Abstract
The classical idea about the function of the mammalian sperm chromatin is that it serves to transmit a highly protected and transcriptionally inactive paternal genome, largely condensed by protamines, to the next generation. In addition, recent sperm chromatin genome-wide dissection studies indicate the presence of a differential distribution of the genes and repetitive sequences in the protamine-condensed and histone-condensed sperm chromatin domains, which could be potentially involved in regulatory roles after fertilization. Interestingly, recent proteomic studies have shown that sperm chromatin contains many additional proteins, in addition to the abundant histones and protamines, with specific modifications and chromatin affinity features which are also delivered to the oocyte. Both gene and protein signatures seem to be altered in infertile patients and, as such, are consistent with the potential involvement of the sperm chromatin landscape in early embryo development. This present work reviews the available information on the composition of the human sperm chromatin and its epigenetic potential, with a particular focus on recent results derived from high-throughput genomic and proteomic studies. As a complement, we provide experimental evidence for the detection of phosphorylations and acetylations in human protamine 1 using a mass spectrometry approach. The available data indicate that the sperm chromatin is much more complex than what it was previously thought, raising the possibility that it could also serve to transmit crucial paternal epigenetic information to the embryo.
Keywords: genomics, male infertility, proteomics, sperm chromatin, sperm epigenetics
INTRODUCTION
The main function of the sperm cell is to transmit to the embryo the paternal genetic message encoded in the DNA sequence together with the presence of appropriate epigenetic information.1,2,3,4 The most well-studied mechanism of epigenetic inheritance is the reversible methylation of cytosine residues in cytosine-guanine dinucleotides at imprinted genes, which is involved in gene expression regulation.5,6 However, additional potential sperm epigenetic information is also constituted by the presence of histone modifications, presence of other chromatin-associated proteins and their modifications, RNAs, a unique chromatin structure (Figure 1), and chromosome territories in the nucleus.7,8,9,10,11,12,13,14,15 As compared to somatic cells, not much is known so far about the potential role of these additional forms of epigenetic information in the sperm, despite that it is an emerging subject of increasing interest.3,10,16,17,18,19 Thus, the present review aims to cover these newer forms of epigenetic information, being focused on the chromatin structure, gene distribution and presence of chromatin proteins and their modifications in the sperm cell (Figure 1). Therefore, it does not aim to cover the topics of sperm DNA methylation and the presence of sperm RNAs, for which the reader is referred to other excellent reviews.5,7,20,21 Furthermore, the present review does not intend to cover related issues concerning the potential origins and consequences of sperm DNA damage, for which recent reviews are also available.22,23,24,25,26
Figure 1.
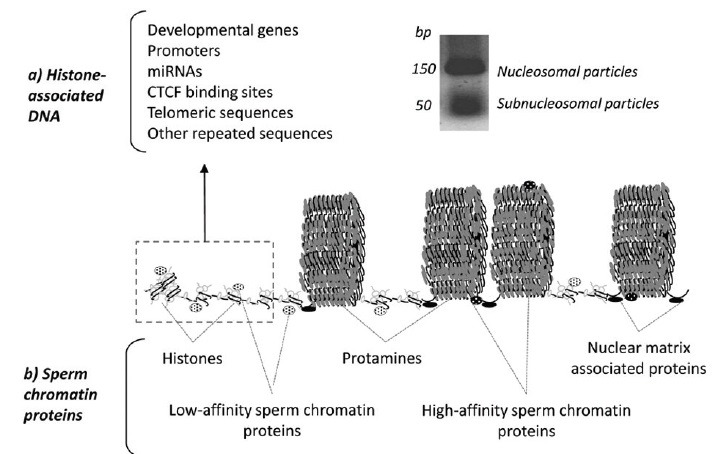
Gene and protein composition of the human sperm chromatin. Hypothetical model of human sperm chromatin showing the histone-associated DNA constitution (a) and protein contents (b). The model is drawn at a scale and inspired by the known nucleo-protamine toroidal structures124 and the high-throughput sequencing data obtained from the analysis of retained human sperm nucleosomes after micrococcal nuclease digestion.43,44,45,51,52
As a source of information, PubMed articles published until submission of this review (November 10, 2014) were considered on the topics of “sperm chromatin,” “sperm chromatin packaging,” “sperm chromatin gene distribution,” “sperm histone retention,” “sperm histone modifications,” “protamine modifications,” “sperm chromatin protein composition,” and “chromatin alterations in infertile patients.” As a complement, we provide here for the first-time experimental evidence for the detection of phosphorylations and acetylations in human protamine 1 using mass spectrometry (MS).
The first part of this review starts with a discussion about the current knowledge of the sequence-specific sperm chromatin distribution, and is followed by a section on the abundant histones and protamines and the additional (less abundant) sperm chromatin proteins recently identified using MS. Finally, the review concludes with a section on the presence of genomic and proteomic alterations detected in the sperm chromatin of infertile patients, both in the gene distribution and in the presence of an altered abundance on sperm chromatin proteins.
The present review complements, expands, and updates other previously published reviews on this topic.3,4,8,9,10,11,12,17,18,21,27,28,29,30,31,32,33,34,35,36
SEQUENCE-SPECIFIC SPERM CHROMATIN DNA DISTRIBUTION
The structure and composition of the inert mammalian sperm chromatin have been extensively studied during the past three decades27,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52 (Table 1). Classical sperm chromatin dissection experiments were based on two main approaches: (1) DNA digestion by endonucleases (typically DNase I and micrococcal nuclease; MNase),39,40,42,43,44,45,46,48,49,50,51,52 and (2) disruption of histone-DNA associations by 0.65 M NaCl followed by the digestion of histone-free DNA (typically by EcoRI and BamHI).37,38,41,43,51 Subsequent DNA analyses using probes, polymerase chain reaction, chromatin immunoprecipitation, microarrays or high-throughput sequencing techniques have provided exciting results (Table 1).
Table 1.
DNA distribution analyses through sperm chromatin in healthy or normozoospermic men and in model species
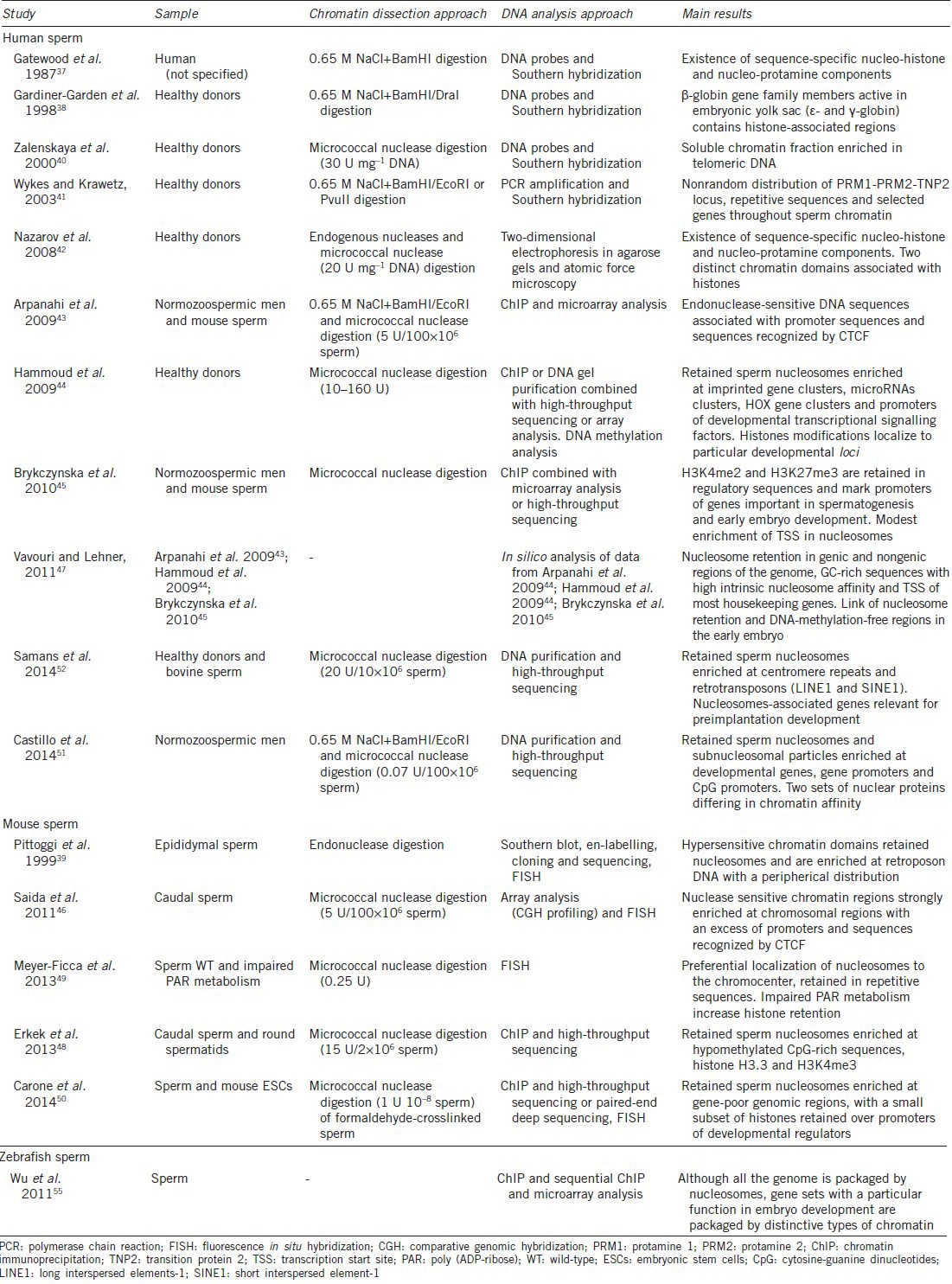
A growing body of evidence suggests the existence of a sequence-specific packaging by histones and protamines involved in potential epigenetic inheritance.9,12,17,27,37,42,53,54 The first studies in this field were focussed on the analysis of specific gene families or clusters, showing a potential involvement of DNA distribution in postfertilization events (Table 1). Interestingly, members of β-globin gene family active in embryonic yolk sac (ε- and γ-globin) contained histone-associated regions, while no presence of β- and δ-globin (inactive in embryonic yolk sac) was found.38 Furthermore, Wykes and Krawetz reported a nonrandom distribution of PRM1-PRM2-TNP2 loci, repetitive sequences and selected genes throughout the chromatin.41 This programmatic sperm DNA distribution has been confirmed recently by the genome-wide sperm nucleosome profiles generated by other groups (Table 1). Particularly, an enrichment of human and mouse retained sperm nucleosomes at developmental loci was reported, which included imprinted genes, microRNAs, HOX genes, promoters of developmental transcriptional signaling factors, GC-rich sequences, and transcription start sites of most housekeeping genes43,44,47,51 (Figure 1).
Of relevance, we have recently contributed to the knowledge of sperm chromatin reporting the existence in human sperm histone-associated chromatin of not only classical nucleosomes, but also of smaller MNase-sensitive regions.51 Although an association with specific histone variants is still unexplored, these so-called subnucleosomal particles were seen to be enriched in alternative and not overlapping developmental loci.51 These results allowed speculation that additional levels of sperm chromatin packaging might be involved in postfertilization gene regulation. A recent study using mouse sperm has also suggested the presence of two types of footprints obtained after MNase digestion, one corresponding to nucleosomes, and a shorter one (<80 bp) associated with other DNA-binding proteins such as transcription factors.50 Similarly, zebrafish (which do not employ protamines for sperm DNA packaging) have a multivalent chromatin constituted by gene sets implicated in embryo development processes and associated with distinctive types of nucleosome packaging.55 All these data are therefore suggesting an exciting dynamic behavior of the sperm chromatin.
The role of sperm histone-retention in male epigenetic inheritance becomes even more significant considering that regulatory loci are marked by specific histone methylation patterns.3,9,12,17,44,45,56 In fact, human and mice genes related to spermatogenesis and cellular homeostasis seem to be associated with activating modifications (H3K4me2), while developmental gene promoters may be related not only with activating (H3K4me2 and H3K4me3) but also with repressive histone marks (H3K27me3). Interestingly, this bivalent gene marking is showing an overlap with embryonic stem cells, suggesting a role in the establishment of embryonic totipotency.44,45
However, histone-retention constitutes just 5% to 15% of sperm chromatin while the major part is indeed tightly packaged by protamines (Figure 1). This higher level of compaction is required not only to avoid any transcriptional and translational activity, but also to reduce the accessibility of external and internal nucleases28,57,58 (Figure 1). In fact, although protamine-packaging is not necessary for proper embryo development,59,60 it appears to be important for DNA integrity maintenance.61,62,63,64,65 Therefore, if the nucleo-protamine structure is ensuring the transmission of intact male genome to the next generation, an important question remaining to be answered is why crucial male developmental loci are vulnerable by virtue of being associated with nucleosomes.
In this regard, contrasting results with those already mentioned regarding sequence-specific sperm DNA distribution, have been reported by several groups during the past years (Table 1). Interestingly, by using similar strategies for chromatin dissection and sequence analysis in human, mouse and bovine sperm, it has been shown that nucleosomes might be moderately retained at unique DNA sequences and regulatory regions (Table 1). In contrast, a majority of sperm histones seemed to be localized to the nuclear periphery, within distal intergenic regions and introns, and associated with centromere and telomere repeats and retrotransposons (LINE and SINE; Figure 1).39,40,45,48,49,50,52 Obtaining such different results following the same strategies could be due to a technical issue. Carone et al. suggest in their study that promoter nucleosomes, although being less abundant in sperm, are more stable to MNase digestion. In this regard, an extensive nuclear digestion of chromatin would degrade more abundant nucleosomes in gene deserts and thus reveal only those associated with regulatory regions.52 This hypothesis seems to be consistent with the identification of distal DNase I-hypersensitive regions characterized by an enrichment at CTCF-binding sites, depletion in H3K4me3 and presence of H3K9ac and H4K12ac in human and mouse spermatozoa43,46 (Table 1).
Whatever the case may be, the sperm nucleosome association with repetitive sequences would be also in agreement with a potential function of the sequence-specific sperm chromatin DNA distribution in postfertilization processes. For instance, it is known that telomeres are involved in microtubule-guided movement during male pronucleus development.40,66 Furthermore, retrotransposons could be conducting regulatory functions for host genes, by serving as a scaffold for the transcription factor binding repertoire in preimplantational processes.20,52 Interestingly, CTCF has been suggested to be a key mediator of epigenetic chromatin remodeling not only during male germ cell development, but also in embryonic genome activation.43,46,50
Taking all these results together, one could hypothesize a model of sperm chromatin structure with a selected subset of relevant regulatory loci packaged in dynamic nucleosomal structures, which together with repeated sequences would be closely involved in the regulation of early postfertilization processes (e.g., sperm chromatin remodeling in male pronucleus; Figure 1). These epigenetic signatures may be potentially important in preimplantational stages, in order to initiate key processes, and preserve the integrity of other male developmental genes required for later stages of embryogenesis.
ABUNDANT SPERM CHROMATIN-ASSOCIATED PROTEINS – HISTONES AND PROTAMINES
Similarly to the sperm DNA distribution analyses, the study of the protein component of sperm chromatin is also providing information supporting the possibility of a potential sperm epigenetic inheritance.
Although the sperm nucleo-histone domain constitutes a minor part of the total sperm chromatin (Figure 1), up to 46 different histones and histone variants have been detected so far as part of the human sperm nuclear proteome.17 Nucleosomes are dynamic, rather than static, structures, and this aspect may be especially relevant in the male germ line, where different testis-specific histones variants (and their modifications) are expressed throughout mammalian spermatogenesis.67,68 The role of noncanonical histones during sperm differentiation is well-documented and becomes essential for different stages, principally after meiosis. In particular, testicular H1 histone, histone H2A.Bdb, and histone H2B type 1-A (TH2B in mouse) are known to be involved in histone hyperacetylation and nucleosome destabilization prior the incorporation of transition proteins and protamines.69,70,71,72 However, histone roles should not be restricted to spermatogenic processes, as an analogous hyperacetylation-based paternal chromatin remodeling occurs after fertilization. Therefore, it would be logical to think that the sperm-derived histone variants that remain in the zygotic nucleus could be also participating at that stage.17,56,70,71
Histone participation during spermatogenic and postfertilization processes becomes fundamental due to the ability to carry modifications (mainly methylations, acetylations, and ubiquitinations), defining the so-called histone code.4,11,12,17,35,73,74 Besides the specific sperm histone-methylation patterns, already pointed out in the above section, histone acetylations are also noteworthy. In fact, in addition to the acetylation wave that takes place prior to the nucleo-histone-to-nucleo-protamine transition in spermatids,75,76,77,78,79,80 histone H4K8ac, and H4K12ac have been also observed preceding full decondensation in the zygotic nucleus.81 Also, interesting was the discovery of a new histone modification, crotonylation, which seems to mark postmeiotically activated genes on autosomes, as well as specific X/Y-linked genes, enabling their postmeiotic activation despite the general repression of the haploid sex chromosomes in the sex body.82
Similar to histones, it is also worth-considering in detail the protamine amino-acid sequences, as protamines are considered critical in the maintenance of sperm chromatin status. Protamines are the most abundant proteins in the mammalian sperm nucleus (Figure 1) and their distinctive physicochemical characteristics (such as extreme basicity and high proportion of arginines and cysteines; Figure 2) confer elevated protein stability,27,28,57,58 at least in Eutherian mammals. For this reason, the identification of potential posttranslational modification (PTM) sites in the sequence of protamine 1 (P1) or protamine 2 (P2) is an intriguing field of study which could increase our knowledge of the sperm chromatin epigenetic landscape.
Figure 2.
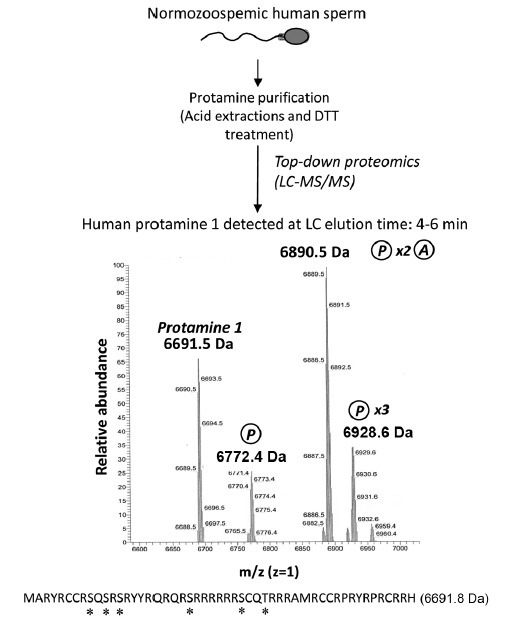
Analysis of the intact human sperm protamine sequences by MS. After following the indicated workflow and LC-MS/MS analysis, the registers obtained demonstrate the presence of the intact unmodified Protamine 1 mass and masses of the protamine 1 containing different combinations of phosphorylation and acetylation. Methods: human sperm cells from normozoospermic sperm donors were collected and processed as described92 following World Health Organization guidelines125 and in accordance with the ethical and internal review board guidelines. Seminal plasma was discarded and sperm were treated with acid and reducing agents,92 in order to obtain a protein extract constituted mostly by a mixture of purified protamine 1 (P1) and protamine 2 (P2) (top). The correct protamine purification was confirmed through acid-urea polyacrylamide gel electrophoresis, as previously described,61 prior to the analysis by LC-MS/MS. Spectra corresponding to the liquid chromatography elution time 4–6 min obtained after MS analysis of nondigested proteins (top-down proteomics) is shown in the middle of the image. Mass peaks corresponding to nonmodified P1, mono-phosphorylated P1, di-phosphorylated and mono-acetylated P1, and tri-phosphorylated P1 were observed (note that each peak is showing a mass/charge (m/z) value corresponding to a specific “m,” since “z” is set as 1). A maximum error of 200 ppm between theoretical and experimental masses was considered to give a match as valid. At the bottom of the figure, the human P1 amino-acid sequence is indicated together with its theoretical “m.” Potential phosphorylation and acetylation sites are highlighted with an asterisk. DTT: dithiothreitol; MS: mass spectrometry; LC-MS/MS: liquid chromatography followed by tandem mass spectrometry.
Reported PTM include the detection of human P1 mono- and di-phosphorylated sites (N-terminal region) and P2 mono-phosphorylated sites (middle region), which seem to be implicated in sperm chromatin remodeling during spermatogenesis.27,83,84,85 The different location of phosphorylated serines between human protamines was suggested to indicate distinct roles for P1 and P2 during sperm maturation.85 Protamines also experience further processing during epididymal maturation, when disulfide bonds and zinc bridges are formed between cysteine residues to stabilize the toroidal structure.25,28,58,86,87
An additional important contribution to our knowledge of protamine PTMs has recently been forthcoming in a study of murine protamine sequences using a novel proteomic workflow based in MS.73 Thus, Brunner et al. were able to detect phosphorylation sites using MS in mouse P1 and P2 corresponding to those previously reported in human sperm using conventional procedures. In addition, although P2 family members were not analyzed separately, P2 residues carrying acetylations and methylations were also identified.73 It is interesting to highlight that while acetylations and methylations were detected in the same sequence, acetylations and phosphorylations seemed to be exclusive, suggesting nonrandom protamine processing in mouse spermatozoa.73
Consistent with the above observations in mice, we report here for the first time the analysis of the intact (not digested) human protamine 1 amino acid sequence through MS (Figure 2). Following a similar strategy to that used in mouse sperm, we were able to detect in human P1 mono-, di- and tri-phosphorylations by MS (Figure 2). In addition, P1 seemed to carry combinations of different PTMs, which included di-acetylations with mono-methylation and, in contrast to mouse P1, di-phosphorylations with mono-acetylation (Figure 1). Concerning human P2 family, the potential presence of PTMs (methylations and acetylations) was only detected in the component P3 (data not shown). Despite this evidence for the presence of phosphorylations and acetylations in mouse (Brunner et al. 2014) and human (Figure 2) protamines, we believe that the existence of a protamine code with a similar function to the histone code (where PTMs are involved in gene expression) is unlikely. The main reasons are the presence of the semicrystalline chromatin status conferred by the nucleo-protamine toroidal packaging and the fact that the sperm chromatin is transcriptionally silent.17,27,28,57,58,88,89,90 Instead, there is substantial evidence for the involvement of protamine phosphorylation in the deposition of recently synthesized protamines to DNA.27,83,84,85,91 What remains still unknown is whether other modifications, such as acetylations of protamines, could also have a role during spermatogenesis. The possibility is thus open to study a potential involvement of protamine PTMs in the mature chromatin structure and in chromatin structural transitions taking place after fertilization (such as the chromatin destabilization in the male pronucleus prior maternal histones incorporation), and how this could be related to gene expression in the early embryo.
These novel protamine PTMs reported here (Figure 2) were obtained following a protamine purification procedure using sperm samples from normozoospermic men as previously described in our lab92 coupled to MS analysis of the nondigested protein extracts. The small size of the protamines (Figure 2) allows the detection of the intact amino acid sequence using an elegant high-throughput technique known as top-down proteomics.73,93 Following this strategy, spectra showing peaks with mass/charge values (m/z) were obtained and used to determine the mass of every protein component of the extract (Figure 2). Because “z” was set as 1, each MS peak provided a unique “m” (Daltons), which was subsequently compared with that corresponding to human P1, or mature and immature individual human P2 family members. Incorporations of “m” to protamines (corresponding to PTMs) were evaluated using information from the Unimod database (http://www.unimod.org/modifications_list.php?), setting 200 ppm as the maximum error acceptable to consider valid an m match.
The gold-standard strategies to detect and quantify protamines have been so far based on protein separation using acid-urea polyacrylamide gel electrophoresis and visualization by Coomassie blue gel staining,21,61,65,92,94,95,96,97,98 the use of specific antibodies for Western blot detection,97,99,100 and determination of the protamine amino-acid sequences using EDMAN cycle protein sequencing.83,85 However, these strategies were quite time-consuming implying a relative limitation. Thus, emerging approaches to identify protamines based in MS strategies73 (Figure 2) seem to have the potential to be more efficient and accurate, in addition to allowing a higher-throughput analysis of patients and model systems. All these reported data are therefore supporting the prospective use of top-down proteomics for protamine PTMs patterns analyses in fertile men, the identification of alterations in infertile patients, and the potential identification of prognostic markers in assisted reproduction.
LOW ABUNDANCE SPERM CHROMATIN-ASSOCIATED PROTEINS
The MS-based study of the male gamete has also largely demonstrated that the sperm chromatin protein composition is more complex that initially considered.17,101,102 In fact, besides histones and protamines, a large number of zinc finger- and bromodomain-containing proteins, transcription factors, histone modifiers, and other DNA-related proteins have been identified as part of the human sperm nucleus17,51,101,103,104,105,106,107 (Figure 1). Interestingly, similar trends were also found in other mammalian species which suggests a conservation of the rich sperm chromatin protein profile.17
Thus, it should be accepted that the sperm nucleus contains a subset of sperm nuclear proteins mainly involved in chromatin organization and gene expression which are delivered to the oocyte upon fertilization.11,17 This fact consequently leads to speculation concerning the purpose of keeping these proteins in the transcriptionally inert male gamete. In this regard, sperm chromatin proteins could be considered at three levels: (1) spermatogenesis residual with no function at all in the mature sperm cell or in the zygote, (2) proteins with a structural or functional role in the mature sperm chromatin, and (3) proteins implicated in future events such as male pronucleus chromatin remodeling or transcriptional regulation of histone-bound paternal genes after fertilization.17,30,51,104,107,108 Of relevance, for many proteins, these three facets would not be necessarily mutually exclusive. For example, this could be the case for the well-studied bromodomain testis-specific protein, the histone-chaperone protein SET, transcription factors involved in differentiation and developmental processes, PHD Finger proteins, effectors of the histone code, or some proteins involved in the regulation of the DNA methylation, replication, repair and transcription.17,51,69,109,110,111
Also interesting is the fact that sperm nuclear proteins can be differentiated according to their chromatin affinity into two subsets with different functional profiles51 (Figure 1). To this extent, high-affinity proteins might be mainly involved in structural roles, which could be related with the nucleo-protamine structural organization and function. In contrast, the low-affinity subset of proteins could conceivably be involved in a higher variety of roles, including protein metabolism, DNA packaging, DNA/RNA binding, and transcription, among others. Of relevance, all those proteins with potential regulatory roles in the sperm nucleus appear to be weakly attached to the chromatin.51 These data suggest that in addition to the sequence-specific distribution of the genes, the spermatozoon is organized such that many different layers of potential epigenetic information are distributed through the chromatin landscape for delivery to the oocyte at fertilization and the subsequent regulation of early embryogenesis.
In order to unravel the sperm chromatin epigenetic potential, it is also necessary to take into account the role of the nuclear matrix and the proteins associated with this structure. Similarly to the somatic cells, sperm chromatin is organized into nuclease sensitive DNA segments attached to the nuclear matrix at 50 kb intervals (Figure 1).8,112 The nuclear matrix itself is thought to incorporate Topoisomerase II-B (TOP2B). TOP2B is a matrix-associated protein reported to have a role in spermiogenesis, reducing nucleosome supercoiling by double-strand DNA breaks and promoting chromatin remodeling.113 The paternal matrix associated regions (MARs), including DNA and protein composition, seem to be inherited by the fertilized embryo and are thought to be essential for paternal pronuclear DNA replication.8,34,59,60 In fact, proper initiation pronuclear formation and DNA synthesis in fertilized mouse oocytes has been demonstrated using spermatozoa exhibiting a high degree of DNA degradation, as long as in situ DNA loop attachments to the nuclear matrix were preserved.59 Therefore, as it happens in somatic cells, the sperm replication machinery is also assembled into the nuclear matrix, which is serving as a scaffold.34 Several lines of evidence suggest that the same MAR sites could also be representing points of DNA cleavage, with a potential role in DNA integrity maintenance, in which TOP2B might also be involved.22,34,114
GENOMIC AND PROTEOMIC ALTERATIONS IN SPERM CHROMATIN OF INFERTILE PATIENTS
As has been discussed in the previous sections, sperm chromatin is characterized by specific genomic and proteomic features, involved in the correct formation of the male gamete as well as the embryo. Therefore, an important question that still remains to be considered is whether alterations in sperm epigenetic signature can lead to male infertility.
Several studies have demonstrated alterations in sperm DNA methylation patterns,115,116,117 RNA content,20,118 and histone retention54,119,120 in infertile/subfertile patients. Alterations in histone retention could affect sperm function at two levels. First, the altered histone retention would directly imply an alteration in protamine content and in the tightly regulated P1/P2 ratio, which is indicative of poor semen quality, increased DNA damage and decreased fertility.9,21,28,31,57,58,61,62,64,65,90,92,95,97,99,100,121 Second, it would lead to rearrangements in chromatin organization of developmental loci and genes, which may have an impact on normal embryo development. In fact, the histone-specific sequence packaging found in healthy men (Figure 1) has been shown to be lost in patients with subfertility, resulting in random sperm DNA chromatin distribution.119 In this study, Hammoud et al. analyzed MNase-sensitive sperm chromatin regions from three patients with poor embryogenesis after in vitro fertilization (IVF) and four men with altered protamination. After genome-wide analysis based on high-throughput sequencing, five of the seven infertile men showed nonprogrammatic retention of histones. Histone modifications were also evaluated, showing a reduction of developmental promoters enriched with H3K4me3 and H3K27me3 in most infertile men, although the localization of modified histones was unaltered.119
Our group has also recently demonstrated the presence of altered histone content in infertile patients using a high-throughput quantitative proteomic approach based on protein isobaric labelling of proteins.120 The significance of sperm nuclear proteomic profiles was recently highlighted when cells from normozoospermic men that were able to perform IVF but whose female partners did not achieve a pregnancy, were compared with those from men with similar semen parameters but with successful pregnancy outcomes (excluding female factor). Interestingly, altered levels of proteins specifically involved in chromatin assembly and metabolism were detected in the so-called “no pregnancy group.” In particular, six histones variants (H2A type 1-A, H2A type 1-C, H2A type 2-C, H2B type 1A, H3 and H4) and a protein involved in protamine 1 phosphorylation (SRSF protein kinase 1; SRPK1) were identified deregulated with abnormally high abundance.120 These results were thus consistent with the hypothesis that an alteration in the sperm chromatin proteome may result in epigenetic errors contributing to failed embryo development.
Additional complementary data have also been obtained in mouse models with impaired poly (ADP-ribose) (PAR) metabolism, which is involved in the nucleo-histone-to-nucleo-protamine transition during spermiogenesis.49,122 Affected PAR mice sperm was shown to carry compromised chromatin with abnormally increased histone retention, although being still associated with repetitive sequences.49,122
Of relevance, Ihara's study went a step further and analyzed for the first time the genome of embryos generated with chromatin affected (by altered PAR metabolism) spermatozoa. Interestingly, it was shown that a statistically significant portion of the differentially expressed genes in mouse embryos corresponded to paternal gene loci showing altered histone retention caused by PAR impairment.122 Despite this and in contrast to the data obtained in human spermatozoa, fertility was not compromised in this model, as embryos developed to term.122
Infertility can be considered a multifactorial disease in which alterations in the epigenetic constitution of the sperm chromatin could be involved. The application of genomic and proteomic high-throughput strategies is thus helping to unravel the potential contribution of sperm chromatin organization to male infertility. Therefore, all these data have the potential in the future to be useful in the identification of putative biomarkers for the diagnosis and prognosis of idiopathic male infertility.107,123
CONCLUSION
About 92% of the human sperm chromatin is formed by highly compact toroidal nucleo-protamine complexes while the remaining 8% is organized with histones. Of importance, there is a nonrandom distribution of the genes, gene sequences, and repetitive elements that has the potential to be involved in the sperm chromatin reorganization in the oocyte and perhaps in the selective activation of key paternal genes in the early embryo. In addition to the protamines and histones, the sperm chromatin also contains many additional chromatin-associated proteins with the potential to provide different layers of epigenetic information or serve in the reorganization of the paternal chromatin after fertilization. Alterations in the distribution of histones and the additional sperm chromatin-associated proteins are also being detected in infertile patients. Altogether the information so far available indicates that sperm chromatin is much more complex than it was previously thought and that it contributes in the transmission of information to the zygote that may be crucial for the paternal pronuclear chromatin remodeling and embryo development.
AUTHOR'S CONTRIBUTIONS
JC and RO analyzed MS data of protamines, drafted the manuscript and approved the final manuscript. JME analyzed MS data of protamines, critically reviewed the manuscript and approved the final version. JLB characterized the clinical samples used for MS detection of protamines, critically reviewed the manuscript and approved the final version.
ACKNOWLEDGMENTS
This work was supported by grants to RO from the Spanish Ministry of Economy and Competitiveness (Ministerio de Economia y Competitividad; FEDER BFU 2009-07118 and PI13/00699). JC was supported by a fellowship from the University of Barcelona (APIF). MS-based proteomic analyses of protamines were conducted at the Proteomic Unit of the Scientific and Technological Centres of the University of Barcelona (CCiTUB), a member of the ProteoRed network (http://www.proteored.org), by Dr. Maria José Fidalgo.
COMPETING FINANCIAL INTERESTS
All authors declare no competing interests.
REFERENCES
- 1.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–83. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 2.Trasler JM. Epigenetics in spermatogenesis. Mol Cell Endocrinol. 2009;306:33–6. doi: 10.1016/j.mce.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 3.Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod. 2010;16:37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- 4.Rando OJ. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–8. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 6.Auclair G, Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94:2202–11. doi: 10.1016/j.biochi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 8.Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–6. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins TG, Carrell DT. The sperm epigenome and potential implications for the developing embryo. Reproduction. 2012;143:727–34. doi: 10.1530/REP-11-0450. [DOI] [PubMed] [Google Scholar]
- 11.Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005;6:633–42. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 12.Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012;97:267–74. doi: 10.1016/j.fertnstert.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Fischer BE, Wasbrough E, Meadows LA, Randlet O, Dorus S, et al. Conserved properties of Drosophila and human spermatozoal mRNA repertoires. Proc Biol Sci. 2012;279:2636–44. doi: 10.1098/rspb.2012.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 15.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loppin B, Lepetit D, Dorus S, Couble P, Karr TL. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 17.Castillo J, Amaral A, Oliva R. Sperm nuclear proteome and its epigenetic potential. Andrology. 2014;2:326–38. doi: 10.1111/j.2047-2927.2013.00170.x. [DOI] [PubMed] [Google Scholar]
- 18.Casas E, Vavouri T. Sperm epigenomics: challenges and opportunities. Front Genet. 2014;5:330. doi: 10.3389/fgene.2014.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. Adv Exp Med Biol. 2014;791:53–66. doi: 10.1007/978-1-4614-7783-9_4. [DOI] [PubMed] [Google Scholar]
- 20.Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA, et al. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update. 2013;19:604–24. doi: 10.1093/humupd/dmt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jodar M, Oliva R. Protamine alterations in human spermatozoa. Adv Exp Med Biol. 2014;791:83–102. doi: 10.1007/978-1-4614-7783-9_6. [DOI] [PubMed] [Google Scholar]
- 22.Shaman JA, Ward WS. Sperm chromatin stability and susceptibility to damage in relation to its structure. In: De Jonge CJ, Barratt CL, editors. Sperm Cell: Production, Maturation, Fertilization, Regeneration. New York: Cambridge University Press; 2006. pp. 31–48. [Google Scholar]
- 23.Lewis SE, Aitken RJ. DNA damage to spermatozoa has impacts on fertilization and pregnancy. Cell Tissue Res. 2005;322:33–41. doi: 10.1007/s00441-005-1097-5. [DOI] [PubMed] [Google Scholar]
- 24.Laberge RM, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod. 2005;73:289–96. doi: 10.1095/biolreprod.104.036939. [DOI] [PubMed] [Google Scholar]
- 25.Aitken RJ, De Iuliis GN. Origins and consequences of DNA damage in male germ cells. Reprod Biomed Online. 2007;14:727–33. doi: 10.1016/s1472-6483(10)60676-1. [DOI] [PubMed] [Google Scholar]
- 26.Aitken RJ, De Iuliis GN. On the possible origins of DNA damage in human spermatozoa. Mol Hum Reprod. 2010;16:3–13. doi: 10.1093/molehr/gap059. [DOI] [PubMed] [Google Scholar]
- 27.Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- 28.Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- 29.Oliva R, Martínez-Heredia J, Estanyol JM. Proteomics in the study of the sperm cell composition, differentiation and function. Syst Biol Reprod Med. 2008;54:23–36. doi: 10.1080/19396360701879595. [DOI] [PubMed] [Google Scholar]
- 30.Oliva R, de Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–17. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- 31.Oliva R, Castillo J. Proteomics and the genetics of sperm chromatin condensation. Asian J Androl. 2011;13:24–30. doi: 10.1038/aja.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane M, Robker RL, Robertson SA. Parenting from before conception. Science. 2014;345:756–60. doi: 10.1126/science.1254400. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins TG, Carrell DT. Dynamic alterations in the paternal epigenetic landscape following fertilization. Front Genet. 2012;3:143. doi: 10.3389/fgene.2012.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamauchi Y, Shaman JA, Ward WS. Non-genetic contributions of the sperm nucleus to embryonic development. Asian J Androl. 2011;13:31–5. doi: 10.1038/aja.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–8. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 36.Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, et al. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction. 2011;141:21–36. doi: 10.1530/REP-10-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–4. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 38.Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. Histone- and protamine-DNA association: conservation of different patterns within the beta-globin domain in human sperm. Mol Cell Biol. 1998;18:3350–6. doi: 10.1128/mcb.18.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, et al. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112(Pt 20):3537–48. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- 40.Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213–8. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- 41.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–7. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 42.Nazarov IB, Shlyakhtenko LS, Lyubchenko YL, Zalenskaya IA, Zalensky AO. Sperm chromatin released by nucleases. Syst Biol Reprod Med. 2008;54:37–46. doi: 10.1080/19396360701876849. [DOI] [PubMed] [Google Scholar]
- 43.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, et al. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–49. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–87. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 46.Saida M, Iles D, Elnefati A, Brinkworth M, Miller D. Key gene regulatory sequences with distinctive ontological signatures associate with differentially endonuclease-accessible mouse sperm chromatin. Reproduction. 2011;142:73–86. doi: 10.1530/REP-10-0536. [DOI] [PubMed] [Google Scholar]
- 47.Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erkek S, Hisano M, Liang CY, Gill M, Murr R, et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20:868–75. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 49.Meyer-Ficca ML, Lonchar JD, Ihara M, Bader JJ, Meyer RG. Alteration of poly (ADP-ribose) metabolism affects murine sperm nuclear architecture by impairing pericentric heterochromatin condensation. Chromosoma. 2013;122:319–35. doi: 10.1007/s00412-013-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, et al. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castillo J, Amaral A, Azpiazu R, Vavouri T, Estanyol JM, et al. Genomic and proteomic dissection and characterization of the human sperm chromatin. Mol Hum Reprod. 2014;20:1041–53. doi: 10.1093/molehr/gau079. [DOI] [PubMed] [Google Scholar]
- 52.Samans B, Yang Y, Krebs S, Sarode GV, Blum H, et al. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Dev Cell. 2014;30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 53.Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–6. [PubMed] [Google Scholar]
- 54.Oliva R, Ballescà JL. Altered histone retention and epigenetic modifications in the sperm of infertile men. Asian J Androl. 2012;14:239–40. doi: 10.1038/aja.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–89. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, et al. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliva R, Castillo J. Sperm nucleoproteins. In: Zini A, Agarwal A, editors. Sperm Chromatin Biological and Clinical Applications in Male Infertility and Assisted Reproduction. New York: Springer; 2011. pp. 45–60. [Google Scholar]
- 59.Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem. 2007;102:680–8. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- 60.Mohar I, Szczygiel MA, Yanagimachi R, Ward WS. Sperm nuclear halos can transform into normal chromosomes after injection into oocytes. Mol Reprod Dev. 2002;62:416–20. doi: 10.1002/mrd.10147. [DOI] [PubMed] [Google Scholar]
- 61.Castillo J, Simon L, de Mateo S, Lewis S, Oliva R. Protamine/DNA ratios and DNA damage in native and density gradient centrifuged sperm from infertile patients. J Androl. 2011;32:324–32. doi: 10.2164/jandrol.110.011015. [DOI] [PubMed] [Google Scholar]
- 62.Simon L, Castillo J, Oliva R, Lewis SE. Relationships between human sperm protamines, DNA damage and assisted reproduction outcomes. Reprod Biomed Online. 2011;23:724–34. doi: 10.1016/j.rbmo.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Ribas-Maynou J, García-Peiró A, Martínez-Heredia J, Fernández-Encinas A, Abad C, et al. Nuclear degraded sperm subpopulation is affected by poor chromatin compaction and nuclease activity. Andrologia. 2015;47:286–94. doi: 10.1111/and.12258. [DOI] [PubMed] [Google Scholar]
- 64.Aoki VW, Moskovtsev SI, Willis J, Liu L, Mullen JB, et al. DNA integrity is compromised in protamine-deficient human sperm. J Androl. 2005;26:741–8. doi: 10.2164/jandrol.05063. [DOI] [PubMed] [Google Scholar]
- 65.Torregrosa N, Domínguez-Fandos D, Camejo MI, Shirley CR, Meistrich ML, et al. Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum Reprod. 2006;21:2084–9. doi: 10.1093/humrep/del114. [DOI] [PubMed] [Google Scholar]
- 66.Dubruille R, Orsi GA, Delabaere L, Cortier E, Couble P, et al. Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr Biol. 2010;20:2090–9. doi: 10.1016/j.cub.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 67.Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenet Genome Res. 2004;105:203–14. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- 68.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 2014;1839:155–68. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–69. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 70.Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol. 2007;176:283–94. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013;27:1680–92. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishibashi T, Li A, Eirín-López JM, Zhao M, Missiaen K, et al. H2A.Bbd: an X-chromosome-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Res. 2010;38:1780–9. doi: 10.1093/nar/gkp1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brunner AM, Nanni P, Mansuy IM. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin. 2014;7:2. doi: 10.1186/1756-8935-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samson M, Jow MM, Wong CC, Fitzpatrick C, Aslanian A, et al. The specification and global reprogramming of histone epigenetic marks during gamete formation and early embryo development in C. elegans. PLoS Genet. 2014;10:e1004588. doi: 10.1371/journal.pgen.1004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oliva R, Mezquita C. Histone H4 hyperacetylation and rapid turnover of its acetyl groups in transcriptionally inactive rooster testis spermatids. Nucleic Acids Res. 1982;10:8049–59. doi: 10.1093/nar/10.24.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oliva R, Bazett-Jones D, Mezquita C, Dixon GH. Factors affecting nucleosome disassembly by protamines in vitro. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J Biol Chem. 1987;262:17016–25. [PubMed] [Google Scholar]
- 77.Oliva R, Bazett-Jones DP, Locklear L, Dixon GH. Histone hyperacetylation can induce unfolding of the nucleosome core particle. Nucleic Acids Res. 1990;18:2739–47. doi: 10.1093/nar/18.9.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurtz K, Martínez-Soler F, Ausió J, Chiva M. Acetylation of histone H4 in complex structural transitions of spermiogenic chromatin. J Cell Biochem. 2007;102:1432–41. doi: 10.1002/jcb.21365. [DOI] [PubMed] [Google Scholar]
- 79.Awe S, Renkawitz-Pohl R. Histone H4 acetylation is essential to proceed from a histone- to a protamine-based chromatin structure in spermatid nuclei of Drosophila melanogaster. Syst Biol Reprod Med. 2010;56:44–61. doi: 10.3109/19396360903490790. [DOI] [PubMed] [Google Scholar]
- 80.Goudarzi A, Shiota H, Rousseaux S, Khochbin S. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J Mol Biol. 2014;426:3342–9. doi: 10.1016/j.jmb.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 81.van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–69. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 82.Montellier E, Rousseaux S, Zhao Y, Khochbin S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays. 2012;34:187–93. doi: 10.1002/bies.201100141. [DOI] [PubMed] [Google Scholar]
- 83.Pruslin FH, Imesch E, Winston R, Rodman TC. Phosphorylation state of protamines 1 and 2 in human spermatids and spermatozoa. Gamete Res. 1987;18:179–90. doi: 10.1002/mrd.1120180208. [DOI] [PubMed] [Google Scholar]
- 84.Chirat F, Arkhis A, Martinage A, Jaquinod M, Chevaillier P, et al. Phosphorylation of human sperm protamines HP1 and HP2: identification of phosphorylation sites. Biochim Biophys Acta. 1993;1203:109–14. doi: 10.1016/0167-4838(93)90043-q. [DOI] [PubMed] [Google Scholar]
- 85.Pirhonen A, Linnala-Kankkunen A, Mäenpää PH. Identification of phosphoseryl residues in protamines from mature mammalian spermatozoa. Biol Reprod. 1994;50:981–6. doi: 10.1095/biolreprod50.5.981. [DOI] [PubMed] [Google Scholar]
- 86.Saowaros W, Panyim S. The formation of disulfide bonds in human protamines during sperm maturation. Experientia. 1979;35:191–2. doi: 10.1007/BF01920608. [DOI] [PubMed] [Google Scholar]
- 87.Björndahl L, Kvist U. Structure of chromatin in spermatozoa. Adv Exp Med Biol. 2014;791:1–11. doi: 10.1007/978-1-4614-7783-9_1. [DOI] [PubMed] [Google Scholar]
- 88.Mezquita C, Teng CS. Studies on sex-organ development. Changes in nuclear and chromatin composition and genomic activity during spermatogenesis in the maturing rooster testis. Biochem J. 1977;164:99–111. doi: 10.1042/bj1640099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ando T, Yamasaki M, Suzuki K. Protamines. Isolation, characterization, structure and function. Mol Biol Biochem Biophys. 1973;12:1–114. [PubMed] [Google Scholar]
- 90.Carrell DT, Emery BR, Hammoud S. The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. Int J Androl. 2008;31:537–45. doi: 10.1111/j.1365-2605.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- 91.Oliva R, Mezquita C. Marked differences in the ability of distinct protamines to disassemble nucleosomal core particles in vitro . Biochemistry. 1986;25:6508–11. doi: 10.1021/bi00369a025. [DOI] [PubMed] [Google Scholar]
- 92.Mengual L, Ballescá JL, Ascaso C, Oliva R. Marked differences in protamine content and P1/P2 ratios in sperm cells from percoll fractions between patients and controls. J Androl. 2003;24:438–47. doi: 10.1002/j.1939-4640.2003.tb02692.x. [DOI] [PubMed] [Google Scholar]
- 93.Moradian A, Kalli A, Sweredoski MJ, Hess S. The top-down, middle-down, and bottom-up mass spectrometry approaches for characterization of histone variants and their post-translational modifications. Proteomics. 2014;14:489–97. doi: 10.1002/pmic.201300256. [DOI] [PubMed] [Google Scholar]
- 94.de Yebra L, Oliva R. Rapid analysis of mammalian sperm nuclear proteins. Anal Biochem. 1993;209:201–3. doi: 10.1006/abio.1993.1104. [DOI] [PubMed] [Google Scholar]
- 95.de Mateo S, Gázquez C, Guimerà M, Balasch J, Meistrich ML, et al. Protamine 2 precursors (Pre-P2), protamine 1 to protamine 2 ratio (P1/P2), and assisted reproduction outcome. Fertil Steril. 2009;91:715–22. doi: 10.1016/j.fertnstert.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 96.de Mateo S, Ramos L, van der Vlag J, de Boer P, Oliva R. Improvement in chromatin maturity of human spermatozoa selected through density gradient centrifugation. Int J Androl. 2011;34:256–67. doi: 10.1111/j.1365-2605.2010.01080.x. [DOI] [PubMed] [Google Scholar]
- 97.de Mateo S, Ramos L, de Boer P, Meistrich M, Oliva R. Protamine 2 precursors and processing. Protein Pept Lett. 2011;18:778–85. doi: 10.2174/092986611795713998. [DOI] [PubMed] [Google Scholar]
- 98.Zatecka E, Castillo J, Elzeinova F, Kubatova A, Ded L, et al. The effect of tetrabromobisphenol A on protamine content and DNA integrity in mouse spermatozoa. Andrology. 2014;2:910–7. doi: 10.1111/j.2047-2927.2014.00257.x. [DOI] [PubMed] [Google Scholar]
- 99.de Yebra L, Ballescá JL, Vanrell JA, Corzett M, Balhorn R, et al. Detection of P2 precursors in the sperm cells of infertile patients who have reduced protamine P2 levels. Fertil Steril. 1998;69:755–9. doi: 10.1016/s0015-0282(98)00012-0. [DOI] [PubMed] [Google Scholar]
- 100.Bench G, Corzett MH, De Yebra L, Oliva R, Balhorn R. Protein and DNA contents in sperm from an infertile human male possessing protamine defects that vary over time. Mol Reprod Dev. 1998;50:345–53. doi: 10.1002/(SICI)1098-2795(199807)50:3<345::AID-MRD11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 101.Baker MA, Naumovski N, Hetherington L, Weinberg A, Velkov T, et al. Head and flagella subcompartmental proteomic analysis of human spermatozoa. Proteomics. 2013;13:61–74. doi: 10.1002/pmic.201200350. [DOI] [PubMed] [Google Scholar]
- 102.Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, et al. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–5. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- 103.Baker MA, Reeves G, Hetherington L, Müller J, Baur I, et al. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin Appl. 2007;1:524–32. doi: 10.1002/prca.200601013. [DOI] [PubMed] [Google Scholar]
- 104.de Mateo S, Castillo J, Estanyol JM, Ballescà JL, Oliva R. Proteomic characterization of the human sperm nucleus. Proteomics. 2011;11:2714–26. doi: 10.1002/pmic.201000799. [DOI] [PubMed] [Google Scholar]
- 105.de Mateo S, Estanyol JM, Oliva R. Methods for the analysis of the sperm proteome. Methods Mol Biol. 2013;927:411–22. doi: 10.1007/978-1-62703-038-0_35. [DOI] [PubMed] [Google Scholar]
- 106.Wang G, Guo Y, Zhou T, Shi X, Yu J, et al. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J Proteomics. 2013;79:114–22. doi: 10.1016/j.jprot.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 107.Amaral A, Castillo J, Ramalho-Santos J, Oliva R. The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Hum Reprod Update. 2014;20:40–62. doi: 10.1093/humupd/dmt046. [DOI] [PubMed] [Google Scholar]
- 108.Chocu S, Calvel P, Rolland AD, Pineau C. Spermatogenesis in mammals: proteomic insights. Syst Biol Reprod Med. 2012;58:179–90. doi: 10.3109/19396368.2012.691943. [DOI] [PubMed] [Google Scholar]
- 109.Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, et al. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol. 2003;23:5354–65. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Steilmann C, Cavalcanti MC, Bartkuhn M, Pons-Kühnemann J, Schuppe HC, et al. The interaction of modified histones with the bromodomain testis-specific (BRDT) gene and its mRNA level in sperm of fertile donors and subfertile men. Reproduction. 2010;140:435–43. doi: 10.1530/REP-10-0139. [DOI] [PubMed] [Google Scholar]
- 111.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39:9061–71. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sotolongo B, Huang TT, Isenberger E, Ward WS. An endogenous nuclease in hamster, mouse, and human spermatozoa cleaves DNA into loop-sized fragments. J Androl. 2005;26:272–80. doi: 10.1002/j.1939-4640.2005.tb01095.x. [DOI] [PubMed] [Google Scholar]
- 113.Roca J, Mezquita C. DNA topoisomerase II activity in nonreplicating, transcriptionally inactive, chicken late spermatids. EMBO J. 1989;8:1855–60. doi: 10.1002/j.1460-2075.1989.tb03581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ribas-Maynou J, Gawecka JE, Benet J, Ward WS. Double-stranded DNA breaks hidden in the neutral Comet assay suggest a role of the sperm nuclear matrix in DNA integrity maintenance. Mol Hum Reprod. 2014;20:330–40. doi: 10.1093/molehr/gat090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 116.Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–9. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 117.Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, et al. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS One. 2011;6:e20280. doi: 10.1371/journal.pone.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A. 2012;109:490–4. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, et al. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26:2558–69. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Azpiazu R, Amaral A, Castillo J, Estanyol JM, Guimerà M, et al. High-throughput sperm differential proteomics suggests that epigenetic alterations contribute to failed assisted reproduction. Hum Reprod. 2014;29:1225–37. doi: 10.1093/humrep/deu073. [DOI] [PubMed] [Google Scholar]
- 121.Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44:52–5. doi: 10.1007/BF01960243. [DOI] [PubMed] [Google Scholar]
- 122.Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, et al. Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet. 2014;10:e1004317. doi: 10.1371/journal.pgen.1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kovac JR, Pastuszak AW, Lamb DJ. The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil Steril. 2013;99:998–1007. doi: 10.1016/j.fertnstert.2013.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hud NV, Allen MJ, Downing KH, Lee J, Balhorn R. Identification of the elemental packing unit of DNA in mammalian sperm cells by atomic force microscopy. Biochem Biophys Res Commun. 1993;193:1347–54. doi: 10.1006/bbrc.1993.1773. [DOI] [PubMed] [Google Scholar]
- 125.Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–45. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]


