice of DNA DSB repair pathways. Our work suggests that CSB facilitates BRCA1-mediated HR repair by repressing the accumulation of NHEJ-promoting factors 53BP1 and Rif1 at sites of DNA DSBs in S and G2 cells (Fig8D). Furthermore, we have demonstrated that CSB is needed for maintaining the ATM- and Chk2-mediated DNA damage checkpoint (Fig8D), preventing premature entry of cells into mitosis following the induction of DNA DSBs.
We observed a large asymmetry in the ratio of targeting versus retargeting in the recovery of null clones. Although a large asymmetry in gene targeting typically is found to be associated with genes whose function is critical to cell viability (Dang et al, 2006; Hucl et al, 2008; Ruis et al, 2008; Oh et al, 2013), homozygous CSB mutations leading to the complete absence of CSB protein have been reported in patients (Horibata et al, 2004; Hashimoto et al, 2008; Laugel et al, 2008). Several lines of evidence strongly argue against the possibility that the observed phenotype of the CSB null clone may be due to a secondary mutation. Firstly, introduction of wild-type CSB rescued the defect of the CSB-knockout cells in the choice of DNA DSB repair pathways as well as the maintenance of G2/M checkpoint. Secondly, dysregulation in the choice of DNA DSB repair pathways was also detected in two independent CSB-deficient cell lines derived from CSB patients. Thirdly, endogenous CSB was found to accumulate at sites of DSBs.
Most patient-derived CSB null cell lines that are available are skin fibroblasts whereas the CSB-KO cells described here are retinal pigment epithelial cells in origin. CS patients are known to exhibit segmental premature aging in certain cell types that are not skin fibroblasts. Therefore, we anticipate that our CSB null clone will provide an added value for understanding the pathology of CS.
CSB has recently been implicated in processing R-loops into DNA DSBs (Sollier et al, 2014). R-loop-dependent DNA DSBs led to a robust DNA damage response including phosphorylation of KAP1, which is sensitive to CSB knockdown (Sollier et al, 2014). We have observed an impaired KAP1 phosphorylation as well as ATM- and Chk2-mediated DNA damage response in CSB-KO cells following ionizing radiation. Conceivably, the fewer R-loop-dependent DNA DSBs resulting from the absence of CSB in the CSB-KO cells may in part account for the reduction in their ATM-dependent damage response (Fig8D). Treatment with the transcription inhibitor DRB or actinomycin D did not abolish the impaired ATM phosphorylation in CSB-KO cells, suggesting that active transcription may not be needed for CSB to regulate ATM activation.
We have reproducibly found that the introduction of wild-type CSB into CSB-KO cells rescued the level of Chk2 phosphorylation at 1 h post-IR but not at 15 min post-IR whereas Chk2 phosphorylation was robust in parental (WT) cells 15 min post-IR. It is possible that the kinetics of Chk2 phosphorylation in CSB-KO cells complemented with wild-type CSB may be different from that in parental (WT) cells. Future studies will be needed to investigate the nature of this difference.
CSB requires its ATPase activity to maintain ATM activation and to regulate DNA DSB repair pathway choice. CSB has been reported to exhibit ATP-dependent chromatin remodeling activity in vitro (Citterio et al, 2000; Cho et al, 2013). Chromatin remodeling is known to influence DNA DSB repair (Goodarzi et al, 2010; Chapman et al, 2012). It is possible that CSB may facilitate DNA DSB repair pathway choice through its ATP-dependent chromatin remodeling activity. Alternatively, CSB might regulate the repair pathway choice through its interactions with chromatin modifying factors. Recently, it has been reported that the chromatin context can influence the choice of DNA DSB repair pathways, especially in S and G2 phases of the cell cycle during which both pathways are available to the cell (Aymard et al, 2014; Carvalho et al, 2014; Jha & Strahl, 2014; Pai et al, 2014; Pfister et al, 2014). For example, in human cells, trimethylation of H3K36 has been suggested to promote DNA end resection and repair of DSBs via HR (Aymard et al, 2014; Pfister et al, 2014) and modification of H3K9 has been implicated in the DSB response in heterochromatin that is preferentially repaired by HR (Chiolo et al, 2011). CSB has been found, in different contexts, to associate with numerous chromatin modifying and remodeling factors such as NuRD (Xie et al, 2012), SMARCA5 (Aydin et al, 2014) as well as histone methyltransferase G9A (Yuan et al, 2007) and acetyltransferase PCAF (Shen et al, 2013), while the yeast homolog of CSB, Rad26, has been reported to genetically interact with the H3K36 methyltransferase SET2 (Jha & Strahl, 2014).
Although transcription inhibition did not abolish the impairment of CSB-KO cells in IR-induced Rad51 foci formation and ATM activation, it abrogated the accumulation of CSB at sites of DNA DSBs. These results suggest that CSB association with DSB-induced damaged chromatin may be regulated distinctively from its role in HR and ATM activation. This notion is further supported by our finding that CSB requires its ATPase activity to facilitate IR-induced Rad51 foci and ATM activation but its ATPase activity is dispensable for its association with DSB-induced damaged chromatin. CSB has previously been reported to possess both ATP-dependent and ATP-independent functions (Wong et al, 2007; Lake et al, 2010).
Mutant CSB-W851R lacking its ATPase activity is competent in DSB-induced chromatin association but defective in UV-induced chromatin association, the latter of which is in line with previous findings (Lake et al, 2010). These results suggest that CSB may use distinct mechanisms to interact with UV-induced DNA damage and DSBs, which is not unprecedented. The chromatin remodeling protein SMARCA5 (also known as SNF2H) does not require its ATPase activity to localize to DSBs, but it does so for its localization to UV-induced DNA damage (Lan et al, 2010; Aydin et al, 2014).
CSB-KO cells exhibit an accumulation of IR-induced 53BP1 foci. Previously, it has been reported that depletion of CSB leads to a decrease in the formation of a specific subset of CPT-induced 53BP1 foci, referred to as type I foci, which are dose dependent and only seen in RPA-negative cells (Sakai et al, 2012). CPT also induces the formation of type II foci of 53BP1, which is not dose dependent and persists after CPT treatment (Sakai et al, 2012). However, whether depletion of CSB might affect CPT-induced type II foci of 53BP1 was not investigated, and therefore, our findings cannot be strictly compared to the previous report. Future studies will be needed to investigate the effect of knockout of CSB on CPT-induced type I and type II foci formation.
CS cells deficient in CSB are known to be sensitive to IR-induced DNA damage as well as to the topoisomerase poisons CPT and Etop (Squires et al, 1993; Elli et al, 1996; Tuo et al, 2002, 2003). However, the multiplicity of the forms of DNA damage generated by these cellular treatments may have contributed to obscure our understanding of CSB as a DNA DSB repair protein. IR, for instance, produces not only DSBs but also oxidative damage and single-strand breaks. Indeed, a defect in repairing oxidative damage may contribute to the sensitivity of CS cells to IR (Stevnsner et al, 2008). Similarly, camptothecin and etoposide generate not only DSBs but also topoisomerase–DNA adducts, which are thought to be removed by base excision repair (Caldecott, 2008). Our finding that CSB accumulates at sites of DSBs and regulates the choice of the DNA DSB repair pathways suggests for the first time that dysregulation in DNA DSB repair may at least in part contribute to the hypersensitivity of CS cells to DSB-inducing agents. In support of this notion, depletion of 53BP1 rescued the formation of BRCA1 damage foci in CSB-knockout cells and fully suppressed their sensitivity to olaparib, a PARP1 inhibitor known to be toxic to cells deficient in HR. Our finding raises a new possibility that targeting 53BP1 might be clinically beneficial to CS patients.
Materials and Methods
Plasmids and antibodies
The retroviral expression constructs for wild-type CSB and the shRNA against CSB or 53BP1 have been described (Batenburg et al, 2012; McKerlie et al, 2013). The QuickChange site-directed mutagenesis kit (Agilent Technologies) was used to generate CSB mutant W851R.
Antibodies used include Rad50, Mre11 and Nbs1 (Zhu et al, 2000) (kindly provided by John Petrini, Memorial Sloan-Kettering Cancer Center); Rif1 (Escribano-Diaz et al, 2013) (generously provided by Daniel Durocher, Samuel Lunenfeld Research Institute); 53BP1 (BD Biosciences); BRCA1 (MS110, Abcam); BRCA1 (Millipore); ATM (clone 2C1, Novus Biologicals); ATM (Ab-3, Calbiochem); ATM-pS1981 (10H11.E12, Cell Signaling); cyclin A (6E6, Abcam); CSB/ERCC6 (A301-354A, Bethyl Laboratories); ERCC6 (553C5a, Fitzgerald); Chk1 (FL-476, Santa Cruz); Chk1-pS317 (A300-163A, Bethyl Laboratories); Chk2 (H300, Santa Cruz); Chk2-pT68 (Cell Signaling); γ-H2AX (Millipore); H3-pS10 (Cell Signaling); KAP1-pS824 (ab70369, Abcam); KAP1 (NB500-158, Novus Biologicals); Rad51 (ab213, Abcam); Rad51 (Santa Cruz); RPA70 (a kind gift from James Ingles, University of Toronto); RPA32 (9H8, Abcam); RPA32-pS4/pS8 (Bethyl Laboratories); SMC1-pS966 (NB100-206, Novus Biologicals); SMC1 (NB100-204, Novus Biologicals); PGBD3 (Fitzgerald); and γ-tubulin (GTU88, Sigma).
Cell culture, retroviral infection and treatments
Cells were grown in DMEM with 10% fetal bovine serum supplemented with non-essential amino acids, l-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin. Phoenix, hTERT-RPE and HeLa-DR-GFP cells were respective gifts from Titia de Lange (Rockefeller University), Prasad Jallepalli (Memorial Sloan-Kettering Cancer Center) and Daniel Durocher (Samuel Lunenfeld Research Institute). GM16095 and the parental line GM10905 for hTERT-GM1095 were obtained from the NIGMS Human Genetic Cell Repository (Coriell Institute for Medical Research). rAAV-293 cells were from Stratagene. Retroviral gene delivery was carried out as described (Wu et al, 2007a, 2008).
To induce DNA DSBs, cells were treated with either 10 μM Etop (Sigma) or 1 μM CPT (Sigma) for 1 h at 37°C. IR was delivered from a Cs-137 source at McMaster University (Gammacell 1000). For UV treatment, cells were exposed to UVC (254 nm) generated by a germicidal lamp (Model G8T5, GE) as described (Wu et al, 2007b). To inhibit transcription, cells were treated with either 50 μM DRB (Cayman Chemical) or 1 μg/ml actinomycin D (Sigma) for 2 h at 37°C except where specified. KU55933 (10 μM, Sigma) and NU7026 (1 μM, Sigma) were used to inhibit ATM and DNA-PKcs, respectively.
Generation of CSB knockout in hTERT-RPE cells
All primers used in the generation of the CSB-knockout cell line are shown in Supplementary Table S1. Construction of targeting vectors was performed as described (Kohli et al, 2004). The primer sets 313/314 and 315/316 were used to amplify the right and left arms flanking exon 5 of the ERCC6 locus, respectively, using genomic DNA harvested from hTERT-RPE cells. The amplified right and left arms of exon 5 were mixed with a 4-kb PvuI fragment derived from the NeDaKO-Neo plasmid, followed by PCR using primers 313 and 316. The resulting fusion PCR product (4.4 kb) was purified, digested with NotI and ligated with the NotI-linearized pAAV-MCS plasmid, giving rise to pAAV-Neo-CSB.
Viral packaging and infection of target cells were done essentially as described (Kohli et al, 2004). Briefly, AAV-293 cells at about 60% confluency were cotransfected with the targeting vector (pAAV-Neo-CSB), pAAV-RC and pHelper plasmids using Lipofectamine 2000 (Invitrogen). Forty-eight hours post-transfection, cells were harvested and subjected to three cycles of freezing and thawing (liquid N2 for 10 min, vortexed for 30 s and then thawed at 37°C for 10 min). The viral supernatant was collected by centrifugation at 14,000 g for 2 min and stored at −80°C.
For infection, the virus was added dropwise to hTERT-RPE cells grown at about 70–80% confluency. Forty-eight hours post-infection, cells were trypsinized and plated in 96-well plates at a density of 2,000 cells per well in media containing 1 mg/ml G418 (Invitrogen). Two weeks later, single colonies were identified and transferred to 24-well plates for expansion.
To screen for CSB targeting events, genomic DNA from cells grown in 24-well plates was harvested using the Qiagen Puregene Cell Kit according to manufacturer’s instructions, followed by PCR reactions with two different sets of primers (364/365 and 366/367). Retargeting was examined by PCR screening for the presence of exon 5 using the primer set 378/367.
Immunofluorescence
Immunofluorescence (IF) was performed as described (Mitchell et al, 2009; McKerlie & Zhu, 2011) except for visualizing Rad51 and CSB. For Rad51 IF, cells grown on coverslips were fixed in PBS-buffered 2% paraformaldehyde at room temperature for 10 min. For CSB IF, cells grown on coverslips were fixed in PBS-buffered 4% paraformaldehyde at room temperature for 10 min. Following three washes in PBS, cells were then permeabilized in 0.5% Triton X-100 for 5 min before proceeding to blocking as described (Zhu et al, 2003; Mitchell & Zhu, 2014) except that the blocking buffer was made with 0.1× PBS. All cell images were recorded on a Zeiss Axioplan 2 microscope with a Hamamatsu C4742-95 camera and processed in Open Lab.
Differential salt extraction of chromatin and immunoblotting
Protein extracts, differential salt extraction of chromatin and immunoblotting were performed as described (Wu et al, 2007a; McKerlie et al, 2012).
Northern analysis of CSB transcripts
Northern analysis was performed as described (Batenburg et al, 2012) except that a PCR product corresponding to CSB nucleotide 1–1,398 was used to generate the radioactively labeled probe.
Random integration assays
For random integration assays, cells were infected with 15 μl of the indicated rAAV adenoviral lysates as described and then plated in media containing 1 mg/ml G418 at 300,000 cells/per 10-cm plate. Following incubation for 12 days, colonies were fixed and stained at room temperature for 10 min with a solution containing 50% methanol, 7% acetic acid and 0.1% Coomassie blue. Colonies consisting of more than 32 cells were scored. To assess plating efficiency, infected cells were plated in media without G418. The number of colonies counted on plates without G418 was normalized to the number of cells seeded to give rise to plating efficiency.
GFP reporter assays and FACS analysis
To assess NHEJ activity, the reporter plasmid pEGFP-Pem1-Ad2 was used as described (Fattah et al, 2010). In brief, Lipofectamine LTX plus reagent (Invitrogen) was used to transfect parental and CSB-KO cells with an I-SceI-expressing plasmid, pCherry and pEGFP-Pem1-Ad2 in a ratio of 1:0.5:1 according to the manufacturer’s instructions. Forty-eight hours post-transfection, cells were harvested and subjected to FACS analysis.
To assess HR activity, HeLa-DR-GFP cells were first transfected with either pRS or shCSB using Lipofectamine 2000 reagents (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours after the first transfection, cells were transfected with an I-SceI-expressing plasmid and pCherry in a ratio of 4:1. Forty-eight hours after the second transfection, cells were harvested and subjected to FACS analysis. For FACS analysis, cells were harvested, washed in 1× PBS and fixed in PBS-buffered 4% paraformaldehyde. FACS analysis was performed using a Becton-Dickinson LSRII located at the McMaster University flow cytometry facility, Hamilton, Canada. The number of cells positive for both GFP and pCherry was normalized to the total number of pCherry-positive cells, giving rise to the percentage of GFP-positive cells.
For cell cycle analysis of parental and CSB-KO cells, two million cells were fixed in 80% ethanol. Fixed cells were then washed twice with PBS, followed by incubation in PBS containing 100 μg/ml RNase A and 50 μg/ml propidium iodide at 37°C for 30 min. FACS analysis was performed on a FACSCalibur instrument and analyzed using FlowJo (vX.0.7).
Clonogenic survival and G2/M checkpoint assays
For clonogenic survival assays, 4 to 6 h prior to Etop or CPT treatment, cells were seeded in triplicate at 200/300 cells (0 to 250 nM CPT and 0 to 5 μM Etop) or 800/2,400 cells (10 μM Etop) for parental and CSB-KO, respectively, per 6-cm plate. After 1 h of CPT or Etop treatment, the drug was washed off with PBS and fresh growth medium was added. For IR treatment, cells were counted, irradiated and seeded in triplicate at 200 or 300 cells for parental and CSB-KO, respectively, per 6-cm plate, followed by replacement with fresh media after a 24-h incubation. For PARP1 inhibitor treatment, cells were seeded in triplicate at 200 and 300 cells for parental and CSB-KO, respectively, except for that 600 knockout cells were seeded for 2 μM olaparib treatment. Twenty-four hours post-seeding, cells were treated with olaparib and allowed to grow in the presence of olaparib for the entirety of the experiments. Ten days later, colonies were fixed and stained at room temperature for 10 min with a solution containing 50% methanol, 7% acetic acid and 0.1% Coomassie blue. Colonies consisting of more than 32 cells were scored.
The G2/M checkpoint assay was performed as described (McKerlie et al, 2013). Briefly, cells seeded on coverslips were treated with 2 Gy IR and allowed to recover in the incubator. Following 1 h, 4 h and 8 h incubations, cells were gently washed with PBS, fixed with paraformaldehyde and then processed for immunofluorescence with anti-H3-pS10 antibody.
Neutral comet assays
Neutral comet assays were carried out as described (Dhawan et al, 2002) with minor modifications. Cells were mixed with 1% agarose, and the mixture was dropped onto slides pre-coated with 1% agarose. Cells on the slides were lysed in comet lysis buffer (100 mM EDTA, 2.5 M NaCl, 10 mM Tris, 1% Triton X-100 pH 10) overnight at 4°C in the dark. The slides were then incubated in 1× TBE buffer (9 mM Tris, 9 mM boric acid, 2 mM EDTA pH 8.0) for 30 min at 4°C in the dark. Following gel electrophoresis run at 0.8 V/cm for 30 min in cold 1× TBE buffer, the slides were dehydrated in 70% ethanol for 30 min, air-dried and stained with SYBR Green I (Invitrogen). ImageJ (v1.49) was used with the Open Comet (v1.3) plugin to analyze at least 200 cells for each sample. The tail moment (TM) represents the product of the tail length (TL) and the fraction of DNA in the comet tail (TM = %DNA in tail × TL/100). The data were plotted using Prism (v5.03) to create a box and whisker graph where the whiskers correspond to the 10–90 percentiles. A non-parametric Mann–Whitney rank-sum t-test was used to derive P-values specifically for comet assays.
Statistical analysis
A Student’s two-tailed unpaired t-test was used to derive all P-values except where specified.
Acknowledgments
We would like to thank Daniel Durocher, James Ingles, Titia de Lange, Prasad Jallepalli and John Petrini for providing cell lines and antibodies. We are indebted to Daniel Durocher and John R. Walker for their insightful suggestions. John R. Walker is also thanked for his critical reading of the manuscript. This work is supported by funding from Canadian Institutes of Health Research to X.-D.Z. Work in the Hendrickson laboratory was supported by the NIH (GM088351) and the NCI (CA15446). N.L.B. is a holder of Ontario Graduate Scholarship.
Author contributions
NLB and ELT performed the experiments. NLB, EAH and X-DZ designed the experiments. NLB and XDZ wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure Legends
Supplementary Table S1
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S5
Source Data for Supplementary Figure S6
Source Data for Supplementary Figure S8
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 8
References
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, 3rd, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin OZ, Marteijn JA, Ribeiro-Silva C, Rodriguez Lopez A, Wijgers N, Smeenk G, van Attikum H, Poot RA, Vermeulen W, Lans H. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res. 2014;42:8473–8485. doi: 10.1093/nar/gku565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, Legube G. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Batenburg NL, Mitchell TR, Leach DM, Rainbow AJ, Zhu XD. Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability. Nucleic Acids Res. 2012;40:9661–9674. doi: 10.1093/nar/gks745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard ME, Randall CL, Larochelle S, Zhang C, Shokat KM, Fisher RP, Jallepalli PV. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci USA. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, Deng CX, Finkel T. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM, Ferreira J, de Almeida SF. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife. 2014;3:e02482. doi: 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. RIF1 Is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Tsai PF, Lake RJ, Basheer A, Fan HY. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013;9:e1003407. doi: 10.1371/journal.pgen.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang LH, Chen F, Ying C, Chun SY, Knock SA, Appelman HD, Dang DT. CDX2 has tumorigenic potential in the human colon cancer cell lines LOVO and SW48. Oncogene. 2006;25:2264–2272. doi: 10.1038/sj.onc.1209247. [DOI] [PubMed] [Google Scholar]
- Dhawan A, Anderson D, de Pascual-Teresa S, Santos-Buelga C, Clifford MN, Ioannides C. Evaluation of the antigenotoxic potential of monomeric and dimeric flavanols, and black tea polyphenols against heterocyclic amine-induced DNA damage in human lymphocytes using the Comet assay. Mut Res. 2002;515:39–56. doi: 10.1016/s1383-5718(01)00347-3. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Arena S, Gallicchio M, Zecchin D, Martini M, Flonta SE, Stella GM, Lamba S, Cancelliere C, Russo M, Geuna M, Appendino G, Fantozzi R, Medico E, Bardelli A. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci USA. 2008;105:20864–20869. doi: 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elli R, Chessa L, Antonelli A, Petrinelli P, Ambra R, Marcucci L. Effects of topoisomerase II inhibition in lymphoblasts from patients with progeroid and “chromosome instability” syndromes. Cancer Genet Cytogenet. 1996;87:112–116. doi: 10.1016/0165-4608(95)00294-4. [DOI] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-Rif1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair. 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Suga T, Kudo E, Ihn H, Uchino M, Tateishi S. Adult-onset neurological degeneration in a patient with Cockayne syndrome and a null mutation in the CSB gene. J Invest Dermatol. 2008;128:1597–1599. doi: 10.1038/sj.jid.5701210. [DOI] [PubMed] [Google Scholar]
- Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M, Tanaka K. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci USA. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, van Steeg H, Berg RJ, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR, Bootsma D, Hoeijmakers JH. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- Hucl T, Rago C, Gallmeier E, Brody JR, Gorospe M, Kern SE. A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res. 2008;68:5023–5030. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Lee SY, Berquist BR, Gileadi O, Bohr VA, Seidman MM, McHugh PJ, Wilson DM., 3rd CSB interacts with SNM1A and promotes DNA interstrand crosslink processing. Nucleic Acids Res. 2015;43:247–258. doi: 10.1093/nar/gku1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha DK, Strahl BD. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun. 2014;5:3965. doi: 10.1038/ncomms4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y, Ruis B, Lin S, Hendrickson EA. The mechanism of gene targeting in human somatic cells. PLoS Genet. 2014;10:e1004251. doi: 10.1371/journal.pgen.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotin RM, Linden RM, Berns KI. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, Yasui A. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell. 2010;40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Laugel V, Dalloz C, Stary A, Cormier-Daire V, Desguerre I, Renouil M, Fourmaintraux A, Velez-Cruz R, Egly JM, Sarasin A, Dollfus H. Deletion of 5′ sequences of the CSB gene provides insight into the pathophysiology of Cockayne syndrome. Eur J Hum Genet. 2008;16:320–327. doi: 10.1038/sj.ejhg.5201991. [DOI] [PubMed] [Google Scholar]
- Leadon SA, Cooper PK. Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active human gene is defective in Cockayne syndrome. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- McKerlie M, Zhu XD. Cyclin B-dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres. Nat Commun. 2011;2:371. doi: 10.1038/ncomms1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerlie M, Lin S, Zhu XD. ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic Acids Res. 2012;40:3975–3989. doi: 10.1093/nar/gks035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerlie M, Walker JR, Mitchell TR, Wilson FR, Zhu XD. Phosphorylated (pT371)TRF1 is recruited to sites of DNA damage to facilitate homologous recombination and checkpoint activation. Nucleic Acids Res. 2013;41:10268–10282. doi: 10.1093/nar/gkt775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TR, Glenfield K, Jeyanthan K, Zhu XD. Arginine methylation regulates telomere length and stability. Mol Cell Biol. 2009;29:4918–4934. doi: 10.1128/MCB.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TR, Zhu XD. Methylated TRF2 associates with the nuclear matrix and serves as a potential biomarker for cellular senescence. Aging (Albany NY) 2014;6:248–263. doi: 10.18632/aging.100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci USA. 2006;103:9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008;4:e1000031. doi: 10.1371/journal.pgen.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Wang Y, Zimbric J, Hendrickson EA. Human LIGIV is synthetically lethal with the loss of Rad54B-dependent recombination and is required for certain chromosome fusion events induced by telomere dysfunction. Nucleic Acids Res. 2013;41:1734–1749. doi: 10.1093/nar/gks1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CC, Deegan RS, Subramanian L, Gal C, Sarkar S, Blaikley EJ, Walker C, Hulme L, Bernhard E, Codlin S, Bahler J, Allshire R, Whitehall S, Humphrey TC. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, Humphrey TC. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan AJ, Squires S, Strutt HL, Johnson RT. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res. 1991;19:3295–3300. doi: 10.1093/nar/19.12.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Sakasai R, Kakeji Y, Kitao H, Maehara Y. PARP and CSB modulate the processing of transcription-mediated DNA strand breaks. Genes Genet Syst. 2012;87:265–272. doi: 10.1266/ggs.87.265. [DOI] [PubMed] [Google Scholar]
- Sakasai R, Teraoka H, Takagi M, Tibbetts RS. Transcription-dependent activation of ataxia telangiectasia mutated prevents DNA-dependent protein kinase-mediated cell death in response to topoisomerase I poison. J Biol Chem. 2010;285:15201–15208. doi: 10.1074/jbc.M110.101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen L, Cassel T, Helleday T. The XPD subunit of TFIIH is required for transcription-associated but not DNA double-strand break-induced recombination in mammalian cells. Mutagenesis. 2010;25:623–629. doi: 10.1093/mutage/geq054. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM, 3rd, Cooper M, Spencer R, de Cabo R, Croteau DL, Bohr VA. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Zhou T, Xie W, Ling T, Zhu Q, Zong L, Lyu G, Gao Q, Zhang F, Tao W. The chromatin remodeling factor CSB recruits histone acetyltransferase PCAF to rRNA gene promoters in active state for transcription initiation. PLoS ONE. 2013;8:e62668. doi: 10.1371/journal.pone.0062668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires S, Ryan AJ, Strutt HL, Johnson RT. Hypersensitivity of Cockayne’s syndrome cells to camptothecin is associated with the generation of abnormally high levels of double strand breaks in nascent DNA. Cancer Res. 1993;53:2012–2019. [PubMed] [Google Scholar]
- Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol Cell Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- Velez-Cruz R, Egly JM. Cockayne syndrome group B (CSB) protein: at the crossroads of transcriptional networks. Mech Ageing Dev. 2013;134:234–242. doi: 10.1016/j.mad.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat Struct Mol Biol. 2007a;14:832–840. doi: 10.1038/nsmb1286. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zacal NJ, Rainbow AJ, Zhu XD. XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair (Amst) 2007b;6:157–166. doi: 10.1016/j.dnarep.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech Ageing Dev. 2008;129:602–610. doi: 10.1016/j.mad.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, Jackson SP, Scully R. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci USA. 2012;109:8161–8166. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure Legends
Supplementary Table S1
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S5
Source Data for Supplementary Figure S6
Source Data for Supplementary Figure S8
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 8