Abstract
The highly condensed chromatin of mammalian spermatozoa is usually considered to be biologically inert before fertilization. However, we have demonstrated that even in this compacted state, sperm chromatin is subject to degradation at open configurations associated with the nuclear matrix through a process we have termed sperm chromatin fragmentation (SCF). This suggests that a mechanism exists to monitor the health of spermatozoa during transit through the male reproductive tract and to destroy the genome of defective sperm cells. The site of DNA damage in SCF, the matrix attachment sites, are the same that we hypothesize initiate DNA synthesis in the zygote. When sperm that have damaged DNA are injected into the oocyte, the newly created zygote responds by delaying DNA synthesis in the male pronucleus and, if the damage is severe enough, arresting the embryo's development. Here we present a model for paternal DNA regulation by the nuclear matrix that begins during sperm maturation and continues through early embryonic development.
We review evidence based on work from several laboratories that the sperm nuclear matrix, a structural element that organizes the DNA, regulates paternal chromatin function. As has been described many times before, the nuclear matrix is a largely proteinaceous structure that organizes the DNA into loops of 5 to 100 kb, depending on the cell type and the chromosomal region by a series of attachments of the linear DNA.1,2,3 In somatic cells, the nuclear matrix has well documented functions in DNA replication,4,5,6 transcription,7,8,9 transcriptional regulation related to cell differentiation,10 and DNA degradation,6,11 to name just a few. The overall function of the nuclear matrix seems to be to provide a functional scaffold on which to organize the six feet of DNA that exists in mammalian cells so that the information encoded in the DNA can be efficiently accessed and utilized by the cell. While sperm chromatin is traditionally viewed as being completely inert, we have argued that it still needs this organizing matrix to keep the DNA functional after fertilization.12,13 Work from our laboratory, described below, suggests that the fully mature spermatozoon does retain some ability to manipulate the compact chromatin with help from the surrounding luminal fluids of the male reproductive tissues, and this requires the sperm nuclear matrix. We have also provided evidence that the sperm nuclear matrix provides the scaffold for DNA replication in the paternal pronucleus after fertilization.14 We have commented previously on the relationship between these two seemingly disparate events in the sperm, both of which seem to occur at the same chromatin sites in these cells.15 Here, we consider new data to develop a model to describe how DNA degradation and DNA replication are related in the sperm cell.
SPERM CHROMATIN FRAGMENTATION (SCF)
When mammalian sperm are incubated with divalent cations, all of the chromatin is fragmented to about 25 kb, in a process we have termed sperm chromatin fragmentation (SCF) (Figure 1, lanes 2, 4 and 6).16 The remarkable aspect of SCF is that when epididymal sperm are stimulated to undergo SCF, subsequent treatment with EDTA appears to reverse most of the breaks (Figure 1, lanes 3, 5 and 7). This is reminiscent of the reversible topoisomerase II (Top2) - induced breaks that occur in somatic cells as the first step of DNA degradation during apoptosis.11,17 Since Top2 is a nuclear matrix associated protein.7,17,18 this first step of apoptotic DNA degradation occurs on the nuclear matrix, and results in the chromatin being digested to loop-sized fragments, about 50 kb in length. As apoptosis progresses, nucleases interact with Top2 to irreversibly continue the DNA degradation.19,20 If SCF is allowed to progress further in epididymal sperm, a similar type of nonreversible degradation occurs.21 We also see this irreversible degradation in sperm cells as they progress through the reproductive tract. When vas deferens sperm are induced to undergo SCF, the degradation is more complete and cannot be reversed as completely (Figure 1, lanes 9, 11 and 13).
Figure 1.
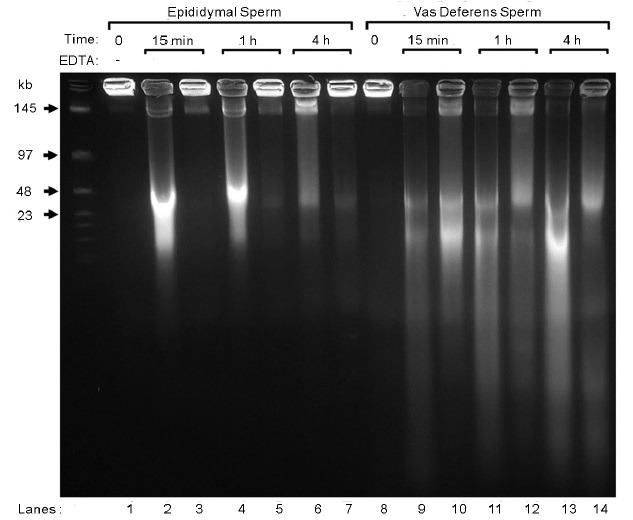
SCF in Epididymal and Vas Deferens Spermatozoa. These data were previously published in Yamauchi, et al.16 Spermatozoa from the epididymis (lanes 1–7) or the vas deferens (lanes 8–14) were embedded in agarose plugs, and incubated in mHCZB (modified Hepes-CZB, CZB medium buffered with HEPES) supplemented with MnCl2 and CaCl2 as described,16 for the times indicated, and treated with or without EDTA to reverse TOP2B-induced breaks. The plugs were then electrophoresed by FIGE.
These two types of DNA degradation in the sperm are closely related. It is possible to induce epididymal sperm to undergo the complete, irreversible DNA degradation that occurs in vas deferens sperm SCF by activating an acidic nuclease in the epididymal lumen.21 In this study, when the complete SCF reaction was allowed to proceed at room temperature, the DNA was degraded to fragments much smaller than the loop-sized 25 kb segments, and this degradation was not reversible. However, when the reaction was slowed by incubation on ice the degradation as limited to the reversible, 25 kb fragmentation. Samples incubated on ice then at room temperature continued to digest the DNA further. This suggests that DNA degradation in the sperm cell follows a similar pattern to that of apoptotic somatic cells. The only difference is that sperm SCF requires the surrounding fluid to supply the nuclease. It is likely that the Top2 required for the first, reversible breaks is already in the sperm chromatin, but is somehow activated to induce the double-strand breaks by an as yet unknown mechanism.
We have recently used the Comet assay to characterize the DNA breaks in SCF in mice. At least two reports used the Comet assay to study DNA breaks in somatic cells with particular attention to whether these breaks remained associated with the nuclear matrix.22,23 Using a modified Comet assay for the sperm cell, we were able to demonstrate that SCF induces both single- and double-stranded DNA breaks.24 The double-stranded DNA breaks in epididymal sperm were largely attached to the sperm nuclear matrix, while those of the vas deferens were released from the nuclear matrix. This is consistent with our model for SCF that double-stranded DNA breaks first occur on the nuclear matrix and subsequent nuclease degradation causes the release of the DNA from this structural scaffold.
The DNA degradation during SCF occurs in the fully condensed chromatin. This suggests that even in this highly condensed state, the sperm cell retains some capability to manipulate the genome. This principle was recently elegantly shown for another enzyme, 8-oxoguanine DNA glycosylase 1 (OGG1).25 When sperm DNA is damaged by oxidative stress, the most common adduct formed is 8-hydroxy-29-deoxyguanosine (8OHdG) and OGG1 is the first enzyme in the normal DNA repair pathway that repairs this defect. OGG1 is present in the sperm nucleus and releases 8OHdG into the extracellular space after oxidative stress. This demonstrates that even in the confined spaces of the condensed chromatin and with the high degree of chromatin packaging resulting from protamine condensation,26,27 the sperm cell has some access to chromatin. In fact, the structure of the highly condensed sperm chromatin may lend itself to breaks at the nuclear matrix attachment sites. When protamines condense sperm chromatin, the DNA is packaged into highly condensed toroids comprised of 25 to 50 kb of DNA.28 This protamine bound DNA is packaged into a highly organized, very compact crystalline-like lattice,26 and is much less sensitive to nuclease degradation than normal chromatin29 and may even be less sensitive to attack by free radicals.30 We, and others, have proposed that these protamine toroids are stacked together in the fully condensed chromatin state similar to a stack of Lifesaver peppermints.12,31 We have also provided evidence that between each protamine toroid, is a nuclease sensitive segment of chromatin we term the toroid linker that is attached to the nuclear matrix.29 According to this model, these toroid linkers represent small sperm chromatin loci that are much more open to nuclease attack than the rest of the protamine bound DNA.
There are two reasons one might consider why this would be important for the sperm cell. During its formation32 and while in transit at fertilization33,34 the sperm cell is susceptible to several types of DNA damage. In the case of OGG1, the sperm cell has the ability to initiate the repair process that may then be completed in the oocyte.25 SCF may be involved in DNA repair when the breaks are restricted to a small number of sites (see below). However, when larger segments of the genome are involved, this mechanism may provide an efficient way to degrade the DNA of dead sperm. SCF requires the luminal fluid of the epididymis or the vas deferens to degrade the DNA fully. Thus, the mechanism to degrade sperm DNA requires both the unique chromatin structure of the sperm cell that protects 90% to 99% of the DNA but leaves a small portion susceptible to attack by nucleases, and the external fluid in which the cells are bathed. This separation of the two components of SCF suggests that only the DNA heavily damaged or dead sperm would be completely degraded when SCF is activated. We hypothesize that SCF is an efficient mechanism for ensuring that so many copies of the complete genome are not floating around either in the male or female reproductive tract when they are no longer needed. The fact that the activation of SCF appears to be outside the cell in the surrounding luminal fluid could be a mechanism for ensuring that the genomes of healthy spermatozoa are protected against premature activation of DNA degradation. Aitken and colleagues have recently proposed that spermatozoa are inherently destined to undergo cell death and must constantly demonstrate their health by producing prosurvival factors.35 SCF may represent the existence of a mechanism to complete the process ensuring that the highly compact DNA of the sperm is not protected when it no longer needs to be.
REPLICATION OF THE PATERNAL GENOME
After fertilization the two maternal and paternal genomes of the zygote replicate independently in two different pronuclei in the same cytoplasm.36 In the mouse, the timing of the initiation of replication in both pronuclei seems to be very similar if not identical, but this is controversial.37,38,39,40,41 We found no differences in the initiation of DNA synthesis in both pronuclei in ICSI-generated mouse zygotes.42 However, it is very clear that it is possible for the zygotic cytoplasm to support asynchronous DNA replication of the two pronuclei if artificially induced to do so. When individual spermatozoa were injected up to 3 h after oocyte activation, the paternal pronucleus was delayed in initiating DNA replication by 2 to 3 h.42,43,44 This is because the sperm nucleus must decondense, and replace all the protamines for histones before replication can begin. The maternal chromatin begins to form its pronucleus as soon as the oocyte is activated. These two events are normally timed to coincide in fully formed pronuclei that are capable of DNA synthesis at the same time. In the case of the artificially delayed paternal DNA replication, shortly after fertilization the zygotic cytoplasm contains two pronuclei in different phases – the paternal pronucleus is at G1 while the maternal pronucleus is in S-phase. The presence of both a G1-phase pronucleus and an S-phase pronucleus in the same cytoplasm raises interesting, and still unanswered, questions about the control that the cytoplasm has on DNA replication in general, with implications for somatic cell DNA synthesis regulation. It also raises questions about how pronuclei communicate with each other in the zygote. This suggests that the pronuclei have independent mechanisms to regulate DNA replication in response to cytoplasmic signals.
A few studies have provided some evidence that the pronuclei do communicate with each other in response to DNA damage, but the mechanisms for this are not known. For example, when the sperm of rats treated with cyclophosphamide were used to fertilize oocytes, the female pronuclei had increased phosphorylation of H2AX, and increased expression of PARP-1.45 When spermatozoa with damaged DNA were used to fertilize oocytes by ICSI, the maternal pronuclei slowed the transition from G2 to M to match that of the paternal pronuclei.46 These studies suggest that the two pronuclei respond to signals from each other during the zygote's progression through its cell cycle.
We have suggested that certain components of the sperm nuclear matrix are inherited by the paternal pronucleus after fertilization that are required for proper DNA replication to occur. The sperm nuclear matrix organizes the DNA into loops that are about 25 kb in size.47,48 The loop organization that is present in the mature sperm cell is the same as that in the round spermatid, so this organization is independent of the protamine condensation that occurs during spermiogenesis.49 We have demonstrated that DNA replication in the paternal pronucleus requires an intact sperm nuclear matrix,14 and have proposed that the origins of paternal DNA replication are determined in part by the attachments sites of DNA to this structure. As described above, this is in keeping with models for DNA replication in somatic cells.4,5,6 Thus, DNA replication initiates at the same chromosomal sites on which degradation of the sperm DNA occurs during SCF.
DNA REPLICATION IS REGULATED BY DNA BREAKS IN THE EMBRYO
If DNA degradation in the sperm cell and DNA replication in the paternal pronucleus are both related to DNA loop attachment sites on the nuclear matrix, it would be reasonable to suggest that these two functions are related. Several laboratories have studied the effects of fertilizing oocytes with spermatozoa that had damaged DNA45,50,51,52 all demonstrating some degree of response to the sperm DNA damage in the zygote. We examined the direct relationship between DNA replication of the paternal pronucleus and damaged sperm DNA. We first reported that when sperm induced to undergo SCF by divalent cations were subsequently used to fertilize oocytes, that the paternal pronuclei failed to replicate in the large majority of zygotes.53 However, when we later followed the zygotes overnight, we noticed that rather than being inhibited, the replication of the paternal pronuclei was severely delayed, even though the maternal pronucleus initiated DNA replication at the normal time.46 As had been noticed previously by other laboratories, we found that sperm with damaged DNA elicited an increased phosphorylation of histone H2AX in the paternal pronucleus.45,46,51
The response of the embryo was related to the level of double-stranded DNA damage in the sperm cell. As mentioned above, epididymal sperm induced to undergo SCF have only reversible double-stranded DNA breaks that remain associated with the nuclear matrix,16,24 and contain a significant amount of single-stranded DNA breaks.24 SCF induced vas deferens sperm also have single-stranded DNA breaks, but the double-stranded breaks are not reversible and are released from the nuclear matrix. We found that when SCF-induced vas deferens sperm were used to fertilize oocytes by ICSI, the paternal DNA replication was severely delayed, but not when SCF-induced epididymal sperm were used.46 In both cases, the formation of the paternal pronucleus was not delayed, so this was not the cause of DNA synthesis delay. Since both types of sperm had similar levels of single-stranded DNA breaks, but different types of double-stranded breaks, it is likely that the delay in DNA replication was a response to the double-stranded breaks being released from the nuclear matrix.
Injection of both epididymal and vas deferens SCF-induced sperm into oocytes resulted in chromosomal aberrations at mitosis.46 As for DNA replication, the severity of the aberrations was also correlated with the degree of DNA damage. Most of the embryos created with SCF-induced epididymal sperm had analyzable chromosome preparations with an average of 5.82 recognizable aberrations per karyotype. However, most (73%) of the embryos created with vas deferens SCF-induced sperm had precondensed chromosomes (PCC) and could not be analyzed. This is characteristic of cells that are forced into mitosis before DNA replication is completed,54,55 and may result because the maternal pronucleus, having completed DNA synthesis, stimulates the zygote to enter M-phase. Finally, the development of the embryos was delayed in both types of embryos, again correlate with the degree of DNA damage. It is likely that if it were possible to measure the initiation of DNA synthesis in both pronuclei more accurately, we would find that even with the lower levels of DNA damage, paternal pronuclei do exhibit some delays.
REGULATION OF PATERNAL CHROMATIN BY THE SPERM NUCLEAR MATRIX
These data demonstrate that paternal pronuclei in zygotes fertilized with sperm that have double-stranded breaks on the sperm nuclear matrix delay the replication of their DNA while maternal DNA in the same cytoplasm replicates normally. There is a clear connection between the presence of DNA breaks in sperm and the regulation of DNA synthesis in the embryo. This is not surprising given that somatic cells contain different mechanisms for handling DNA replication in the presence of double-stranded DNA breaks.56,57,58 In developing a model to hypothesize what our experimentally induced SCF can tell us about true chromatin function it is important to remember that in SCF the entire genome is fragmented to 25 kb or less. SCF-induced spermatozoa are not motile and would not normally fertilize an oocyte. The fact that the zygote can progress through the first cell cycle even with such severely damaged DNA, but that it is delayed, suggests that the zygote does respond to DNA breaks and that it contains a mechanism (s) for progressing through S-phase with the damage. As described above, when the DNA damage is less severe, as in SCF-induced epididymal sperm, there is no detectable delay in DNA synthesis but chromosomal breaks are detected at mitosis demonstrating that DNA synthesis is possible in the zygote with some breaks. Thus, this mechanism does not require the full repair of the DNA breaks because most of the zygotes fertilized with SCF-induced sperm contained broken paternal chromosomes at metaphase even though the maternal chromosomes were intact.46
We hypothesize that the sperm nuclear matrix serves as a checkpoint at multiple stages for chromatin integrity. The unique structure of sperm chromatin results in 90% to 99% of the chromatin being inert and protected against nuclease and physical degradation. The small amount of chromatin that remains susceptible to external influences is made up of the attachment regions to the nuclear matrix. It is not surprising, therefore, that the matrix attachment regions (MARs) regulate the chromatin structure of the mature spermatozoa because this is the only part of the sperm chromatin that is susceptible to any type of manipulation. The sperm MARs appear to contain Top2, an enzyme that is involved in both DNA degradation11,17 and DNA replication,59,60,61 and therefore represent potentially active sites of chromatin modification in mature spermatozoa. As stated above, we hypothesize that these active sites of the sperm chromatin are inherited by the paternal pronucleus after fertilization. We have also suggested that sperm MARs are the sites of initiation of DNA synthesis in the paternal pronucleus. This would structurally link the DNA degradation that occurs in SCF with DNA replication in the zygote.
In this model, the sperm nuclear matrix functions to regulate aspects of the paternal genome related to chromatin integrity. As mentioned above, we propose that SCF functions to deactivate the paternal genome in defective spermatozoa during maturation. There are two factors involved, one in the surrounding fluid and the other the components of the sperm nuclear matrix at the sites of DNA attachment. We have previously suggested that a similar type of chromatin surveillance might function to eliminate sperm that are damaged or infected with pathogens during fertilization.62 SCF, however, is the most severe form of sperm chromatin degradation, and is probably activated only upon sperm death. Even though embryos injected with SCF-induced sperm do eventually progress through the first cell cycle, they do not develop to the blastocyst stage in vitro.46 Therefore, it is not likely that SCF as we have described it is directly related to normal physiological function after fertilization. Our data indicate that when SCF is activated genome-wide, embryos that are injected with the DNA-damaged sperm exhibit some delay of paternal DNA replication initiation in the pronuclei, suggesting that there is a mechanism that halts replication forks in the presence of double-stranded breaks at or near the MARs. This suggests that the same mechanism that deactivates the genome in sperm cell death also signals a zygotic response to the DNA damage. We propose that when a limited number of double-stranded breaks occur in the sperm genome below a certain threshold during spermiogenesis or fertilization due to mechanical stress, the nuclear matrix mediated double-strand breaks are activated at these loci only Figure 2. It is possible, for example, that when spermatozoa undergo single-stranded breaks from exposure to reactive oxygen species (ROS) that the OGG1 repair mechanism also signals the nuclear matrix to induce double-stranded breaks to slow DNA replication enough to allow the DNA to be repaired. After fertilization, these breaks might be repaired following replication delay. Somatic cells have a DNA damage tolerance (DDT) mechanism that allows DNA replication to continue in the presence of double-strand DNA breaks so that the entire cell cycle is not stalled.63,64 We would not expect DDT to be activated in zygotes because this might propagate mutations in the template genome of the embryo. Rather, we would expect the DNA to be repaired.65
Figure 2.
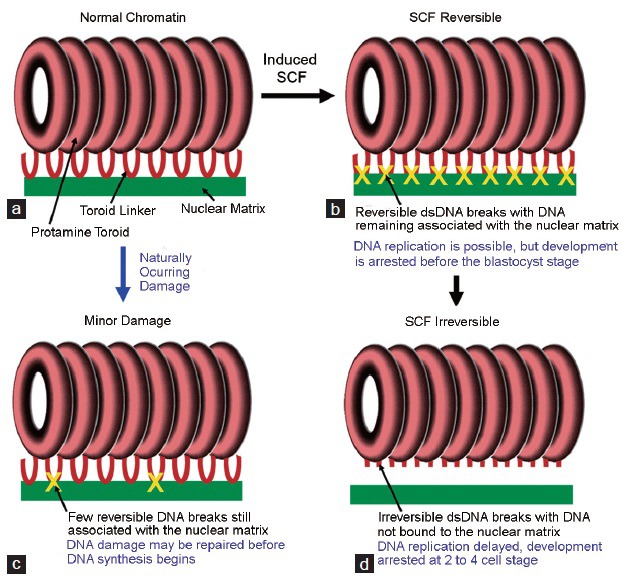
A Model for the nuclear matrix as a regulator for the paternal genome. (a) In normal sperm chromatin most of the DNA is protected by protamines that condense the DNA into tightly compacted toroids. Nuclease sensitive “toroid linkers” connect the toroids to each other, and these are bound to the nuclear matrix. (b) When SCF is induced by treating the sperm with divalent cations, double-stranded DNA breaks are induced at all matrix attachment sites, and the DNA remains bound to the matrix. At this stage, the DNA can still be reversed with EDTA. Zygotes created by injecting sperm at this stage into oocytes can support DNA replication with no evidence for DNA replication delays, but they do not develop to the blastocyst stage. (c) As SCF progresses, the breaks are no longer associated with the nuclear matrix. Zygotes created with these sperm have severe replication and developmental delays. (d) We hypothesize that this signaling mechanism helps the zygote repair minor DNA damage by slowing DNA replication at damaged sites to allow for repair.
It is important to note that the zygote responds to this particular type of DNA damage, double-stranded breaks on the nuclear matrix, more than others. As we have noted before, SCF-induced sperm from both the epididymis and vas deferens have a large amount of single-stranded DNA breaks,24 but only embryos from SCF-induced vas deferens sperm have significant developmental delays.46 Furthermore, when sperm that have significant levels of double-stranded breaks distal from the nuclear matrix but with intact MARs are injected into oocytes, there is no significant replication delay in either pronucleus.14 It appears that the zygote is particularly sensitive to DNA damage associated with the nuclear matrix.
In conclusion, our work on SCF suggests that the sperm nuclear matrix regulates chromatin structure by alerting the zygote to the presence of DNA damage. Strand breaks associated with the nuclear matrix delay DNA replication until either the damage can be repaired, or until development with damaged DNA is no longer possible. We propose that the sperm nuclear matrix is a checkpoint for the integrity of the sperm chromatin.
ACKNOWLEDGEMENTS
This work was supported by a grant from the U.S. National Institutes of Health (NIH) number HD060722-01.
COMPETING FINANCIAL INTERESTS
There are no competing financial interests.
REFERENCES
- 1.Getzenberg RH. Nuclear matrix and the regulation of gene expression: tissue specificity. J Cell Biochem. 1994;55:22–31. doi: 10.1002/jcb.240550105. [DOI] [PubMed] [Google Scholar]
- 2.Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–74. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WG, Pienta KJ, Barrack ER, Coffey DS. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–75. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- 4.Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19:527–36. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- 5.Dijkwel PA, Hamlin JL. Origins of replication and the nuclear matrix: the DHFR domain as a paradigm. Int Rev Cytol. 1995;162A:455–84. doi: 10.1016/s0074-7696(08)61236-x. [DOI] [PubMed] [Google Scholar]
- 6.Wilson RH, Coverley D. Relationship between DNA replication and the nuclear matrix. Genes Cells. 2013;18:17–31. doi: 10.1111/gtc.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–82. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 8.Choi J, Yang ES, Cha K, Whang J, Choi WJ, et al. The nuclear matrix protein, NRP/B, acts as a transcriptional repressor of E2F-mediated transcriptional activity. J Cancer Prev. 2014;19:187–98. doi: 10.15430/JCP.2014.19.3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty S, Das K, Saha S, Mazumdar M, Manna A, et al. Nuclear matrix protein SMAR1 represses c-Fos-mediated HPV18 E6 transcription through alteration of chromatin histone deacetylation. J Biol Chem. 2014;289:29074–85. doi: 10.1074/jbc.M114.564872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–88. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 11.Li TK, Chen AY, Yu C, Mao Y, Wang H, et al. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev. 1999;13:1553–60. doi: 10.1101/gad.13.12.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–6. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamauchi Y, Shaman JA, Ward WS. Non-genetic contributions of the sperm nucleus to embryonic development. Asian J Androl. 2011;13:31–5. doi: 10.1038/aja.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem. 2007;102:680–8. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- 15.Shaman JA, Yamauchi Y, Ward WS. Function of the sperm nuclear matrix. Arch Androl. 2007;53:135–40. doi: 10.1080/01485010701329378. [DOI] [PubMed] [Google Scholar]
- 16.Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal pronuclear DNA degradation is functionally linked to DNA replication in mouse oocytes. Biol Reprod. 2007;77:407–15. doi: 10.1095/biolreprod.107.061473. [DOI] [PubMed] [Google Scholar]
- 17.Solovyan VT, Bezvenyuk ZA, Salminen A, Austin CA, Courtney MJ. The role of topoisomerase II in the excision of DNA loop domains during apoptosis. J Biol Chem. 2002;277:21458–67. doi: 10.1074/jbc.M110621200. [DOI] [PubMed] [Google Scholar]
- 18.Earnshaw WC, Halligan B, Cooke CA, Heck MM, Liu LF. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J Cell Biol. 1985;100:1706–15. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem. 2005;94:1078–87. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 20.Varecha M, Potešilová M, Matula P, Kozubek M. Endonuclease G interacts with histone H2B and DNA topoisomerase II alpha during apoptosis. Mol Cell Biochem. 2012;363:301–7. doi: 10.1007/s11010-011-1182-x. [DOI] [PubMed] [Google Scholar]
- 21.Boaz SM, Dominguez K, Shaman JA, Ward WS. Mouse spermatozoa contain a nuclease that is activated by pretreatment with EGTA and subsequent calcium incubation. J Cell Biochem. 2008;103:1636–45. doi: 10.1002/jcb.21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson D, Laubenthal J. Analysis of DNA damage via single-cell electrophoresis. Methods Mol Biol. 2013;1054:209–18. doi: 10.1007/978-1-62703-565-1_14. [DOI] [PubMed] [Google Scholar]
- 23.Afanasieva K, Zazhytska M, Sivolob A. Kinetics of comet formation in single-cell gel electrophoresis: loops and fragments. Electrophoresis. 2010;31:512–9. doi: 10.1002/elps.200900421. [DOI] [PubMed] [Google Scholar]
- 24.Ribas-Maynou J, Gawecka JE, Benet J, Ward WS. Double-stranded DNA breaks hidden in the neutral Comet assay suggest a role of the sperm nuclear matrix in DNA integrity maintenance. Mol Hum Reprod. 2014;20:330–40. doi: 10.1093/molehr/gat090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith TB, Dun MD, Smith ND, Curry BJ, Connaughton HS, et al. The presence of a truncated base excision repair pathway in human spermatozoa that is mediated by OGG1. J Cell Sci. 2013;126:1488–97. doi: 10.1242/jcs.121657. [DOI] [PubMed] [Google Scholar]
- 26.Hud NV, Downing KH. Cryoelectron microscopy of lambda phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc Natl Acad Sci U S A. 2001;98:14925–30. doi: 10.1073/pnas.261560398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilfan ID, Conwell CC, Hud NV. Formation of native-like mammalian sperm cell chromatin with folded bull protamine. J Biol Chem. 2004;279:20088–95. doi: 10.1074/jbc.M312777200. [DOI] [PubMed] [Google Scholar]
- 28.Hud NV, Downing KH, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc Natl Acad Sci U S A. 1995;92:3581–5. doi: 10.1073/pnas.92.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sotolongo B, Lino E, Ward WS. Ability of hamster spermatozoa to digest their own DNA. Biol Reprod. 2003;69:2029–35. doi: 10.1095/biolreprod.103.020594. [DOI] [PubMed] [Google Scholar]
- 30.De Iuliis GN, Thomson LK, Mitchell LA, Finnie JM, Koppers AJ, et al. DNA damage in human spermatozoa is highly correlated with the efficiency of chromatin remodeling and the formation of 8-hydroxy-2’-deoxyguanosine, a marker of oxidative stress. Biol Reprod. 2009;81:517–24. doi: 10.1095/biolreprod.109.076836. [DOI] [PubMed] [Google Scholar]
- 31.Zalensky A, Zalenskaya I. Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans. 2007;35:609–11. doi: 10.1042/BST0350609. [DOI] [PubMed] [Google Scholar]
- 32.Sakkas D, Alvarez JG. Sperm DNA fragmentation: mechanisms of origin, impact on reproductive outcome, and analysis. Fertil Steril. 2010;93:1027–36. doi: 10.1016/j.fertnstert.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 33.Wright C, Milne S, Leeson H. Sperm DNA damage caused by oxidative stress: modifiable clinical, lifestyle and nutritional factors in male infertility. Reprod Biomed Online. 2014;28:684–703. doi: 10.1016/j.rbmo.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Aitken RJ, Bronson R, Smith TB, De Iuliis GN. The source and significance of DNA damage in human spermatozoa; a commentary on diagnostic strategies and straw man fallacies. Mol Hum Reprod. 2013;19:475–85. doi: 10.1093/molehr/gat025. [DOI] [PubMed] [Google Scholar]
- 35.Pujianto DA, Curry BJ, Aitken RJ. Prolactin exerts a prosurvival effect on human spermatozoa via mechanisms that involve the stimulation of Akt phosphorylation and suppression of caspase activation and capacitation. Endocrinology. 2010;151:1269–79. doi: 10.1210/en.2009-0964. [DOI] [PubMed] [Google Scholar]
- 36.Sirlin JL, Edwards RG. Timing of DNA synthesis in ovarian oocyte nuclei and pronuclei of the mouse. Exp Cell Res. 1959;18:190–4. doi: 10.1016/0014-4827(59)90308-8. [DOI] [PubMed] [Google Scholar]
- 37.Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote. 1999;7:165–72. doi: 10.1017/s0967199499000532. [DOI] [PubMed] [Google Scholar]
- 38.Bouniol-Baly C, Nguyen E, Besombes D, Debey P. Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp Cell Res. 1997;236:201–11. doi: 10.1006/excr.1997.3708. [DOI] [PubMed] [Google Scholar]
- 39.Ferreira J, Carmo-Fonseca M. Genome replication in early mouse embryos follows a defined temporal and spatial order. J Cell Sci. 1997;110(Pt 7):889–97. doi: 10.1242/jcs.110.7.889. [DOI] [PubMed] [Google Scholar]
- 40.Luthardt FW, Donahue RP. Pronuclear DNA synthesis in mouse eggs. An autoradiographic study. Exp Cell Res. 1973;82:143–51. doi: 10.1016/0014-4827(73)90256-5. [DOI] [PubMed] [Google Scholar]
- 41.Howlett SK, Bolton VN. Sequence and regulation of morphological and molecular events during the first cell cycle of mouse embryogenesis. J Embryol Exp Morphol. 1985;87:175–206. [PubMed] [Google Scholar]
- 42.Yamauchi Y, Ward MA, Ward WS. Asynchronous DNA replication and origin licensing in the mouse one-cell embryo. J Cell Biochem. 2009;107:214–23. doi: 10.1002/jcb.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishigami S, Wakayama S, Nguyen VT, Wakayama T. Similar time restriction for intracytoplasmic sperm injection and round spermatid injection into activated oocytes for efficient offspring production. Biol Reprod. 2004;70:1863–9. doi: 10.1095/biolreprod.103.025171. [DOI] [PubMed] [Google Scholar]
- 44.Maleszewski M, Borsuk E, Koziak K, Maluchnik D, Tarkowski AK. Delayed sperm incorporation into parthenogenetic mouse eggs: sperm nucleus transformation and development of resulting embryos. Mol Reprod Dev. 1999;54:303–10. doi: 10.1002/(SICI)1098-2795(199911)54:3<303::AID-MRD11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 45.Barton TS, Robaire B, Hales BF. DNA damage recognition in the rat zygote following chronic paternal cyclophosphamide exposure. Toxicol Sci. 2007;100:495–503. doi: 10.1093/toxsci/kfm242. [DOI] [PubMed] [Google Scholar]
- 46.Gawecka JE, Marh J, Ortega M, Yamauchi Y, Ward MA, et al. Mouse zygotes respond to severe sperm DNA damage by delaying paternal DNA replication and embryonic development. PLoS One. 2013;8:e56385. doi: 10.1371/journal.pone.0056385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nadel B, de Lara J, Finkernagel SW, Ward WS. Cell-specific organization of the 5S ribosomal RNA gene cluster DNA loop domains in spermatozoa and somatic cells. Biol Reprod. 1995;53:1222–8. doi: 10.1095/biolreprod53.5.1222. [DOI] [PubMed] [Google Scholar]
- 48.Kramer JA, Krawetz SA. Nuclear matrix interactions within the sperm genome. J Biol Chem. 1996;271:11619–22. doi: 10.1074/jbc.271.20.11619. [DOI] [PubMed] [Google Scholar]
- 49.Klaus AV, McCarrey JR, Farkas A, Ward WS. Changes in DNA loop domain structure during spermatogenesis and embryogenesis in the Syrian golden hamster. Biol Reprod. 2001;64:1297–306. doi: 10.1095/biolreprod64.5.1297. [DOI] [PubMed] [Google Scholar]
- 50.Grenier L, Robaire B, Hales BF. Paternal exposure to cyclophosphamide affects the progression of sperm chromatin decondensation and activates a DNA damage response in the prepronuclear rat zygote. Biol Reprod. 2010;83:195–204. doi: 10.1095/biolreprod.109.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mu XF, Jin XL, Farnham MM, Li Y, O’Neill C. DNA damage-sensing kinases mediate the mouse 2-cell embryo's response to genotoxic stress. Biol Reprod. 2011;85:524–35. doi: 10.1095/biolreprod.110.089334. [DOI] [PubMed] [Google Scholar]
- 52.Adiga SK, Toyoshima M, Shiraishi K, Shimura T, Takeda J, et al. p21 provides stage specific DNA damage control to preimplantation embryos. Oncogene. 2007;26:6141–9. doi: 10.1038/sj.onc.1210444. [DOI] [PubMed] [Google Scholar]
- 53.Yamauchi Y, Shaman JA, Ward WS. Topoisomerase II-mediated breaks in spermatozoa cause the specific degradation of paternal DNA in fertilized oocytes. Biol Reprod. 2007;76:666–72. doi: 10.1095/biolreprod.106.057067. [DOI] [PubMed] [Google Scholar]
- 54.Schlegel R, Pardee AB. Caffeine-induced uncoupling of mitosis from the completion of DNA replication in mammalian cells. Science. 1986;232:1264–6. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- 55.Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci U S A. 2001;98:9092–7. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castel SE, Ren J, Bhattacharjee S, Chang AY, Sánchez M, et al. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell. 2014;159:572–83. doi: 10.1016/j.cell.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziv O, Zeisel A, Mirlas-Neisberg N, Swain U, Nevo R, et al. Identification of novel DNA-damage tolerance genes reveals regulation of translesion DNA synthesis by nucleophosmin. Nat Commun. 2014;5:5437. doi: 10.1038/ncomms6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson WG, Liu LF, Coffey DS. Newly replicated DNA is associated with DNA topoisomerase II in cultured rat prostatic adenocarcinoma cells. Nature. 1986;322:187–9. doi: 10.1038/322187a0. [DOI] [PubMed] [Google Scholar]
- 60.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol. 2003;160:645–55. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–45. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 62.Ward MA, Ward WS. A model for the function of sperm DNA degradation. Reprod Fertil Dev. 2004;16:547–54. doi: 10.10371/RD03072. [DOI] [PubMed] [Google Scholar]
- 63.Saugar I, Ortiz-Bazán MÁ, Tercero JA. Tolerating DNA damage during eukaryotic chromosome replication. Exp Cell Res. 2014;329:170–7. doi: 10.1016/j.yexcr.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Derijck A, van der Heijden G, Giele M, Philippens M, de Boer P. DNA double-strand break repair in parental chromatin of mouse zygotes, the first cell cycle as an origin of de novo mutation. Hum Mol Genet. 2008;17:1922–37. doi: 10.1093/hmg/ddn090. [DOI] [PubMed] [Google Scholar]


