Key Points
Elevation of HbF in 3 patients heterozygous for distinct 2p15-p16.1 syndrome microdeletions affecting BCL11A.
Identification of novel, putative regulatory elements downstream of BCL11A that govern its expression in erythroid cells.
Abstract
Elevated fetal hemoglobin (HbF) ameliorates the clinical severity of hemoglobinopathies such as β-thalassemia and sickle cell anemia. Currently, the only curative approach for individuals under chronic transfusion/chelation support therapy is allogeneic stem cell transplantation. However, recent analyses of heritable variations in HbF levels have provided a new therapeutic target for HbF reactivation: the transcriptional repressor BCL11A. Erythroid-specific BCL11A abrogation is now actively being sought as a therapeutic avenue, but the specific impact of such disruption in humans remains to be determined. Although single nucleotide polymorphisms in BCL11A erythroid regulatory elements have been reported, coding mutations are scarcer. It is thus of great interest that patients have recently been described with microdeletions encompassing BCL11A. These patients display neurodevelopmental abnormalities, but whether they show increased HbF has not been reported. We have examined the hematological phenotype, HbF levels, and erythroid BCL11A expression in 3 such patients. Haploinsufficiency of BCL11A induces only partial developmental γ-globin silencing. Of greater interest is that a patient with a downstream deletion exhibits reduced BCL11A expression and increased HbF. Novel erythroid-specific regulatory elements in this region may be required for normal erythroid BCL11A expression, whereas loss of separate elements in the developing brain may explain the neurological phenotype.
Introduction
Chromosome 2p15-p16.1 microdeletions constitute a contiguous gene deletion syndrome characterized by shared phenotypic traits including intellectual disability, growth retardation, and distinct dysmorphisms. To date, 11 microdeletions of discrete lengths have been described, each associated with overlapping subsets of phenotypic features.1 These deletions collectively encompass approximately 17 protein-coding genes and are accordingly associated with a complex spectrum of physical and mental traits. The smallest microdeletion yet has recently been reported to solely contain the BCL11A gene and is associated with hypotonia, mild intellectual delay, speech disorder, and gross motor impairments.1
BCL11A is a transcription factor that is highly expressed in the brain, B-lymphocytes, and adult erythroid lineage. It has emerged from several genome-wide association studies as a negative modulator of fetal hemoglobin expression.2-4 Indeed, BCL11A knockout disrupts developmental silencing of human fetal γ-globin in transgenic mice.5 Moreover, BCL11A knockdown in primary human erythroid cells results in increased fetal hemoglobin (HbF),6 whereas conditional knockout in adult sickle cell disease transgenic mice leads to reactivation of γ-globin and amelioration of symptoms.7 Erythroid-specific abrogation of BCL11A has thus been pursued as an attractive therapeutic for β-hemoglobinopathies.8 Although HbF-associated single-nucleotide polymorphisms (SNPs) have been shown to disrupt an erythroid enhancer in the second intron of BCL11A, until recently, coding BCL11A mutations had not been reported.9-11 Here we have investigated the hematopoietic parameters of 3 patients with distinct de novo, heterozygous deletions encompassing, or proximal to, BCL11A. All display modestly reduced BCL11A expression and considerable HbF elevation. In one patient, a downstream deletion that leaves the BCL11A coding gene intact results in reduced BCL11A transcripts in erythroblasts, alluding to the existence of novel, erythroid regulatory elements within this region.
Methods
See the supplemental Data, available on the Blood Web site.
Results and discussion
The 3 patients herein have been described previously.12,13 The first is a 15-year-old female with a 3.5-Mb deletion downstream of BCL11A (Figure 1A).13 The second and third are an 8-year-old female and 7-year-old male with 642-kb and 2.5-Mb deletions, respectively, covering the entire BCL11A gene (Figure 1A).12 Although their neurological and physical traits have been extensively characterized, their hematopoietic profiles have not been previously assessed.
Figure 1.
Patients with 2p15-16.1 microdeletions display depleted BCL11A expression in erythroblasts and markedly increased fetal γ-globin in peripheral blood. (A) Schematic of the 2p15-16.1 region showing segments that are deleted in the 3 patients. In addition to BCL11A, the following deleted genes are expressed at 1 or more stages during erythroid maturation:25 VRK2, FANCL, PAPOLG, REL, PUS10, PEX13, KIAA1841, C2orf74, AHSA2, USP34, XPO1, FAM161A, CCT4, COMMD1, and B3GNT2. (B) Real-time reverse transcription-polymerase chain reaction quantification of BCL11A mRNA isoforms in erythroblasts expanded in vivo derived from patients (n = 3) and matched parents (n = 5). Shown are total BCL11A transcripts, isoforms previously implicated in γ-globin repression (BCL11A-XL and BCL11A-L) and short variants (BCL11A-S). All levels have been normalized to GAPDH mRNA. Error bars represent standard deviation and P values signify analysis of variation comparison. (C) HPLC determination of hemoglobin tetramer levels for the 3 patients and their parents. HbF peaks are shown in red. Note that HbA2 (α2δ2) levels are relatively constant across all samples (2.3%, 2.6%, and 1.6% for the patients, and 2.2%, 2.8% and 2.6%, 2.4% and 2.2%, respectively, for the parents).
We first determined the extent to which these heterozygous deletions influence BCL11A expression in erythroblasts derived from the 3 patients. In all cases, BCL11A transcripts were significantly reduced by approximately twofold relative to parental expression (Figure 1B). We also specifically analyzed the abundance of different BCL11A messenger RNA (mRNA) isoforms, and found that those implicated in γ-globin repression (BCL11A-XL and BCL11A-L)5,6 were significantly downregulated in the patients’ erythroblasts, whereas short transcript variants (BCL11A-S) were not (Figure 1B). Expression of KLF1 and GATA1, erythroid transcription factors that regulate globin expression, was unaltered (data not shown).
Complete blood counts revealed that hematological parameters were largely normal in the 3 individuals (supplemental Table 1). The third patient displayed mildly reduced hematocrit and increased variability in red blood cell size at 5 months of age. These findings were recapitulated in a follow-up count at 7 years of age. This patient also exhibited modest lymphocytosis at this age. BCL11A is highly expressed in B cells and has been implicated in human B-lymphopoiesis.7,14 It should be noted, however, that the first 2 patients showed normal lymphocyte levels, suggesting that these hypomorphic BCL11A mutations have minimal impact on B-cell numbers.
We next assayed relative amounts of hemoglobin isoforms by high-performance liquid chromatography (HPLC). All 3 patients had markedly elevated fetal γ-globin compared with their unaffected parents (Figure 1C; supplemental Figure 1A). This was reflected both as a fraction of total fetal plus adult β-like globin as well as α-globin (supplemental Figure 1B,C). HPLC analysis of hemoglobin tetramers determined the patients’ HbF levels to be 7.3%, 4.8%, and 6.2%, respectively, compared with that of their parents (0.5%, 0.7% and 0.2%, and 0.2% and 0.6%) (Figure 1C). Quantification of individual globin chains revealed that fetal γ-globin represented 11.5%, 8.2%, and 10.0% of total β-like globin in the 3 patients compared with parental levels of 1.5%, 1.9% and 1.3%, and 1.9% and 2.0% (supplemental Figure 1A). In all individuals, a balanced ratio of β-like globin to α-globin was sustained (supplemental Figure 1D).
These results demonstrate that BCL11A haploinsufficiency has an appreciable effect on HbF expression. This mirrors findings of the master erythroid regulator KLF1, an upstream activator of BCL11A.15,16 In the short time since a nonsense mutation in KLF1 was initially linked to hereditary persistence of fetal hemoglobin,15 a raft of additional mutations in the locus have been described resulting from focused sequencing efforts,17-20 culminating in the recent discovery of a KLF1-null individual.21 Thus far, BCL11A coding mutations have not been frequently encountered. Given its implicated B-lymphopoietic and neurological roles, it is possible that BCL11A mutations with erythroid-restricted consequences (ie, elevated HbF) will largely lie in regulatory elements with erythroid activity (as in Bauer et al9).
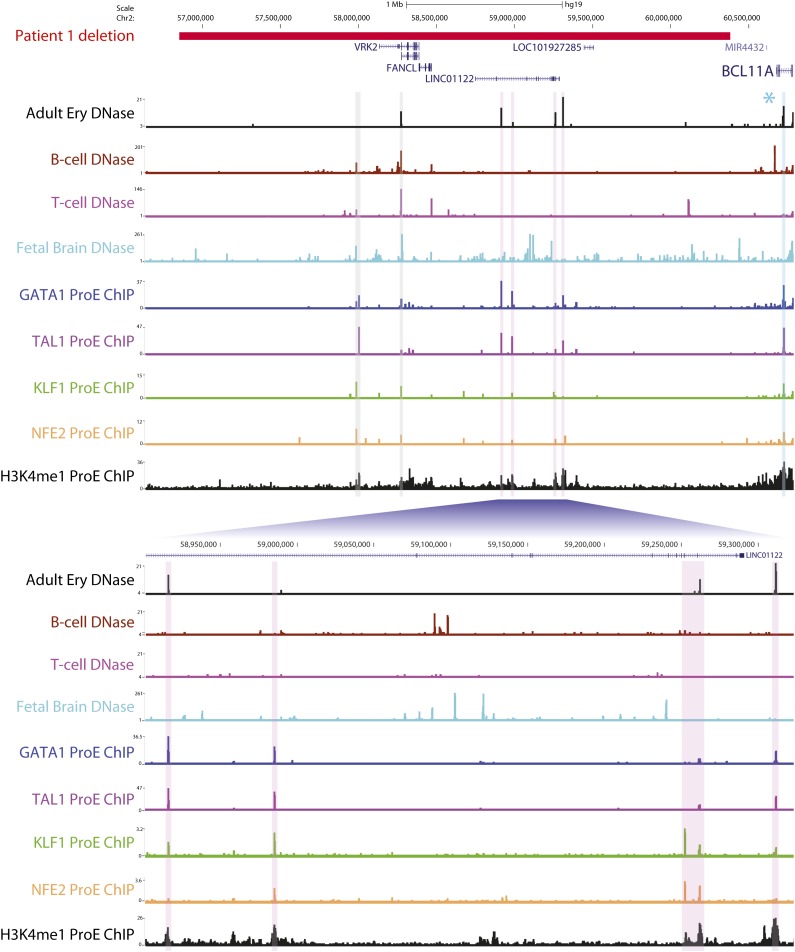
The approximately twofold downregulation of BCL11A mRNA in patients 2 and 3 can likely be attributed to monoalleleic expression arising from BCL11A heterozygosity. However, in the first patient, both BCL11A alleles are intact (Figure 1A). This raises the possibility that downstream regulatory elements within the deleted region are required for full erythroid expression of BCL11A. Inspection of this region revealed several prominent DNase-hypersensitive sites in adult erythroid cells that are cobound by various combinations of key erythroid transcription factors (GATA1, TAL1, KLF1, and NFE2) and are enriched for the active enhancer-associated marker H3K4me1 (Figure 2). These elements represent potential erythroid enhancers, although their functional characterization remains to be determined. It is also plausible that the deletion in patient 1 might result in reduced BCL11A expression through alternative molecular mechanisms, such as downstream heterochromatic regions being brought into the vicinity of BCL11A.
Figure 2.
A downstream microdeletion associated with reduced BCL11A expression contains several putative erythroid enhancers. The region deleted in patient 1 is shown in red. Shown are DNase sequencing tracks for adult erythroblasts (derived from peripheral blood CD34+ cells), other hematopoietic cells in which BCL11A is expressed (B cells) or not expressed (T cells), and the developing fetal brain. Sequencing tracks have been emboldened for clarity. Potential erythroid regulatory elements are highlighted with vertical bars and are bound in human pro-erythroblasts by different combinations of noted erythroid transcription factors (GATA1, TAL1, KLF1, and NFE2) as evidenced by chromatin immunoprecipitation (ChIP) sequencing. These regions are also marked by H3K4me1, typically associated with active enhancer elements. Of note, regions highlighted in pink display erythroid DNase hypersensitivity that is not present in the other hematopoietic cells, suggesting that they are erythroid-specific sites. Regions in gray signify DNase-hypersensitive sites at an alternative VRK2 promoter and upstream element, which are present in other tissues. The segment in blue indicates a known erythroid enhancer in BCL11A intron 2, to which SNPs associated with HbF levels are localized.9 A second cluster of high HbF-associated SNPs (marked by an asterisk) lies downstream of BCL11A but outside the deleted region. The fetal brain exhibits widespread DNase hypersensitivity across the deleted region, which might account for the neurological phenotype of this patient.
The extent to which BCL11A need be disrupted to induce clinically protective HbF levels in sickle cell and thalassemic sufferers is unknown. The patients here exhibit approximately 5% to 10% HbF and twofold downregulated BCL11A mRNA. In sickle cell sufferers, such HbF levels are associated with reduced major organ failure, but higher amounts (>20%) are required to deter recurrent clinical crises.22 Further, because of limited availability of tissue samples, we have been unable to ascertain whether the increased HbF in these patients is pancellular or heterocellular. This is an important consideration because pancellular upregulation is predominantly associated with clinical amelioration. Indeed, studies using transgenic, humanized sickle cell mice found that HbF levels of approximately 10% or greater were clinically beneficial when expressed in at least two-thirds of peripheral erythrocytes23 or higher.24
During the revision of this manuscript, an additional 3 patients were described, each with heterozygous 2p15-p16.1 deletions distinct from those reported here. Each of these patients exhibited a similar reduction in BCL11A expression to the cases here as well as persistence of fetal hemoglobin and unperturbed lymphocytes. However, the reported patients exhibited markedly higher levels of HbF (16.1-29.7%) than those described here. The reason for this discrepancy is unclear, but may be due to modifiers, genetic or otherwise, that influence ethnic or regional variation in HbF levels.11
Last, it is worth noting that several of the genes deleted in the 3 patients are expressed during erythroid differentiation (Figure 1A), and their deletion could thus conceivably contribute to the elevated HbF observed. However, the striking phenotypic similarities of the patients (in terms of both BCL11A and HbF levels) might suggest that the HbF modifier effects of these genes, which are variably deleted in different combinations, are minimal.
Taken together, these findings suggest that modest downregulation of erythroid BCL11A expression results in markedly increased HbF, supporting the utility of erythroid-specific disruption of BCL11A as a therapeutic for β-hemoglobinopathies. That these hypomorphic BCL11A mutations are associated with a spectrum of neurodevelopmental defects further emphasizes the need for such abrogation to be erythroid-specific. We suggest that candidate erythroid regulatory elements, such as those described previously9 and here, should be inspected during routine screening of HbF modifiers, and may represent targets for therapeutic intervention.
Acknowledgments
The authors sincerely thank the patients and their families for their willingness to contribute to this research.
This study was supported by grants from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK101328), National Heart, Lung, and Blood Institute (HL116329-01), National Cancer Institute (P01-CA108671), and Italian Ministry of Health (Ricerca Corrente 2013-2015).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.P.W.F. analyzed data and performed the bioinformatics; P.P., V.O., M.P., A.G., L.V., and A.M. provided important hematologic data; M.M., F.C., L.V., and F.M. performed cell culture and analyzed BCL11A mRNA expression levels; S.S.-S. conducted the HPLC analysis; T.P., A.R.M., and J.A.S. oversaw project design; T.P. and A.R.M. conceived the study and contributed equally to this work; and A.P.W.F., A.R.M., and T.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thalia Papayannopoulou, Division of Hematology, Department of Medicine, University of Washington, Box 357710, Health Sciences Building, Room K243, Seattle, WA 98195; e-mail: thalp@uw.edu; and Anna Rita Migliaccio, Department of Medicine, Mount Sinai School of Medicine, One Gustave L Levy Pl, Box #1079, New York, NY 10029; e-mail: annarita.migliaccio@mssm.edu.
References
- 1.Peter B, Matsushita M, Oda K, Raskind W. De novo microdeletion of BCL11A is associated with severe speech sound disorder. Am J Med Genet A. 2014;164A(8):2091–2096. doi: 10.1002/ajmg.a.36599. [DOI] [PubMed] [Google Scholar]
- 2.Menzel S, Garner C, Gut I, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39(10):1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 3.Lettre G, Sankaran VG, Bezerra MA, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA. 2008;105(33):11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uda M, Galanello R, Sanna S, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA. 2008;105(5):1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankaran VG, Xu J, Ragoczy T, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460(7259):1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Peng C, Sankaran VG, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334(6058):993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankaran VG, Weiss MJ. Anemia: progress in molecular mechanisms and therapies. Nat Med. 2015;21(3):221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer DE, Kamran SC, Lessard S, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet. 2010;42(12):1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basak A, Hancarova M, Ulirsch JC, et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations [published online ahead of print May 4, 2015]. J Clin Invest. doi: 10.1172/JCI81163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccione M, Piro E, Serraino F, et al. Interstitial deletion of chromosome 2p15-16.1: report of two patients and critical review of current genotype-phenotype correlation. Eur J Med Genet. 2012;55(4):238–244. doi: 10.1016/j.ejmg.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Prontera P, Bernardini L, Stangoni G, et al. Deletion 2p15-16.1 syndrome: case report and review. Am J Med Genet A. 2011;155A(10):2473–2478. doi: 10.1002/ajmg.a.33875. [DOI] [PubMed] [Google Scholar]
- 14.Liu P, Keller JR, Ortiz M, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4(6):525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 15.Borg J, Papadopoulos P, Georgitsi M, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet. 2010;42(9):801–805. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet. 2010;42(9):742–744. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 17.Satta S, Perseu L, Moi P, et al. Compound heterozygosity for KLF1 mutations associated with remarkable increase of fetal hemoglobin and red cell protoporphyrin. Haematologica. 2011;96(5):767–770. doi: 10.3324/haematol.2010.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallienne AE, Dréau HM, Schuh A, Old JM, Henderson S. Ten novel mutations in the erythroid transcription factor KLF1 gene associated with increased fetal hemoglobin levels in adults. Haematologica. 2012;97(3):340–343. doi: 10.3324/haematol.2011.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T, He Y, Zhou JY, et al. KLF1 gene mutations in Chinese adults with increased fetal hemoglobin. Hemoglobin. 2013;37(5):501–506. doi: 10.3109/03630269.2013.805304. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Zhang X, Yu L, et al. KLF1 mutations are relatively more common in a thalassemia endemic region and ameliorate the severity of β-thalassemia. Blood. 2014;124(5):803–811. doi: 10.1182/blood-2014-03-561779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magor GW, Tallack MR, Gillinder KR, et al. KLF1-null neonates display hydrops fetalis and a deranged erythroid transcriptome. Blood. 2015;125(15):2405–2417. doi: 10.1182/blood-2014-08-590968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powars DR, Weiss JN, Chan LS, Schroeder WA. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63(4):921–926. [PubMed] [Google Scholar]
- 23.Perumbeti A, Higashimoto T, Urbinati F, et al. A novel human gamma-globin gene vector for genetic correction of sickle cell anemia in a humanized sickle mouse model: critical determinants for successful correction. Blood. 2009;114(6):1174–1185. doi: 10.1182/blood-2009-01-201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blouin MJ, Beauchemin H, Wright A, et al. Genetic correction of sickle cell disease: insights using transgenic mouse models. Nat Med. 2000;6(2):177–182. doi: 10.1038/72279. [DOI] [PubMed] [Google Scholar]
- 25.An X, Schulz VP, Li J, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123(22):3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]