Abstract
Controlled gene regulation during gamete development is vital for maintaining reproductive potential. During the complex process of mammalian spermatogenesis, male germ cells experience extended periods of the inactive transcription despite heavy translational requirements for continued growth and differentiation. Hence, spermatogenesis is highly reliant on mechanisms of posttranscriptional regulation of gene expression, facilitated by RNA binding proteins (RBPs), which remain abundantly expressed throughout this process. One such group of proteins is the Musashi family, previously identified as critical regulators of testis germ cell development and meiosis in Drosophila, and also shown to be vital to sperm development and reproductive potential in the mouse. This review describes the role and function of RBPs within the scope of male germ cell development, focusing on our recent knowledge of the Musashi proteins in spermatogenesis. The functional mechanisms utilized by RBPs within the cell are outlined in depth, and the significance of sub-cellular localization and stage-specific expression in relation to the mode and impact of posttranscriptional regulation is also highlighted. We emphasize the historical role of the Musashi family of RBPs in stem cell function and cell fate determination, as originally characterized in Drosophila and Xenopus, and conclude with our current understanding of the differential roles and functions of the mammalian Musashi proteins, Musashi-1 and Musashi-2, with a primary focus on our findings in spermatogenesis. This review highlights both the essential contribution of RBPs to posttranscriptional regulation and the importance of the Musashi family as master regulators of male gamete development.
Keywords: gene regulation, Musashi, Musashi-1, Musashi-2, posttranscriptional control, RNA binding proteins, spermatogenesis, splicing, testis, translation
INTRODUCTION
Spermatogenesis
Spermatogenesis defines the maturation process of male gametes and is one of the most complex differentiation events that occur within developmental biology, necessitating the controlled regulation of gene expression (Figure 1). Within the male testis, spermatogenesis commences shortly after birth with the differentiation of prespermatogonial gonocytes to spermatogonia. These spermatogonia provide the pool of stem cells essential for the continual production of spermatozoa throughout postpubertal life. Spermatogonial stem cells undergo a series of mitotic amplifications to produce primary spermatocytes. These spermatocytes then go through two rounds of meiosis to form haploid round spermatids. Postmeiotic spermatid differentiation (spermiogenesis) defines the profound morphological changes that mark transition into elongating spermatids and the progressive development of immature spermatozoa.
Figure 1.
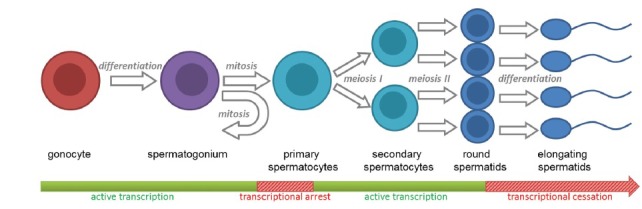
Important stages of spermatogenesis relative to the transcription. Here, we commence with the differentiation of prespermatogonial gonocytes in spermatogonia. Spermatogonial stem cells then undergo mitotic self-maintenance and mitotic amplifications to produce primary spermatocytes. These spermatocytes are transcriptionally arrested during homologous recombination and then continue through two rounds of meiosis to form haploid round spermatids. Postmeiotic spermatid differentiation (spermiogenesis) defines the profound morphological changes that mark transition into elongating spermatids and the progressive development of immature spermatozoa which occurs independently of RNA synthesis. The bar describes the transcriptional status relative to the stage of germ cell development.
During the process of spermatogenesis, two phases of the inactive transcription have been described. The first of these phases takes place during homologous recombination of spermatocytes entering early meiosis,1 whereby the genome is damaged and repaired for crossing over and transcription is blocked.2 The second phase marks the cessation of mRNA synthesis, and takes place in late elongating spermatids, at the time of chromatin condensation in spermiogenesis, denoted by the mass degradation of mRNAs and the gradual decline of translation.3,4 These extended periods of transcriptional cessation that occur during the continued differentiation program of spermatogenesis necessitate extensive regulation of translation.5 Consequently, the mechanisms of posttranscriptional regulation, controlled primarily by RNA binding proteins (RBPs), become of utmost importance. In instances where the mechanisms for posttranscriptional control function abnormally, spermatogenesis can fail resulting in the production of nonviable gametes.6,7
RNA binding proteins
RBPs are an extensive class of proteins defined by their ability to recognize and bind to specific sequences of RNA, and regulate the function utilizing an array of mechanisms. They contain at least one RNA recognition site responsible for identifying a particular motif within target sequences.
The sub-cellular level at which RBPs are expressed is important in determining the function. Consequently, RBPs can be divided into nuclear RBPs and cytoplasmic RBPs. Nuclear RBPs localize to the nucleus where they primarily regulate nascent mRNA (pre-mRNA) processing events, including capping, polyadenylation, and splicing.7 Cytoplasmic RBPs bind mature mRNA sequences in the cytoplasm as they are released from the nucleus. Cytoplasmic RBPs operate more directly in the translation via: directing mRNA transport, competitive or co-operative interactions with translation machinery, and regulating mRNA stability. A number of RBPs are both nuclear and cytoplasmic and can, therefore, be involved in a combination of the previously mentioned processes, as well as uniquely in mRNA nuclear export.
Importantly RBPs are highly expressed throughout spermatogenesis and have been well documented as being essential to posttranscriptional control during all stages of germ cell development. Characterization studies using transgenic models of testis-expressed RBP disruption commonly identify an irregular spermatogenesis phenotype and often exhibit various stages of spermatogenic arrest and resultant sterility.7 Piecing together the mechanisms that these RBPs utilize to make them fundamental during testis germ cell development is vital to our overall understanding of spermatogenesis and male factor infertility.
FUNCTIONAL MECHANISMS OF RNA BINDING PROTEINS
Within the cell, RBPs function at all levels of RNA metabolism. Assembling on nascent and processed mRNAs, RBPs have the ability to govern gene regulation at the posttranscriptional level in both health and disease. Here, we divide the mechanisms utilized by RBPs into six sub-categories in terms of posttranscriptional regulatory processes (Figure 2).
Figure 2.
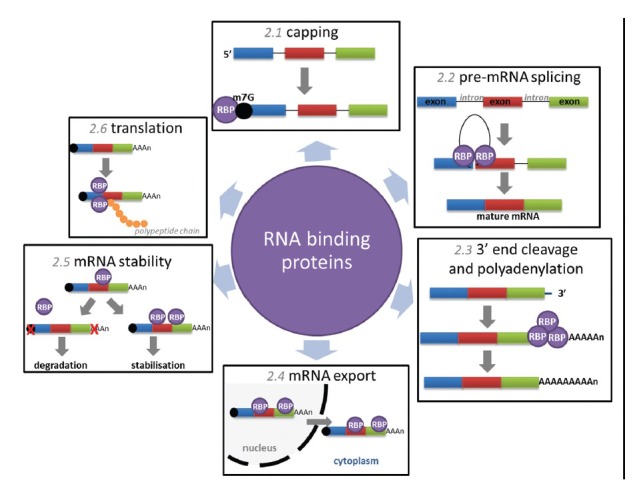
Mechanisms of posttranscriptional control regulated by RNA binding proteins. Capping (section in this article) describes the addition of a 7-methylguanosine to the 5’ end of nascent mRNA, RBPs bind to the cap and promote mRNA stability. Pre-mRNA splicing (section in this article) describes the excision of noncoding introns from nascent mRNA regulated by numerous RBPs within the macromolecular spliceosome. 3’-end cleavage and polyadenylation (section in this article) involves cleavage at a defined site and the 3’-end of fully transcribed pre-mRNA followed by the addition of 150–200 adenosine residues, facilitated by a complex of RBPs. mRNA export (section in this article) refers to the shuttling of mature mRNAs through the nuclear pore complex to the cytoplasm, mediated by the association of RBPs with specific transcripts. MRNA stability (section in this article) can be modulated by transcript associations with specific RBPs, poly(A) tail alterations and decapping often precede rapid degradation. Translation (section in this article) is orchestrated by a complex of RBPs, known as polysomes, RBPs can also modulate translation via exonuclease degradation or sequestering of transcripts in protective cytoplasmic compartments. RBP: RNA binding proteins.
Capping
Messenger RNA production requires synthesis of a pre-mRNA and processing of the nascent precursor by 5’ capping, splicing of introns, and 3’ cleavage/polyadenylation to make mature mRNA. Although these processes are interconnected: the 5’ cap can enhance splicing of the first intron and 3’ processing,8 while 3’ processing depends on splicing of the last intron,9 it is important to assess each step individually. Capping describes the addition of a 7-methylguanosine linked via a triphosphate bridge, to the first nucleotide of the 5’ end of pre-mRNA and occurs co-transcriptionally. Capping is essential for the growth of eukaryotic cells: the guanylate protects the mRNA from untimely degradation, while the methyl group facilitates translation.10
Capping of transcripts confers stability. One main function of the 5’ cap is to prevent Xrn1 and Xrn2 exoribonuclease-mediated digestion of mRNAs.11 In the cytoplasm, the cap is also used as a signal to direct translational machinery to the 5’ end of protein-encoding RNAs. The eukaryotic translation initiation factor eIF4E recognizes the cap-structure and recruits the 40S ribosome subunit to the 5’ end of the mRNA to signal translation initiation.12
In mammalian cells, the caps of pre-mRNAs and newly synthesized mRNAs are bound by the nuclear cap binding complex (CBC) heterodimer of RBP cap binding protein 20 (CBP20) and auxiliary protein CBP80. This complex promotes nuclear cap function during pre-mRNA processing, but also functions in the cytoplasm in the pioneer round of translation, responsible for monitoring mRNA quality via the nonsense-mediated mRNA decay pathway.13
It is important to note that within the CBC, CBP20 is responsible for cap binding through its ribonucleoprotein domain while CBP80 causes a conformational change in CBP20 and is required for CBC to bind with high affinity. In mammalian cells, siRNA-mediated depletion of CBP80 results in deregulation of ~400 genes and a significant reduction in the cell proliferation rate.14 Deletion of the CBC in vitro prevents spliceosome assembly at an early step in complex formation.15
Pre-mRNA splicing
Splicing describes the precise removal of noncoding intervening sequences (introns) from pre-mRNAs and is essential for proper protein expression. This process of intron removal is efficient and precise, constituting the majority of constitutive splicing events in the cell. However, most transcripts in higher eukaryotic cells also contain regions that are subjected to selection of alternative exons, resulting in the production of different mRNA isoforms. This process, known as alternative splicing, has the capacity to alter gene coding and affect RNA stability and has been recognized as a mechanism for increasing the functional diversity of the proteome, altering the expression of a protein 3- to 4-fold.16,17 Both constitutive and alternative splicing can occur co- and post-transcriptionally and is catalyzed by the spliceosome, a macromolecular complex comprised of the U1-U6 family of small nuclear ribonucleic particles (RNPs) in conjunction with >100 different additional proteins.18 Unsurprisingly, mutations in spliceosome components and subsequent splicing errors underlie a large number of human diseases.19
Spliceosome proteins are a well-studied class of RBPs. There are also a number of RBPs involved in alternative splicing that bind pre-mRNA either to encourage or block specific splicing events. DExD/H-type RNA-dependent ATPases/helicases have long been implicated in rearrangements within the spliceosome and act at discrete stages of splicing including single-strand RNA translocation, strand annealing, and protein displacement.20
Despite being composed of a large number of RBPs, very few mutations in core spliceosome components have been found, suggesting that such mutations are nonviable either at the cellular level or in early development.21 A diverse set of diseases is also associated with even moderate changes in expression of any number of RBPs involved in splicing and splicing regulation.22,23 Recently, an unbiased genetic screen for essential male fertility genes in the mouse identified the RBP, RBM5, as an essential regulator of haploid male germ cell pre-mRNA splicing and fertility. Mice carrying a missense mutation in the second RNA recognition motif of RBM5, affecting pre-mRNA splicing of putative targets, exhibited spermatid differentiation arrest, which led to azoospermia and male sterility.24
Cleavage and polyadenylation
Endonucleolytic cleavage marks the final stage of transcription and is followed by addition of a poly(A) tail at the 3’-end. Similar to the 5’ cap, the poly(A) tail is important for the stability and translational efficiency of the mRNA transcript.25 And like splicing, transcripts can be alternatively polyadenylated, altering stability, localization, and transport. More than half of the genes in the human genome are estimated to be subject to alternative 3’-end processing, generating isoforms that differ in 3’ UTR length or encoding different proteins.26
With the exception of replication-dependent histone genes, all protein encoding mRNAs contain a uniform 3’-end consisting of around 200 adenosine residues. The formation of this poly(A) tail is directed by sequences present on the pre-mRNA and the polyadenylation machinery. In mammals this consists of six multimeric proteins which come together to firstly mediate cleavage of the nascent mRNA 3’-end, and secondly facilitate coupled polyadenylation, namely: cleavage and polyadenylation specificity factor, cleavage stimulation factor (CstF), cleavage factors I and II (CFI and CFII), poly(A) polymerase, and poly(A)-binding protein II.27
A number of reports suggest that there are testis-specific mechanisms which support nuclear polyadenylation in male germ cells. One example of this is the testis-specific CstF paralog, τCstF-64 (gene name: Cstf2t), considered necessary for germ-cell polyadenylation and gene expression during spermatogenesis.28 Targeted deletion of Cstf2t resulted in male infertility due to aberrant meiotic and postmeiotic development.29
mRNA export
Transport through the nuclear pore complex (NPC) represents the link between the nucleus and cytoplasm. With nuclear export of mature mRNAs likely to involve distinct docking, translocation and release steps from the NPC.30 Export is mediated by protein factors associated with the mRNA, and mRNAs without the necessary adaptor and export factors remain held in the nucleus. The mRNA export machinery includes numerous RBPs, ATPase/RNA helicases, and NPC-associated proteins. Most of these are essential, with conditional mutations for many these genes in yeast showing rapid and strong defects in mRNA export.31
Following mRNA export from the nucleus, interacting RBPs either remain in the nucleus or accompany the transcript into the cytoplasm, where the transcript either remains bound to the same RBPs or is recruited by others. This ultimately determines the cytoplasmic compartment localisation.7
mRNA stability
Maintenance of mRNA stability is essential for the translation of essential proteins and also the proteosomal-mediated degradation of unwanted transcripts. Stability of mRNAs can be rapidly modulated to alter the expression of specific genes, providing flexibility in affecting changes in patterns of protein synthesis. Degradation of transcripts is highly variable and thought to be controlled mainly by RBPs. An important step commonly observed in the regulation of stability is an alteration in the length of the poly(A) tail, which often precedes decapping, and 5ingth of the poly(A).32 It is often the case that RBPs that accompany the transcript from the nucleus to the cytoplasm, with the assistance of newly assembled translation initiation factors, aid in the recruitment of the translation machinery.33 In contrast, mRNA binding of the miRNA-induced silencing complex brings about decapping, deadenylation, and translational repression of the mRNA.34
Translation
Perhaps the best-described and also most important event that RBPs control is translation. Regulation of translation is controlled through three main mechanisms: poly(A) tail modification, association with RNPs, and competition with translation machinery. Lengthening the poly(A) tail of a transcript can significantly increase translation rates. This is achieved through the regulation of binding of the RBP poly(A)-binding protein (PABP), which recruits factors essential for translation initiation.35 The most significant mechanism involves directing the transcripts to either cytoplasmic compartments - correlated with translational inhibition, or polyribosomes - known sites of translation.36 Cytoplasmic compartmentalization of transcripts in RNPs represents a targeted location for processing, sorting, storage, or degradation. Generally associated with translational arrest (stress granules), some RNPs are associated with exonucleases (exosome complexes) and are thus sites for mRNA degradation, while others represent locations where transcripts can be protected and remain translationally silent until they are needed (processing bodies, intermitochondrial cement/nuage, and chromatoid bodies).7 Polyribosomes or polysomes are groups of ribosomes clustered around a single mRNA transcript, allowing for simultaneous translation of the transcript, resulting in rapid protein production.7 Polysome aggregates represent the main sites of translation, controlled to correspond with cell needs.
STAGE-SPECIFIC EXPRESSION OF RNA BINDING PROTEINS DURING SPERMATOGENESIS
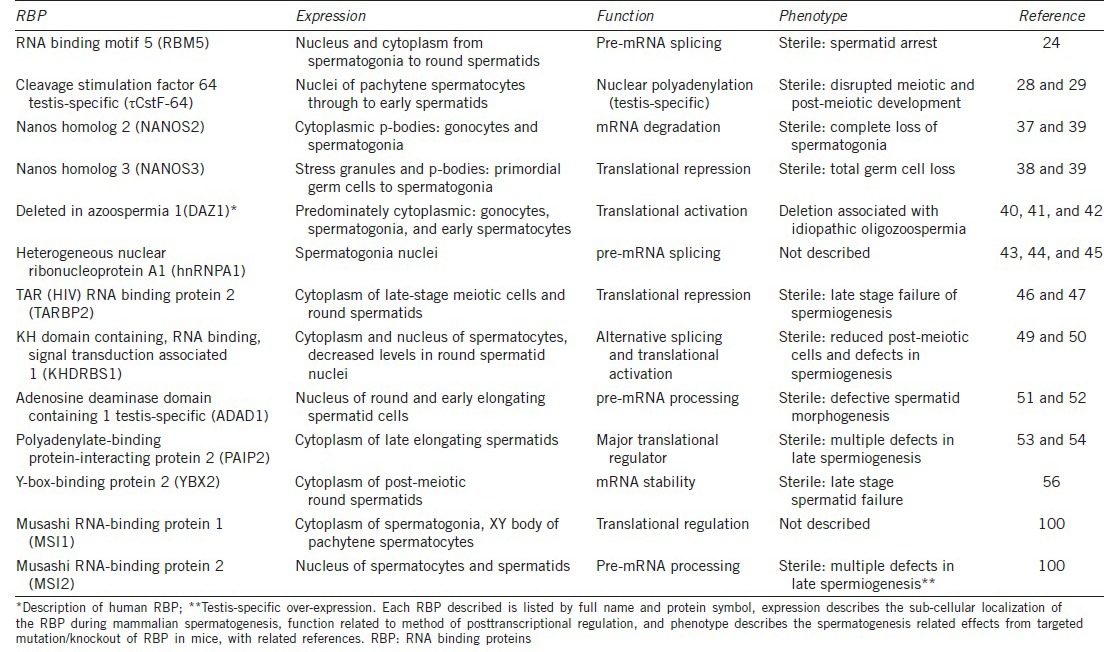
The stage at which RBPs are expressed during spermatogenesis is also essential in determining their role. Given the sequential process of germ cell differentiation, defining the expression patterns of testis-expressed RBPs in terms of the differentiation status is relatively straightforward. We can sub-divide these RBPs into three major germ cell categories: mitotic, meiotic, and postmeiotic and in the following section explore all three groups in depth, providing examples of each (Table 1). It is important to note that some RBPs are expressed throughout all stages of spermatogenesis, quite often serving a variety of functions.
Table 1.
RBP expressed during mammalian spermatogenesis, function, and phenotype
Mitotic RNA binding proteins
Mitotic RBPs consist of those expressed during early stage spermatogenesis, primarily in gonocytes and spermatogonia. Transcription remains active during this period and, as a result, the cell is not as heavily reliant upon posttranscriptional gene regulation. Fewer RBPs have been identified during this stage; however those that are expressed appear to be essential.
Two well-characterized RBPs expressed during mitosis are Nanos2 and Nanos3. Nanos2 is predominantly expressed in early male germ cells, and the elimination of this gene results in a complete loss of spermatogonia.37 Nanos3 is expressed slightly earlier, in migrating gonocytes,38 with deletion of this factor resulting in complete germ cell loss.39
Similarly, the RBP DAZ1, present only in higher primates, is known to be expressed in gonocytes, with increasingly high levels in observed spermatogonial stem cells plateauing at the first wave of meiosis.40 This unique pattern of expression indicates that DAZ1 participates in differentiation, proliferation, or maintenance of germ cells during early spermatogenesis. Deletion of DAZ1 is associated with azoospermia, strongly suggesting that DAZ1 plays a critical role in normal spermatogenesis,41 with a predicted function in translational activation.42
HnRNP proteins are a class of nuclear pre-mRNA binding proteins with important roles in the biogenesis of mRNA. In the testis, hnRNP proteins are abundant, yet their expression remains tightly regulated. Specifically, hnRNPA1 is highly expressed only in early spermatogonia and absent in later stages.43 Although no hnRNPA1 mutations have been described, it is predicted based on its role in splicing regulation in other cell systems,44,45 that this protein would serve a similar essential function during spermatogenesis.
Meiotic RNA binding proteins
These RBPs are present in cells undergoing either of the two meiotic divisions that occur during spermatogenesis. These cells, spermatocytes, initially undergo homologous recombination to differentiate their genome from the parental ones. During this period, the genome is damaged and repaired for crossing over and transcription is blocked.2 Following this, pachytene spermatocytes begin to synthesize large amounts of stored mRNAs into proteins that will allow them to sustain two consecutive rounds of cell divisions without a real interphase.4 Round spermatids are the resultant germ cells. Given the importance of posttranscriptional regulation during this period, it is not surprising that there is an abundance of RBPs expressed in meiotic spermatocytes.
TRBP2 (formerly PRBP) is a protamine-1 RBP present in the cytoplasmic compartment of late-stage meiotic cells and haploid round spermatids. Recombinant TRBP2 protein inhibits the translation of multiple mRNAs, suggesting that TRBP2 acts as a translational repressor during spermatogenesis.46 Mice that carry a targeted disruption of Tarbp2 are sterile and severely oligospermic due to failure of late-stage spermiogenesis.47
Khdrbs1 (formerly SAM68) belongs to the STAR family of RBPs, which regulate a range of processes, including RNA stability, export, splicing, and mRNA translation.48 The expression of Khdrbs1 protein during spermatogenesis peaks in spermatocytes as the meiotic cells prepare for division. Khdrbs1 is a nuclear protein during most of spermatogenesis but is found within the cytoplasm in meiosis.49 Analysis of the reproductive phenotype of Khdrbs1 knockout mice revealed that males are completely infertile due to azoospermia,50 indicating the requirement of Khdrbs1 expression in fertility.
Postmeiotic RNA binding proteins
Postmeiotic RBPs describe those expressed in both round and elongating spermatids during the differentiation process known as spermiogenesis. The cessation of mRNA synthesis during spermatid elongation necessitates extensive posttranscriptional regulation as a number of new proteins appear during the later stages of this process.5
Adad1 (formerly TENR) is a testis-specific nuclear RBP, detected in postmeiotic cells, primarily in round and early elongating spermatids, and in association with the nuclear scaffold.51 Adad1 is predicted to function in either in pre-mRNA editing or transport. Targeted mutation of the Adad1 gene causes male sterility, through a combination of reduced sperm number, decreased motility, and increased malformed heads in remaining spermatozoa, indicating an essential function in spermatid morphogenesis.52
The RBP Paip2 is a PABP binding partner expressed late elongated spermatids and acts in the translational repression of poly(A)-containing mRNAs.53 Paip2-KO mice exhibit male infertility via translational inhibition of essential development proteins, resulting in defective elongated spermatids.54
Ybx2 (also MSY2) is a germ cell-specific member of the Y-box family of proteins with broad DNA and mRNA binding capacity. Largely localized in the cytoplasm, Ybx2 exhibits maximal expression in postmeiotic round spermatids and is predicted to function in mRNA storage and translational delay.55 The inactivation of Ybx2 results in spermatogenic arrest and infertility, with incomplete nuclear condensation prominent in later-stage spermatids at the time of massive spermatid loss.56 This occurs due to increased mRNA instability, as indicated by polysomal redistribution resulting in decreases to the abundance of numerous mRNAs normally expressed in these germ cells.56
MUSASHI FAMILY OF RNA BINDING PROTEINS
Historically, the Musashi family of RBPs has well-established roles in stem cell function and cell fate determination. All Musashi family members contain two tandem RNA recognition motifs located at the N-terminal of the protein, each composed of two highly conserved motifs; RNP-1 and RNP-2, that bind to target mRNAs through a Musashi binding element (MBE, (G/A) U1–3AGU) in the mRNA 3’ untranslated region.57,58,59,60 Originally described in Drosophila, Musashi has also been well-characterized in Xenopus. Both of these model species have provided keen insights into elucidating the pivotal functions of Musashi in the mammalian system, specifically in stem cell maintenance, nervous system development, and tumorigenesis.
Musashi in Drosophila
The original Drosophila Musashi (dMsi), was discovered to function in the nucleus of adult external sensory organs. Loss-of-function dMsi mutants exhibit a phenotype of extra outer support cells.61 Consequently, dMsi was shown to play an essential role in the asymmetric division of the external sensory organs progenitor cells; the sensory organ precursor cells. This was later found to be achieved through dMsi-mediated mRNA binding and subsequent translational repression of the neural differentiation inhibitory factor Tramtrack69 (ttk69).62 Similarly, dMsi is required to negatively regulate ttk69 expression in the photoreceptor cells of the developing eye.63 dMsi is also expressed in the central nervous system (CNS) in proliferating neural stem/progenitor cells within the Drosophila larvae brain with overexpression resulting in the proliferation of undifferentiated cells.64
More recently, a second dMsi protein has been identified. Termed RNA-binding protein 6 (Rbp6), it is considered more closely related to the vertebrate Musashi proteins due to sequence homology.65 Rbp6 is expressed in multiple tissues throughout development, but interestingly, Rbp6 mutants are viable and fertile, exhibiting only a delay in the timing of larval development.65 Furthermore, this work showed no overlap in function between Rbp6 and dMsi.
Musashi in Xenopus
Studies in Xenopus laevis have uncovered evidence for the additional roles of vertebrate Musashi in posttranscriptional regulation, both in retinal development and oocyte maturation. The Xenopus Musashi-1 homolog (xMsi1) is a heterogeneous nuclear RBP specifically expressed in the nervous system.66 During Xenopus retina development xMsi1 is expressed in retinal stem cells, mitotically active neural precursors, postmitotic photoreceptors, and retinal pigment epithelium.67 In Xenopus, Musashi function is also considered essential to establishing the temporal order of maternal mRNA translation during the meiotic cell cycle in developing oocytes.68 Here, xMsi1 directs the activation of the mRNAs required for MAP kinase- and CDK-mediated promotion of cell cycle progression.69 In to oocyte, it has been demonstrated that Xenopus Musashi proteins auto-regulate xMsi1 translation,70 while Musashi-mediated translational activation of the proto-oncogene Mos is considered a necessary event for meiotic cell cycle progression.68
Musashi-1
Mammalian Musashi-1 (Msi1) is strongly expressed in fetal and adult CNS and brain, with functional roles in the maintenance of stem-cell state, differentiation, and tumorigenesis.64,71 Msi1 null mice develop obstructive hydrocephalus and suffer early postnatal lethality.72 This indicates a vital role for Msi1 in the normal development of ependymal stem cells and highlights the importance of Msi1 expression in the proliferation and maintenance of CNS stem cell population.
In mouse neural stem cells, Msi1 inhibits translation by binding to consensus sequences of transcripts encoding Numb,73 the cell cycle regulator Cdkn1a,73,74 and Dcx.75 The mechanisms described for Msi1 translational repression competition with eIF4G for PABP binding demonstrates an essential mechanism for preventing the formation of the 80S ribosome.76 Conversely, in the hindbrain, Msi1 has been shown to facilitate the translational activation of Robo3, required for axonal midline crossing of precerebellar neurons.77
Interestingly, Msi1 is often up-regulated in tumor cells, specifically in malignant gliomas and astrocytomas where Msi1 is highly enriched when compared with nonneoplastic brain tissue. Furthermore, the level of increased expression was positively correlated with the aggressiveness of the tumour.78 Considered a putative marker of intestinal stem cells,79 Msi1 expression is frequently detected at elevated levels in both premalignant gastric lesions and invasive gastric cancer.80,81 Up-regulation of Msi1 has also been linked with endometrial carcinoma82 and of lymph node metastases.83,84
Musashi-2
Musashi-2 (Msi2) shares high sequence homology to mammalian Msi1, appearing to have arisen following gene duplication.85 Msi2-deficient mice demonstrate 50% embryonic lethality, and subfertility when crossed among themselves.86
In neural precursor cells, Msi2 and Msi1 are strongly co-expressed and share similar RNA-binding target specificity, as well as being predicted to be co-operatively involved in the proliferation and maintenance of CNS stem cell population.72 Indeed, extensive studies of Musashi proteins in the CNS unequivocally favor a redundant and compensatory role for Msi2 that is indistinct from Msi1.72,87,88,89 In instances involving primary regulation via Msi2, it is common for Msi1 expression is effectively undetectable.90,91 However, a recent study in pancreatic islet (endocrine) cells did identify distinctive roles for Msi1 and Msi2.92
Like Msi1, Msi2 has also been linked with tumorigenesis. Specifically, up-regulation of Msi2 has been implicated in brain tumor growth,93 leukemia progression,94,95 and the differentiation of hematopoietic stem cells (HSCs).96 Further studies in HSC utilizing a Msi2 conditional ablation model revealed failure of HSC maintenance and engraftment resulting from a loss of quiescence and increased commitment divisions.97
MUSASHI IN SPERMATOGENESIS
Genetic screening of Drosophila genes involved in germ cell biology identified Musashi as a critical regulator of testis stem cell maintenance and meiosis.98 Using the fly testis as a model system, it has been demonstrated that loss of Musashi function disrupts the balance between germ-line stem cell renewal and later-stage differentiation, resulting in the premature differentiation of germ-line stem cells and meiotic defects.98 Studies in the mouse have identified differential expression of the two primary mammalian orthologs: Msi1 and Msi2, both at a developmental stage and sub-cellular level.98,99
Recent work has by our group detailed the distinctive expression of mammalian Msi1 and Msi2 and explored the outcomes of aberrant expression of both RBPs during the process of male gamete development.100 The unique spatial and temporal expression patterns of the Musashi proteins throughout spermatogenesis indicate individual roles of both RBPs throughout during gamete development in the mammalian testis (Figure 3). Specifically, Msi1 predominately localizes to the cytoplasmic compartments of mitotic gonocytes and spermatogonia, while Msi2 shows nuclear expression in meiotic spermatocytes and differentiating spermatids. The two novel transgenic mouse models utilized, with germ cell-specific overexpression of full-length isoforms of Msi1 or Msi2, demonstrated that aberrant expression of either gene was deleterious to normal spermatogenesis and detrimental to cell health. In addition to this, preliminary studies performed on human testicular seminoma tumors have provided further insights into the relevance of Msi1 and Msi2 over-expression as diagnostic markers to human stem cell cancers.100
Figure 3.
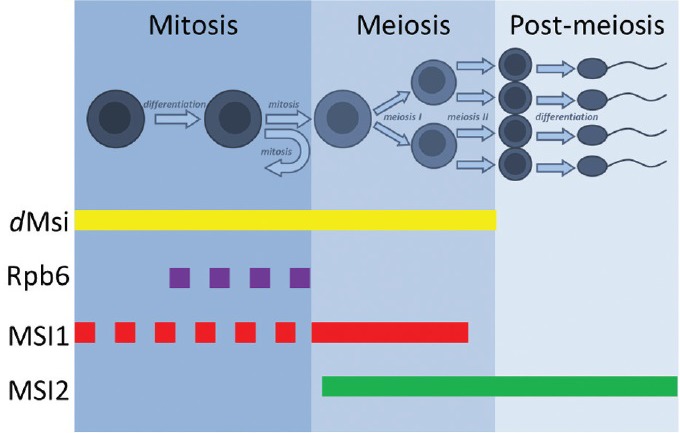
Musashi RBP expression during spermatogenesis. dMsi paralogs dMsi and Rbp6 are differentially expressed in the fly testis: dMsi is nuclear and expressed in early germ cells and spermatocytes while Rbp6 is cytoplasmic and localized to spermatogonial cyst cells. Mammalian Msi1 is expressed in the cytoplasm of early mitotic germ cells before translocating to the nucleus upon transition to meiosis. Mammalian Msi2 is entirely nuclear, expressed throughout meiosis and during spermatid differentiation. The dotted line refers to cytoplasmic localization while the solid line refers to nuclear-specific expression. RBP: RNA binding proteins; Msi1: Musashi-1; Msi2: Musashi-2; dMsi: Drosophila Musashi; Rbp6: RNA-binding protein 6.
SUMMARY
Herein, we have highlighted various mechanisms utilized by RBPs within the cell and emphasized the importance of sub-cellular localization and stage-specific expression on the function of these master regulators of posttranscriptional control. Through the detailed description of the Musashi family of RBPs, we acknowledge their fundamental role in stem cell maintenance, nervous system development, and tumorigenesis, and observe their major contributions to our general knowledge of RBPs. Finally, by focusing on the expression and function of Musashi in spermatogenesis, we have provided new evidence for the novel and unique roles of both Msi1 and Msi2 during male germ cell development.
REFERENCES
- 1.Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J Cell Biol. 1964;22:521–32. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monesi V, Geremia R, D’Agostino A, Boitani C. Biochemistry of male germ cell differentiation in mammals: RNA synthesis in meiotic and postmeiotic cells. Curr Top Dev Biol. 1978;12:11–36. doi: 10.1016/s0070-2153(08)60592-x. [DOI] [PubMed] [Google Scholar]
- 3.Bettegowda A, Wilkinson MF. Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1637–51. doi: 10.1098/rstb.2009.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paronetto MP, Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int J Androl. 2010;33:2–12. doi: 10.1111/j.1365-2605.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- 5.Venables JP, Eperon I. The roles of RNA-binding proteins in spermatogenesis and male infertility. Curr Opin Genet Dev. 1999;9:346–54. doi: 10.1016/s0959-437x(99)80052-5. [DOI] [PubMed] [Google Scholar]
- 6.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin Cell Dev Biol. 1998;9:483–9. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 7.Idler RK, Yan W. Control of messenger RNA fate by RNA-binding proteins: an emphasis on mammalian spermatogenesis. J Androl. 2012;33:309–37. doi: 10.2164/jandrol.111.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3’ processing. Proc Natl Acad Sci U S A. 1997;94:11893–8. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baurén G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3’-end formation and excision of the 3’-terminal intron. Genes Dev. 1998;12:2759–69. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat Rev Mol Cell Biol. 2002;3:619–25. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 11.Schwer B, Mao X, Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998;26:2050–7. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 13.Schoenberg DR, Maquat LE. Re-capping the message. Trends Biochem Sci. 2009;34:435–42. doi: 10.1016/j.tibs.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, et al. NELF interacts with CBC and participates in 3’ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–65. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–68. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011;21:336–43. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark H, Lührmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct. 2006;35:435–57. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 19.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 21.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–93. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–25. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: the splicing regulatory machinery. Cell. 2008;133:192.e1. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 24.O’Bryan MK, Clark BJ, McLaughlin EA, D'sylva RJ, O’Donnell L, et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet. 2013;9:e1003628. doi: 10.1371/journal.pgen.1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond DR, Armstrong J, Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–94. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proudfoot N. New perspectives on connecting messenger RNA 3’ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–8. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 28.MacDonald CC, McMahon KW. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond. Wiley Interdiscip Rev RNA. 2010;1:494–501. doi: 10.1002/wrna.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, et al. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci U S A. 2007;104:20374–9. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–30. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 2008;582:1987–96. doi: 10.1016/j.febslet.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 32.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta. 2012;1819:593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–68. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 35.Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3’-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–38. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 36.Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci U S A. 2006;103:7712–7. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci U S A. 2010;107:3594–9. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaji M, Tanaka T, Shigeta M, Chuma S, Saga Y, et al. Functional reconstruction of NANOS3 expression in the germ cell lineage by a novel transgenic reporter reveals distinct subcellular localizations of NANOS3. Reproduction. 2010;139:381–93. doi: 10.1530/REP-09-0373. [DOI] [PubMed] [Google Scholar]
- 39.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, et al. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–41. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 40.Reijo RA, Dorfman DM, Slee R, Renshaw AA, Loughlin KR, et al. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–6. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- 41.Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8:286–98. doi: 10.1093/molehr/8.3.286. [DOI] [PubMed] [Google Scholar]
- 42.Vangompel MJ, Xu EY. The roles of the DAZ family in spermatogenesis: more than just translation? Spermatogenesis. 2011;1:36–46. doi: 10.4161/spmg.1.1.14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamma H, Portman DS, Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp Cell Res. 1995;221:187–96. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- 44.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–9. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zearfoss NR, Johnson ES, Ryder SP. hnRNP A1 and secondary structure coordinate alternative splicing of Mag. RNA. 2013;19:948–57. doi: 10.1261/rna.036780.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K, Fajardo MA, Braun RE. A testis cytoplasmic RNA-binding protein that has the properties of a translational repressor. Mol Cell Biol. 1996;16:3023–34. doi: 10.1128/mcb.16.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22:171–4. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 48.Li LJ, Zhang FB, Liu SY, Tian YH, Le F, et al. Decreased expression of SAM68 in human testes with spermatogenic defects. Fertil Steril. 2014;102:61–67.e3. doi: 10.1016/j.fertnstert.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 49.Ehrmann I, Elliott DJ. Expression and functions of the star proteins Sam68 and T-STAR in mammalian spermatogenesis. Adv Exp Med Biol. 2010;693:67–81. doi: 10.1007/978-1-4419-7005-3_5. [DOI] [PubMed] [Google Scholar]
- 50.Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol. 2009;185:235–49. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schumacher JM, Lee K, Edelhoff S, Braun RE. Distribution of Tenr, an RNA-binding protein, in a lattice-like network within the spermatid nucleus in the mouse. Biol Reprod. 1995;52:1274–83. doi: 10.1095/biolreprod52.6.1274. [DOI] [PubMed] [Google Scholar]
- 52.Connolly CM, Dearth AT, Braun RE. Disruption of murine Tenr results in teratospermia and male infertility. Dev Biol. 2005;278:13–21. doi: 10.1016/j.ydbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 53.Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, et al. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci U S A. 2006;103:9494–9. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanagiya A, Delbes G, Svitkin YV, Robaire B, Sonenberg N. The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J Clin Invest. 2010;120:3389–400. doi: 10.1172/JCI43350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu W, Tekur S, Reinbold R, Eppig JJ, Choi YC, et al. Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol Reprod. 1998;59:1266–74. doi: 10.1095/biolreprod59.5.1266. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Morales CR, Medvedev S, Schultz RM, Hecht NB. In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic arrest. Biol Reprod. 2007;76:48–54. doi: 10.1095/biolreprod.106.055095. [DOI] [PubMed] [Google Scholar]
- 57.Good P, Yoda A, Sakakibara S, Yamamoto A, Imai T, et al. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics. 1998;52:382–4. doi: 10.1006/geno.1998.5456. [DOI] [PubMed] [Google Scholar]
- 58.Nagata T, Kanno R, Kurihara Y, Uesugi S, Imai T, et al. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J Mol Biol. 1999;287:315–30. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- 59.Ohyama T, Furukawa A, Mashima T, Sugiyama T, Ohgara S, et al. Structural analysis of Musashi-RNA complex on the basis of long-range structural information. Nucleic Acids Symp Ser (Oxf) 2008;52:193–4. doi: 10.1093/nass/nrn098. [DOI] [PubMed] [Google Scholar]
- 60.MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10:39–44. doi: 10.4161/cc.10.1.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 62.Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–8. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 63.Hirota Y, Okabe M, Imai T, Kurusu M, Yamamoto A, et al. Musashi and seven in absentia downregulate Tramtrack through distinct mechanisms in Drosophila eye development. Mech Dev. 1999;87:93–101. doi: 10.1016/s0925-4773(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 64.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–56. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 65.Siddall NA, Kalcina M, Johanson TM, Monk AC, Casagranda F, et al. Drosophila Rbp6 is an orthologue of vertebrate Msi-1 and Msi-2, but does not function redundantly with dMsi to regulate germline stem cell behaviour. PLoS One. 2012;7:e49810. doi: 10.1371/journal.pone.0049810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richter K, Good PJ, Dawid IB. A developmentally regulated, nervous system-specific gene in Xenopus encodes a putative RNA-binding protein. New Biol. 1990;2:556–65. [PubMed] [Google Scholar]
- 67.Amato MA, Boy S, Arnault E, Girard M, Della Puppa A, et al. Comparison of the expression patterns of five neural RNA binding proteins in the Xenopus retina. J Comp Neurol. 2005;481:331–9. doi: 10.1002/cne.20387. [DOI] [PubMed] [Google Scholar]
- 68.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arumugam K, Wang Y, Hardy LL, MacNicol MC, MacNicol AM. Enforcing temporal control of maternal mRNA translation during oocyte cell-cycle progression. EMBO J. 2010;29:387–97. doi: 10.1038/emboj.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arumugam K, Macnicol MC, Macnicol AM. Autoregulation of Musashi1 mRNA translation during Xenopus oocyte maturation. Mol Reprod Dev. 2012;79:553–63. doi: 10.1002/mrd.22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002;115:1355–9. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 72.Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A. 2002;99:15194–9. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Horisawa K, Imai T, Okano H, Yanagawa H. 3’-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009;583:2429–34. doi: 10.1016/j.febslet.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 76.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–53. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuwako K, Kakumoto K, Imai T, Igarashi M, Hamakubo T, et al. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron. 2010;67:407–21. doi: 10.1016/j.neuron.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 78.Kong DS, Kim MH, Park WY, Suh YL, Lee JI, et al. The progression of gliomas is associated with cancer stem cell phenotype. Oncol Rep. 2008;19:639–43. [PubMed] [Google Scholar]
- 79.Yuqi L, Chengtang W, Ying W, Shangtong L, Kangxiong L. The expression of Msi-1 and its significance in small intestinal mucosa severely damaged by high-dose 5-FU. Dig Dis Sci. 2008;53:2436–42. doi: 10.1007/s10620-007-0155-0. [DOI] [PubMed] [Google Scholar]
- 80.Murata H, Tsuji S, Tsujii M, Nakamura T, Fu HY, et al. Helicobacter pylori infection induces candidate stem cell marker Musashi-1 in the human gastric epithelium. Dig Dis Sci. 2008;53:363–9. doi: 10.1007/s10620-007-9858-5. [DOI] [PubMed] [Google Scholar]
- 81.Nagata H, Akiba Y, Suzuki H, Okano H, Hibi T. Expression of Musashi-1 in the rat stomach and changes during mucosal injury and restitution. FEBS Lett. 2006;580:27–33. doi: 10.1016/j.febslet.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 82.Götte M, Wolf M, Staebler A, Buchweitz O, Kelsch R, et al. Increased expression of the adult stem cell marker Musashi-1 in endometriosis and endometrial carcinoma. J Pathol. 2008;215:317–29. doi: 10.1002/path.2364. [DOI] [PubMed] [Google Scholar]
- 83.Wang XY, Yin Y, Yuan H, Sakamaki T, Okano H, et al. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol Cell Biol. 2008;28:3589–99. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, et al. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akindahunsi AA, Bandiera A, Manzini G. Vertebrate 2xRBD hnRNP proteins: a comparative analysis of genome, mRNA and protein sequences. Comput Biol Chem. 2005;29:13–23. doi: 10.1016/j.compbiolchem.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 86.de Andrés-Aguayo L, Varas F, Kallin EM, Infante JF, Wurst W, et al. Musashi 2 is a regulator of the HSC compartment identified by a retroviral insertion screen and knockout mice. Blood. 2011;118:554–64. doi: 10.1182/blood-2010-12-322081. [DOI] [PubMed] [Google Scholar]
- 87.Kawase S, Imai T, Miyauchi-Hara C, Yaguchi K, Nishimoto Y, et al. Identification of a novel intronic enhancer responsible for the transcriptional regulation of musashi1 in neural stem/progenitor cells. Mol Brain. 2011;4:14. doi: 10.1186/1756-6606-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishimoto Y, Okano H. New insight into cancer therapeutics: induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res. 2010;20:1083–5. doi: 10.1038/cr.2010.122. [DOI] [PubMed] [Google Scholar]
- 89.Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21:8091–107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–8. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–8. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szabat M, Kalynyak TB, Lim GE, Chu KY, Yang YH, et al. Musashi expression in ß-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2011;2:e232. doi: 10.1038/cddis.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cox JL, Wilder PJ, Gilmore JM, Wuebben EL, Washburn MP, et al. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. PLoS One. 2013;8:e62857. doi: 10.1371/journal.pone.0062857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thol F, Winschel C, Sonntag AK, Damm F, Wagner K, et al. Prognostic significance of expression levels of stem cell regulators MSI2 and NUMB in acute myeloid leukemia. Ann Hematol. 2013;92:315–23. doi: 10.1007/s00277-012-1637-5. [DOI] [PubMed] [Google Scholar]
- 95.Mu Q, Wang Y, Chen B, Qian W, Meng H, et al. High expression of Musashi-2 indicates poor prognosis in adult B-cell acute lymphoblastic leukemia. Leuk Res. 2013;37:922–7. doi: 10.1016/j.leukres.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 96.de Andrés-Aguayo L, Varas F, Graf T. Musashi 2 in hematopoiesis. Curr Opin Hematol. 2012;19:268–72. doi: 10.1097/MOH.0b013e328353c778. [DOI] [PubMed] [Google Scholar]
- 97.Park SM, Deering RP, Lu Y, Tivnan P, Lianoglou S, et al. Musashi-2 controls cell fate, lineage bias, and TGF-ß signaling in HSCs. J Exp Med. 2014;211:71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A. 2006;103:8402–7. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gunter KM, McLaughlin EA. Translational control in germ cell development: a role for the RNA-binding proteins Musashi-1 and Musashi-2. IUBMB Life. 2011;63:678–85. doi: 10.1002/iub.499. [DOI] [PubMed] [Google Scholar]
- 100.Sutherland JM, Fraser BA, Sobinoff AP, Pye VJ, Davidson TL, et al. Developmental expression of Musashi-1 and Musashi-2 RNA-binding proteins during spermatogenesis: analysis of the deleterious effects of dysregulated expression. Biol Reprod. 2014;90:92. doi: 10.1095/biolreprod.113.115261. [DOI] [PubMed] [Google Scholar]


