Abstract
Short-term increases in oxidative stress and decreases in motor function, including debilitating effects on balance and motor control, can occur following primary mild traumatic brain injuries (mTBI). However, the long-term effects on motor unit impairment and integrity as well as the molecular mechanisms underlying secondary injuries are poorly understood. We hypothesized that changes in central nervous system-specific protein (CSP) expression might correlate to these long-term effects. To test our hypothesis, we longitudinally assessed a closed-skull mTBI mouse model, vs. sham control, at 1, 7, 30, and 120 days post-injury. Motor impairment was determined by rotarod and grip strength performance measures, while motor unit integrity was determined using electromyography. Relative protein expression was determined by microwave & magnetic (M2) proteomics of ipsilateral brain tissue, as previously described. Isoprostane measurements were performed to confirm a primary oxidative stress response. Decoding the relative expression of 476 ± 56 top-ranked proteins for each specimen revealed statistically significant changes in the expression of two well-known CSPs at 1, 7 and 30 days post-injury: P < 0.001 for myelin basic protein (MBP) and P < 0.05 for myelin associated glycoprotein (MAG). This was confirmed by Western blot. Moreover, MAG, αII-spectrin (SPNA2) and neurofilament light (NEFL) expression at 30 days post-injury were directly related to grip strength (P < 0.05). While higher-powered studies of larger cohorts merit further investigation, this study supports the proof-of-concept that M2 proteomics is a rapid method to quantify putative protein biomarkers and therapeutic targets of mTBI and suggests the feasibility of CSP expression correlations to long-term effects on motor impairment.
Keywords: microwave, magnetic, proteomics, mild traumatic brain injury, proteins, biomarkers, motor impairment, MBP, MAG, central nervous system-specific protein
INTRODUCTION
Traumatic brain injury (TBI) is caused by sudden and violent trauma, including: vehicle accidents, falls, sport related injuries, and acts of violence such as those occurring in combat situations. The CDC has recently reported that nearly one third (30.5%) of deaths associated with injury include a diagnosis of TBI and there are an estimated 5.3 million U.S. residents living with TBI-related disabilities [1]. Economic costs resulting from TBI were estimated at $76.5 billion for 2010, including $11.5 billion for direct medical costs and $64.8 billion for indirect costs (e.g., lost wages, lost productivity, and nonmedical expenditures) [2, 3]. TBI has been described as the “signature” injury of veterans from the conflicts in Iraq and Afghanistan, where repetitive and multiple combat injures are common [4]. Consequently, the DOD alone has invested more than $374.9 million to increase the quality and access to care for these veterans [5]. Approximately 20% of the 1,348,405 citizens that were deployed since September 11, 2001, were diagnosed with TBI, where 82% of these injuries were considered mild (mTBI) [5].
The devastating physical and emotional effects of TBI are compounded by a lack of FDA-approved diagnostic, prognostic or predictive biomarkers. The acute effects of TBI (primary injuries) have been the focus of most biomarker studies, while sub-acute and long-term effects of TBI (secondary injuries) have not been received as much attention. Secondary injuries due to mTBI are expected to be particularly subtle at the molecular level, posing a profound challenge for the discovery of clinically relevant biomarkers. Primary injuries are characterized by short-term increases in oxidative stress and decreases in motor function [6–9]. These initial events are followed by a poorly understood secondary response characterized by long-term effects associated with neuronal degeneration and functional and cognitive deficits, including deficits in memory, coordination, judgment, balance and fine motor skills [7]. While the importance of investigating these long-term changes is becoming more appreciated due to strengthening links between TBI and multiple age-associated neurodegenerative diseases [10–15], few pre-clinical studies have examined the long-term functional and biochemical changes associated with mTBI [11, 16–19].
The most sensitive (most true-positive) and specific (least false-positive) biomarkers are expected to be proteins. More than 24,000 genes are translated into an estimated 2 million protein isoforms in humans, encoding far more molecular diversity than the relatively static genome or transcriptome. Paradoxically, less than 100 proteins are routinely quantified in blood today [20, 21]. Proteins must be measured directly due to the poor correlation between the transcriptome and proteome due to alternative splicing, post-translational modifications, single nucleotide polymorphisms, limiting ribosomes available for translation, mRNA and protein stability, and other actors (e.g., microRNA). Central nervous system-specific proteins (CSPs), transported across the damaged blood-brain-barrier to cerebral spinal fluid (CSF) or blood, are attractive protein biomarkers for TBI because they are not expected at appreciable levels in the circulation of healthy controls. However, amino acid sequence specific tandem mass spectrometry (MS/MS)-based proteomic analysis of low abundance CSPs can be confounded by masking effects due to high abundance proteins, particularly in CSF or blood where protein abundance can span up to 12 orders of magnitude. For these reasons and others, proteomic analysis of CSPs in brain tissue is a sound strategy for prioritizing putative protein biomarkers for future immunoassay (e.g., ELISA) measurements in CSF or blood.
We hypothesized that changes in CSP expression might correlate to these long-term secondary effects. To test our hypothesis, we longitudinally assessed a closed-skull mTBI mouse model, vs. sham control, at 1, 7, 30, and 120 days post-injury. Motor impairment was determined by rotarod and grip strength performance measures, while motor unit integrity was determined by electromyography. Relative protein expression was determined by microwave & magnetic (M2) proteomics of brain tissue as previously described [22, 23], where confirmation for selected proteins was provided with Western blotting. Isoprostane measurements were performed to confirm a primary oxidative stress response. Decoding the relative protein expression for each specimen for 476 ± 56 top-ranked proteins revealed statistically significant changes in the expression of two well-known CSPs at 1, 7 and 30 days post-injury: P < 0.001 for myelin basic protein (MBP) and P < 0.05 for myelin associated glycoprotein (MAG). This was confirmed by Western blot. Moreover, MAG, αII-spectrin (SPNA2) and neurofilament light (NEFL) expression at 30 days post-injury were significantly correlated to grip strength (P < 0.05).
MATERIAL & METHODS
Closed-Skull Impact Model
mTBI was induced at 60 days with the TBI 0310 impact device (Percision Systems LLC). TBI was administered as a closed cortical injury (CCI) using pneumatic force. The mortality rate was less than 5%; there were no overt structural abnormalities, intracranial bleeds, or edema observed with MRI, indicating that TBI severity was mild. Prior to surgery mice were anesthetized in a chamber with 2–4% isoflurane in 100% oxygen. Anesthesia was maintained at 1% for the remaining procedures. During surgery the mean arterial pressure was monitored with a transducer, and mice were fixed to a pad in the prone position under a heating lamp to maintain body temperature. A midline incision in the scalp was made and the skin and periosteum retracted. A stainless steel disc (7mm in diameter and 3mm thick) was glued to the skull between the coronal and lambdoid sutures over the somatosensory cortex using super glue. TBI was induced using a CCI device calibrated to deliver a blow at 4.5 m/s, 100ms dwell time and a depth of 2mm directly to the disc. Following injury, the disc and glue were removed and the incision sutured. Antibiotic ointment was applied to wounds. Animals were allowed to wake in a warm/dry cage with a sterile liner and monitored for at least 1 hour. Sham animals were subjected to all procedures except that the impact device was calibrated to a level just above the disk resulting in no impact. All animals were observed and weighed daily until completion of experimentation. At selected survival times, mice were anesthetized under isoflurane, sacrificed, and brain tissue (and plasma) specimens were snap frozen in liquid nitrogen prior to storage at −80C.
Imaging
For Nissl staining, standard procedures were used for the detection of Nissl bodies found within neurons. Briefly, brains were harvested as described above and sectioned at 20µm and placed on plus slides. The slides were dried at 37C overnight, hydrated with distilled water, 0.1% cresyl violet was applied for 7 minutes, and washed with distilled water. The slides were then hydrated, cleared in xylene and coverslipped. Images were captured with a Nikon Eclipse TE2000-U fluorescence microscope (Nikon Inc.). For immunofluoroescence, Mice were perfused with 4% paraformaldehyde and brains dissected and placed in PFA overnight. Tissue was then transferred to glucose for 48 hours. Following cryosectioning, slices were permeabilized (0.1% Tween-20 in PBS, 5 min), and non-specific binding of antibodies was blocked with PBS/5.0% BSA for 1 h. Slices were probed with primary antibodies and incubated overnight at 4°C. The following antibodies were used: (1:500, Neuron Signaling, Danvers, Mass., USA): Anti-GFAP, Anti-NeuN. After a washing step (PBS, 5 min), slices were counterstained with AlexaFluor-conjugated secondary antibodies (1:1000, 1h, RT, PBS/5% BSA; Molecular Probes, Eugene, Oregon, USA), washed again and mounted onto slides with Prolong Gold Antifade reagent containing DAPI (Molecular Probes). Stained slides were visualized with a Nikon Eclipse TE2000-U fluorescence microscope (Nikon Inc.). For MRI, experiments were conducted at the Research Imaging Institute using a horizontal 7T Biospec system (BrukerBioSpin, Ettlingen, Germany) and ParaVision 5 software. A small circular surface coil (ID = 1.1 cm) was placed on top of the head. Mice were imaged under 1.2% isoflurane with spontaneous breathing after placement in a custom-made animal holder with ear and mouth bars. Respiration rate (80–130 bpm) and rectal temperature (37 ± 0.5° C) were continuously monitored and maintained within normal physiological ranges unless otherwise perturbed. High-resolution (isotropic 100 µm), T1-weighted images were acquired using 3D FLASH sequence (scan parameters: with echo time (TE) 5.1 ms, repetition time (TR) 50 ms, flip angle of 30 degrees, field of view (FOV) of 11×11×11 mm, matrix size 1024×1024×1024). Preprocessing consisted of removing non-brain tissues and global spatial normalization. The GM and WM were separated using FMRIB Software Library (FSL) packet (EPSRC, UK). The GM and WM volumes were determined using the Multi-Image Analysis GUI (MANGO) software (http://ric.uthscsa.edu/mango).
Rotarod
Neuromuscular function was tested using Rotamex 4/8 (Columbus Instruments, Columbus, OH). Each mouse was trained for five consecutive days (six trails/day) where the speed of the rotor was accelerated from 4 to 40rpm with an acceleration of 0.2 rpm/sec. Twenty-four hours after the last training session, the mice were tested in a probe trial consisting of six trials as previously described. The latency to fall was then recorded.
Grip Strength
Forelimb muscle strength was determined by measuring peak force (in pounds) using the Digital Grip Strength meter equipped with a Hind Limb Pull Bar Assembly (Columbus Instruments, Columbus, OH). Mice are allowed to grip the metal grids of a grip meter with their forepaws, and gently pulled backwards by the tail until they could no longer hold the grids. The peak grip force observed in 10 trials was recorded [24].
Electromyography
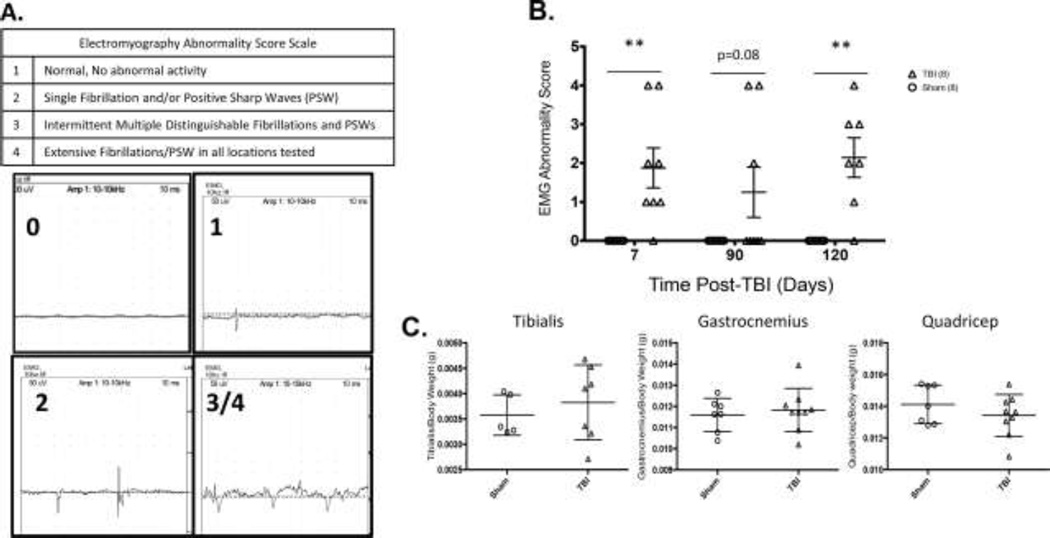
Bilateral hind limb muscle groups were examined with a Nicolet Viking Quest portable EMG apparatus (CareFusion San Diego, CA. USA). A monopolar electrode (active) was inserted into the muscle of interest. An identical electrode (reference) was inserted subcutaneously into the lateral and distal-most tendinous portion of the gastroc-soleus complex, ipsilateral to the muscle studied. A subdermal needle (ground) was inserted subcutaneously into tendinous tissue posterior to and near the reference electrode. Abnormal spontaneous activity in the form of denervation potentials (positive sharp waves and fibrillations) was recorded using an electromyography abnormality score scale, Figure 5A.
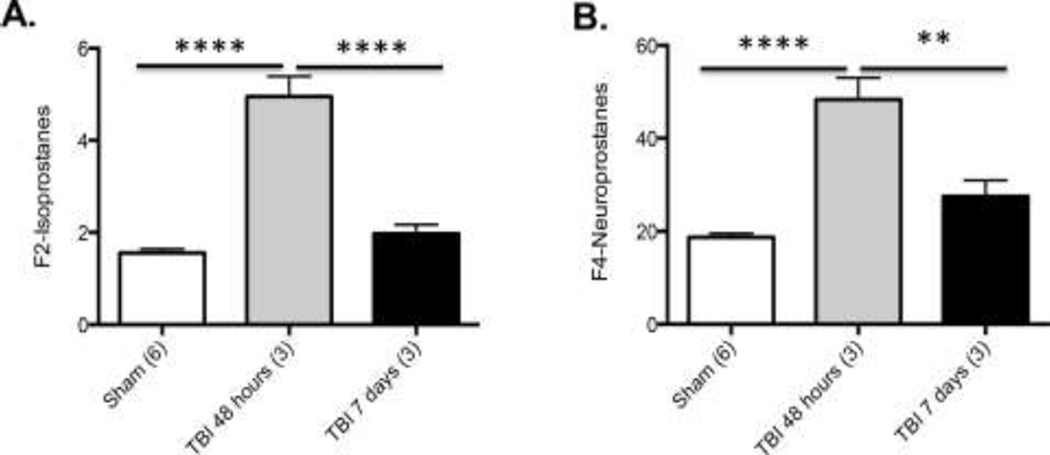
Figure 5. Isoprostane measures of mTBI (****p<0.0001, **p<0.001).
Significantly increased A: F2-Isoprostanes and B: F4-Isoprostanes 48 hours following mTBI as compared to Sham.
Isoprostanes
F2-isoprostanes and F4-neuroprostanes were measured in ipsilateral brain using the gas chromatography–mass spectrometry method of Morrow and Roberts [25]. Tissue was collected and homogenized in chloroform:methanol containing 0.005% butylatedhydroxytoluene (BHT) to prevent auto-oxidation, dried under a stream of nitrogen, and re-suspended in methanol containing BHT. Esterified F2-isoprostanes in phospholipids were saponified, to free fatty acids from lipids, by adding aqueous potassium hydroxide. Then, the sample was acidified and diluted with water. Next, deuterated-F2-isoprostane internal standard was added to the mixture. For the measurement of free F2-isoprotanes/F2-isofuranes in plasma, the extraction and hydrolysis steps were omitted, and the sample was simply acidified, diluted, and the internal standard added. The mixture was subsequently run on a silica column to separate isoprostanes/isofuranes from bulk fatty acids. The eluate was converted to pentafluorobenzyl esters, by treatment with pentafluorobenzyl bromide to improve separation efficiency. The mixture was then subjected to thin layer chromatography to remove the excess pentafluorobenzyl bromide and unreacted fatty acids. The F2-isoprotane/isofurane fraction was extracted using ethyl acetate, and analyzed. F2-isoprostanes were quantified by peak height, the data was corrected with the internal standard, and results were calculated as nanogram of F2-isoprostanes per mL of plasma or per gram tissue. F4-neuroprostanes, a lipid peroxidation product of docosahexaenoic acid was also determined some modifications of the F2-isoprostane method. Briefly, 100–200mg tissue was homogenized in ice-cold Folch solution containing BHT. Lipids were then extracted and chemically hydrolyzed with 15% KOH. After acidification with HCl and addition of a stable isotope-labeled internal standard, 8-iso-prostaglandin F2α-d4, ,F4-neuroprostanes were applied to a C18 Sep-Pak cartridge and a silica Sep-Pak column for further purification. Unlike the F2-isoprostane assay, the washing step for silica columns used an ethyl acetate / heptane (75:25) mixture instead of pure ethyl acetate because of the polarity difference between F2-isoprostanes and F4-neuroprostanes. The eluted compounds are dried under N2, converted to pentafluororobenzyl esters, and purified by thin-layer chromatography. Instead of a 1.5cm narrow cut for isoprostane measurement, the scraped area was extended to 4cm above and 1cm below the PGF2a methyl ester migration. The purified F4-neuroprostanes were derivatized to trimethysilyl ether derivatives then dissolved in undecane that was dried over a bed of calcium hydride. Negative ion chemical ionization MS was performed by Agilent 6890 GC and Model 5975 MSD instruments with selected ions monitored for [2H4]15-F2α-IsoP internal standard (m/z 573) and F4-NeuroPs (m/z 593).
Sham Control vs. mTBI Mouse Specimens
Cryropreserved ipsilateral C57Bl6 mouse brain specimens were obtained at various post-injury time points following closed skull mTBI. All mice used were 60 days of age at the time of primary brain injury.
Microwave & Magnetic (M2) Sample Preparation
Protein was pooled from all specimens by protein amount as reference material. For isobaric TMT labeling, 50 mg of C8 magnetic beads (BcMg, Bioclone Inc.) were suspended in 1 mL of 50% methanol. Immediately before use, 100 µL of the beads were washed 3 times with equilibration buffer (200 mM NaCl, 0.1% trifluoroacetic acid (TFA)). Whole cell protein lysate (25–100 µg at 1µg/µL) was mixed with pre-equilibrated beads and 1/3rd sample binding buffer (800 mM NaCl, 0.4% TFA) by volume. The mixture was incubated at room temperature for 5 min followed by removing the supernatant. The beads were washed twice with 150 µL of 40 mM triethylammonium bicarbonate (TEAB), and then 150 µL of 10 mM dithiolthreitol (DTT) was added. The bead-lysate mixture underwent microwave heating for 10 s. DTT was removed and 150 µL of 50 mM iodoacetamide (IAA) added, followed by a second microwave heating for 10 s. The beads were washed twice and re-suspended in 150 µL of 40 mM TEAB. In vitro proteolysis was performed with 4 µL of trypsin in a 1:25 trypsin-to-protein ratio (stock = 1µg/µL in 50mM acetic acid) with microwave-assisted heating for 20 s in triplicate. The supernatant was used immediately or stored at −80°C. Released tryptic peptides from digested protein lysates, including the reference materials described above, were modified at the N-terminus and at lysine residues with the tandem mass tagging (TMT)-6plex isobaric labeling reagents (Thermo scientific, San Jose, CA). Each individual specimen was encoded with one of the TMT-126-130 reagents, while reference material was encoded with the TMT-131 reagent: 41 µL of anhydrous acetonitrile was added to 0.8 mg of TMT labeling reagent for 25µg of protein lysate and microwave-heated for 10s. To quench the reaction, 8 µL of 5% hydroxylamine was added to the sample at room temperature. To normalize across all specimens, TMT-encoded cell lysates from individual specimens, labeled with the TMT-126-130 reagents, were mixed with the reference material encoded with the TMT-131 reagent in 1126:1-127:1128:1129:1130:1131 ratios. These sample mixtures, including all TMT-encoded specimens, were stored at −80°C until further use.
Capillary Liquid Chromatography-Fourier-Transform-Tandem Mass Spectrometry (LC/FT/MS/MS) with Protein Database Searching
Capillary LC/FT/MS/MS was performed with a splitless nanoLC-2D pump (Eksigent, Livermore, CA), a 50 µm-i.d. column packed with 7 cm of 3 µm-o.d. C18 particles, and a hybrid linear ion trap-Fourier-transform tandem mass spectrometer (LTQ-ELITE; ThermoFisher, San Jose, CA) operated with a lock mass for calibration. The reverse-phase gradient was 2 to 62% of 0.1% formic acid (FA) in acetonitrile over 60 min at 350 nL/min. For unbiased analyses, the top 6 most abundant eluting ions were fragmented by data-dependent HCD with a mass resolution of 120,000 for MS and 15,000 for MS/MS. For isobaric TMT labeling, probability-based protein database searching of MS/MS spectra against the Trembl_mouse protein database (release 2012_dec29; 59,862 sequences) was performed with a 10-node MASCOT cluster (v. 2.3.02, Matrix Science, London, UK) with the following search criteria: peak picking with Mascot Distiller; 10 ppm precursor ion mass tolerance, 0.8 Da product ion mass tolerance, 3 missed cleavages, trypsin, carbamidomethyl cysteines as a static modification, oxidized methionines and deamidated asparagines as variable modifications, an ion score threshold of 20 and TMT-6-plex for quantification.
Western Blot
Western Blot Analysis was performed on lysates from ipsilateral brain samples in order to confirm our proteomics results. Equivalent amounts of protein from each sample were subjected to sodium dodecyl sulfate-polyacrylamide electrophoresis using 4–12% Bis–Tris precast gels (Invitrogen, California, USA) under reducing and non reducing conditions (1h, RT) and electro-blotted onto a nitrocellulose membrane (18h, Overnight, Bio Rad). Following a blocking step (0.1% Tween-20/5% nonfat milk in PBS, 1h, RT) membranes were incubated with primary antibodies overnight (12–14 h, 4°C) with gentle agitation. The following primary antibodies were used (1:1000): Anti-MBP (Millipore), Anti-MAG (AbCam), Anti-Beta Actin (Cell Signaling). Membranes were washed, incubated with secondary antibody (RT, 1h, Cell Signaling) and developed with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific).
Statistical Analysis
M2 proteomics technical replicates estimated protein expression for individual specimens, TMT- -encoded in sample mixtures, relative to pooled reference materials. Relative protein expression levels were transformed to log base 2 for quantile normalization. We tested the association between relative protein expression with rotarod, grip strength and motor unit integrity measures (EMG) using linear regression or ANOVA. All statistical analysis was performed with GraphPad Prism software (GraphPad Software Inc.) and R v3.0+ (R Project, Vienna, Austria).
RESULTS
Imaging measures of mTBI
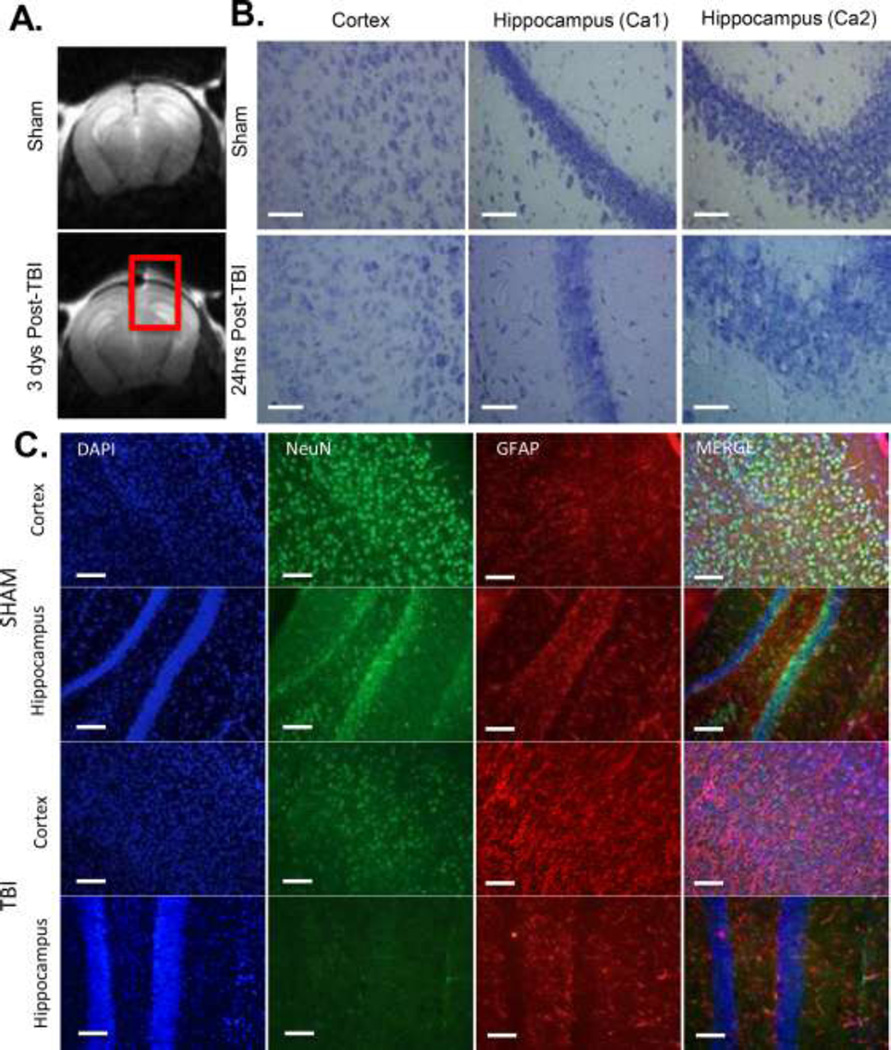
Anatomical images of the mouse brain after mTBI, obtained with 7T MRI show no signs of herniation, midline shift, overt edema or hemorrhaging (Figure 1A), consistent with the clinical diagnosis of mTBI and supporting our closed-skull mTBI mouse model. For analysis of cell loss and edema in brain cryosections, the mice were perfused 48 hours post-TBI, and brains from wild-type sham and TBI mice were collected and cryosectioned. Sections were stained with Nissl in order to visualize edema (Figure 1B). Staining was more diffuse in the brains of TBI animals with visible decreases in cell number and increases in cellular size (edema). Immunoflourescence double-labeling for neurons and astrocytes indicates a loss of neurons (NeuN, Green) in the cortex and hippocampus under the site of injury and an increase in astrogliosis as indicated by increases in GFAP (Red) 24 hours following injury (Figure 1C).
Figure 1. Imaging measures of mTBI.
A) Structural MRI of WT mouse brain 3 days post-TBI. Red Square encompasses area of direct impact. N=3 B) Nissl stain(40× magnification, line=500µm) of sham and TBI treated WT mouse cortex and hippocampus (Ca1 and Ca2 regions) 24 hours post-TBI. N=4 C) DAPI (Blue), NeuN (Green) and GFAP (Red) (20× magnification, line=100µm) used to label nuclei, neurons and astrocytes in the ipsilateral (Injured) cortexand hippocampus of Sham and TBI animals 24 hours post-TBI. N=3
Body weight, apnea and righting reflex measures of mTBI
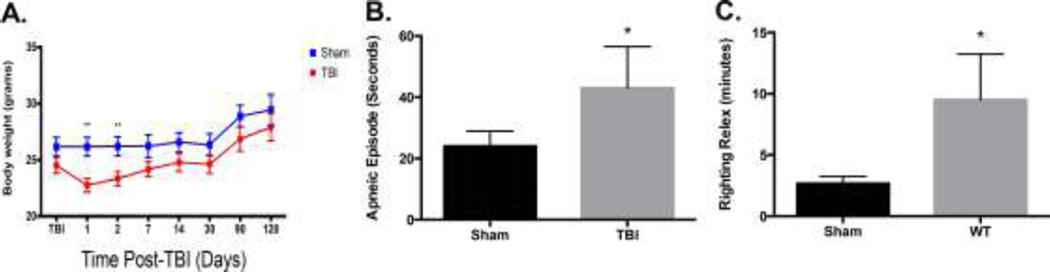
Following mTBI, animals experienced a significant loss of body weight at 1 and 2 days post TBI that returned to non-significant levels by 7 days postinjury (p=0.01, Figure 2A). After mTBI, mice experienced an apneic episode averaging 45 seconds, significantly higher than sham controls (p=0.02, Figure 2B). Animals also experienced a significant increase righting reflex following recovery from anesthesia when compared to sham controls (Figure 2C, p=0.02).
Figure 2. Body weight, apnea and righting reflex measures of mTBI (**p<0.01, *p<0.02).
A) mTBI causes a significant reduction in bodyweight loss up to 7 days post-injury B) Time spent apneic following mTBI is significantly increased as compared to sham controls C) Increased time to right self (righting reflex) following TBI is also significantly increased.
Motor impairment measures of mTBI
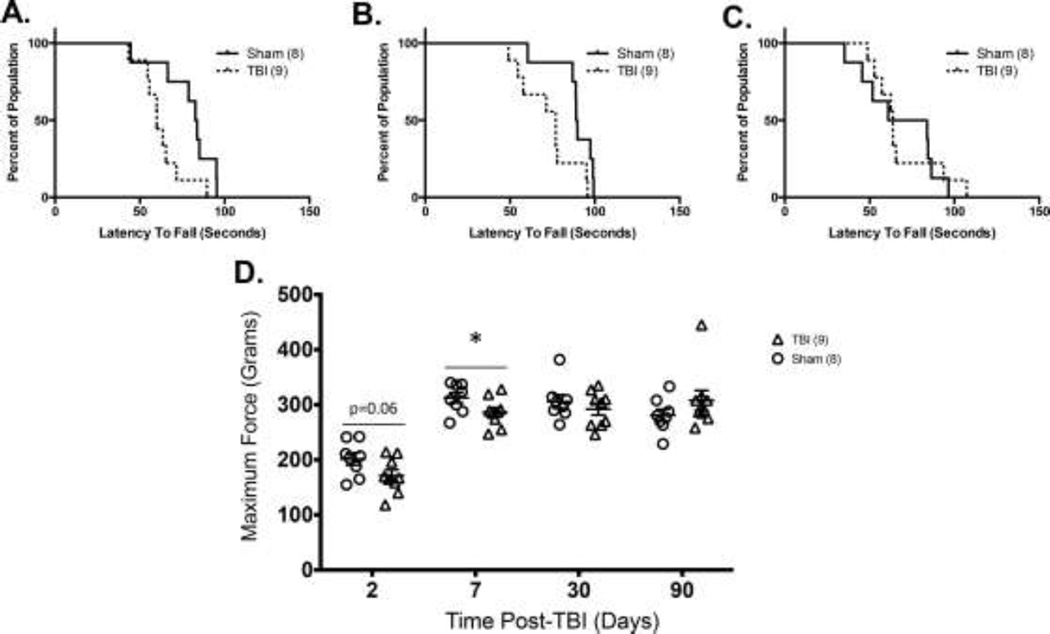
As indicated in Figures 3A and 3B, mTBI is capable of initiating a significant decrease in rotarod performance in WT mice at 7 and 30 days post-injury (mTBI vs. sham, p=0.05) but not at 90 days post-injury, Figure 3C. TBI animals had a trend toward lower maximum grip strength at 2 days post-injury (p=0.06), which became significant by 7 days post-injury (p=0.05) as compared to sham controls (Figure 3D).
Figure 3. Motor impairment measures of mTBI (*p<0.05).
A) Significantly reduced rotarod performance at 7 days B) 30 days and C) 90 days post-injury (p<0.05) D) Grip strength was decreased at 2 days post-injury and significantly reduced at 7 days post-injury.
Motor unit integrity measures of mTBI
Further, there is a marked increase in EMG abnormalities in mTBI mice as early as 7 days post-injury (Two Way ANOVA, p=0.001, Figure 4B). These significant abnormalities in motor unit integrity persist up to 120 months after mTBI. EMG abnormalities are not accompanied by loss of muscle mass as shown in Figure 4C.
Figure 4. Motor unit integrity measures of mTBI (**p<0.01).
A) Representative tracings of Electromyography Abnormality Scoring used in the current study B) Electromyography of hind limb muscles indicate significant abnormality in the form of denervation potentials at all time points tested post-injury. C) Changes in muscle mass at 4 months post-injury D) Muscle mass normalized to body weight.
Isoprostane measures of mTBI
We sought to determine if our closed-skull mTBI mouse model (primary injury) led to increases in oxidative stress. To address this question, we examined levels of F2-isoprostanes (Figure 5A) and F4-Neuorprostanes (Figure 5B) following mTBI. Our results are consistent with the literature: we observed significant increases in F2-isoprostances and F4-neuroprostanes in the ipsilateral cortex 48 hours post-injury mTBI (p=0.0001) that returned to sham levels by 7 days post-injury, supporting our closed-skull mTBI mouse model.
M2 proteomics of mTBI
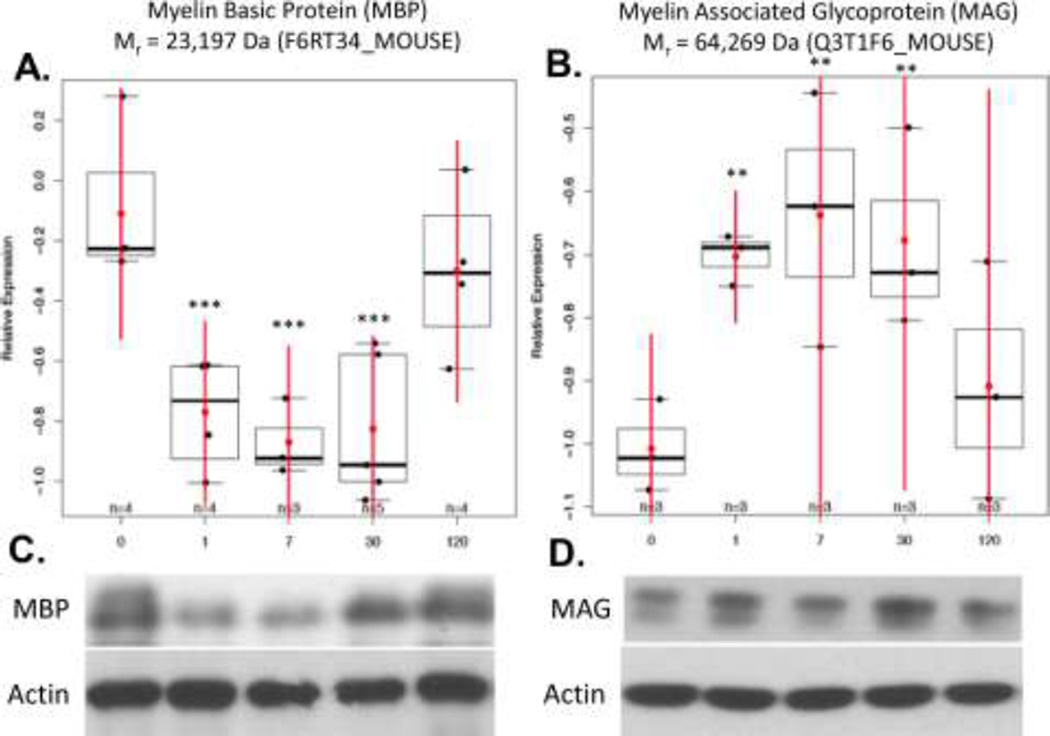
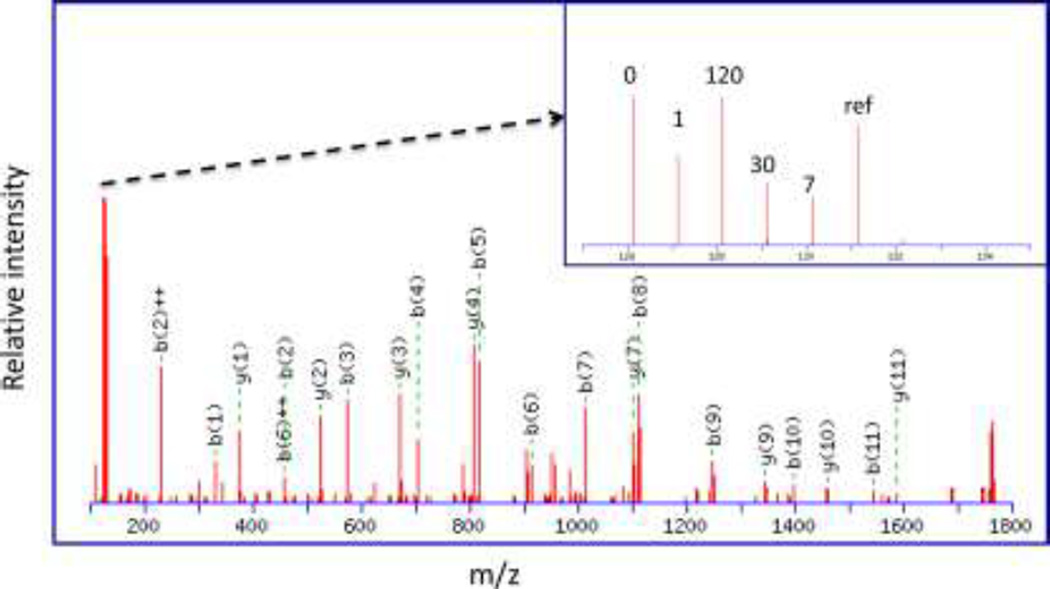
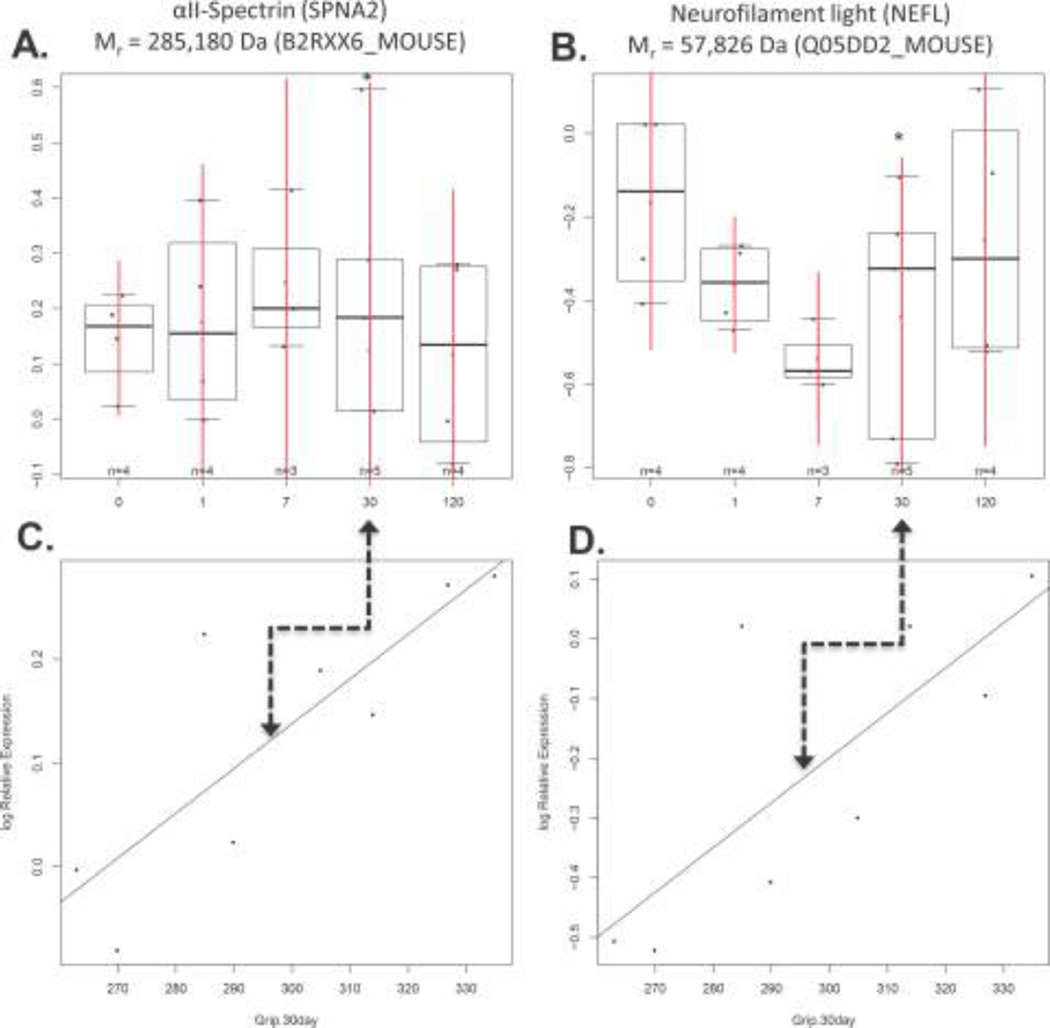
Decoding the relative expression of 476 ± 56 top-ranked proteins for each specimen revealed statistically significant changes in the expression of two well-known CSPs at 1, 7 and 30 days post-injury: P < 0.001 for myelin basic protein (MBP) and P < 0.05 for myelin associated glycoprotein (MAG) (Figure 6A–B, and Supplementary Table 1). This was confirmed with Western blotting (Figure 6C). MBP and MAG protein expression was inferred from the following top-scoring TMT-labeled tryptic peptides generated in vitro as part of our M2 proteomics procedure: MBP50-59 (DTGILDSIGR); MBP60-65 (FFSGDR); MBP121-132 (TQDENPVVHFFK); and MBP155-171 (FSWGAEGQKPGFGYGGRASDYK). Likewise, the following top-scoring TMT-labeled peptides from MAG were quantified: MAG78-90 (TQVVHESFQGRSR) and MAG189-208 (LREDEGTWVQVSLLHFVPTR). A representative MS/MS spectrum for MBP121–132 (TQDENPVVHFFK) shows the probability-based protein database search assignment of 19/23 amino acid sequence-specific b- and y-type ions (expectation = 5.8E-7), with a zoomed-in view of the TMT126–131 reporter ions used for quantification of this peptide in specimens from individual mice (inset) (Figure 7). The post-injury time point (0,1,7,30 and 120 days) for each reporter ion is also shown, where ref = the pooled reference used to normalize relative expression across all specimens. The trajectory of MBP121–132 expression is evident in the trajectory inferred for MBP expression (Figure 6A). Other differentially expressed proteins, including other well-known CSPs, were also revealed by M2 proteomics. For example, decreased expression of αII-spectrin (SPNA2) and neurofilament light (NEFL) were directly correlated to decreased grip strength (P < 0. 05) (Figure 8 and Supplementary Table 2). The majority of the remaining proteins did not exhibit statistically significant correlations to post-injury time and/or grip strength, as expected. However, some of these proteins are known to be important to TBI, including: glutathione S-transferase µ (GSTM5) and glucose-6-phosphate isomerase (GPI) (see Table 1 showing top-ranked correlations from Supplementary Table 1).
Figure 6. M2 proteomics measures of mTBI (***p<0.001, **p<0.05).
A) Significantly decreased MBP expression up to 30 days post-injury as compared to sham controls at 180 days of age. B) Significantly increased MAG expression up to 30 days post-injury. Western blots of C) MBP (18–23 kDa) and D) MAG (63 kDa) with Beta-Actin (45 kDa) as a loading control were used to confirm M2 proteomics results.
Figure 7. A representative MS/MS spectrum for MBP121-132 (TQDENPVVHFFK).
This spectrum shows the probability-based protein database search assignment of 19/23 amino acid sequence-specific b- and y-type ions (expectation = 5.8E-7), with a zoomed-in view of the TMT126-131 reporter ions used for quantification of this peptide in specimens from individual mice (inset). The post-injury time point (0,1,7,30 and 120 days) for each reporter ion is also shown, where ref = the pooled reference used to normalize relative expression across all specimens.
Figure 8. M2 proteomics measures of mTBI correlated to grip strength (*p<0.05).
Decreased expression of A) SPNA2 and B) NEFL directly correlated to decreased grip strength at 30 days post-injury, as shown in C) and D), respectively.
Table 1.
Top-ranked associations for relative protein expression correlated to post-injury time (vs. sham controls). The post-injury time point (vs. sham), log2 fold change in protein expression (vs. sham), p-value for post-injury time point (vs. sham), overall p-value for all time-points (vs. sham), Tembl protein symbol, and protein description are shown with examples in bold and italics.
| post- injury time point (vs. sham) |
log2 fold change in protein expression (vs. sham) |
p- value for post- injury time point (vs. sham) |
overall p- value for all time- points (vs. sham) |
Trembl protein symbol |
protein description |
|---|---|---|---|---|---|
| Day30 | −0.7162 | 0.0004 | 0.0007 | F6RT34_MOUSE | Myelin basic protein (Fragment) OS=Mus musculus GN=Mbp PE=4 SV=1 |
| Day7 | −0.7605 | 0.0007 | 0.0007 | F6RT34_MOUSE | Myelin basic protein (Fragment) OS=Mus musculus GN=Mbp PE=4 SV=1 |
| Day1 | −0.6614 | 0.0011 | 0.0007 | F6RT34_MOUSE | Myelin basic protein (Fragment) OS=Mus musculus GN=Mbp PE=4 SV=1 |
| Day7 | −0.3883 | 0.0095 | 0.0712 | D5MR34_MOUSE | Tubulin beta 3 (Fragment) OS=Mus musculus GN=Tubb3 PE=3 SV=1 |
| Day7 | −0.6319 | 0.0098 | 0.0690 | Q8R1B2_MOUSE | Actr1b protein (Fragment) OS=Mus musculus GN=Actr1b PE=2 SV=1 |
| Day7 | 0.3705 | 0.0117 | 0.0450 | Q3T1F6_MOUSE | Myelin-associated glycoprotein OS=Mus musculus GN=Mag PE=2 SV=1 |
| Day7 | −0.2128 | 0.0122 | 0.0779 | E9PVM7_MOUSE | Glutathione S-transferase Mu 5 (Fragment) OS=Mus musculus GN=Gstm5 PE=3 SV=1 |
| Day120 | 0.4588 | 0.0141 | 0.0602 | B2RTM0_MOUSE | Histone H4 OS=Mus musculus GN=Gm11275 PE=3 SV=1 |
| Day7 | −0.3137 | 0.0166 | 0.0652 | Q80ZV2_MOUSE | Tubb5 protein (Fragment) OS=Mus musculus GN=Tubb5 PE=2 SV=1 |
| Day30 | 0.3310 | 0.0205 | 0.0450 | Q3T1F6_MOUSE | Myelin-associated glycoprotein OS=Mus musculus GN=Mag PE=2 SV=1 |
| Day1 | −0.1867 | 0.0231 | 0.0779 | E9PVM7_MOUSE | Glutathione S-transferase Mu 5 (Fragment) OS=Mus musculus GN=Gstm5 PE=3 SV=1 |
| Day7 | −0.3895 | 0.0273 | 0.1359 | B1AQZ2_MOUSE | Kinesin family member 3A OS=Mus musculus GN=Kif3a PE=3 SV=1 |
| Day7 | 0.3047 | 0.0296 | 0.2298 | Q3TJH1_MOUSE | Putative uncharacterized protein OS=Mus musculus GN=Gnai3 PE=2 SV=1 |
| Day1 | 0.3050 | 0.0297 | 0.0450 | Q3T1F6_MOUSE | Myelin-associated glycoprotein OS=Mus musculus GN=Mag PE=2 SV=1 |
| Day30 | −0.1733 | 0.0322 | 0.0779 | E9PVM7_MOUSE | Glutathione S-transferase Mu 5 (Fragment) OS=Mus musculus GN=Gstm5 PE=3 SV=1 |
| Day120 | −0.2409 | 0.0322 | 0.2271 | E9Q0U7_MOUSE | Heat shock protein 105 kDa OS=Mus musculus GN=Hsph1 PE=3 SV=1 |
| Day30 | −0.4112 | 0.0346 | 0.2477 | Q3UMG4_MOUSE | Putative uncharacterized protein (Fragment) OS=Mus musculus GN=Ina PE=2 SV=1 |
| Day7 | −0.4627 | 0.0382 | 0.1140 | Q3ULW0_MOUSE | Putative uncharacterized protein OS=Mus musculus GN=Ran PE=2 SV=1 |
| Day30 | 0.6178 | 0.0528 | 0.3263 | Q9Z1R9_MOUSE | MCG124046 OS=Mus musculus GN=Prss1 PE=2 SV=1 |
| Day7 | 0.3261 | 0.0539 | 0.2529 | D3Z2Y8_MOUSE | D-beta-hydroxybutyrate dehydrogenase, mitochondrial (Fragment) OS=Mus musculus GN= |
| Day1 | −0.2217 | 0.0587 | 0.2989 | Q3TWL6_MOUSE | Putative uncharacterized protein OS=Mus musculus GN=Psmd2 PE=2 SV=1 |
| Day7 | −0.3719 | 0.0592 | 0.2837 | Q05DD2_MOUSE | Nefl protein (Fragment) OS=Mus musculus GN=Nefl PE=2 SV=1 |
| Day120 | −0.4108 | 0.0601 | 0.1140 | Q3ULW0_MOUSE | Putative uncharacterized protein OS=Mus musculus GN=Ran PE=2 SV=1 |
| Day30 | 0.2755 | 0.0607 | 0.1185 | A4FUS1_MOUSE | MCG123443 OS=Mus musculus GN=Rps16 PE=2 SV=1 |
| Day1 | 0.2431 | 0.0616 | 0.2378 | H3BK84_MOUSE | cAMP-dependent protein kinase type II-beta regulatory subunit OS=Mus musculus GN=Prkar2b |
| Day120 | 0.2345 | 0.0621 | 0.3348 | Q3TIU7_MOUSE | Putative uncharacterized protein OS=Mus musculus GN=Ndufs1 PE=2 SV=1 |
| Day1 | −0.6893 | 0.0646 | 0.3239 | Q3UJ20_MOUSE | Putative uncharacterized protein OS=Mus musculus GN=Eno2 PE=2 SV=1 |
| Day120 | −0.5581 | 0.0666 | 0.2934 | F6SAC3_MOUSE | Glucose-6-phosphate isomerase (Fragment) OS=Mus musculus GN=Gm1840 PE=3 SV=1 |
DISCUSSION
The goal of the current study was to investigate whether changes in CSP expression correlate to long-term secondary effects on motor unit impairment and integrity, as well as to investigate potential underlying molecular mechanisms for these lasting effects, with M2 proteomics. Our imaging and isoprostane measures were consistent with the clinical diagnosis of mild TBI (mTBI) and are support for our closed-skull mTBI mouse model. Decoding the relative protein expression for each specimen revealed statistically significant changes in the expression of the CSPs known as MBP and MAG. MBP expression was rapidly reduced, by 24 hours, in the ipsilateral brain following mTBI and was significantly down-regulated for up to 30 days post-injury. Decreased MBP expression was mirrored by increased MAG expression during the same time period. Moreover, increased grip strength revealed that increased MAG expression was directly related to motor impairment at 30 days post-injury (Supplementary Table 2). A brief discussion of previous work on MBP, MAG and other CSP biomarkers of mTBI are provided below.
MBP is the second most abundant protein in CNS myelin, comprising ~ 30% of the total protein in the myelin sheath [26–28]. It is a positively charged membrane bound protein that binds to negatively charged lipids, present at the cytosolic surface of myelin, and alternative splicing and post-translational modifications generate numerous isoforms [26, 29–34]. MBP has most often been associated with pediatric mTBI [35–37]. For example, an increase in MBP levels in the CSF was observed in a small sample of 14 pediatric mTBI patients [38]. It is also believed to be a potential biomarker of traumatic axonal injury (TAI) [39]. TAI represents a mechanism of secondary (long-term) injury, resulting from increased oxidative stress due to calcium accumulation and mitochondrial failure in injured axons [40–42]. Increases in CSF levels of MBP have been seen in multiple models of mTBI also, including pediatric TBI and blast-induced TBI [28, 35]. MBP is cytotoxic and promotes inflammation by activating the release of histamine and it is present at extracellular sites of pathological fibrotic lesions in several disease models [43, 44]. Therefore, the transport of MBP across the blood-brain-barrier and into the periphery could be contributing to the motor impairment observed in this animal model of mTBI.
MAG is a member of the immunoglobulin-like family and provides a source of inhibition for growing neuritis after CNS injury [45, 46]. Most of the current studies on injury-induced growth inhibition have been performed in spinal cord injury. Few studies have investigated the role of MAG in the pathogenesis of mTBI [46–49]. Intriguingly, a rodent model of fluid percussion injury has been employed to show that treatment with an anti-MAG monoclonal antibody can improve neurologic motor, sensory and cognitive function for up to 8 weeks post-injury [49]. Likewise, central and systemic administration of anti-MAG antibody significantly reduced lesion volume, improved motor function and reduced oxidative stress responses in a rat model of middle cerebral artery occlusion [50]. These studies support the involvement of MAG in CNS injury pathology as well as its use as a potential therapeutic target for future studies.
We also observed that MAG, SPNA2 and NEFL expression at 30 days post-injury were directly correlated to grip strength (P < 0.05) (Figure 8 and Supplementary Table 2). Breakdown products (i.e., cleavage or proteolytic processing) of the cytoskeletal protein SPNA2 (e.g., SBDP145) that is abundant in axons and pre-synaptic terminals of neurons, are generated by calcium-dependent cysteine protease(s) (e.g., calpains and caspases) during necrosis (and/or apoptosis) following TBI [51, 52]. The nominal mass of the MBP isoform measured by M2 proteomics herein is 23,197 Da, while an 18kDa isoform was the most abundant band measured with Western blotting (Figure 6A and 6C). However, since M2 proteomics did not achieve 100% sequence coverage (i.e., the C-terminus is missing) for this (or any other CSP) and the antibody employed was not isoform-specific, we cannot unambiguously assign our results solely to this isoform or its breakdown products [53].
Neurofilament proteins are major cytoskeletal structural proteins of neurons and are found heavily concentrated in axons [54–56]. NEFL shows some promise as an indicator of acute axonal damage [57]. Unfortunately, NEFL is degraded relatively rapidly in the CSF or blood, thus potentially confounding NEFL measurements [58]. While our data on linking MAG, SPNA2 and NEFL expression to grip strength are preliminary, epidemiological data suggest significant correlations of TBI to the development of CNS pathologies with long-term motor dysfunction, including; Amyotrophic Lateral Sclerosis (ALS), Parkinson’s Disease, Alzheimer’s Disease and Chronic Traumatic Encephalopathy [10–13]. For example, TBI has been linked to a ~ 3-fold increased risk of ALS [13, 59], and the genes and environmental exposures in veterans with ALS (GENEVA) case-control study revealed significantly increased risk of ALS in veterans with a TBI [12].
Lastly, we briefly discuss two of the proteins shown in Table 1 that did not exhibit statistically significant correlations to post-injury time and/or grip strength: GSTM5 and GPI. GSTM5 is a member of the glutathione s-transferase superfamily: a major group of detoxification enzymes that alleviates the damage from lipid peroxidation by-products (e.g., 4-HNE and acrolein) by catalyzing their conjugation with glutathione [60, 61]. Our results indicate increased lipid peroxidation during mTBI, which could be exacerbated by decreased levels of GSTM5 [62]. GPI is a dimeric enzyme that catalyzes the reversible isomerization of glucose-6-phosphate and fructose-6-phosphate. In the cytoplasm, GPI is involved in glycolysis and gluconeogenesis, while outside the cell it functions as a neurotrophic factor for spinal and sensory neurons. Interestingly, we also observed that GPI levels were decreased in mTBI vs. sham. In contrast, we observed increased levels of GPI in our previous work on a mouse model of multiple sclerosis [22].
Significance of M2 Proteomics
M2 proteomics was developed to provide the following advantages: to improve sample throughput by decreasing lengthy sample preparation times, to improve sensitivity by decreasing adsorptive losses, and to improve statistical power by enabling quantitative MS/MS-based proteomic studies with relatively large numbers of specimens. One disadvantage is the greater computational burden that comes with larger numbers of specimens and MS/MS spectra. A rigorous comparison of the analytical figures of merit for M2 proteomics vs. other proteomics methods is beyond the scope of this work, including: M2 proteomics vs. conventional approaches for quantitative MS/MS-based proteomics, bead-based immunoassays, and gel-based proteomic methods such as two-dimensional electrophoresis. However, since our initial publications on M2 proteomics [22, 23], we have successfully applied M2 proteomics to a variety of preclinical and clinical studies. Moreover, there is a growing number of reports of high-throughput sample preparation by microwave- assisted digestion [63, 64] or by magnetic beads [65], high-sensitivity on-bead digestions [66], and isobaric labeling reagents for multiplexed protein quantification by MS/MS-based proteomics [67, 68].
Conclusions
While higher-powered studies of larger cohorts merit further investigation, this study supports the proof-of-concept that M2 proteomics is a rapid method to quantify putative CSP biomarkers and therapeutic targets of mTBI and suggests the feasibility of correlations to long-term effects on motor impairment. Based on our own results and previous work, we posit that a decrease in MBP expression and/or an increase in MAG expression might contribute to impaired motor function and neuronal regeneration in mTBI patients. From these preliminary studies, we also hypothesize that M2 proteomics can reveal subtler changes in CSP expression than those observed herein, such as those reflecting long-term secondary effects on motor impairment and unit integrity, as well as underlying molecular mechanisms, at 180 days post-injury and beyond. For these reasons and others, M2 proteomics is expected to become increasingly important to accurately predict clinical outcome and improve risk group stratification and therapy for mTBI patients.
Supplementary Material
Prioritization of relative protein expression associated with post-injury time relative to sham controls at 180 days of age. The post-injury time point (vs. sham), log2 fold change in protein expression (vs. sham), p-value for post-injury time point (vs. sham), overall p-value for all time-points (vs. sham), Tembl protein symbol, and protein description are shown with examples in bold and italics.
Prioritization of relative protein expression regressed onto grip strength at day 30. The Trembl protein symbol, protein description, log2 fold change in protein expression due to unit increase in grip strength, and p-value for day 30 grip are shown with examples in bold.
Highlights.
M2 proteomics is a rapid method to study mild traumatic brain injury.
M2 proteomics can reveal changes in CNS-specific protein (CSP) expression.
CSP expression can be related to long-term effects on motor impairment.
Acknowledgments
We acknowledge the RCMI and RTRN grants from the National Institute on Minority Health and Health Disparities (G12MD007591 and U54MD008149, respectively) for funding (Haskins WE). This research was funded in part by an independent National Research Service Award, National Institute for Neurological Diseases and Stroke (1F31NS080508-01; Evans TM) and the Hartford Foundation/American Federation for Aging Research Scholars in Geriatric Medicine Program (Jaramillo CA). We would also like to acknowledge the support of the Sam and Ann Barshop institute for Longevity and Aging Studies. Lastly, we thank the dedicated patients, physicians and researchers in the TBI community for their strong support of protein biomarker research for TBI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to report.
Contributor Information
Teresa M. Evans, Email: EvansTM@uthscsa.edu.
Holly Van Remmen, Email: Holly-Van Remmen@omrf.org.
Anjali Purkar, Email: anjali.purkar@utsa.edu.
Swetha Mahesula, Email: swetha.mahesula@utsa.edu.
J AL Gelfond, Email: gelfondJAL@uthscsa.edu.
Marian Sabia, Email: sabia@uthscsa.edu.
Wenbo Qi, Email: Qi@uthscsa.edu.
Ai-Ling Lin, Email: lina3@uthscsa.edu.
Carlos A. Jaramillo, Email: jaramilloc3@uthscsa.edu.
William E. Haskins, Email: WEH.scholar@gmail.com.
References Cited
- 1.Dankers AC, et al. Hyperuricemia influences tryptophan metabolism via inhibition of multidrug resistance protein 4 (MRP4) and breast cancer resistance protein (BCRP) Biochim Biophys Acta. 2013;1832(10):1715–1722. doi: 10.1016/j.bbadis.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Coronado VG, et al. Trends in Traumatic Brain Injury in the U.S. and the public health response: 1995–2009. J Safety Res. 2012;43(4):299–307. doi: 10.1016/j.jsr.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein ES, et al. Are baby boomers who care for their older parents planning for their own future long-term care needs? J Aging Soc Policy. 2012;24(1):29–45. doi: 10.1080/08959420.2012.630905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer H. U.S. Military Casualty Statistics: Operation New Dawn, Operation Iraqi Freedom, and Operation Enduring Freedom. Congressional Research Service. 2010 http://www.crs.gov.
- 5.Logan BW, et al. Concussive Brain Injury in the Military: September 2001 to the Present. Behav Sci Law. 2013 doi: 10.1002/bsl.2092. [DOI] [PubMed] [Google Scholar]
- 6.Kochanek PM, et al. Screening of biochemical and molecular mechanisms of secondary injury and repair in the brain after experimental blast-induced traumatic brain injury in rats. J Neurotrauma. 2013;30(11):920–937. doi: 10.1089/neu.2013.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis AE. Mechanisms of traumatic brain injury: biomechanical, structural and cellular considerations. Crit Care Nurs Q. 2000;23(3):1–13. doi: 10.1097/00002727-200011000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzbold ML, et al. Effects of traumatic brain injury of different severities on emotional, cognitive, and oxidative stress-related parameters in mice. J Neurotrauma. 2010;27(10):1883–1893. doi: 10.1089/neu.2010.1318. [DOI] [PubMed] [Google Scholar]
- 9.Malkesman O, et al. Traumatic Brain Injury - Modeling Neuropsychiatric Symptoms in Rodents. Front Neurol. 2013;4:157. doi: 10.3389/fneur.2013.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson VE, Stewart W, Smith DH. Traumatic brain injury and amyloid-beta pathology: a link to Alzheimer's disease? Nat Rev Neurosci. 2010;11(5):361–370. doi: 10.1038/nrn2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uryu K, et al. Age-dependent synuclein pathology following traumatic brain injury in mice. Exp Neurol. 2003;184(1):214–224. doi: 10.1016/s0014-4886(03)00245-0. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt S, et al. Association of ALS with head injury, cigarette smoking and APOE genotypes. J Neurol Sci. 2010;291(1–2):22–29. doi: 10.1016/j.jns.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKee AC, et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J Neuropathol Exp Neurol. 69(9):918–929. doi: 10.1097/NEN.0b013e3181ee7d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bigler ED. Traumatic brain injury, neuroimaging, and neurodegeneration. Front Hum Neurosci. 2013;7:395. doi: 10.3389/fnhum.2013.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith DH, Johnson VE, Stewart W. Chronic neuropathologies of single and repetitive TBI: substrates of dementia? Nat Rev Neurol. 2013;9(4):211–221. doi: 10.1038/nrneurol.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breunig JJ, Guillot-Sestier MV, Town T. Brain injury, neuroinflammation and Alzheimer's disease. Front Aging Neurosci. 2013;5:26. doi: 10.3389/fnagi.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acosta SA, et al. Long-term upregulation of inflammation and suppression of cell proliferation in the brain of adult rats exposed to traumatic brain injury using the controlled cortical impact model. PLoS One. 2013;8(1):e53376. doi: 10.1371/journal.pone.0053376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan V, et al. Older adults with acquired brain injury: outcomes after inpatient rehabilitation. Can J Aging. 2013;32(3):278–286. doi: 10.1017/S0714980813000317. [DOI] [PubMed] [Google Scholar]
- 19.Corrigan JD, Hammond FM. Traumatic brain injury as a chronic health condition. Arch Phys Med Rehabil. 2013;94(6):1199–1201. doi: 10.1016/j.apmr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 21.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Raphael I, et al. Microwave and magnetic (M(2)) proteomics of the experimental autoimmune encephalomyelitis animal model of multiple sclerosis. Electrophoresis. 2012;33(24):3810–3819. doi: 10.1002/elps.201200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahesula S, et al. Immunoenrichment microwave and magnetic proteomics for quantifying CD47 in the experimental autoimmune encephalomyelitis model of multiple sclerosis. Electrophoresis. 2012;33(24):3820–3829. doi: 10.1002/elps.201200515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin PT, et al. Overexpression of Galgt2 in skeletal muscle prevents injury resulting from eccentric contractions in both mdx and wild-type mice. Am J Physiol Cell Physiol. 2009;296(3):C476–C488. doi: 10.1152/ajpcell.00456.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrow JD, Roberts LJ., 2nd Mass spectrometric quantification of F2-isoprostanes in biological fluids and tissues as measure of oxidant stress. Methods Enzymol. 1999;300:3–12. doi: 10.1016/s0076-6879(99)00106-8. [DOI] [PubMed] [Google Scholar]
- 26.Boggs JM. Myelin basic protein: a multifunctional protein. Cellular and molecular life sciences : CMLS. 2006;63(17):1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas DG, Palfreyman JW, Ratcliffe JG. Serum-myelin-basic-protein assay in diagnosis and prognosis of patients with head injury. Lancet. 1978;1(8056):113–115. doi: 10.1016/s0140-6736(78)90415-4. [DOI] [PubMed] [Google Scholar]
- 28.Gyorgy A, et al. Time-dependent changes in serum biomarker levels after blast traumatic brain injury. J Neurotrauma. 2011;28(6):1121–1126. doi: 10.1089/neu.2010.1561. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M, et al. Overexpression of a minor component of myelin basic protein isoform (17.2 kDa) can restore myelinogenesis in transgenic shiverer mice. Brain research. 1998;785(2):245–252. doi: 10.1016/s0006-8993(97)01383-8. [DOI] [PubMed] [Google Scholar]
- 30.Nakajima K, et al. Novel isoforms of mouse myelin basic protein predominantly expressed in embryonic stage. Journal of neurochemistry. 1993;60(4):1554–1563. doi: 10.1111/j.1471-4159.1993.tb03321.x. [DOI] [PubMed] [Google Scholar]
- 31.Ottens AK, et al. Proteolysis of multiple myelin basic protein isoforms after neurotrauma: characterization by mass spectrometry. Journal of neurochemistry. 2008;104(5):1404–1414. doi: 10.1111/j.1471-4159.2007.05086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Givogri MI, Bongarzone ER, Campagnoni AT. New insights on the biology of myelin basic protein gene: the neural-immune connection. Journal of neuroscience research. 2000;59(2):153–159. [PubMed] [Google Scholar]
- 33.Akiyama K, et al. Study of expression of myelin basic proteins (MBPs) in developing rat brain using a novel antibody reacting with four major isoforms of MBP. Journal of neuroscience research. 2002;68(1):19–28. doi: 10.1002/jnr.10188. [DOI] [PubMed] [Google Scholar]
- 34.Boggs JM, et al. Effect of posttranslational modifications to myelin basic protein on its ability to aggregate acidic lipid vesicles. Biochemistry. 1997;36(16):5065–5071. doi: 10.1021/bi962649f. [DOI] [PubMed] [Google Scholar]
- 35.Su E, et al. Increased CSF concentrations of myelin basic protein after TBI in infants and children: absence of significant effect of therapeutic hypothermia. Neurocrit Care. 2012;17(3):401–407. doi: 10.1007/s12028-012-9767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger RP, et al. Trajectory analysis of serum biomarker concentrations facilitates outcome prediction after pediatric traumatic and hypoxemic brain injury. Dev Neurosci. 2010;32(5–6):396–405. doi: 10.1159/000316803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papa L, et al. Systematic review of clinical research on biomarkers for pediatric traumatic brain injury. J Neurotrauma. 2013;30(5):324–338. doi: 10.1089/neu.2012.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger RP, et al. Identification of inflicted traumatic brain injury in well-appearing infants using serum and cerebrospinal markers: a possible screening tool. Pediatrics. 2006;117(2):325–332. doi: 10.1542/peds.2005-0711. [DOI] [PubMed] [Google Scholar]
- 39.Kochanek PM, et al. The potential for bio-mediators and biomarkers in pediatric traumatic brain injury and neurocritical care. Front Neurol. 2013;4:40. doi: 10.3389/fneur.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong KA, et al. Diffuse axonal injury in children: clinical correlation with hemorrhagic lesions. Ann Neurol. 2004;56(1):36–50. doi: 10.1002/ana.20123. [DOI] [PubMed] [Google Scholar]
- 41.Galloway NR, et al. Diffusion-weighted imaging improves outcome prediction in pediatric traumatic brain injury. J Neurotrauma. 2008;25(10):1153–1162. doi: 10.1089/neu.2007.0494. [DOI] [PubMed] [Google Scholar]
- 42.Smith C, et al. The neuroinflammatory response in humans after traumatic brain injury. Neuropathol Appl Neurobiol. 2013;39(6):654–666. doi: 10.1111/nan.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piliponsky AM, et al. Human eosinophils induce histamine release from antigen-activated rat peritoneal mast cells: a possible role for mast cells in late-phase allergic reactions. J Allergy Clin Immunol. 2001;107(6):993–1000. doi: 10.1067/mai.2001.114656. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi H, et al. Tissue eosinophilia and eosinophil degranulation in syndromes associated with fibrosis. Am J Pathol. 1992;140(2):521–528. [PMC free article] [PubMed] [Google Scholar]
- 45.Lenzlinger PM, et al. Delayed inhibition of Nogo-A does not alter injury-induced axonal sprouting but enhances recovery of cognitive function following experimental traumatic brain injury in rats. Neuroscience. 2005;134(3):1047–1056. doi: 10.1016/j.neuroscience.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 46.Chytrova G, Ying Z, Gomez-Pinilla F. Exercise normalizes levels of MAG and Nogo-A growth inhibitors after brain trauma. Eur J Neurosci. 2008;27(1):1–11. doi: 10.1111/j.1460-9568.2007.05982.x. [DOI] [PubMed] [Google Scholar]
- 47.Dubreuil CI, et al. Activation of Rho after traumatic brain injury and seizure in rats. Exp Neurol. 2006;198(2):361–369. doi: 10.1016/j.expneurol.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Marklund N, et al. Selective temporal and regional alterations of Nogo-A and small proline-rich repeat protein 1A (SPRR1A) but not Nogo-66 receptor (NgR) occur following traumatic brain injury in the rat. Exp Neurol. 2006;197(1):70–83. doi: 10.1016/j.expneurol.2005.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson HJ, et al. Tissue sparing and functional recovery following experimental traumatic brain injury is provided by treatment with an anti-myelin-associated glycoprotein antibody. Eur J Neurosci. 2006;24(11):3063–3072. doi: 10.1111/j.1460-9568.2006.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irving EA, et al. Identification of neuroprotective properties of anti-MAG antibody: a novel approach for the treatment of stroke? J Cereb Blood Flow Metab. 2005;25(1):98–107. doi: 10.1038/sj.jcbfm.9600011. [DOI] [PubMed] [Google Scholar]
- 51.Newcomb JK, et al. Immunohistochemical study of calpain-mediated breakdown products to alpha-spectrin following controlled cortical impact injury in the rat. J Neurotrauma. 1997;14(6):369–383. doi: 10.1089/neu.1997.14.369. [DOI] [PubMed] [Google Scholar]
- 52.Pike BR, et al. Accumulation of non-erythroid alpha II-spectrin and calpain-cleaved alpha II-spectrin breakdown products in cerebrospinal fluid after traumatic brain injury in rats. J Neurochem. 2001;78(6):1297–1306. doi: 10.1046/j.1471-4159.2001.00510.x. [DOI] [PubMed] [Google Scholar]
- 53.Ottens AK, et al. Proteolysis of multiple myelin basic protein isoforms after neurotrauma: characterization by mass spectrometry. J Neurochem. 2008;104(5):1404–1414. doi: 10.1111/j.1471-4159.2007.05086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teunissen CE, Khalil M. Neurofilaments as biomarkers in multiple sclerosis. Multiple sclerosis. 2012;18(5):552–556. doi: 10.1177/1352458512443092. [DOI] [PubMed] [Google Scholar]
- 55.Khalil M, et al. CSF neurofilament and N-acetylaspartate related brain changes in clinically isolated syndrome. Multiple sclerosis. 2012 doi: 10.1177/1352458512458010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mateen FJ, et al. Neurological disorders in the 11th revision of the International Classification of Diseases: now open to public feedback. Lancet neurology. 2012;11(6):484–485. doi: 10.1016/S1474-4422(12)70125-4. [DOI] [PubMed] [Google Scholar]
- 57.Gunnarsson M, et al. Axonal damage in relapsing multiple sclerosis is markedly reduced by natalizumab. Annals of neurology. 2011;69(1):83–89. doi: 10.1002/ana.22247. [DOI] [PubMed] [Google Scholar]
- 58.Koel-Simmelink MJ, et al. The neurofilament light chain is not stable in vitro. Annals of neurology. 2011;69(6):1065–1066. doi: 10.1002/ana.22438. author reply 1066–7. [DOI] [PubMed] [Google Scholar]
- 59.Gavett BE, Stern RA, McKee AC. Chronic traumatic encephalopathy: a potential late effect of sport-related concussive and subconcussive head trauma. Clin Sports Med. 30(1):179–188. doi: 10.1016/j.csm.2010.09.007. xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 61.Hayes JD, Strange RC. Potential contribution of the glutathione S-transferase supergene family to resistance to oxidative stress. Free Radic Res. 1995;22(3):193–207. doi: 10.3109/10715769509147539. [DOI] [PubMed] [Google Scholar]
- 62.Ansari MA, Roberts KN, Scheff SW. Dose- and time-dependent neuroprotective effects of Pycnogenol following traumatic brain injury. J Neurotrauma. 2013;30(17):1542–1549. doi: 10.1089/neu.2013.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandoval WN, et al. Applications of microwave-assisted proteomics in biotechnology. Combinatorial chemistry & high throughput screening. 2007;10(9):751–765. doi: 10.2174/138620707783018504. [DOI] [PubMed] [Google Scholar]
- 64.Lill JR, et al. Microwave-assisted proteomics. Mass spectrometry reviews. 2007;26(5):657–671. doi: 10.1002/mas.20140. [DOI] [PubMed] [Google Scholar]
- 65.Wong MY, et al. A magnetic bead-based serum proteomic fingerprinting method for parallel analytical analysis and micropreparative purification. Electrophoresis. 2010;31(10):1721–1730. doi: 10.1002/elps.200900571. [DOI] [PubMed] [Google Scholar]
- 66.Labugger R, et al. Strategy for analysis of cardiac troponins in biological samples with a combination of affinity chromatography and mass spectrometry. Clinical chemistry. 2003;49(6 Pt 1):873–879. doi: 10.1373/49.6.873. [DOI] [PubMed] [Google Scholar]
- 67.Thompson A, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical chemistry. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 68.Dayon L, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Analytical chemistry. 2008;80(8):2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prioritization of relative protein expression associated with post-injury time relative to sham controls at 180 days of age. The post-injury time point (vs. sham), log2 fold change in protein expression (vs. sham), p-value for post-injury time point (vs. sham), overall p-value for all time-points (vs. sham), Tembl protein symbol, and protein description are shown with examples in bold and italics.
Prioritization of relative protein expression regressed onto grip strength at day 30. The Trembl protein symbol, protein description, log2 fold change in protein expression due to unit increase in grip strength, and p-value for day 30 grip are shown with examples in bold.