Abstract
Gene disruption technology has long been beneficial for the study of male reproductive biology. However, because of the time and cost involved, this technology was not a viable method except in specialist laboratories. The advent of the CRISPR/Cas9 system of gene disruption has ushered in a new era of genetic investigation. Now, it is possible to generate gene-disrupted mouse models in very little time and at very little cost. This Highlight article discusses the application of this technology to study the genetics of male fertility and looks at some of the future uses of this system that could be used to reveal the essential and nonessential genetic components of male reproductive mechanisms.
Mice are one of the most ideal organisms to study mammalian reproduction. This is because of their relatively fast reproductive cycle coupled with their similarity to the human genome.1 However, reproduction remains one of the most complex yet poorly understood biological processes, despite decades of dedicated research. Numerous genes have been thought to play essential roles in fertilization, because of their localization or specific expression in the male and/or female gonads, yet analysis of their specific roles has been problematic due to the difficulty in maintaining gametes and embryos in vitro.2
Thus, gene manipulation experiments in animal models have played an essential role in the investigation of reproductive processes. Reproduction is arguably one of the best-suited biological systems to which gene knockout (KO) can be applied, for several reasons. First, genes essential for fertility are most often highly specific to the gonads, eliminating the need for conditional gene knockout models to be utilized. Often spermatogenic or haploid male germ cell genes are comprised of a single exon, eliminating the issues of alternate splicing. Another benefit of utilizing gene-disrupted mice is the incidental discovery of genes involved in reproduction by groups investigating other body systems.3,4 The breeding schemes involved in producing homozygous genetic manipulations for any target gene automatically highlight genes involved in gametogenesis, fertilization and pregnancy. Whatever the reason, there is a higher level of specific homologues in reproductively related genes than in those coding for somatic cell phenotypes, making the use of gene-disruption essential to uncover the truly essential factors of fertility. This review will look at the history of gene manipulation techniques for the study of mammalian reproduction, with a focus on discoveries in the male system and analyze the application of the latest in these technologies, the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR associated (Cas) protein number 9) system.
GENE-DISRUPTION DISCOVERIES IN MALE FACTOR FERTILITY
Disruption of genes involved in male gamete development has shown how highly complex male fertility really is. For example, specific factors involved in premeiotic stages were found to be essential for fertility, such as Pi3K (the KO of which lead to impaired spermatogonia proliferation and increased apoptosis of spermatogonia) and Ddx4 (leading to impaired premeiotic germ cell differentiation and increased apoptosis of spermatogonia) have been shown to contribute to male infertility.5,6
Meiotic defects are well known to result in aneuploidy, leading to embryonic death or developmental issues in offspring.1 KO studies of genes thought to be involved in meiosis also revealed several contributors to male infertility such as Spo11 (impaired double-stranded break (DSB) formation and initiation of recombination), Prdm9 (impaired chromosomal synapsis and sex body formation), and Eif4g3 (meiotic arrest, spermatocytes failure to exit prophase via G2/M1 transition).1,7,8,9
As more has been discovered about postmeiotic and posttesticular maturation processes, KO mouse models have played a significant role in clarifying the complexities of the many mechanisms involved. In terms of capacitation, there is a multitude of evidence supporting the role of calcium and the calcium channel CATSPER in onset of hyperactivation10,11 but there is also evidence suggesting calcium release from intracellular stores may be induced by NO- or cAMP-mediated pathways.11,12 Structurally, in vitro and microscopic analyses have shown various abnormalities in the axonemes of spermatozoa from infertile men and mouse KO studies have revealed the mechanisms behind them.13 An example of this is the KOs of the genes encoding the Dynein heavy chain (Dnahc) family of proteins. Dyneins are ubiquitously expressed in all ciliated cells and are linked to respiratory conditions such as primary ciliary dyskinesia (PCD). Male patients with PCD often present with fertility issues and a specific KO for Dnahc7; a dynein heavy chain protein found in the axoneme of the principal piece of the sperm flagellum, resulted in mice with an infertility phenotype.13,14
One of the main impairments found in the Dnach7 KO mouse was the inability to move from the uterus to the oviduct, despite displaying normal fertilizing ability in vitro.13 A similar phenotype has been found in at least 13 other genes, where KO studies demonstrate an inability to cross the uterotubal junction (UTJ).15 Unlike the dynein mutation (which leads to direct impairment of motility by affecting the structure of the flagellum), these mutations where all found to have one thing in common: the loss or inactivation of Adam3 (for a full review of this phenomenon see Okabe 2014). Adam3 is a sperm surface protein that appears to be essential for sperm migration into the oviduct, yet Adam3 is only a pseudogene in humans. Another KO mouse model might hold the answer to this conundrum, namely the lymphocyte antigen 6 complex, locus K (Ly6K) KO mouse.15,16 This mouse line demonstrated the same phenotype of impaired UTJ migration yet Adam3 expression was not affected, indicating that there must be another mechanism exists that drives this particular part of the fertilization process.
What happens when the spermatozoon meets the egg is still not fully understood, despite years of dedicated research. Only one sperm-specific factor has thus far been identified as being completely essential for sperm-egg fusion: Izumo1.17 Recently the binding partner of Izumo1 on the oocyte was discovered and named Juno.18 Both of these proteins were initially discovered using in vitro techniques but were only considered confirmed once the KO mouse models had been produced and shown to have the predicted phenotype.
Over the past couple of decades over 10 000 genes out of the 23–25 000 have been disrupted in mice (IMPC website accessed 24/10/14). In 2003, it was estimated that over 2300 genes are involved in spermatogenesis alone – not to mention sperm maturation, capacitation and sperm-egg interaction.19,20
NEW TECHNOLOGY – THE CRISPR/CAS9 SYSTEM
CRISPR/Cas9 development and adaptation
Since early 2013, gene-disruption technology has advanced to a new level, allowing the mutation or ablation of the mouse genome to be achieved faster and easier than ever.21 Until, the main method of gene modification in mammals was achieved through manipulation of the target cells own homologous recombination machinery, via microinjection of embryonic stem (ES) cells.22 This had been the most efficient and often used method of generating gene-disrupted mice.23 The most current technology for creating genetically modified organisms is the “clustered regulatory interspaced short palindromic repeats” or CRISPR/Cas9 system of gene-disruption. This system is faster and just as efficient (if not more so) than the traditional use of ES cells.21,24,25
This novel method is a genome editing system adapted from the Streptococcus pyrogenes bacterium, and has been adapted to work in a similar manner on the mammalian genome.21 CRISPRs work in tandem with an endonuclease; CRISPR-associated (Cas) proteins.26 There are three types of CRISPR systems, with the most commonly used known as CRISPR type II, as this system associates with a single Cas endonuclease, Cas9.27,28,29 In the original bacterial system, the chimeric CRISPR RNAs contain a “guide” sequence homologous to the genome of the invading pathogen.21 The system works by Cas9 unfolding and running along the host DNA until reaching a region complementary to the CRISPR guide sequence, which then binds to and cleaves the target site by Watson–Crick base pairing and the endonuclease activity of the Cas9 protein.24,30 This system has been adapted to introduce a genetic manipulation in mammalian cells by designing CRISPR “guide” sequences from the organisms own genome.27,29 These “guide” sequences (originally named “protospacer” sequences) follow a specific pattern; 20 nucleotides (N) followed by NGG (20N-NGG).29 This motif appears frequently in the genome of the mouse – theoretically every eight nucleotides – allowing CRISPR signal sequences to be selected at the start, middle, or end of a gene sequence. The final “NGG” sequence is known as the protospacer adjacent motif (PAM) and is important as Cas9 can only cleave if the correct PAM sequence is present at the 3′ end of the target.29
CRISPR/Cas9 basic functions and benefits
The CRISPR/Cas9 system is able to target the mammalian genome following either microinjection of a plasmid vector expressing the guide sequence, a specific promoter and the humanized Cas9 endonuclease (e.g., the px330 vector available from Addgene: http://www.addgene.org/42230/) or co-injection of the CRISPR guide RNA (containing the guide sequence) and the mRNA of the Cas9 vector separately into the cytoplasm or pronucleus of a fertilized oocyte.21,27
Following cleavage of the target region the double-stranded break (DSB) is repaired by error-prone nonhomologous end joining (NHEJ) or, if the vector is co-injected with an oligonucleotide with a sequence of high homology to the targeted gene, homology directed repair (HDR) occurs.21,27
The first step, NHEJ, achieves gene knockout whereas the second HDR step results in very specific mutations being introduced into the host genome (Figure 1).21 The many benefits of CRISPR/Cas9 include cutting out a significant portion of the time to make a knockin/KO animal.21 The usual process of creating a vector, transfecting ES cells, growing and aggregation of ES cells with preimplantation embryos, transplanting to a pseudopregnant female and waiting for pups to be born, then confirming the germline transmission by mating chimeric mice31,32 can be lessened as CRISPR/Cas9 is used on fertilized eggs that can be implanted directly into a recipient female mouse.21,27,29 The conventional method can take up to 1–2 years, whereas the CRISPR/Cas9 system takes only 1–2 months to produce homozygous and heterozygous mutant mouse lines (Figure 2).
Figure 1.
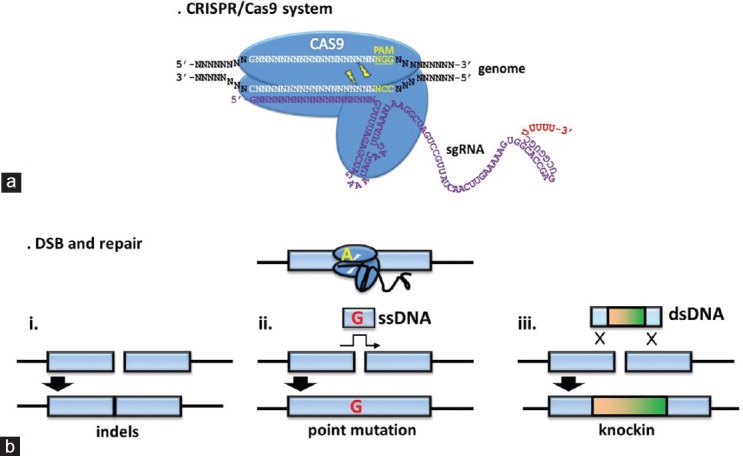
The CRISPR/Cas9 system. (a) The CRISPR sequence and the associated Cas9 endonuclease guide sequence (white) consisting of 20N followed by the PAM sequence (yellow) of NGG. (b) Different uses of CRISPR/Cas9: (i) NHEJ resulting in an insertion/deletion (indel); (ii) HDR when co-injected with a single-stranded DNA (ssDNA) resulting in a point mutation; and (iii) HDR when co-injected with a double-stranded DNA (dsDNA) results in the insertion of a small DNA sequence.
Figure 2.
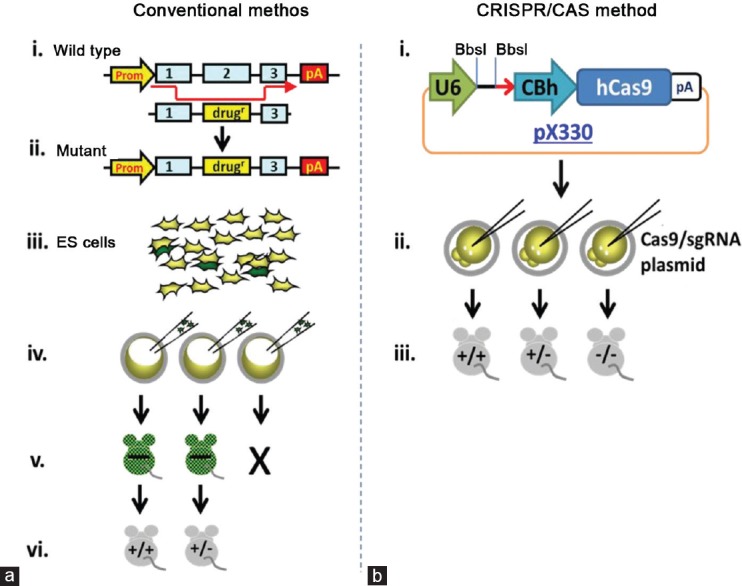
Conventional versus CRISPR/Cas9 method of gene-disruption. (a) (i and ii): design and construction of a vector; (iii) transfection of ES cells with a vector, followed by multiple rounds of positive and negative selection; (iv) injection of ES cells carrying the mutation into fertilized oocytes; (v) chimeric mice born and bred; and (vi) generation of heterozygous/homozygous mice for analysis. This process takes 1–2 years. (b) (i): design and production of a CRISPR/Cas9 plasmid (the guide sequence is inserted between BbsI restriction enzyme sites); (ii) the CRISPR/Cas9 plasmid is either injected alone if NHEJ is desired, or co-injected with an oligonucleotide if HDR is desired; (iii) plasmid or RNA is injected into the fertilized oocyte; and (iv) the founder generation contains heterozygous/homozygous mice for analysis and proliferation. This process takes only 1–2 months.
This system also has the potential to allow for the creation of specific mutations, again shortening the time for mutational experimentation. This, coupled with the ability to generate a homozygous mutant in the founder generation also reduces time and costs (both financial and in terms of animal lives). Germline transmission is guaranteed. The sensitivity of this system also allows researchers to develop mouse models with a “scalpel” rather than a “hammer”, investigating subtle genetic factors such as point mutations, and gene clusters, as well as allowing the specific targeting of the maternal or paternal contributing genomes rather than just total gene deletion or up-regulation.21,33
CRISPR AND REPRODUCTION
Current uses and benefits for reproduction
An important point to consider when utilizing this new technology for studying reproduction is that it is estimated that more than 50% of candidate genes for fertility will produce no discernible phenotype when disrupted.15 When using the conventional method of gene-disruption this often meant months or years of work resulting in the dreaded conclusion “no phenotype”. With the speed and ease of the CRISPR/Cas9 system this has turned around, with the ability to discover whether a gene product is worth pursuing further within a matter of months (i.e., produces a fertility/infertility phenotype), as well as introducing the ability to generate point mutations quickly to investigate the finer detail of potential candidate genes for fertility.
A straight KO of genes has been achieved in a large number of mice in a short amount of time (approximately 200 genes in the past year in our laboratory alone34). However, a large issue in reproduction-related genes is the high level of redundancy that is apparent.35 Current theory posits that this redundancy is an evolutionary advantage, as animals need to be able to produce offspring to proliferate the species, therefore requiring multiple fall-back mechanisms in case of damage to the genome or reproductive systems.35 The use of CRISPR/Cas9 can circumvent this issue by allowing one to create mutations on multiple sites simultaneously.21,36 This was previously impossible to achieve in a single round of conventional methods. Utilizing these conventional methods required the production of independent mutant mouse lines that required multiple crossbreeding to obtain mice carrying multiple mutations for analysis. CRISPR/Cas9 can target multiple sites simultaneously, as demonstrated by Wang et al. where they targeted the Tet1 and Tet2 genes simultaneously. As CRISPR/Cas9 can produce homozygous mice in the founder generation: this not only speeds up analysis considerably, but also allows researchers to expand their list of target genes, generating more data than ever before.
In terms of reproduction, this allows targeting of multiple genes to elucidate the combined effects or roles of genes on fertility, or to make connections between seemingly unrelated genes and analyzing their combined effect on reproduction. An example of this exists in the study of growth differentiation factor 9 (Gdf9) and bone morphogenetic protein 15 (Bmp15) in the female reproductive system. The balance in the products of these genes is important for normal folliculogenesis as well as other important preovulatory events.37 This was discovered through the use of gene-disrupted mice.37 However, in the case of human disease conditions, they are rarely caused by a complete gene deletion. More often than not it is a mutation in one or more genes that causes or contributes to human phenotypes. Advances in proteomic studies allows us to determine where these point mutations may be occurring, providing a guide for the generation of mutant mice.38 Injection of the CRISPR/Cas9 plasmid along with an oligonucleotide containing a desired mutation allows the generation of mice carrying this mutation that can pass it on to their offspring.39 There is no need for the transgenic integration steps of the conventional methods of gene manipulation, which have always been problematic because of the difficulty in controlling the site of integration and the number of gene copies inserted into the host genome.40
In summary, the CRISPR/Cas9 system is currently used to generate KO mice to study the roles of individual genes as well as clarifying how multiple genes act together. This is done via the basic NHEJ repair mechanisms triggered by cleavage of DNA. It is also used to generate mouse models of human conditions by introducing point mutations via the HDR mechanism. This second protocol is also being used to analyze more complex aspects of genetics, such as posttranslational modifications. Nevertheless, what complications are there with this system?
Complications
While having many advantages, there are some cases in which use of the CRISPR/Cas9 system cannot achieve the desired genetic disruption. One such outcome is conditional gene-disruption, which at the time of writing has been achieved via the CRISPR/Cas9 system but is inefficient.41,42 This is desirable when investigating reproductive genes that are also expressed in somatic cells. For example, many proteins involved in the structure and function of the sperm flagellum are often also expressed in other ciliated cells. One such example is the Dynein family of proteins. A complete KO of a Dynein family member (dynein axonemal intermediate chain 1; Dnaic1) in the mouse produced mice with severe hydrocephaly, sinus abnormalities, and developmental defects.43 To analyze the effects of this gene on ciliary (and flagella) movement a conditional KO was required that allowed mice to develop to adulthood.43
Conventional methods of gene-disruption allow conditional mutations via use of the Cre/LoxP system.20,33 The problem with adapting these systems to CRISPR/Cas9 is the need to insert LoxP sites surrounding the target gene on the same allele.44 Yang et al. attempted this by integrating LoxP sites into the Mecp2 gene. This was done via co-injection of an oligonucleotide containing the LoxP sequences and four guide sequences targeting intron 2 and exon 3. The integration was successful (as was subsequent conditional deletion via Cre-mediated recombination) demonstrating that the theory is sound; however, the rate of successful integration of both LoxP sites on the same allele was low.41 There was also a high incidence of mice lacking the LoxP insertion that still had mutations produced by NHEJ. However, a conditionally disrupted mouse was achieved in a single generation, and with continued research a simpler method should be available very soon.
As with any genetic manipulation, researchers must be careful that the phenotype observed is indeed caused by disruption of the target region. The risk of off-target cleavage from the CRISPR/Cas9 system is high enough that researchers have warned against its use (the current methods) in treating human patients.45 Nevertheless, for the creation of mouse models there are several ways in which the incidence of off-target cleavage can be limited, such as utilizing a different guide sequence or attempting transgenic rescue following backcrossing.29,34,46 Indeed for studying male reproduction, many genes involved in spermatogenesis are highly specific to the testis, further limiting the risk of off-target cleavage.
FUTURE APPLICATIONS AND CONCLUSIONS
The CRISPR/Cas9 system has been used on mammalian cells since early 2013. Since then, the technology has progressed from introducing a single gene-disruption to targeting multiple genes across multiple sites on the genome.36 It has been used to insert specific point mutations, and research has begun on creating conditional KO models (see above). There is no sign of slowing down. Different research groups have modified the Cas9 endonuclease to allow the system to perform various tasks, previously unable to be done by conventional methods. By deactivating this nuclease (dCas9), the targeting machinery can guide the CRISPR/dCas9 complex to bind to a specific region of the genome, blocking binding of transcription factors and effectively halting gene transcription.25 This has been expanded by the addition of an effector protein to the dCas9 – an activator or repressor.47 This allows researchers even more control of gene action. Another expansion is the addition of a fluorescent tag to the dCas9, permitting the real-time visualization of gene location and movement in live cells for the first time.48
These modifications have great potential for the study of reproduction. In analyzing genes whose role is suspected in spermatogenesis, use of the CRISPR/dCas9 transcriptional blockage allows in-depth analysis of what roles – if any – candidate genes play. The visualization of sperm-specific genes allows their chromosomal location to be determined. Utilizing the CRISPR/dCas9 with a fluorescent tag might allow the location of candidate genes in spermatogenesis, which in turn could highlight genes of importance to reproductive mechanisms.
As mentioned above, it is estimated that at least 50% of gene-disrupted mouse models show no clear phenotype. The strategies discussed here might help clarify what genes and/or gene components are important for reproduction. However, there is still difficulty in controlling double-stranded DNA (dsDNA) integration. The conventional method of ES cell manipulation remains the most efficient way of inducing these knock-in models. CRISPR/Cas9 is currently being combined with the conventional method to disrupt genes in ES cells, allowing confirmation of mutations before transplantation to recipient female mice (unpublished data). The advantage of combining these technologies is the ability to analyze the chimeric mice produced from ES cells (where the homozygous KO of a gene is confirmed) for a fertility phenotype in much less time than conventional ES cell mutations.
Overall, the CRISPR/Cas9 methodology will allow the investigation of reproductive mechanisms to be achieved much faster and more easily than before. With the ability to target multiple genes in multiple ways (by blocking transcription and fluorescent tagging to name a few) and by the creation of specific point mutations, the genetic mysteries of male reproduction will be rapidly revealed.
COMPETING FINANCIAL INTERESTS
The authors declare there are no competing financial interests.
REFERENCES
- 1.Jamsai D, O’Bryan MK. Mouse models in male fertility research. Asian J Androl. 2011;13:139–51. doi: 10.1038/aja.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy A, Matzuk MM. Deconstructing mammalian reproduction: using knockouts to define fertility pathways. Reproduction. 2006;131:207–19. doi: 10.1530/rep.1.00530. [DOI] [PubMed] [Google Scholar]
- 3.Ikawa M, Inoue N, Benham AM, Okabe M. Fertilization: a sperm's journey to and interaction with the oocyte. J Clin Invest. 2010;120:984–94. doi: 10.1172/JCI41585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue N, Yamaguchi R, Ikawa M, Okabe M. Sperm-egg interaction and gene manipulated animals. Soc Reprod Fertil Suppl. 2007;65:363–71. [PubMed] [Google Scholar]
- 5.Blume-Jensen P, Jiang G, Hyman R, Lee KF, O’Gorman S, et al. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3’-kinase is essential for male fertility. Nat Genet. 2000;24:157–62. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, et al. The mouse homolog of Drosophila vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–53. [PMC free article] [PubMed] [Google Scholar]
- 7.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–98. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 9.Sun F, Palmer K, Handel MA. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development. 2010;137:1699–707. doi: 10.1242/dev.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung JJ, Navarro B, Krapivinsky G, Krapivinsky L, Clapham DE. A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nat Commun. 2011;2:153. doi: 10.1038/ncomms1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitken RJ, Nixon B. Sperm capacitation: a distant landscape glimpsed but unexplored. Mol Hum Reprod. 2013;19:785–93. doi: 10.1093/molehr/gat067. [DOI] [PubMed] [Google Scholar]
- 12.Alasmari W, Costello S, Correia J, Oxenham SK, Morris J, et al. Ca2+ signals generated by CatSper and Ca2+ stores regulate different behaviors in human sperm. J Biol Chem. 2013;288:6248–58. doi: 10.1074/jbc.M112.439356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escalier D. Knockout mouse models of sperm flagellum anomalies. Hum Reprod Update. 2006;12:449–61. doi: 10.1093/humupd/dml013. [DOI] [PubMed] [Google Scholar]
- 14.Vernon GG, Neesen J, Woolley DM. Further studies on knockout mice lacking a functional dynein heavy chain (MDHC7).1. Evidence for a structural deficit in the axoneme. Cell Motil Cytoskeleton. 2005;61:65–73. doi: 10.1002/cm.20066. [DOI] [PubMed] [Google Scholar]
- 15.Okabe M. Mechanism of fertilization: a modern view. Exp Anim. 2014;63:357–65. doi: 10.1538/expanim.63.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujihara Y, Okabe M, Ikawa M. GPI-anchored protein complex, LY6K/TEX101, is required for sperm migration into the oviduct and male fertility in mice. Biol Reprod. 2014;90:60. doi: 10.1095/biolreprod.113.112888. [DOI] [PubMed] [Google Scholar]
- 17.Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–8. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- 18.Bianchi E, Doe B, Goulding D, Wright GJ. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–7. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–6. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamowski S, Aston KI, Carrell DT. The use of transgenic mouse models in the study of male infertility. Syst Biol Reprod Med. 2010;56:260–73. doi: 10.3109/19396368.2010.485244. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–12. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 23.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, et al. The knockout mouse project. Nat Genet. 2004;36:921–4. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 31.Mashiko D, Young SA, Muto M, Kato H, Nozawa K, et al. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes. Dev Growth Differ. 2014;56:122–9. doi: 10.1111/dgd.12113. [DOI] [PubMed] [Google Scholar]
- 32.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menke DB. Engineering subtle targeted mutations into the mouse genome. Genesis. 2013;51:605–18. doi: 10.1002/dvg.22422. [DOI] [PubMed] [Google Scholar]
- 34.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klinovska K, Sebkova N, Dvorakova-Hortova K. Sperm-egg fusion: a molecular enigma of mammalian reproduction. Int J Mol Sci. 2014;15:10652–68. doi: 10.3390/ijms150610652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Archambeault DR, Matzuk MM. Disrupting the male germ line to find infertility and contraception targets. Ann Endocrinol (Paris) 2014;75:101–8. doi: 10.1016/j.ando.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–66. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 38.Baker MA, Hetherington L, Weinberg A, Naumovski N, Velkov T, et al. Analysis of phosphopeptide changes as spermatozoa acquire functional competence in the epididymis demonstrates changes in the post-translational modification of Izumo1. J Proteome Res. 2012;11:5252–64. doi: 10.1021/pr300468m. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Wang J, Shen B, Chen L, Su Y, et al. Dual sgRNAs facilitate CRISPR/Cas9-mediated mouse genome targeting. FEBS J. 2014;281:1717–25. doi: 10.1111/febs.12735. [DOI] [PubMed] [Google Scholar]
- 40.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 41.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AY, Lloyd KC. Conditional targeting of Ispd using paired Cas9 nickase and a single DNA template in mice. FEBS Open Bio. 2014;4:637–42. doi: 10.1016/j.fob.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostrowski LE, Yin W, Rogers TD, Busalacchi KB, Chua M, et al. Conditional deletion of dnaic1 in a murine model of primary ciliary dyskinesia causes chronic rhinosinusitis. Am J Respir Cell Mol Biol. 2010;43:55–63. doi: 10.1165/rcmb.2009-0118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young SA, Baker MA, Ikawa M. Genome editing in mice using CRISPR/Cas. In: Yamamoto T, editor. Targeted Genome Editing Using Engineered Nucleases: ZFNs, TALENs and the CRISPR/Cas System. Ch. 11. Tokyo: Springer Japan; 2015. pp. 151–66. [Google Scholar]
- 45.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013;41:e187. doi: 10.1093/nar/gkt772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


