Abstract
N-Glycans of human proteins possess both α2,6- and α2,3-linked terminal sialic acid (SA). Recombinant glycoproteins produced in Chinese hamster overy (CHO) only have α2,3-linkage due to the absence of α2,6-sialyltransferase (St6gal1) expression. The Chinese hamster ST6GAL1 was successfully overexpressed using a plasmid expression vector in three recombinant immunoglobulin G (IgG)-producing CHO cell lines. The stably transfected cell lines were enriched for ST6GAL1 overexpression using FITC-Sambucus nigra (SNA) lectin that preferentially binds α2,6-linked SA. The presence of α2,6-linked SA was confirmed using a novel LTQ Linear Ion Trap Mass Spectrometry (LTQ MS) method including MSn fragmentation in the enriched ST6GAL1 Clone 27. Furthermore, the total SA (mol/mol) in IgG produced by the enriched ST6GAL1 Clone 27 increased by 2-fold compared to the control. For host cell engineering, the CHOZN® GS host cell line was transfected and enriched for ST6GAL1 overexpression. Single-cell clones were derived from the enriched population and selected based on FITC-SNA staining and St6gal1 expression. Two clones (“ST6GAL1 OE Clone 31 and 32”) were confirmed for the presence of α2,6-linked SA in total host cell protein extracts. ST6GAL1 OE Clone 32 was subsequently used to express SAFC human IgG1. The recombinant IgG expressed in this host cell line was confirmed to have α2,6-linked SA and increased total SA content. In conclusion, overexpression of St6gal1 is sufficient to produce recombinant proteins with increased sialylation and more human-like glycoprofiles without combinatorial engineering of other sialylation pathway genes. This work represents our ongoing effort of glycoengineering in CHO host cell lines for the development of “bio-better” protein therapeutics and cell culture vaccine production.
Keywords: Chinese hamster ovary cells, host cell engineering, α2, 6-linked sialic acid, St6gal1, glycoengineering
Introduction
Sialic acid modulation remains one of the most focused areas in glycoengineering due to the critical role that sialic acid plays in therapeutic glycoprotein half-life and efficacy. The hepatocyte asialoglycoprotein receptor recognizes terminal galactose and mediates protein uptake prior to degradation. Sialylated therapeutic proteins are slower to degrade and have longer serum half-life, as the galactose moiety is linked to terminal sialic acid, therefore, not exposed.1–3 On the other hand, sialylation contributes to heterogeneity in the core N-glycan in IgG and non-IgG therapeutic proteins.4,5 Hence, it is one of the critical product quality parameters in therapeutic protein production. Terminal sialic acid can be attached to galactose in α2,6-, α2,3-, or α2,8-linkages. Six β-galactoside α2,3-sialyltransferases (St3gal1–6) and two β-galactoside α2,6-sialyltransferases (St6gal1–2) are responsible for forming these terminal sialic acids in mammalian cells.6 Human proteins have all linkages, predominantly α2,6-linkage, whereas CHO cells have incomplete sialylation with only α2,3-linked sialic acid.7,8 Therapeutic proteins possessing both sialic acid linkages are more “human-like” and are less likely to be immunogenic. Furthermore, Anthony et al. reported that α2,6-linked sialic acid residues play a critical role in anti-inflammatory activity of human intravenous immunoglobulin.9,10 Despite that half-life may not be significantly improved for low sialic acid containing IgG molecules, enhanced anti-inflammatory activity may also be achieved by producing recombinant IgG therapeutics that contain α2,6-linked sialic acid, which is lacking in current CHO host cells.
Approaches to modulate sialylation of recombinant glycoproteins in CHO and other host cell lines have been reviewed previously.11,12 In general, media composition (glycosylation substrate feeding or media supplements) and process optimization (pH, temperature, and osmolality) can be employed to increase sialic acid content. Cell engineering efforts have identified various gene targets for enhancing sialic acid biosynthesis and transfer or reducing sialic acid degradation.13,14 The employed approaches include overexpression of UDP-GlcNAc2-epimerase15 or CMP-sialic acid transporter,16 RNA interference of sialidases17,18 and overexpression of sialyltransferases. The pathways involved are schematically illustrated in Figure 1 in Supporting Information. Overexpression of human or rat sialyltransferases in CHO cells, either alone or in combination with other glycosylation genes, was explored by numerous groups.19–24 Bragonzi et al. overexpressed rat α2,6-sialyltransferase in a CHO DuxB11 host cell line to produce interferon-γ with more human-like sialylation patterns.25 Fukata et al. utilized an interferon-γ-producing CHO cell line to co-overexpress the following three genes: the glycan-branching enzyme UDP-N-acetylglucosamine: α1,6-D-mannoside β1,6-N-acetylglucosaminyltransferase (Mgat4 or GnT-V); mouse α2,3-sialyltransferase (St3gal4); and/or rat St6gal1.3 Their cell engineering resulted in increased sialylation, particularly tri- and tetra-sialic acid glycan structures. Similarly, Jeong and colleagues co-overexpressed α2,3-sialyltransferase and β1,4-galactosyltransferase and achieved increase in tri-sialylated glycans in erythropoietin (EPO).26 Combinatorial engineering with multiple gene targets can become a lengthy and laborintensive process; multiple selection reagents have to be present in culture media to ensure stability of overexpression; and multiple rounds of selection can result in loss of desired phenotypes that are fundamental for protein expression. Recently, Onitsuka and colleagues first demonstrated enhanced sialylation in a recombinant CHO line producing a bi-specific IgG-like protein by overexpressing Chinese hamster St6gal1 alone.27 They reported altered glycan-type ratios in the recombinant cell line. Maï et al. reported a host CHO cell line with a St6gal1 minigene integrated to overexpress ST6GAL1, and they generated adherent clones secreting hypersialylated proteins.28
The main objective of this study was to create a versatile CHO host cell line with enhanced human-like sialylation that can be used to express various therapeutic glycoproteins. The host line has to retain the desired phenotypes for biopharmaceutical production, such as high transfectability, robust growth, and high productivity in chemically defined culture media. To prove this concept, we successfully overexpressed the Chinese hamster ST6GAL1, first in three recombinant CHO lines of different cell types that produce industry-relevant levels of humanized or human IgG. When α2,6-linkage was confirmed in the recombinant CHO lines, we proceeded to host cell engineering and overexpressed ST6GAL1 in a CHO K1-derived GS (−/−) host cell line (“CHOZN® GS”). A clonal cell line (“ST6GAL1_OE_32”) with the highest St6gal1 expression and highest α2,6-linked sialic acid levels was selected to express a recombinant IgG as a model protein (Supporting Information Figure 1). The recombinant IgG produced in this novel host cell line demonstrated not only α2,6-linkage but also increased total sialic acid content.
Materials and Methods
Cell culture
The glutamine synthetase (−/−) SAFC CHOZN® GS (−/−) host cell line (Sigma Aldrich, St. Louis, MO) and its derivative, a recombinant SAFC human IgG1 cell line (“Clone 27”), were used. Two other recombinant cell lines used in this study, “Clone N22” and “Clone N46,” were derived previously from CHOZN® DHFR (−/−) host cell line and produced a proprietary humanized antibody (“IgG#2B”). The sialidase 3 (Neu3) locus was disrupted using zinc-finger nuclease (ZFN) technology in Clones N22 and N46 (unpublished data). All cell culture media, supplements, and other reagents used were obtained from Sigma-Aldrich unless otherwise specified. Cells were maintained as shaken suspension cultures at 37°C and 5% CO2 in EX-CELL®CHO CD Fusion containing no hypoxanthine and thymidine and supplemented with 6 mM L-glutamine for the host line, 25 μM methionine sulfoximine (MSX) for Clone 27, or 500 nM methotrexate (MTX) for Clones N22 and N46. Dextran sulfate was used at a concentration of 160 μg/mL to prevent clumping to the shaken cultures of ST6GAL1 overexpressing clonal cell lines described in the following section.
Overexpression of ST6GAL1
The coding sequence of Chinese hamster St6gal1 was obtained from GenBank (AB492855) and chogenome.org (AFTD01061789 and AFTD01061790). The open reading frame was synthesized by DNA2.0 (Menlo Park, CA) with a Kozak sequence (5′-GCCGCCACCAatg-3′) added to the 5′-untranslated region (UTR). The synthesized fragment was cloned into expression vector pJ602 (DNA2.0). Both IgG expressing and the CHOZN® GS host cell line were transfected by this construct. Cells were seeded at 106 cells/mL in 50 mL TPP TubeSpin® Bioreactors (Trasadingen, Switzerland) one day prior to transfection. 5 × 106 cells in 800 μL growth media and 30 μg plasmid DNA were used for each transfection. Transfections were conducted by electroporation at 300 V and 950 μF in 0.4-cm cuvettes using a Gene Pulser XCell™ system (Bio-Rad, Hercules, CA). Electroporated cells were subsequently placed in 6 mL growth media in a suspension 25-cm2 tissue culture flask (Greiner Bio-One, Monroe, NC) for 24 h prior to 250 μg/mL Zeocin® (Life Technologies, Carlsbad, CA) selection. Once the stably transfected pool was > 95% viable, cells were collected for quantitative reverse transcription polymerase chain reaction (qRT-PCR) for overexpression validation.
Quantitative RT-PCR
Exponential phase cultures (Day 4 after inoculation) were used for total RNA isolation as previously described.29 Relative expression levels of St6gal1 and St3gal4 and St3gal6 were determined using qRT-PCR and normalized to ActR5. M-MLV reverse transcriptase was used for the RT reactions according to the manufacturer’s protocol. Quantitative PCR was carried out using TaqMan® Universal PCR Master Mix (Life Technologies). he primers and probes are listed in Table 1 in Supporting Information.
FACS enrichment using 2,6-sialic acid-specific lectin
Fluorescein labeled Sambucus nigra lectin (FITC-SNA, EY Laboratories, San Mateo, CA) that preferentially binds to α2,6-linked sialic acid structure was used to enrich the ST6GAL1 overexpressed pools (both IgG expressing and host cell lines). About 5 × 106 cells were incubated with 100 μg/mL FITC-SNA in 1 mL phosphate buffered saline (PBS) for 15 min at 25°C, washed three times with PBS. Cells were resuspended in 1 mL PBS and sorted using a FACSAria™ III cell sorter (Becton-Dickinson) based on FITC fluorescence intensity. Cells were collected in 6-well plates containing chemically defined culture media as described in 2.1 for recovery and expansion. When the enriched cultures fully recovered (3–3.5 weeks postsort), they were subjected to postsort analysis using FITC-SNA stain.
Single-cell cloning and clone selection for ST6GAL1 overexpression in host cell lines
The enriched Zeocin resistant stable pool derived from CHOZN® GS host cell line that overexpressed ST6GAL1 was single-cell cloned using a FACSAria™ III cell sorter. The cells with top 5% FITC-SNA fluorescence was plated at 1 cell/well in 96-well plates, with 200 μL per well of Ham’s F-12 supplemented with 6 mM L-glutamine and 10% fetal bovine serum (FBS). One plate was plated using unstained cells based on forward and side scatter and no fluorescence as control for clonal outgrowth.
Biotinylated Maackia amurensis lectin II (MALII, Vector Labs, Burlingame, CA) at a final concentration of 5 μg/mL for 15 min at 25°C was incubated with 5 × 105 cells to stain for α2,3-linked sialic acid. Cells were washed twice with 1 × PBS supplemented with 0.1% bovine serum albumin (BSA) and incubated for 15 min at 25°C with 0.5 μg/mL streptavidin-Alexa Fluor647 (Life Technologies, Eugene, OR) and 10 μg/mL FITC-SNA. The cells were washed twice with 1 × PBS supplemented with 0.1% BSA before responded for two-color FACS analysis on a MACSQuant® Analyzer (Miltenyi Biotec, San Diego, CA).
Sialic acid linkage analysis by LTQ linear ion trap mass spectrometry including MSn fragmentation
IgG was purified as previously described. Total cellular protein was extracted using CelLytic M according to manufacturer’s protocol. IgG, or total cellular protein extracts, was reduced and carboxyamidomethylated according to standard procedures prior to trypsinization at 37°C overnight (12–16 h). Trypsin was deactivated by heating at 100°C for 5 min. Purification of fragments was carried out with a C18 SPE cartridge (Waters, 300 mg packing). After a wash with 5% acetic acid (AcOH), the peptides/glycopeptides were eluted sequentially in 20% isopropanol/5% AcOH, 40% isopropanol/5% AcOH, and 100% isopropanol. The eluent was dried down, reconstituted in phosphate buffer containing PNGase F, and incubated at 37°C overnight. The released glycans were purified using a C18 cartridge, permethylated31 and diluted into 1 mM lithium carbonate/50% MeOH and infused directly into an LTQ Orbitrap Discovery Mass Spectrometer (Thermo Scientific) at a flow rate of 0.5 μL/min for nanospray ionization. A full Fourier transform mass spectrometry (FTMS) spectrum was obtained at a 30,000 resolution for each sample to determine which glycans contained sialic acids. The sialic acid linkage of the sialylated glycans was determined by MSn analysis as described by Anthony et al.9 The sample was subjected to multiple ion selection and fragmentation steps within the ion trap to break down a complex-type glycan down to a single galactose and then, the fragmentation pattern was observed. This process is outlined in Figure 2 in Supporting Information.
Total sialic acid content analysis
Purified IgG samples were prepared for sialic acid analysis as follows: Briefly, 30 μL of 10 M acetic acid was added to a 120 μL aliquot of each sample and triplicates of control sample (25 μg of fetuin in 120 μL of water) and incubated at 80°C for 1.5 h. Samples were centrifuged and cooled to 25°C. Supernatant was removed and a 5 μL aliquot of each sample was added to 100 μL of the 1,2-diamino-4,5-methylenedioxybenzene (DMB) labeling solution. The labeling solution consisted of 7 mM DMB dissolved in 18 mM sodium hydrosulfite, 0.75 M 2-mercaptoethanol in 2 M acetic acid. Similarly, a standard mixture of two sialic acids, N-acetylneuraminic acid (NANA) and N-glycolylneuraminic acid (NGNA) were also prepared and treated with the DMB labeling solution. The labeling reaction was allowed to continue at 50°C for 3 h and quenched with 500 μL water. The resulting sample solutions were analyzed by HPLC with fluorescence detection according to the following parameters. Column: express RPA 100 × 2.1 mm (Supelco); column temperature: 30°C; sample temperature: 4°C; mobile phase: A: water, B: ACN (both with 0.1% formic acid); flow rate: 0.2 mL/min; injection volume: 10 μL; FExcitation = 373 nm; FEmission = 448 nm.
Glycoform and sialylated glycan analysis of IgG stable expression in the ST6GAL1 overexpression host cell lines
ST6GAL1 OE clone 32 was used for generating stable pools expressing a model human IgG using GS selection as described elsewhere.30 Recombinant proteins were collected from 7-day fed-batch cultures (fed with 2 g/L glucose on Day 4), quantified for volumetric productivity as described previously, purified using protein A affinity chromatography and analyzed for glycoform profiles by intact protein mass as previously described.30 Glycoform relative abundance (% of total glycoforms) was calculated for all significant peaks. A second independent HPLC-fluorescence mass spectrometry method was employed to characterize the released N-glycans of these biomolecules.32 Approximately, 200 μg of glycoprotein was PNGase F-treated in a filter-aided sample prep (FASP) format33 after urea-mediated denaturation and buffer exchange. Reduction and alkylation steps were not employed. The glycosidase reaction progressed at 37°C for 18 h and released glycans captured in the filtrate and dried by vacuum centrifugation. Dried glycans were procainamide-labeled by 3 h 65°C reductive amination using a reaction solution of dimethyl sulfoxide/acetic acid/water/procainamide hydrochloride/sodium cyanoborohydride in a 24:13:41:1:2.4 molar ratio. About 6.6 μL reaction solutions were diluted with 100 μL 70% acetonitrile 30% water (v:v) and transferred to autosampler vials. UPLC/FLR/MS was accomplished on a Waters (Milford, MA) Acquity UPLC-Waters FLR fluorescence detector-Thermo (Waltham, MA) LTQ mass spectrometer system. Ten microliters of each sample was injected onto an Ascentis® Express OH5 15 cm × 2.1 mm, 2.7 μm column (Sigma-Aldrich Supelco, St. Louis MO) with 0.3 mL/min, 26 min 75%/25% to 52%/48% acetonitrile/ammonium formate pH 4.4 gradient elution. Fluorescence was monitored at 308 nm excitation/359 nm emission. The IonMax ESI-MS source was plumbed in series after the FLR detector, and positive-mode scans acquired from 500–2000 m/z.
In order to verify the sialic acid linkage, western blotting using biotinylated SNA lectin (Vector Labs) was performed according to manufacturer’s protocol. Briefly, 2 μg purified IgG and 1 μg human transferrin were resolved on SDS-PAGE and blotted on nitrocellulose membranes. Biotinylated SNA at 1.7 μg/mL was incubated with the membrane for 30 min at 25°C, and streptavidin-horse radish peroxidase (Thermo Scientific) at 2 μg/mL was used for detection.
Results
ST6GAL1 overexpression in IgG-producing cell lines and lectin-based enrichment in IgG-producing cell lines overexpressing ST6GAL1
The IgG-producing lines Clone 27, Clone N22, and Clone N46 (described in Materials and Methods) were stably transfected with the St6gal1 expression vector under the same selection conditions. However, the three cell lines demonstrated different levels of cell surface FITC-SNA staining. Clone 27 had the strongest pre- and postenrichment staining (37.7% and 64.6% positive, respectively, Figure 1). Clone N46 was 14.7% positive prior to enrichment and sustained enrichment as well (39.1% positive, Figure 1). Clone N22 ST6GAL1 pool was zeocin-resistant but did not have positive FITC-SNA staining and did not proceed to enrichment (data not shown). Interestingly, St6gal1-relative mRNA levels by qRT-PCR indicated that the overexpression stable pools from Clone 27 and Clone N22 had similar levels of St6gal1, Clone N46 had over 5-fold higher St6gal1 mRNA than Clones 27 and N22, whereas the nontransfected controls did not have detectable St6gal1 mRNA (data not shown).
Figure 1. FACS analysis and enrichment of cell lines stably transfected with St6gal expression vector.
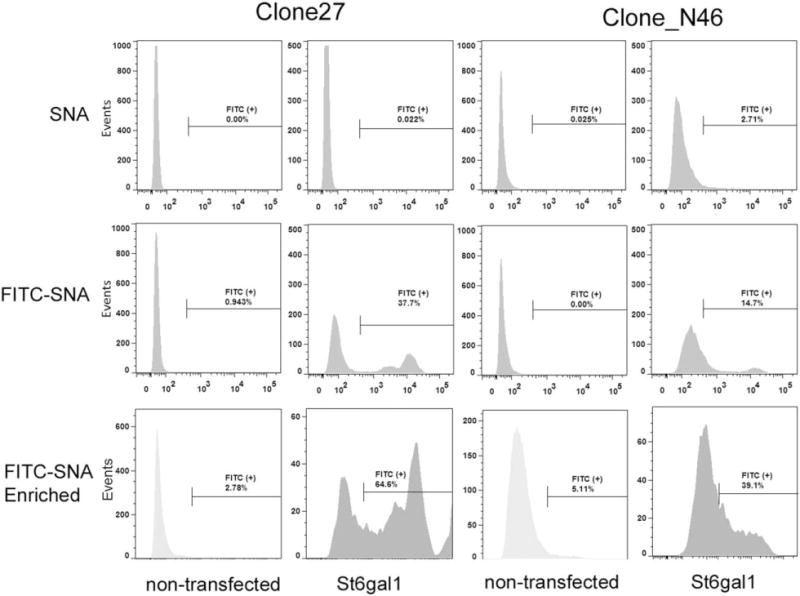
IgG-producing Clone 27 and Clone N46 demonstrated positive FITC-SNA lectin staining (specific for α2,6-linked sialic acid, middle row of histograms). Cells that were positive for FITC-SNA were collected, expanded, and reanalyzed for FITC-SNA staining, with nontransfected cells as control (bottom row of histograms). Nonconjugated SNA was used to stain the cells as background control (top row of histograms).
Three glycans were identified to contain sialic acid from IgG produced by Clone 27 St6gal1 OE and the nontransfected control (Supporting Information Figure 2). The sialylated glycans were further fragmented down to a single galactose for observing the fragmentation pattern (Figure 2). The fragmentation patterns of all three glycans indicated that the sialylation of nontransfected control to be primarily α2,3-linked, while Clone 27 St6gal1 OE gave a combination of α2,3- and α2,6-linked.
Figure 2. Sialic acid linkage analysis of the enriched stable pool overexpressing ST6GAL1.
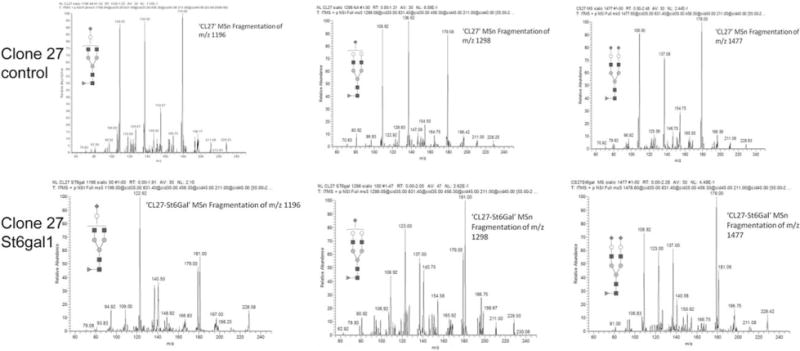
The three sialylated glycans were subjected to MSn analysis (Supporting Information Figures 1 and 2) for sialic acid linkage analysis. The fragmentation patterns obtained by this analysis were compared to α2,3- and α2,6-sialyllactose standards.9
IgG harvested and purified from late-exponential phase batch cultures (Day 7 of culture) demonstrated increased total sialic acid content in the enriched stable pool overexpressing ST6GAL1 derived from Clone 27, but not in Clone N46 (Figure 3). This is consistent with the higher percentage of FITC-SNA (+) population in the former enriched stable pool, which indicated a higher overall content of α2,6-linked sialic acid.
Figure 3. Total sialic acid content analysis of the enriched stable pool overexpressing ST6GAL1.
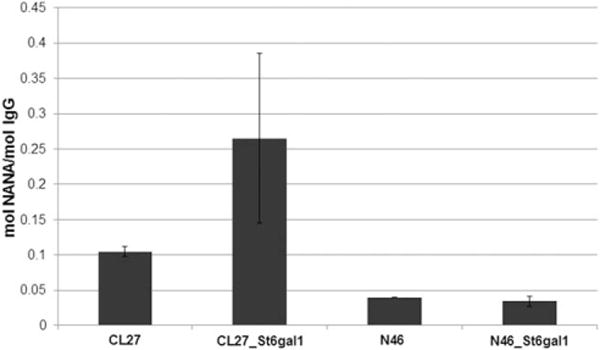
CL27: Clone 27 stably transfected with the zeocin control vector. CL27-St6gal1: enriched stable pool derived from Clone 27 overexpressing ST6GAL1. N46: Clone N46 stably transfected with the zeocin control vector. N46-St6gal1: enriched stable pool derived from Clone N46 overexpressing ST6GAL1. Error bars represent standard deviation of biological duplicates. NANA: N-acetylneuraminic acid. No N-glycolylneuraminic acid (NGNA) was detected.
Significant enrichment of St6gal1 overexpression in the clonal cell lines derived from the CHOZN® GS host cell line
The CHOZN® GS host cell line was transfected and enriched for FITC-SNA (+) population as previously described for the IgG-producing cell lines. The enriched ST6GAL1 stable pool demonstrated 25% FITC-SNA (+) compared to pre-enrichment (12%, data not shown here). The clonal outgrowth of the FITC-SNA stained and nonstained cells was equal after FACS plating (data not shown), indicating no growth inhibition or cytotoxicity from lectin staining. Thirty-seven single-cell clones were selected based on their percentage of FITC-SNA (+) populations and expanded to repeat the FACS analysis. Figure 4A gave FITC-SNA histograms of two representative clones. The majority of the clones that outgrew from the enriched population demonstrated increased FITC-SNA fluorescence in 96-well culture (Panel B), and the 35 clones selected sustained high FITC-SNA fluorescence in 24-well cultures (Panel C). Twelve clones with the highest FITC-SNA fluorescence in 24-well were expanded to TPP tube cultures for further characterization. Interestingly, FITC-SNA stain (% positive) reduced after adapted in shaken cultures for some clones, while the others maintained the same. This resulted a poor linear correlation (R2 = 0.5) for the % FITC-SNA (+) for the shaken versus static cultures (data not shown).
Figure 4. FITC-SNA staining of single-cell clones derived from the CHOZN® GS enriched ST6GAL1 stable pool.
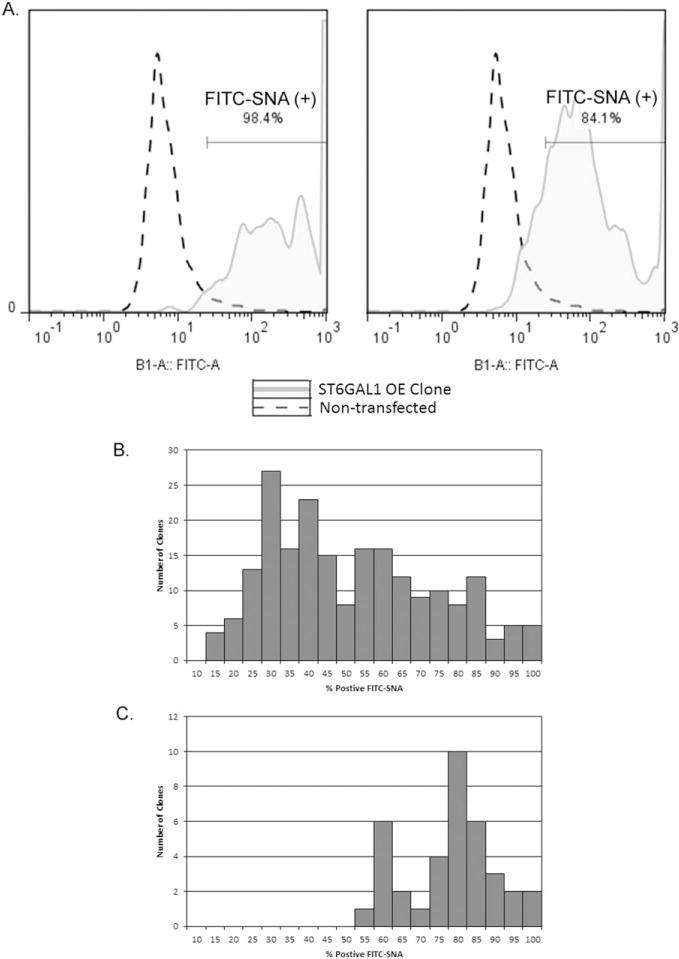
A. FACS histograms of two representative single-cell clones overexpressing ST6GAL1 stained with FITC-SNA lectin. B. Histogram of % FITC-SNA (+) cells of the clones isolated from the FITC-SNA enriched pool overexpressing ST6GAL1 (N=208) in static 96-well cultures. C. Histogram of % FITC-SNA (+) cells of the clones with the highest % FITC (+) selected from the clones depicted in Panel B (N=35) in static 24-well cultures.
Two-color FACS analysis indicated the presence of both α2,6- and α2,3-linked sialic acid in the ST6GAL1 overexpression clones
A two-color staining method was optimized to detect surface sialic acid with both linkage types simultaneously. Figure 4 in Supporting Information illustrates that there was no competition between MALII and SNA using this protocol, as indicated by the excellent correlation with a slope of 1.02 between the two- and single-color staining. All 12 ST6GAL1 overexpression clones are 100% MALII (+) with various percentages of SNA (+) cells (Figure 5). ST6GAL1 Clone 32 is notably the highest % FITC-SNA among the 12 clones in all stages of expansion. The nontransfected CHOZN® GS host cell line is also 100% MALII (+), but no FITC-SNA stain above background. This implies that the two sialyltransferases are independent in forming terminal sialic acid residues when the substrate is abundant.
Figure 5. Two-color FACS analysis of SNA and MALII lectin staining of ST6GAL1 overexpression clones.
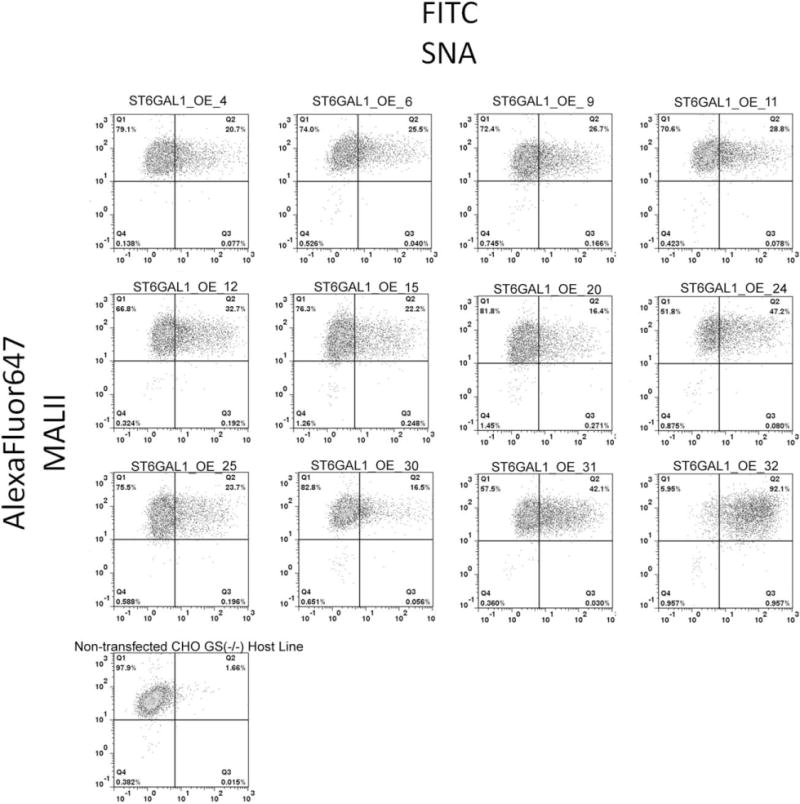
Representative scatter plots with quadrant analysis from the 12 clones overexpressing ST6GAL1 are shown here. The X-axis is fluorescence of FITC-SNA. The Y-axis is fluorescence of AlexaFluor647, representing MALII lectin staining. Each quadrant is labeled with % of total population. The gating was set based on streptavidin-AlexaFluor647 only of each OE clone (scatter plots not shown here) and based on FITC of the nontransfected host cell line, respectively, to exclude background fluorescence.
Expression assessment of endogenous ST3 sialyltransferases in the ST6GAL1 overexpression clones
Cells were harvested during exponential growth phase of TPP cultures and analyzed using qRT-PCR for the recombinant St6gal1 and three ST3 sialyltransferases (St3gal3, St3gal4, and St3gal6) in the 12 ST6GAL1 overexpression clones. St3gal3, 4, and 6 has been reported in literature to be expressed in CHO cells,3,34 whereas the absence of St6gal1 transcript is reported in CHO cells.35,36 All the ST6GAL1 overexpression clones express detectable levels of mRNA of St6gal1 (Figure 6). Interestingly, no linear correlation was observed between the surface SNA stain and St6gal1 mRNA levels. Western blot results were unavailable (Materials and Methods in Supporting Information) due to lack of commercial antibodies to Chinese hamster ST6GAL1. The expression levels of the ST3 sialyltransferases appear to be independent of the recombinant St6gal1 expression in these clones, with St3gal4 being the highest expressing sialyltransferase, followed by St3gal6 and St3gal3 nondetected (data not shown). All clones have similar levels of surface α2,3-linked sialic acid as detected by MALII lectin stain (data not shown).
Figure 6. Expression assessment of St6gal1.
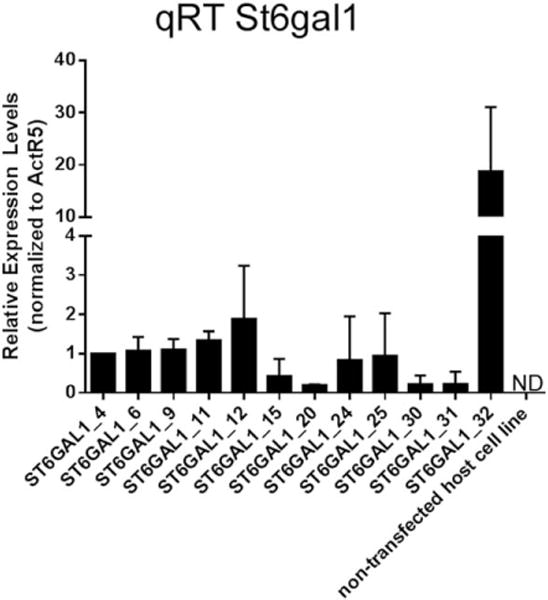
Quantitative RT-PCR was used to assess the relative expression of St6gal1 in 12 overexpression cell lines and the nontransfected CHOZN®GS host cell line. ActR5 was used for normalization. Error bars represent standard deviation of two independent qRT-PCR experiments. Relative expression was reported as fold increase to clone ST6GAL1_4. ND: not detected.
Sialic acid linkage analysis of total cell protein
There is striking difference between the N-glycan profiles between GS (−/−) host cell line and the two ST6GAL1 OE cell lines (ST6GAL1_OE_31 and 32). As illustrated in Figure 7, there are no sialylated glycans identified in “2E3” due to their low abundance. By contrast, mono-, bi-, tri-, and tetrasialylated N-glycans are identified in both ST6GAL1 OE cell lines. ST6GAL1_OE_32 demonstrated all but one sialylated population to contain both α2,3- and α2,6-linkages. ST6GAL1_OE_31, on the other hand, has several populations that cannot be confirmed to have α2,6-linked sialic acid (Figure 8). The most abundant sialylated N-glycan for both ST6GAL1 OE cell lines is the bisialylated, fucosylated (GlcNAc)4(Man)3(Gal)2(Fuc)(NeuAc)2. ST6GAL1_OE_32 has 9% of this glycan, a 1.8-fold increase compared to 31. ST6GAL1_OE_31 contains higher mono-sialylated glycans, albeit some only contain α2,3-linkage. The complete glycan identification results are included in Table 2 in Supporting Information. The linkage analysis results indicated that overexpression of ST6GAL1 led to increase in complex glycans. Higher levels of St6gal1 expression in ST6GAL1_OE_32 also resulted in higher relative content α2,6-linked sialylated glycans compared to ST6GAL1_OE_31.
Figure 7. FTMS (800–2000 m/z) of permethylated glycans found in total cellular protein extracts from two host cell lines with ST6GAL1 overexpressed and the nontransfected CHOZN® GS host cell line.
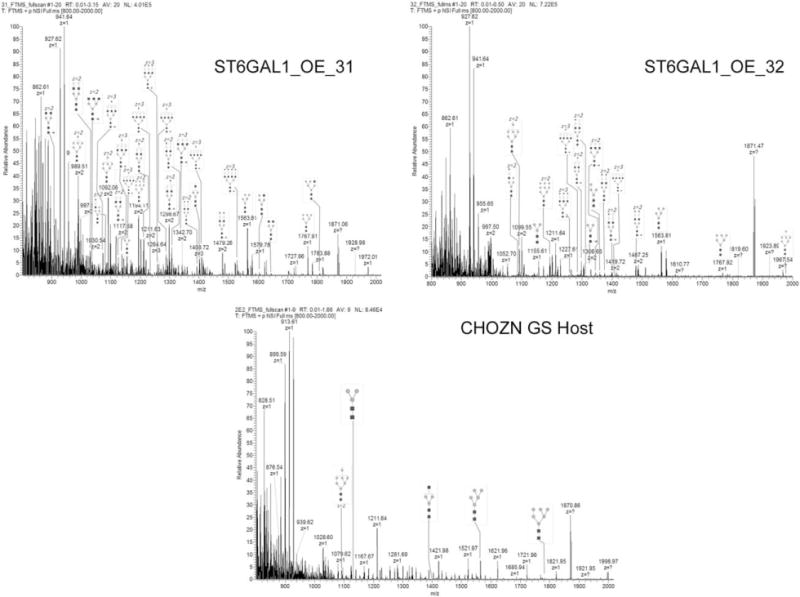
The identity of each glycan was observed based on mass and confirmed by fragmentation following total ion mapping conducted on all samples. Full MS-spectra including glycan peak identification are shown here. Tables listing each of the glycans detected are found in Table 2 in Supporting Information. The peaks are labeled by their observed m/z and the glycoform structure.
Figure 8. Semi-quantitative analysis of sialylated N-glycans.
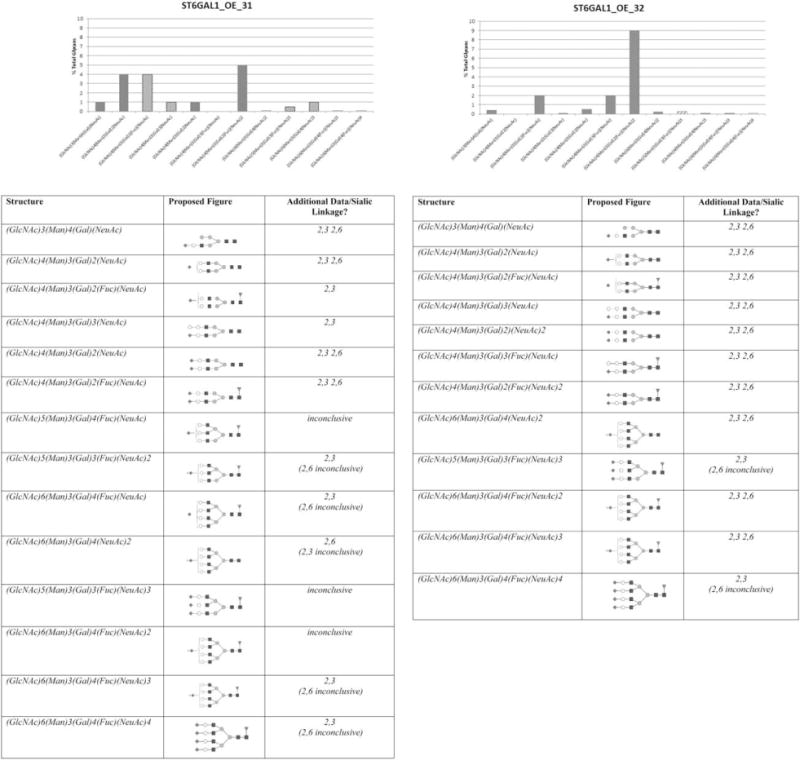
The tables summarize the glycoform structure and the sialic acid linkage of all N-glycans containing sialic acid. The percentage of each glycan was calculated by totaling intensities for each peak and calculating percentages from the sum. The solid columns in the graphs indicate both α2,3- and α2,6-linked sialic acid. The dashed columns indicate α2,3-linkage.
Productivity, glycoform profiles, sialylated glycan, and linkage analysis of IgG expressed in the selected St6gal1 OE host cell line
In seven-day simple fed-batch cultures, the two stable pools generated from ST6GAL1_OE_32 host cell line produced significantly more IgG than the nonengineered host cell line using minus glutamine selection (Figure 9A). Viability was above 90% when harvested for all cultures. Intact protein mass analyses of the two IgG stable pools derived from two transfections using ST6GAL1_OE_32 host cell line revealed three significant peaks of 50850, 51011, and 51303 Da, all larger than the G2F glycoform (Figure 9B). The peaks indicated complex glycoforms that are likely sialylated and consist of 4.5%–9.1% of total glycoforms. By contrast, these peaks were not detected in the control IgG stable pools derived from CHOZN® GS host cell line (Figure 9C). The IgG produced in ST6GAL1_OE_32 host cell line also appeared to have a lower relative abundance of the G0F and G2F glycoforms than IgG produced in CHOZN® GS host cell line (Figure 9B), which may contribute to the increased relative content of more complex glycoforms (G1FS1, G2FS1, and G2FS2). G2FS2 peaks were identified, but they did not meet the cutoff for % total glycoforms therefore only noted presence but not quantified.
Figure 9. Stable pool productivity and glycoform profiles of IgG produced in ST6GAL1 OE host cell line by intact protein mass.
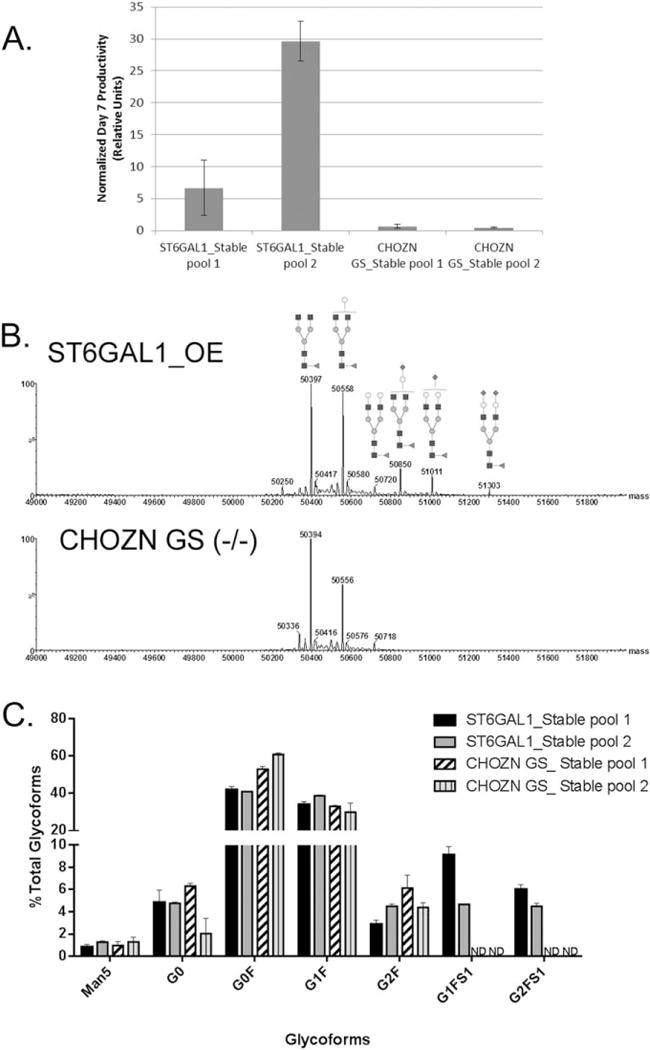
A. Day 7 IgG productivity from two stable pools generated from different transfections. The IgG concentrations were normalized by that of the CHOZN GS_Stable pool 1 collected from the first experiment. Error bars represent three independent cell culture experiments. B. The representative mass spectra of the IgG glycoform profiles of a stable IgG-producing “bulk” pool derived from ST6GAL1_OE_32 and a stable pool from the CHOZN® GS host cell line. The top pane shows the de-charged mass spectrum of IgG heavy chain from ST6GAL1_OE_32. Annotated mass peaks are consistent with glycoforms bearing the indicated glycans. Species 50850, 51011, and 51303 are sialyated. The bottom pane shows the mass spectrum of IgG heavy chain from the CHOZN® GS host cell line. C. The IgG glycoform distribution (% of total glycoforms) of two stable pools derived from ST6GAL1_OE_32 and two stable pools from the CHOZN® GS host cell line. G2SA2F (51303) is below quantification limit. Error bars represent three independent cell culture experiments.
Released glycan HPLC analysis indicated that IgG produced in ST6GAL1_OE_32 host cell line had more sialic acid per molecule (Figure 10A). This assessment is based on the mass spectral identification of the features eluting in the normal phase HILIC chromatography (Figure 10B). Also corroborating the MS-based ID is the late elution of the hydrophilic sialylated glycans, which matches the trend seen with the released glycans of transferrin that was used as an external control (data not shown). Only terminal N-acetylneuraminic acid (NANA) was detected. N-glycolylneuraminic acid (NGNA) was not detected in this a model human IgG, nor in previous studies using our CHO cell lines (data not shown here). Western blotting using biotinylated SNA showed strong positive bands at 50 kDa, indicating the glycosylated heavy chain, for IgG produced in both ST6GAL1 stable pools, and no bands were detected for the IgG produced in the control CHOZN® GS-transfected pools (Figure 10B). A duplicate SDS-PAGE gel stained with Coomassie Blue was included in Figure 6 in Supporting Information. This validated the presence of α2,6-linked sialic acid in the IgG produced in both ST6GAL1 stable pools.
Figure 10. Released glycan HPLC-fluorescence-MS analysis and SNA lectin western blotting of IgG produced in ST6GAL1 OE and the control host cell lines.
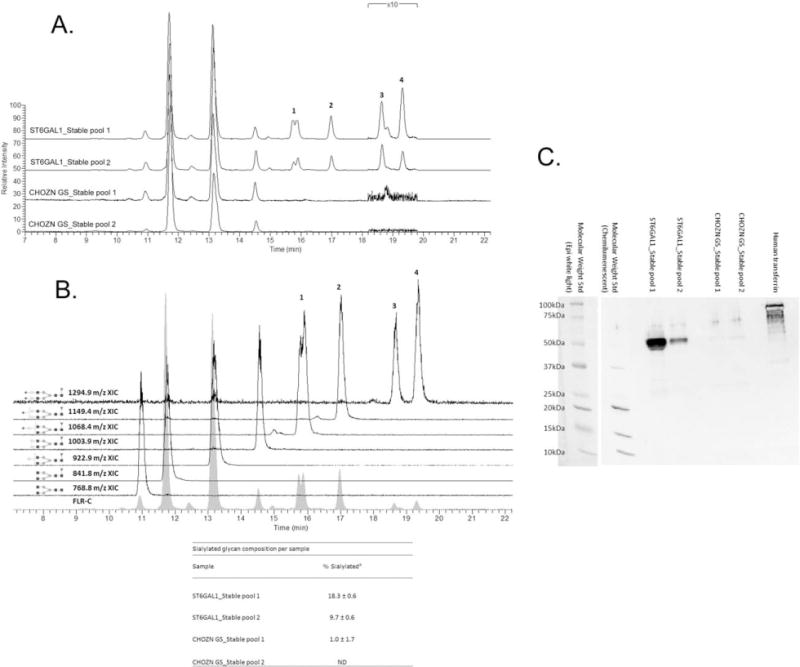
A. Fluorescence HPLC-based glycoprofiles for IgG expressed in transfected and control cell lines. Released N-glycans were labeled with procainamide and resolved by HILIC normal phase chromatography coupled to a fluorescence detector (308 nm excite, 359 nm emission) and the ESI source of a linear trap mass spectrometer. Each feature has been identified by MS as a procainamide-labeled glycan; peaks 1–4 correspond to sialylated glycans. B. Fluorescence and extracted ion chromatograms (XICs) for the glycoprofile of IgG expressed in a ST6Gal1-transfected cell line pool. Each fluorescence peak corresponded to a characteristic doubly charge ion with a mass consistent with the annotated glycan plus procainamide label (+219 Da). The XICs show the elution of these ions values, ±0.5 m/z. Squares are N-acetylglucosamine, grey circles mannose, triangles fucose, light circles galactose, and diamonds N-acetylneuraminic (sialic) acid. Linkage and anomericity is not conveyed by these data. Peaks 1–4 are glycans with atleast one sialic acid residue. The inset table: % sialylated glycan composition based on fluorescence (Mean±SD from three independent cell culture experiments). C. Western blotting using biotinylated SNA. Each lane was loaded with 2 μg purified IgG from one of the three cell culture experiments, and the control lane was loaded with 1 μg human transferrin as the positive control for α2,6-linked sialic acid. The molecular weight standard was imaged with both chemiluminescence and epi-white light due to the short exposure (0.5 and 1 s) required for adequate chemiluminescence imaging of the high-intensity heavy chain bands.
Discussion
We are the first to report sialic acid enhancement in a GS host cell line that is increasingly employed in biopharmaceutical production. We successfully applied FACS enrichment and single-cell selection for α2,6-linked sialic acid host cell engineering, resulting in cell lines overexpressing functional ST6GAL1. FITC-SNA stain did not seem to exhibit cytotoxicity nor affect clonal outgrowth in presence of 10% FBS. FACS enrichment started from a stably transfected pool in chemically defined suspension culture and single-cell clones with top 5% FITC-SNA fluorescence were plated in media containing 10% FBS. The serum-containing static cultures (96 well and 24 well, Figure 4) demonstrated similar relative FITC-SNA fluorescence intensity. After adaptation into chemically defined suspension cultures, some ST6GAL1 overexpression clones (such as clone 32) maintain very stable FITC-SNA fluorescence, whereas others showed reduction in relative FITC-SNA fluorescence and less % positive with similar FACS gating strategy. Indeed, a recent publication reported sialylation changes during the serum-free adaptation process.37 Suspension adaptation may also alter surface sialic acid by physical sheer or physiological mechanism such as accelerated sialic acid degradation. Similarly, the role of surface sialic acid has been well studied in cellular adhesion process.38–40 All clones exhibited clumping in suspension culture, which was corrected after incorporating dextran sulfate in the media, implying that increased surface sialic acid may alter cell adhesion properties. On a practical note, the FACS-enriched stable pools can certainly be used for serum-free or chemically defined single-cell selection for biopharmaceutical production. Likewise, 96-well shaken cultures can be introduced in the clone selection process to reduce the risk of selecting clones that tend to lose α2,6-linked sialic acids during adaptation to shaken cultures.
The clones have varied levels of St6gal1 expression, which may be attributed to position effect of random genomic integration. The lack of linear correlation between recombinant St6gal1 mRNA and surface α2,6-linked sialic acid may imply a threshold-type dose response. Clone 32 has the highest FITC-SNA stain and mRNA level among the clones screened. Some clones have increased FITC-SNA stains, indicating the desired phenotype (α2,6-linked sialic acid), but much lower mRNA levels than Clone 32. One should start with sufficient number of candidate clones and use phenotypic screen (SNA binding) as the primary criteria, in order to ensure high levels of α2,6-linked sialic acid in the recombinant therapeutic proteins produced by the engineered host cell lines.
The N-glycan analyses using total cellular proteins demonstrated that ST6GAL1 overexpression in Clone 31 and 32 was sufficient to generate complex sialylated glycans with both linkages, while the CHOZN® GS host line with no ST6GAL1 had lower abundant complex glycans in cellular proteins (and nondetectable sialylated glycans). Despite the drastically different St6gal1 mRNA levels, the difference between the two clones is that ST6GAL1_OE_32 has 10 peaks fully determined 2,6 linkage and ST6GAL1_31 has 5, but not a significant difference in % total glycans (Figure 8). This also appears to support the threshold theory for St6gal expression.
Previous work reported altered proportions of sialylation but no report of net increase in total sialic acid.27 One limiting factor for sialylation is sufficient transgolgi pool of CMP-sialic acid12. Overexpression of ST6GAL1 in a cell line with no endogenous expression may lead to potential substrate competition between the two classes of sialyltransferases and even a decrease of α2,3-linked sialic acid. Another limiting factor for sialylation is feedback regulatory mechanisms based on concentrations of CMP-sialic acid or UDP-GlcNAc.39,41 The net increase in sialic acid content that we reported may indicate that either CHO K1 host cells provide enough substrate to accommodate sialyltransferase OE or can override these potential negative regulatory mechanisms to achieve net increase of N-linked sialylation. Additionally, based on the qRT-PCR results of the closely related genes of the N-glycan biosynthesis pathway, it was found that there was no occurrence of up- or down-regulation of the ST3 transferase genes or the β1,4-galactosyltransferase (B4galt1) in the ST6GAL1_OE clones (data not shown).
As previously reported, “minipool” selection has demonstrated significant advantage over bulk selection. It is fascinating that the ST6GAL1 OE host cell line demonstrated higher producing stable pools after minus glutamine selection, albeit about a week longer for recovery time than the CHOZN GS host line. We speculate that the observed decreased specific growth rate in the ST6GAL1_OE clones that is similar to Onitsuka et al description27 may allow for sufficient expression of recombinant GS for the transfectants to overcome selection and hence led to higher expressing stable pools. In static culture, the altered surface adhesion could also contribute to better recovery in a bulk stable pool. This gained characteristic may enable bypass of “minipool” selection and evaluation and proceed to single-cell cloning directly from bulk pool selection, which would save at least four weeks in cell line development according to our current protocols.
Cell engineering sometimes requires reasonable compromise and balance between existing characteristics and the gained function. In simple fed-batch, the peak viable cell density (VCD) of ST6GAL1_OE_32 is 4−5×106/mL, whereas CHOZN® GS host cell line is routinely 8−9×106/mL (data not shown). Although the growth was much improved from previously reported, it can be further improved through media and process optimization, particularly in producing clones. For host cell engineering, selecting clones with similar ST6GAL1 overexpression but higher growth from more candidates can also solve this. ST6GAL1 OE host cell lines can be very well suitable for pharmaceutical production after optimizing growth during clone selection or upstream process development. Furthermore, combinatorial overexpression approaches, despite the well-known challenges, may provide further increase in sialylation. Our St6gal1 OE cell lines aim for an improved baseline for sialylation development than wild-type CHO host cells.
Additionally, this work may contribute to host cell engineering for cell culture vaccine production. Membrane protein sialylation is critical in the mechanism of viral entry to mammalian cells.42,43 Influenza hemagglutinin interacts with sialylated membrane receptors that mediate membrane fusion during infection.44 α2,6-Linked sialic acid is considered essential for the infectivity; the ratio of the linkages may alter species preference, as the H7N9 influenza viruses in the most recent epidemic in Asia was reported to have preference with α2,3-linked over α2,6-linked sialic acid.45 Consistent with these mechanisms, Bilsel demonstrated that overexpression of human ST6GAL1 led to influenza infectivity to CHO cells.46 CHO cells are easier to culture in animal component free and chemically defined media compared with other current cell culture vaccine host cell lines such as Vero, MDCK, and PER.C6® cells. Host cell engineering through sialic acid modulation may provide a viable approach for vaccine production in CHO cells.
In conclusion, this work represents our ongoing effort of glycoengineering in CHO host cell lines for the development of “bio-better” protein therapeutics and cell culture vaccine production.
Supplementary Material
Acknowledgments
The authors thank Drs. Kevin Ray, Ben Cutak, and Pegah Jalili for their analytical support, discussions, and editing of this manuscript, Ms. Kelly Keys for flow cytometry support and discussions and Ms. Laura Lauck for technical support in cell culture.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Contributor Information
Nan Lin, Cell Sciences and Development, SAFC/Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103.
Joaquina Mascarenhas, Cell Sciences and Development, SAFC/Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103.
Natalie R. Sealover, Cell Sciences and Development, SAFC/Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103
Henry J. George, Cell Sciences and Development, SAFC/Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103
Jeanne Brooks, Cell Sciences and Development, SAFC/Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103.
Kevin J. Kayser, Cell Sciences and Development, SAFC/Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103
Brian Gau, Analytical R&D, Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103.
Isil Yasa, Analytical R&D, Sigma-Aldrich, 2909 Laclede Avenue, Saint Louis, MO 63103.
Parastoo Azadi, The Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602.
Stephanie Archer-Hartmann, The Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602.
Literature Cited
- 1.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 2.Byrne B, Donohoe GG, O’Kennedy R. Sialic acids: carbohydrate moieties that influence the biological and physical properties of biopharmaceutical proteins and living cells. Drug Discov Today. 2007;12:319–326. doi: 10.1016/j.drudis.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Fukuta K, Yokomatsu T, Abe R, Asanagi M, Makino T. Genetic engineering of CHO cells producing human interferon-gamma by transfection of sialyltransferases. Glycoconj J. 2000;17:895–904. doi: 10.1023/a:1010977431061. [DOI] [PubMed] [Google Scholar]
- 4.Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19:936–949. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins N, Parekh RB, James DC. Getting the glycosylation right: implications for the biotechnology industry. Nat Biotechnol. 1996;14:975–981. doi: 10.1038/nbt0896-975. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol. 2012;94:887–905. doi: 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santell L, Ryll T, Etcheverry T, Santoris M, Dutina G, Wang A, Gunson J, Warner TG. Aberrant metabolic sialylation of recombinant proteins expressed in Chinese hamster ovary cells in high productivity cultures. Biochem Biophys Res Commun. 1999;258:132–137. doi: 10.1006/bbrc.1999.0550. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi M, Takasaki S, Miyazaki H, Kato T, Hoshi S, Kochibe N, Kobata A. Comparative study of the asparagine-linked sugar chains of human erythropoietins purified from urine and the culture medium of recombinant Chinese hamster ovary cells. J Biol Chem. 1988;263:3657–3663. [PubMed] [Google Scholar]
- 9.Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG antiinflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthony RM, Ravetch JV. A novel role for the IgG Fc glycan: the anti-inflammatory activity of sialylated IgG Fcs. J Clin Immunol. 2010;30:S9–S14. doi: 10.1007/s10875-010-9405-6. [DOI] [PubMed] [Google Scholar]
- 11.Bork K, Horstkorte R, Weidemann W. Increasing the sialylation of therapeutic glycoproteins: the potential of the sialic acid biosynthetic pathway. J Pharm Sci. 2009;98:3499–3508. doi: 10.1002/jps.21684. [DOI] [PubMed] [Google Scholar]
- 12.Butler M. Optimisation of the cellular metabolism of glycosylation for recombinant proteins produced by mammalian cell systems. Cytotechnology. 2006;50:57–76. doi: 10.1007/s10616-005-4537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickramasinghe S, Hua S, Rincon G, Islas-Trejo A, German JB, Lebrilla CB, Medrano JF. Transcriptome profiling of bovine milk oligosaccharide metabolism genes using RNA-sequencing. PLoS one. 2011;6:e18895. doi: 10.1371/journal.pone.0018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong DC, Wong NS, Goh JS, May LM, Yap MG. Profiling of N-glycosylation gene expression in CHO cell fed-batch cultures. Biotechnol Bioeng. 2010;107:516–528. doi: 10.1002/bit.22828. [DOI] [PubMed] [Google Scholar]
- 15.Son YD, Jeong YT, Park SY, Kim JH. Enhanced sialylation of recombinant human erythropoietin in Chinese hamster ovary cells by combinatorial engineering of selected genes. Glycobiology. 2011;21:1019–1028. doi: 10.1093/glycob/cwr034. [DOI] [PubMed] [Google Scholar]
- 16.Wong NS, Yap MG, Wang DI. Enhancing recombinant glycoprotein sialylation through CMP-sialic acid transporter over expression in Chinese hamster ovary cells. Biotechnol Bioeng. 2006;93:1005–1016. doi: 10.1002/bit.20815. [DOI] [PubMed] [Google Scholar]
- 17.Ngantung FA, Miller PG, Brushett FR, Tang GL, Wang DI. RNA interference of sialidase improves glycoprotein sialic acid content consistency. Biotechnol Bioeng. 2006;95:106–119. doi: 10.1002/bit.20997. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Koskie K, Ross JS, Kayser KJ, Caple MV. Enhancing glycoprotein sialylation by targeted gene silencing in mammalian cells. Biotechnol Bioeng. 2010;105:1094–1105. doi: 10.1002/bit.22633. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins N, Buckberry L, Marc A, Monaco L. Genetic engineering of alpha 2,6-sialyltransferase in recombinant CHO cells. Biochem Soc Trans. 1998;26:S115. doi: 10.1042/bst026s115. [DOI] [PubMed] [Google Scholar]
- 20.Lee EU, Roth J, Paulson JC. Alteration of terminal glycosylation sequences on N-linked oligosaccharides of Chinese hamster ovary cells by expression of beta-galactoside alpha 2,6-sialyltransferase. J Biol Chem. 1989;264:13848–13855. [PubMed] [Google Scholar]
- 21.Minch SL, Kallio PT, Bailey JE. Tissue plasminogen activator coexpressed in Chinese hamster ovary cells with alpha(2,6)-sialyltransferase contains NeuAc alpha(2,6)Gal beta(1,4)Glc-N-AcR linkages. Biotechnol Prog. 1995;11:348–351. doi: 10.1021/bp00033a015. [DOI] [PubMed] [Google Scholar]
- 22.Monaco L, Marc A, Eon-Duval A, Acerbis G, Distefano G, Lamotte D, Engasser JM, Soria M, Jenkins N. Genetic engineering of alpha 2,6-sialyltransferase in recombinant CHO cells and its effects on the sialylation of recombinant interferon-gamma. Cytotechnology. 1996;22:197–203. doi: 10.1007/BF00353939. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Lok SH, Kon OL. Stable expression of human alpha-2,6-sialyltransferase in Chinese hamster ovary cells: functional consequences for human erythropoietin expression and bioactivity. Biochim Biophys Acta. 1998;1425:441–452. doi: 10.1016/s0304-4165(98)00095-6. [DOI] [PubMed] [Google Scholar]
- 24.Warner TG. Enhancing therapeutic glycoprotein production in Chinese hamster ovary cells by metabolic engineering endogenous gene control with antisense DNA and gene targeting. Glycobiology. 1999;9:841–850. doi: 10.1093/glycob/9.9.841. [DOI] [PubMed] [Google Scholar]
- 25.Bragonzi A, Distefano G, Buckberry LD, Acerbis G, Foglieni C, Lamotte D, Campi G, Marc A, Soria MR, Jenkins N, Monaco L. A new Chinese hamster ovary cell line expressing alpha 2,6-sialyltransferase used as universal host for the production of human-like sialylated recombinant glycoproteins. Biochim Biophys Acta. 2000;1474:273–282. doi: 10.1016/s0304-4165(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 26.Jeong YT, Choi O, Lim HR, Son YD, Kim HJ, Kim JH. Enhanced sialylation of recombinant erythropoietin in CHO cells by human glycosyltransferase expression. J Microbiol Biotechnol. 2008;18:1945–1952. [PubMed] [Google Scholar]
- 27.Onitsuka M, Kim WD, Ozaki H, Kawaguchi A, Honda K, Kajiura H, Fujiyama K, Asano R, Kumagai I, Ohtake H, Omasa T. Enhancement of sialylation on humanized IgG-like bispecific antibody by overexpression of alpha 2,6-sialyltransferase derived from Chinese hamster ovary cells. Appl Microbiol Biotechnol. 2012;94:69–80. doi: 10.1007/s00253-011-3814-1. [DOI] [PubMed] [Google Scholar]
- 28.Maï N, Donadio-Andréi S, Iss C, Calabro V, Ronin C. Engineering a Human-Like Glycosylation to Produce Therapeutic Glycoproteins Based on 6-Linked Sialylation in CHO Cells. In: Beck A, editor. Glycosylation Engineering of Biopharmaceuticals. Vol. 988. Humana Press; 2013. pp. 19–29. [DOI] [PubMed] [Google Scholar]
- 29.Bahr SM, Borgschulte T, Kayser KJ, Lin N. Using microarray technology to select housekeeping genes in Chinese hamster ovary cells. Biotechnol Bioeng. 2009;104:1041–1046. doi: 10.1002/bit.22452. [DOI] [PubMed] [Google Scholar]
- 30.Sealover NR, Davis AM, Brooks JK, George HJ, Kayser KJ, Lin N. Engineering Chinese hamster ovary (CHO) cells for producing recombinant proteins with simple glycoforms by zinc-finger nuclease (ZFN)-mediated gene knockout of mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (Mgat1) J Biotechnol. 2013;167:24–32. doi: 10.1016/j.jbiotec.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem. 1992;203:101–108. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- 32.Klapoetke S, Zhang J, Becht S, Gu X, Ding X. The evaluation of a novel approach for the profiling and identification of N-linked glycan with a procainamide tag by HPLC with fluorescent and mass spectrometric detection. J Pharm Biomed Anal. 2010;53:315–324. doi: 10.1016/j.jpba.2010.03.045. [DOI] [PubMed] [Google Scholar]
- 33.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 34.Zhang P, Tan DL, Heng D, Wang T, Mariati, Yang Y, Song Z. A functional analysis of N-glycosylation-related genes on sialylation of recombinant erythropoietin in six commonly used mammalian cell lines. Metab Eng. 2010;12:526–536. doi: 10.1016/j.ymben.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O’Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C, Xie M, Chen W, Li N, Baycin-Hizal D, Latif H, Forster J, Betenbaugh MJ, Famili I, Xu X, Wang J, Palsson BO. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31:759–765. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- 36.Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa AR, Withers J, Rodrigues ME, McLoughlin N, Henriques M, Oliveira R, Rudd PM, Azeredo J. The impact of cell adaptation to serum-free conditions on the glycosylation profile of a monoclonal antibody produced by Chinese hamster ovary cells. Nat Biotechnol. 2013;30:563–572. doi: 10.1016/j.nbt.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Hedlund M, Ng E, Varki A, Varki NM. Alpha 2–6-linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 2008;68:388–394. doi: 10.1158/0008-5472.CAN-07-1340. [DOI] [PubMed] [Google Scholar]
- 39.Valley U, Nimtz M, Conradt HS, Wagner R. Incorporation of ammonium into intracellular UDP-activated N-acetylhexosamines and into carbohydrate structures in glycoproteins. Biotechnol Bioeng. 1999;64:401–417. [PubMed] [Google Scholar]
- 40.Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pels Rijcken WR, Overdijk B, Van den Eijnden DH, Ferwerda W. The effect of increasing nucleotide-sugar concentrations on the incorporation of sugars into glycoconjugates in rat hepatocytes. Biochem J. 1995;305:865–870. doi: 10.1042/bj3050865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, Qi Y, Zhang FY, Yu XH, Wu YG, Chen Y, Jiang CL, Kong W. Overexpression of alpha-2,6 sialyltransferase stimulates propagation of human influenza viruses in Vero cells. Acta Virol. 2011;55:147–153. doi: 10.4149/av_2011_02_147. [DOI] [PubMed] [Google Scholar]
- 43.Nam HJ, Gurda-Whitaker B, Gan WY, Ilaria S, McKenna R, Mehta P, Alvarez RA, Agbandje-McKenna M. Identification of the sialic acid structures recognized by min virus of mice and the role of binding affinity in virulence adaptation. J Biol Chem. 2006;281:25670–25677. doi: 10.1074/jbc.M604421200. [DOI] [PubMed] [Google Scholar]
- 44.Schwarzer J, Rapp E, Hennig R, Genzel Y, Jordan I, Sandig V, Reichl U. Glycan analysis in cell culture-based influenza vaccine production: influence of host cell line and virus strain on the glycosylation pattern of viral hemagglutinin. Vaccine. 2009;27:4325–4336. doi: 10.1016/j.vaccine.2009.04.076. [DOI] [PubMed] [Google Scholar]
- 45.Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, Garcia-Sastre A, Fernandez-Sesma A. H7N9 influenza viruses interact preferentially with alpha 2,3-linked sialic acids and bind weakly to alpha 2,6-linked sialic acids. J Gen Virol. 2013;94:2417–2423. doi: 10.1099/vir.0.056184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bilsel P. Cell-Based Systems for Producing Influenza Vaccines. WO Patent. 2009
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


