Abstract
The currently used biomarkers for acute myocardial infarction (AMI) are blood creatinine phosphokinase-muscle band (CPK-MB), troponin-T (TnT), and troponin I (TnI). However, no good biomarkers are identified in urine after AMI, because these blood protein biomarkers are difficult to be filtered into urine. In this study, the role of urine microRNAs in the diagnosis of AMI and the mechanism involved were determined. We found that urine miR-1 was quickly increased in rats after AMI with peak at 24 h after AMI, in which an over 50-fold increase was demonstrated. At 7 days after AMI, the urine miR-1 level was returned to the basal level. No miR-208 was found in normal urine. In urine from rats with AMI, miR-208 was easily detected. To determine the mechanism involved, we determined the levels of heart-released miR-1 in the liver, spleen and kidney after AMI in rats and found that the kidney was an important metabolic organ. To determine the renal elimination of blood miRNAs, we isolated serum exosomes from rats after AMI and injected these exosomes into the circulating blood of normal rats. We found that the urine miR-1 was significantly increased in exosome-injected animals. Moreover, PKH67-labeled exosomes injected into circulating blood could enter into the kidney tissues and cells, as well as urine. Furthermore, the levels of urine miR-1 were significantly increased in patients with AMI. The results suggest that urine miRNAs such as miR-1 could be novel urine biomarkers for AMI.
Keywords: Acute myocardial infarction, microRNA, miR-1, miR-208, Urine, Biomarker
1. Introduction
The currently used diagnostic biomarkers for acute myocardial infarction (AMI) are blood creatinine phosphokinase-muscle band (CPK-MB), troponin-T (TnT), and troponin I (TnI) [1]. However, no good biomarkers are identified in urine after AMI, because these blood protein biomarkers are difficult to be filtered into urine. Indeed, these blood protein biomarkers can only be detected in urine from patients with some special circumstances such as impaired kidney function [2].
MicroRNAs (miRNAs) have emerged as a novel class of endogenous, small (<24 nucleotide), noncoding RNAs with strong biological functions in cardiovascular system [3–5]. Recent studies from us and other groups have revealed that the heart-specific or heart-enriched miRNAs such as miR-1 and miR-208 can be quickly released into circulating blood after AMI [6–13]. In contrast to our original thought, the heart-released circulating cell-free miRNAs are relatively stable in blood due to the protection from degradation by their inclusion in exosomes and by the formation of protein–miRNA complexes. Thus, these heart-released blood miRNAs may be novel biomarkers for AMI [6–13].
Recent studies have demonstrated that miRNAs are identified in urine [14–16]. These urine miRNAs could be released locally by the urinary system. Another potential source of these miRNAs might be from the circulating blood miRNAs via renal elimination [17,18]. Although blood miRNAs have no obvious superiority compared with these well-established blood protein biomarkers such as TnT, and TnI based on the current qRT-PCR determination, they may have the potential to enter the urine. If this is true, these heart-derived urine miRNAs may represent a novel group of biomarkers of AMI in urine. In this translational study, we attempt to determine the role of urine miRNAs in the diagnosis of AMI and the detailed mechanisms involved by using molecular, cell, animal, and human approaches.
2. Methods
2.1. Rat model of AMI
AMI in rats was induced by LAD ligation as described in our recent studies [6,19,20]. In brief, 10-week-old female Sprague–Dawley rats (weighing 250–300 g) were anesthetized with ketamine (80 mg/kg i.p.) and xylazine (5 mg/kg i.p.). (Note: our preliminary data demonstrated that male rats could release prostatic fluid and sperm into urine during the anesthesia. Thus, male rats were excluded in this study). Under sterile conditions, an anterior transmural AMI was created by occlusion of the LAD with a silk suture. Sham-operated rats served as controls. Sham operation involved an identical procedure, except the suture was passed around the vessel without LAD occlusion. The animals were divided into two study groups. Group 1 is for the time course study of urine and serum miR-1. In this group, the urine and blood samples were collected from rats before (0 h) and at 1 h, 3 h, 6 h, 12 h, 24 h, 3 day, 7 day, and 14 day after AMI. Eight rats finished the time course study. In addition, 8 sham-opened rats were used as sham control for the study. Group 2 had 12 rats that were used to study the relationship between urine miR-1 and serum miR-1. All protocols were approved by the Institutional Animal Care and Use Committee, and were consistent with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 85-23, revised 1985).
2.2. Measurement of infarct size
Myocardial infarct size was determined by pathological staining as described [6,19,20]. At the end of experiments, rats were anesthetized and 6 mL of 10% Evans blue dye was injected into the vena cava to define the area that was not supplied by LAD. The myocardial ischemic area at risk (IAR) was identified as the region lacking blue staining. The ventricles of the hearts were sliced transversely into 2-mm thick slices. The slices were incubated in 1% triphenyltetrazolium chloride (TTC) at 37 °C for 10 min to identify the noninfarcted and infarcted areas. TTC staining was displayed as red color. The infarcted area was defined as TTC unstained area. Infarct size was expressed as a percentage of the IAR (% IAR).
2.3. Blood and urine sample collection, miRNA isolation and measurement
Before and at different times after AMI or sham surgery, the blood samples were collected via tail vein. Urine samples were collected in anesthetized rats using the following noninvasive method. Briefly, a tube was placed at the urethral orifice. After the collection of about 30 μL of initial urine, a new tube was used to replace the old one and the experimental urine sample was then collected. The samples were placed at 4 °C for 1 h and then centrifuged at 12,000 ×g for 20 min at 4 °C. Aliquot urine and serum samples were stored at −70 °C until miRNA isolation. Blood and serum miRNAs were isolated from 250 μL urine or serum samples using Trizol LS based isolation kit (RNA bioscience, NJ). miR-1, mir-208, and miR-122 (a control liver-specific miRNA which is not expressed in heart) were measured using the qRT-PCR based solution miRNA quantitative kit developed by our group as described [6] (RNA bioscience, NJ). Briefly, miR-1 and miR-208 was measured by qRT-PCR with a Roche Lightcycler 480 Detection System using the primers provided by the Applied Biosystems. The same isolation and assay were performed using a series of concentrations of standard miR-1 and miR-208 (synthesized by IDT, Coralville, IA, U.S.A.) to make a standard curve. The absolute amount of miR-1 or miR-1 was calculated by software based on sample qRT-PCR numbers and the standard curve, and are expressed as pmol/L.
2.4. Tissue sample collection, miRNA isolation and measurement
In an effort to determine which organ is related to the metabolism of the heart-released miRNAs in blood after AMI, we carefully perfused the AMI rats at 24 h or sham-opened surgery rats with RNA-free saline. This was done at a physiological pressure for a period of at least 15 min to remove all the circulating blood in the rat body. Then, the liver, spleen and kidney were isolated to determine the miR-1 levels. miR-1 was isolated and measured using qRT-PCR as described in our previous studies [6,19,20].
2.5. Exosome isolation from serum or urine
Exosomes from serum and urine were isolated by ultracentrifugation. Briefly, the samples were filtered through a 0.22 μm Millex-GS Filter Unit (Millipore), followed by ultracentrifugation at 100,000 ×g for 2 h to pellet the exosomes. The pellet exosomes were resuspended in PBS to perform the experiments.
2.6. Electron microscopy
Exosomes isolated from serum and urine were washed in PBS to further purify the sample, filtered, and ultracentrifuged again at 100,000 ×g for 2 h to re-pellet the exosomes. The exosome pellet was resuspended and fixed in phosphate buffer containing 2% glutaraldehyde and then loaded onto formar/carbon-coated electron microscopy grids. The samples were contrasted with uranyl acetate to visualize membrane and viewed with a JEOL 1200EX electron microscope.
2.7. Exosome labeling
Serum exosomes were labeled with PKH67 Green Fluorescent Cell Linker Kit (Sigma-Aldrich) according to the manufacturer's protocol. Briefly, the exosomes were diluted in PBS. One microliter of PKH67 dye was added to 250μL of experimental solution before being added to the exosomes. The exosomes without PKH67 dye were used as the control. The samples were mixed gently for 4 min before 500 μL of 1% BSA was added to remove the excess dye.
2.8. Renal elimination of exosomes and exosome-carried miRNAs
To determine the potential transrenal release of exosomes and exosome-carried miRNAs, serum exosomes from AMI rats at 6 h were isolated. These exosomes were labeled with PKH67 or vehicle. PKH67-labeled exosomes (20 μg in 500 μL of PBS), unlabelled exosomes (20 μg in 500 μL of PBS) or vehicle (500 μL of PBS) was injected into the aorta above the renal artery level. The urine was collected before and at 3 h, 6 h, and 24 h after injection for urine miRNA and urine exosome isolation. In some animals, the unlabeled exosomes (20 μg in 500 μL of PBS) or vehicle (500 μL of PBS) were injected into circulation via external jugular vein and the urine samples were collected at 6 h after injection. The potential renal elimination of exosomes and exosome-carried miRNAs was determined by the following three experiments. First, to determine whether the exosomes injected to circulating blood could increase the level of miR-1 in urine. Second, to determine whether PKH67-labeled exosomes could enter the kidney tissues and kidney cells. In this experiment, the labeled exosomes were detected in 5 μm frozen sections of kidney with a Nikon Ti fluorescence microscope. Third, to determine whether the labeled exosomes could be released into urine. The labeled individual exosome cannot be directly observed by the fluorescence microscope due to its small size. However, the labeled exosomes in the cells could be easily identified via a fluorescence microscope. In this experiment, the isolated urine exosomes from animals treated with PKH67-labeled exosomes or unlabeled exosomes were added to the culture medium of HEK293 cells and incubated for 4 h at 37 °C. The cells were washed twice with PBS. Then, the PKH67-labeled exosomes in HEK-293 cells were analyzed with a Nikon Ti fluorescence microscope.
2.9. Clinical study
AMI was defined as 1) characteristic chest pain of myocardial ischemia for 30 min or more, 2) ST segment elevation within 6 h of chest pain at least 0.1 mV in at least two leads of the echocardiography (ECG), and 3) confirmation of the diagnosis of AMI by elevated CK-MB in serum which was at least twice the normal range. The blood samples from patients without the confirmation of AMI were not used in this study. In total, 20 patients with AMI were used in this study (11 men and 9 women; mean age 56.1+/−13.9). Nine of them had coronary angiography data showing at least one vessel disease. Among the patients studied, three had hypertension on antihypertetive medicines, angiotensin II inhibitor and /or beta-blocker; five patients had hyperlipidemia with simvastatin therapy. No patients with diabetes were included in this study. Blood and urine samples were obtained within 24 h of AMI. The time interval between the onset of typical chest pain and urine samples was 12.5±5.3 h. Urine samples obtained from age and sex matched 20 healthy volunteers were used as the controls. The protocol was approved by the Institutional Review Board of the Luzhou Hospital and Guangdong Hospital. All the samples were stored for 1 h at room temperature (26 °C) and were centrifuged at 1600 ×g for 20 min at 4 °C. Urine miR-1 and miR-208 were determined by qRT-PCR.
2.10. Bioinformatical analysis
The putative targets of miR-1 and miR-208 were analyzed by using the online software TargetScan 5.1 and published database as described [21].
2.11. Statistics
All data is presented as mean±standard error. For relative gene expression, the mean value of the vehicle control group was defined as 100% or 1. Differences in miRNA levels were analyzed using one-way ANOVA with Bonferroni's multiple comparisons post hoc test. Linear regression analysis was used to determine the relationship between serum TnI and urine miR-1. Normality tests of all parameters were performed using D'Agostino-Pearson omnibus method. Sigma stat statistical analysis program was used for data analysis. A p value<0.05 was considered significant.
3. Results
3.1. Urine miR-1 and miR-208 are increased in rats after AMI
As shown in Figs. 1A and B, AMI was induced by the left anterior descending coronary artery (LAD) ligation. In normal rats or sham-opened rats, the heart-enriched miR-1 is almost undetectable in urine by the high sensitive qRT-PCR. However, an over 50-fold increase in miR-1 level was demonstrated in urine from rats at 24 h after AMI (p<0.0001, Fig. 1C). In contrast, no difference was found in the levels of urine miR-122, which is a liver specific miRNA, between AMI group and the control group (Fig. 1C). There is no miR-208, the heart-specific miRNA, in normal urine. However, miR-208 was easily identified in urine from AMI rats. As shown in Fig. 1D, qRT-PCR showed that no miR-208 signal was found even after 60 cycles in urine from normal and sham-opened rats. In contrast, a significant amount of miR-208 was identified in urine from rats at 24 h after AMI (Fig. 1D).
Fig. 1.
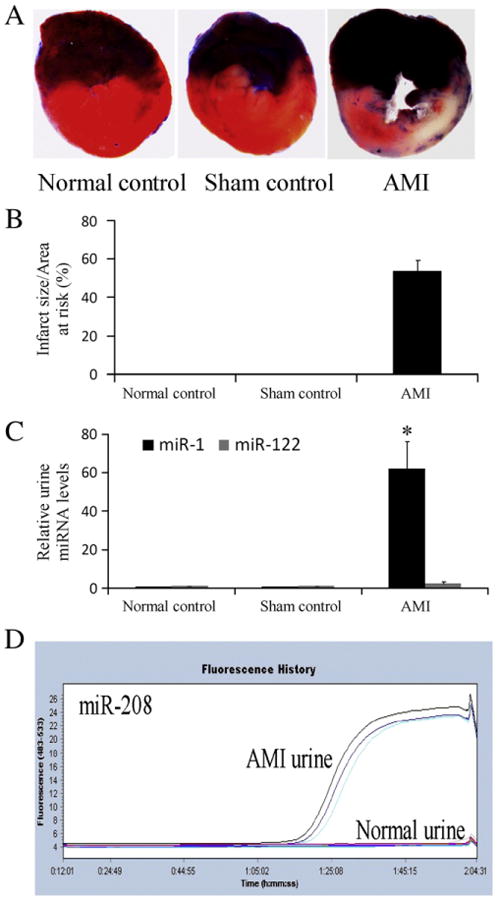
Urine miRNAs from rats with or without acute AMI. (A) AMI was induced by left anterior descending coronary artery ligation and the infarct size was determined by pathological staining in heart slices. Note: Color blue is Evans blue staining. The region without Evans blue staining is myocardial ischemic area at risk (IAR). Color red is the triphenyltetrazolium chloride (TTC) staining. TTC unstained area within IAR was the infarcted area. (B) Infarct size is expressed as a percentage of the IAR (% IAR). (C) The relative urine miR-1 and miR-122 levels in 8 rats without surgery, 8 sham-opened control rats, and 8 rats at 24 h after AMI. n=8, *P<0.001 compared with the control group. (D) The urine levels of miR-208 in rats with and without AMI. Note: The 3 curves above the baseline after 1:05:02 represent the miR-208 signals in urine from 3 AMI rats. In contrast, 3 lines that are still at baseline after 1:05:02 represent 3 normal urine samples without miR-208 signal.
3.2. Time course of urine miR-1 in rats after AMI
To determine the time course changes of urine miR-1, rat urine samples were collected before (0 h), and at 3 h, 6 h, 12 h, 24 h, 48 h, 3 days, 7 days, and 14 days after AMI. As shown in Fig. 2A, compared with that in normal urine (0 h group), urine miR-1 level was quickly increased with its peak at 24 h after AMI, in which an over 50-fold increase in miR-1 level was demonstrated. At 7 days after AMI, the urine miR-1 level was returned to baseline (0 h) level.
Fig. 2.
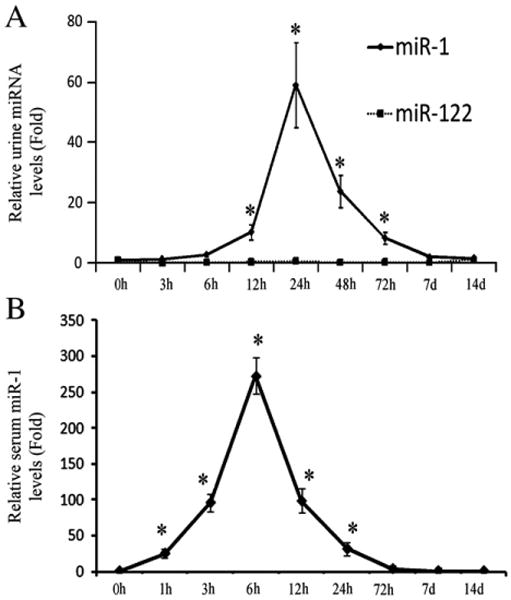
Time-course changes of urine and serum miR-1 in rats after AMI. (A) The time course of urine miR-1 in rats after AMI. (B) The time course of serum miR-1 in rats after AMI. Note: n=8, *P<0.05 compared with the group before AMI (0 h group).
3.3. The putative targets of miR-1 and miR-208
Bioinformatical analysis of the putative targets of miRNAs using the online software TargetScan 5.1 revealed both miRNAs have multiple target genes that may be involved in mechanistic pathways relevant to the pathophysiology of AMI. For example, transgelin 2 (TAGLN2), purine nucleoside phosphorylase (PNP), prothymosin alpha (PTMA), Connexin 43 (Cx43), stromal cell derived factor-1 (SDF-1), transgelin 2 (TAGLN2), fibronectin 1, and Kruppel-like factor 4 (KLF4) are the targets of miR-1 that are related to cell proliferation, apoptosis, death, and migration. Thyroid hormone-associated protein 1 and myostatin are two putative target genes of miR-208 that are related to heart disease [22].
3.4. Relationship of urine miR-1 and serum miR-1 in rats with AMI
The time course of serum miR-1 in rats after AMI was shown in Fig. 2B. After AMI, the serum miR-1 level was quickly increased within 1 h, peaked at 6 h, and was return to basal (0 h) level at 3 days. Clearly, there was a delay in the increase of miR-1 in urine compared with that in serum. However, the period of the increased miR-1 in urine was longer compared with that in serum. In addition, the absolute peak value of urine miR-1 (<0.1 pmol/L) was much lower than that of serum miR-1 (3–5 pmol/L).
3.5. Stability and distribution of urine miR-1 in rats with AMI
To determine the stability of urine miR-1, we collected the urine samples from AMI rats at 24 h. The urine miR-1 levels were determined immediately after collection and at 24 h following collection. As shown in Fig. 3A, the urine miR-1 was partially degraded during the 24 h period. To identify the distribution of urine miR-1, the urine was divided into two parts, exosome part and supernatant part by ultracentrifugation. Representative urine exosomes were displayed in Fig. 3B. We found about 51% of urine miR-1 was located in urine exosomes. The remaining 49% of miR-1 was at urine supernatant (Fig. 3C). However, the exosomes occupy only a very small part of the urine volume. Thus, miRNAs are relatively concentrated in urine exosomes. The urine miR-1 levels in both exosome part and supernatant part were increased in rats after AMI (Figs. 3D and E). The miR-1 in exosome part and supernatant part was partially degraded during the 24 h period at 4 °C (Figs. 3F and G).
Fig. 3.
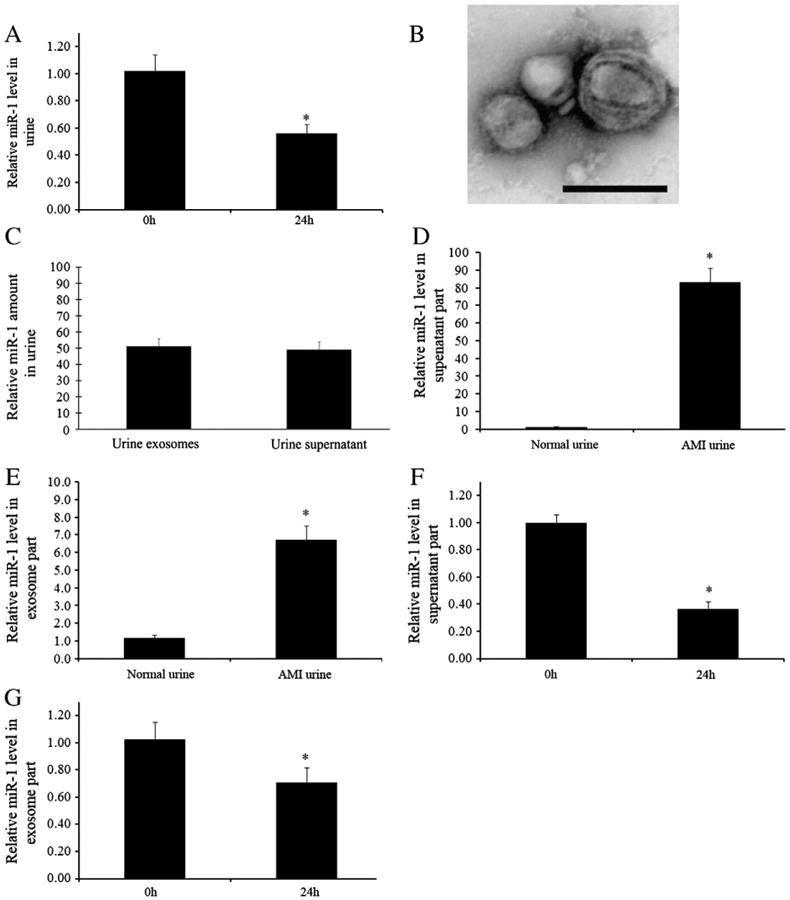
Stability and distribution of urine miR-1 in rats with AMI. (A) Urine samples were collected from rats at 24 h after AMI and miR-1 levels were determined immediately after urine collection and 24 h later at 4 °C. n=5, *P<0.01 compared with 0 h group. (B) Representative urine exosomes isolated by ultracentrifugation and detected by electron microscopy. (C) The distribution of urine miR-1 in exosome part and supernatant part (100% of total urine miR-1). (D) miR-1 levels in supernatant part of urine in rats with and without AMI. (E) miR-1 levels in exosome part of urine in rats with and without AMI. (F) miR-1 levels in freshly isolated supernatant part of AMI urine and in supernatant part kept for 24 h at 4 °C. (G) miR-1 levels in freshly isolated exosome part of AMI urine and in exosome part kept for 24 h at 4 °C. Note: n=5, *P<0.01 compared with 0 h group.
3.6. The kidney is an important metabolic organ for heart-released miR-1 after AMI
To determine which organ is related to the metabolism of the heart-released miRNAs in blood, the rats at 24 h after AMI were carefully perfused with RNA-free saline at physiological pressure to remove all the circulating blood in the rats. Sham-opened rats were used as control animals. Then, the liver, spleen and kidney were isolated to determine the miR-1 levels. Interestingly, as shown in Figs. 4A–C, the miR-1 levels in the kidneys from infarcted rats were markedly higher than those in kidneys from rats without AMI. In contrast, in the liver and spleen, no difference in miR-1 levels was found between the AMI and control group.
Fig. 4.
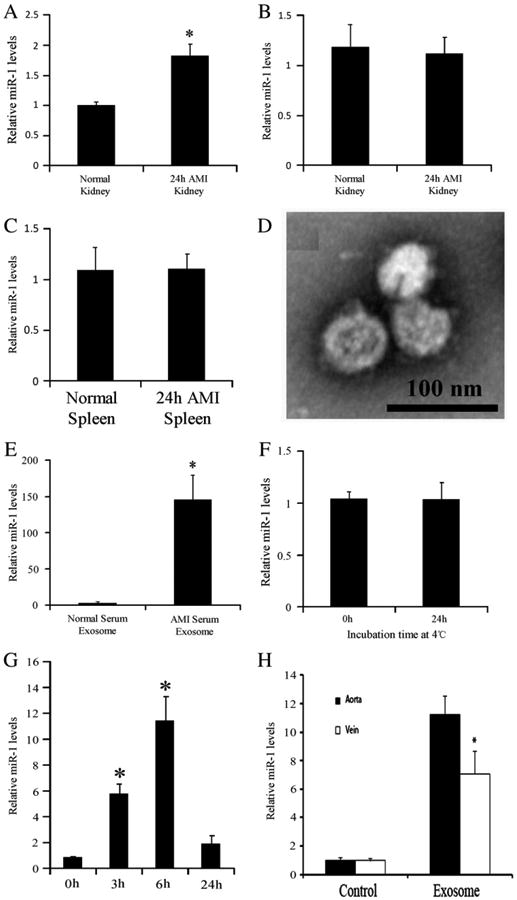
Transrenal release of exosome-carried miRNAs. (A)–(C) The miR-1 levels in liver, spleen and kidney of Sham-opened rats and of the rats at 24 h after AMI. (D) Representative serum exosomes isolated by ultracentrifugation and detected by electron microscopy. (E) miR-1 levels in serum exosomes from normal rats and from rats with AMI. (F) miR-1 levels in freshly isolated exosomes of AMI serum and in these exosomes kept for 24 h at 4 °C. G. Administration of the serum exosomes from rats with AMI to aortas of normal rats resulted in the time-dependent increase in urine miR-1. H. Comparison of urine miR-1 levels exosomes-treated rats between the delivery via aortas and the delivery via external jugular veins. Note: n=5, *P<0.05 compared with the control group.
3.7. Renal elimination of exosomes and exosome-carried miRNAs
To determine the renal elimination of exosomes and exosome-carried miRNAs, we isolated serum exosomes from rats at 6 h after AMI. Fig. 4D illustrates the representative serum exosomes. The miR-1 level in serum exosomes was significantly increased after AMI (Fig. 4E). Unlike the miR-1 in urine exosomes, the miR-1 in serum exosomes was relatively stable (Fig. 4F). Interestingly, administration of the serum exosomes from AMI rats to the aortas of normal rats could result in the time-dependent increase in urine miR-1 (Fig. 4G). The result indicated that the exosome-carried miRNAs could enter the urine via transrenal release. Injection of exosomes via external jugular vein also resulted in the increase in urine miR-1. However, the increase in urine miR-1 was less compared with that in animals injected via aortas (Fig. 4H). The result revealed that that exosomes may be partially cleared by other organs.
To provide direct evidence that the exosome may have the ability of transrenal release, we delivered the PKH67-labeled exosomes to rat blood [23]. As in Fig. 5A, there was no fluorescence signal (green color) in the kidney from unlabeled exosomes-injected animals. In contrast, the PKH67-labeled exosomes (green color) were identified in the kidney section from PKH67-labelled exosomes-injected rats (Fig. 5B). The result suggested that the circulating exosomes could enter the kidney tissues and kidney cells. The PKH67-labeled exosomes cannot be seen directly in urine and isolated exosomes cannot be observed directly by the fluorescence microscope. To provide direct evidence that the injected circulating exosomes could enter the urine, we isolated the urine exosomes at 12 h after injection of PKH67-labeled exosomes. The isolated urine exosomes were added to the culture medium of the HEK293 cells. As shown in Fig. 6, the PKH67-labeled exosomes (green color) were indeed demonstrated in HEK293 cells. The results clearly showed that the circulating exosomes could enter urine via transrenal release.
Fig. 5.
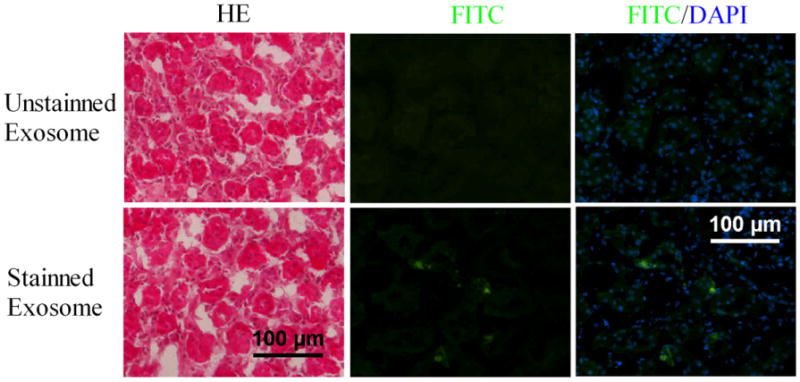
The capability of circulating exosomes into kidney tissue. PKH67-labeled exosomes or unlabeled exosomes were injected into circulating blood of rats. The kidneys were isolated to determine the PKH67-labeled exosomes in the kidney tissue sections (A). HE-staining and fluorescence signal detection (green color) in kidney sections from unlabeled exosomes-injected animals. No fluorescence signal (green color) is demonstrated. (B) HE-staining and fluorescence signal detection (green color) in kidney sections from PKH67-labeled exosomes-injected animals. Fluorescence signal (green color) is identified.
Fig. 6.
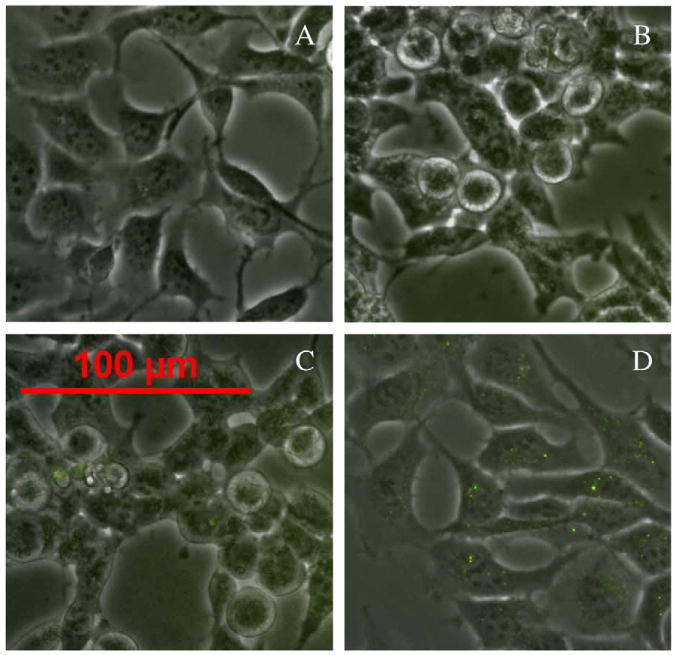
Transrenal release of exosomes. PKH67-labeled exosomes or unlabeled exosomes were injected into circulating blood of rats. The urine exosomes from these rats were isolated and were added to the culture medium of HEK293 cells to detect to PKH67-labeled exosomes in cells. HEK293 cells cultured with vehicle are used as a negative control and HEK293 cells cultured directly with PKH67-labeled exosomes were used as a positive control. Fluorescence signal (green color) was detected by a fluorescence microscopy. (A) Negative control. (B) Cells cultured with urine exosomes from unlabeled exosomes-injected animals. (C) Cells cultured with urine exosomes from PKH67-labeled exosomes-injected animals. (D) Positive control.
3.8. The results from clinical study on urine miRNAs in patients with AMI
As shown in Fig. 7, the urine miR-1 was significantly increased in patients with AMI compared with that in urine from age and sex-matched normal controls. To investigate the potential relationship between urine miR-1 levels and myocardial infarct sizes in humans, serum TnI levels in these patients were also determined. As shown in Fig. 7B, a positive correlation was demonstrated between serum TnI and urine miR-1 levels (r=0.70; p<0.05). There was no miR-208 in normal urine. In contrast to urine miR-1, only a few of patients (5 out of 20 detected patients) were found to have very low levels of miR-208 in urine.
Fig. 7.
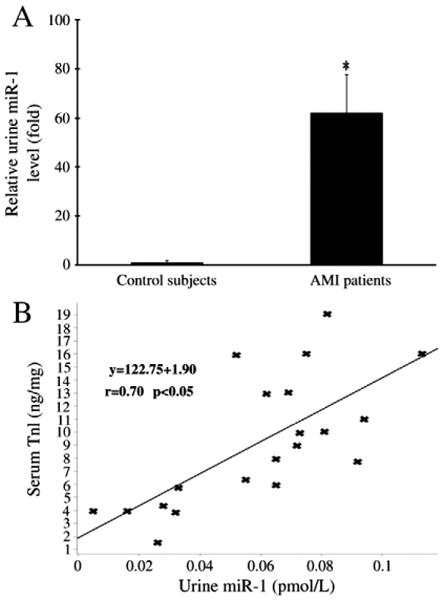
Urine miR-1 in patients with AMI. (A) Urine miR-1 in patients with AMI and their age and sex-matched normal controls. n=20, *P<0.01 compared with the control group. (B) The relationship between serum TnI levels and urine miR-1 in these patients. A positive correlation was demonstrated between serum TnI and urine miR-1 levels (r=0.70; p<0.05).
4. Discussion
AMI is still one of the leading causes of death in the world. An early and accurate diagnosis is the prerequisite to facilitate rapid decision making and treatment and therefore improves outcomes in patients with AMI [22]. Testing of blood protein markers such as CPK-MB, TnT, and TnI has become the standard procedure and plays an important role in the diagnosis of AMI, especially in patients without typical symptoms [1]. However, these large molecular protein biomarkers are difficult to enter the urine. Thus, there are no good urine biomarkers available for the diagnosis of AMI.
MicroRNAs (miRNAs) are small (<24 nucleotide) RNAs with strong biological functions [4,5]. Recent studies have reported that miRNAs are found in urine [14–18]. miR-122, a liver-specific miRNA is among the identified miRNAs in urine [18], suggesting that miRNA may be filtered into urine from the circulating blood. More recently, our studies and those of other groups have demonstrated that the heart-specific or heart-enriched miRNAs such as miR-1 and miR-208 can be quickly released into circulating blood after AMI [6–13]. These heart-released blood miRNAs may represent a novel class of biomarkers for AMI. Blood miRNAs may have no obvious superiority compared with these well-established blood protein biomarkers at current stage due to their time consuming of qRT-PCR. However, these heart-released miRNAs may have the potential to enter the urine. If this is true, miRNAs might be a novel group of biomarkers for AMI in urine.
The metabolic approaches of the heart-released miRNAs in blood after AMI are still unknown. In this study, we first determined the levels of heart-released miR-1 in the liver, spleen and kidney in AMI rats. The results revealed that the kidney is an important metabolic organ for heart-released miR-1 after AMI. We found that the heart-enriched specific miR-1 in urine was significantly increased in animals after AMI. The heart-specific miR-208 cannot be found in normal rat urine. However, in urine from animals with AMI, a significant amount of miR-208 is easily identified. Our time course study has demonstrated an obvious delay in the increase of miR-1 in urine compared with that in blood, but the increase of miR-1 in urine was sustained longer than that in blood. These results suggested that a metabolic process may occur in the kidney. In addition, the standard deviation or standard error of miR-1 in urine was larger compared with that in blood. This finding may reflect the different rate of urine formation and kidney metabolism among the rats after AMI.
The urine was separated into exosome part and supernatant part by ultracentrifugation. The miR-1 levels were measured in two parts to determine the distribution of miR-1 in urine from animals with AMI. We identified that miR-1 was located in both parts. Previous studies have reported that miRNAs in serum exosomes are pretty stable [6–13]. We thus compared the stability of miR-1 in serum exosomes and urine exosomes. Consistent with previous reports, miR-1 was stable in serum exosomes. However, a slow degradation of urine exosome-carried miR-1 was found in our study. A slow degradation of miR-1 in the supernatant part was also identified. The results suggested that an unknown degradation mechanism may exist in urine.
To determine the capability of renal elimination of exosomes and exosome-carried miRNAs, we isolated serum exosomes from rats after AMI and injected into aortas in normal rats. We found that the urine miR-1 level in these exosome-injected animals was significantly increased. In addition, injection of these exosomes intravenously also increase the urine miR-1 level, but the increase is smaller than that induced by arterial injection. The result suggests that exosomes may also be cleared by other organs. To further provide direct evidence that circulating exosomes can enter into urine by transrenal release, we injected the exosomes with fluorescence label into the circulating blood. We found that the injected exosomes indeed could be identified in kidney tissues and cells. To further verify the discovery, we isolated urine exosomes in animals after the injection with fluorescence-labeled exosomes and added them to the culture medium of HEK293 cells. The result showed that the HEK293 cells indeed had the fluorescence-labeled exosomes.
One interesting question is that, in addition to the transrenal release of miRNAs via exosomes, whether the circulating free miRNAs are able to enter urine directly. It is possible, because about half of urine miRNAs are not in exosomes. However, it is difficult to perform the experiment to provide direct evidence to verify this hypothesis due to the following two reasons: First, it is difficult to obtain the real-free miRNAs in blood, because most of the miRNAs are binding to exosomes or, at least, to other proteins. Second, the synthesized mature free miRNAs are not stable in blood.
Finally, in the clinical study, we found that urine miR-1 in patients with AMI were also quickly increased within hours. In patients within 24 h of AMI, there is over 60-fold increase in miR-1 urine level. The heart-specific miR-208 cannot be found in normal human urine. In contrast to urine miR-1, only a few of patients (5 out of 20 detected patients) were found to have very low levels of urine miR-208. We think the potential reason for the difference between human urine miR-1 and urine miR-208 is that the level of miR-208 is much lower than that of miR-1. Thus, the low level of human urine miR-208 cannot be easily detected by the regular qRT-PCR. Improvement of the detective method for urine miR-208 is thus needed in future studies.
The search for new biomarkers of cardiovascular diseases including AMI remains a large and growing enterprise [24]. The new surrogate markers for AMI should have a higher sensitivity, higher specificity and need to be quickly detected. There is no doubt that urine miRNAs are a novel class of AMI biomarkers with high sensitivity and specificity. However, they are not the surrogate markers for AMI using the current qRT-PCR technology because of the time consuming in detection compared with the method to determine serum CK-MB or troponin (TnT or TnI). However, the development of high sensitive, quick assay of RNA is a highly pursued research topic. In fact, progress has been made such as fluoresce and ELISA methods in RNA array. Any breakthroughs in this research area may overcome the pitfall in the future. We understand that it still has a long way to go to bring the discovery of this study to clinical practice.
During the writing of this article, we are happy to see that another group also reported the urine miR-1 in AMI urine samples [25]. It provided an additional support to our observations. However, they did not find miR-208 in urine from animals and patients. As we discussed above, it might be induced by the low level of urine miR-208. Increasing of the cycle number of qRT-PCR or urine RNA samples may provide help to detect urine miR-208 correctly.
In summary, in the current study, we identified that the heart-released miRNAs could enter the urine by their transrenal release and provided a detailed molecular mechanism about the heart-released urine miRNAs in AMI. The urine miRNAs such as miR-1 and miR-208 may represent a novel group of urine biomarkers for ischemic heart disease such as AMI.
Acknowledgments
Role of funding source: This work was supported by NIH Grants HL080133, HL095707, HL109651 and a grant from the American Heart Association 09GRNT2250567 to C. Zhang. This project was also supported in part by the Projects of International Cooperation, Department of Science & Technology of Sichuan Province (2011HH0031) to X. Wang.
Footnotes
Disclosure statement: C.Z., Y.Z., J.Y., and X.L. are named as co-inventors on a patent application to the US pertaining to the Therapeutic and Diagnostic MIRNA Products (Patent Application#61476974).
References
- 1.Howie-Esquivel J, White M. Biomarkers in acute cardiovascular disease. J Cardiovasc Nurs. 2008;23:124–31. doi: 10.1097/01.JCN.0000305072.49613.92. [DOI] [PubMed] [Google Scholar]
- 2.Ziebig R, Lun A, Hocher B, Priem F, Altermann C, Asmus G, et al. Renal elimination of troponin T and troponin I. Clin Chem. 2003;49:1191–3. doi: 10.1373/49.7.1191. [DOI] [PubMed] [Google Scholar]
- 3.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–95. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meder B, Keller A, Vogel B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol. 2011;106:13–23. doi: 10.1007/s00395-010-0123-2. [DOI] [PubMed] [Google Scholar]
- 9.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 10.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 11.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–9. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 12.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;39:73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 14.Melkonyan HS, Feaver WJ, Meyer E, Scheinker V, Shekhtman EM, Xin Z, et al. Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci. 2008;1137:73–81. doi: 10.1196/annals.1448.015. [DOI] [PubMed] [Google Scholar]
- 15.Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–61. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC, et al. Serum and urinary cell-free MiR-146a and MiR-155 in patients with systemic lupus erythematosus. J Rheumatol. 2010;37:2516–22. doi: 10.3899/jrheum.100308. [DOI] [PubMed] [Google Scholar]
- 17.Neal CS, Michael MZ, Pimlott LK, Yong TY, Li JY, Gleadle JM. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant. 2011;26:3794–802. doi: 10.1093/ndt/gfr485. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11:2221–7. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 19.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–25. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, et al. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431–9. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–86. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balk EM, Ioannidis JP, Salem D, Chew PW, Lau J. Accuracy of biomarkers to diagnose acute cardiac ischemia in the emergency department: a meta-analysis. Ann Emerg Med. 2001;37:478–94. doi: 10.1067/mem.2001.114905. [DOI] [PubMed] [Google Scholar]
- 25.Gidlöf O, Andersson P, van der Pals J, Götberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology. 2011;118:217–26. doi: 10.1159/000328869. [DOI] [PubMed] [Google Scholar]


