Circulating tumor cells (CTCs) are cells shed from the primary tumor and metastatic sites, and can be found at very low frequencies in the peripheral blood of cancer patients.[1] Enumeration of CTCs has been shown to correlate with disease progression in metastatic cancer patients,[2] and recent studies suggest that CTCs can serve as a less invasive biomarker for therapeutic assays.[3] However, the heterogeneity inherent in CTCs[4] suggests that molecular profiling at a single cell level may be necessary to capture the evolution of tumor genotypes during treatment and disease progression.[5] To successfully achieve in-depth interrogation of individual CTCs and identify tumor drivers or secondary mutations,[6] the isolated cells must be intact, viable and free of contaminating cells. On the other hand, bulk analysis of the same CTC population may be desirable for other clinical applications, such as the establishment of CTC cell lines through cell culture for in-vitro drug screening. CTC isolation technologies with such dynamic versatility could serve as a powerful clinical tool.
The growing demand for molecular profiling of CTCs has led to the development of microfluidic devices patterned with stimuli-responsive coatings that can release captured CTCs in response to a coating-specific degradation mechanism. Previously, our group has developed the herringbone CTC-Chip (HBCTC-Chip),[7] a microfluidic device used to isolate CTCs from patient blood,[8] identify signaling pathways in CTCs through RNA sequencing,[9] study dynamic changes in epithelial and mesenchymal composition in CTCs[4] and most recently, to explore the role of CTC clusters in the metastatic process.[10] Removal of viable CTCs isolated on the HBCTC-Chip and other positive selection-based microfluidic devices has been extremely challenging due to the fragile nature of CTCs and the non-specific interactions that can occur between whole blood components (e.g., white blood cells, plasma proteins) and the microchannel substrate. Proteolytic enzymes such as trypsin have been combined with surfactants for the release of captured cells,[11] but when applied to other device architectures, this strategy has resulted in poor release (< 10%) with limited viability.[12] Ionically cross- linked sacrificial hydrogels have been used for microfluidic capture and release of endothelial progenitor cells without the need for enzymatic digestion,[13] with the principles of calcium chelation driving the substrate degradation. This approach is limited to the use of heparin blood tubes, preventing the use of common blood anti-cogulants (e.g., EDTA, citrates). Using a similar approach but applied to nanoprobes, Xie et al. reported 65% release efficiency using EDTA-assisted cell release, with 70% of the released cells being viable after five minutes of 50mM EDTA exposure.[14] Our group has previously presented a photo-cross-linkable alginate-based degradable biopolyer that is compatible with all anti-cogulants and degrades under exposure to alginate lyase.[15] However, the excessive thickness of the alginate coating prevented its precise integration with the complex three-dimensional microfluidic features present in the HBCTC-Chip.
DNA-based[16] and Aptamer-based[12a, 17] coatings that rely upon DNAse[16] and endonucleases to digest all forms of DNA and RNA have been used to capture and release cell lines.[12a, 17b] The use of DNAse has a detrimental impact on cell viability with Zhao et al. reporting that 66% of the cells were viable post-release.[16] Thermally responsive polymer brushes have been grafted onto silicon nanowire substrates for the capture of cancer cell lines from buffer and serum.[12b] While promising, this system may be challenging to implement clinically due to the limited volumes processed (< 1ml), and the requirement to run the samples at 37°C.[18] Laser microdissection systems (LMDs) allow for the release of individual CTCs from disassembled microfluidic devices, but LMDs can be cost-prohibitive and the released cells are non-viable.[18-19] The exposure of individual cells to UV-light during the release process may cause cell DNA and RNA degradation.[20]
We present a dual-mode gelatin-based nanostructured coating that can achieve temperature-responsive release (for bulk-population recovery) or mechano-sensitive release (for single cell recovery) of CTCs from peripheral blood. Both release mechanisms are non-fouling and extremely sensitive for isolating these rare, delicate cells. The coating is formed by a layer-by-layer (LbL) deposition of biotinylated gelatin and streptavidin, in which the complementary binding of biotin with streptavidin are the interactions that drive coating assembly. For bulk-population release of CTCs, raising the device temperature to physiologic temperature (37°C) deconstructs the nanocoating from the surface within minutes. For single cell release of CTCs, mechanical stress from a frequency-controlled microtip was used to dissolve localized regions of the nanocoating (mimicking thixotropic hydrogel behaviors). This dual-mode recovery strategy was successfully used to characterize CTCs in bulk or at the single cell level such that driver mutations in the PIK3CA and EGFR oncogenes were identified.
Gelatin-biotin molecules were directly deposited onto plasma-activated PDMS surfaces with alternating layers of streptavidin applied to increase and stabilize the nanocoating architecture (Figure 1a). For gelatin molecules near the surface, intermolecular forces between the substrate and gelatin will dominate and inhibit the inherent temperature responsiveness of gelatin. As such, the first layer was physisorbed onto the surface of the device, likely due to hydrogen bonding as well as electrostatic and hydrophobic interactions. This initial layer then served as the foundation upon which the remaining layers were built. Gelatin substrate adsorption has been shown to be dependent upon solution concentration,[21] and we confirmed this with the nanocoating by varying the concentration of gelatin and measuring the resulting thickness of the physisorbed layer (ε). For a 0.1% (w/v) solution at 20°C, ε was 10.7 ± 6.3 nm; while for a 1 % (w/v) solution at 20°C, the thickness of the physisorbed layer increased to 18.2 ± 7.5 nm (Figure S1). The efficiency of biotinylation of the gelatin coating was analyzed using a standard HABA assay (Supplementary Table 1).
Figure 1. Nanocoating characterization.
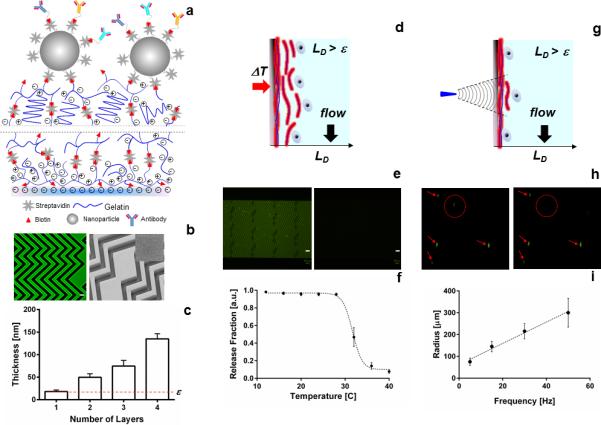
(a) Schematic of the modified LBL nanocoating at the surface of a microfluidic device. Lines indicate the process is repeated four times. (b) Left, confocal micrograph of the nanocoating using streptavidin-FITC; right, electron microscopy image of the coating with the insert showing a high-resolution imaging of the surface, scale bar represents 50 μm. (c) Thickness-growth curve of the deposited layers. (d) Cartoon of the bulk release mechanism for the nanocoating. (e) Fluorescence microscopy images of the coating before (left) and after degradation, scale bar represents 200 μm. (f) Quantification of the release fraction of microbeads immobilized on the surface of the nanocoating. (g) Cartoon of the single cell / selective release mechanism. (h) Micrographs of a single cell being released from the nanocoating. In the left image, the cell targeted for release is identified with a dotted red circle; all other cells that should remain are marked with red arrows. The right image was taken after applying localized shear stress with our microtip, releasing only the target cell. (i) Size of the release radius based on the magnitude of the frequency of vibration of the microtip.
Surface heterogeneity of any coating can result in poor reproducibility and performance. We evaluated the heterogeneity of the nanocoating across the device length using confocal and electron microscopy techniques. Micrographs of the microchannel surface (Figure 1b) revealed the uniformity achieved using the LbL process, even on complex 3-D microfluidic structures with high aspect ratio grooves.[7] The incremental deposition of each layer (gelatin-biotin and streptavidin) was verified with fluorescence microscopy (Figure S1). Additionally, we used electron microscopy to visually inspect the deposited gelatin for surface impurities (Figure S1). Streptavidin-FITC labeling of the nanocoating surface revealed a fluorescence signal intensity that was continuous and uniform along the microchannel length and at different planes of the device boundaries (Figure S1). This qualitative information was combined with quantitative profilometry measurements to characterize the thickness of the nanocoating. After the deposition of four layers, the total substrate thickness was LD = 135.0 ± 11.2 nm (Figure 1c). We tested the lifespan of the microfluidic devices by evaluating the binding capacity of streptavidin nanocoating using fluorescently tagged biotinylated molecule (Biotin-R-Phycoerythrin). Devices with the nanocoating were stored at 4°C for 5, 30 and 45 days (Figure S2). The biotinylated molecule was added and the average fluorescence intensity was recorded across the surface of the device. Statistically similar intensities were observed for the time span evaluated (p = 0.54).
The resulting nanocoating has two distinct, biocompatible mechanisms of dissolution: (1) thermal and (2) shear-responsive dissolution. The first mechanism of substrate solubilization occurs when the surface temperature is increased from room temperature to physiologic temperatures (37°C), resulting in the bulk release of the entire nanocoating (Figure 1d-e). To have this level of responsiveness within the nanocoating, gelatin molecules form entangled intermolecular alpha-helix structures with reversible hydrogen bonds between gelatin molecules and surrounding water.[21] We determined the minimum thickness LD at which the temperature responsiveness of the nanocoating was restored to be 135.0 ± 11.2 nm. At LD thickness, we found negligible detachment of the coating at temperatures below 30°C with a retention fraction of 0.96 ± 0.01 (Figure 1f). Statistical analysis between the first five points revealed no significant difference (p = 0.69). However, an increase in temperature over 30°C produced the detachment of the external layers of the coating. Upon its release, the nanocoating was removed from the device using pressure-driven flow at flow rates of 2.0 to 3.5 ml/h. A residual fraction of 0.11 ± 0.05 nm of the coating remained on the surface of the device (Figure 1f). Changes in the flow rate in the range of 2.0 to 3.5 ml/h did not produce significant variations in the amount of degradation of the nanocoating (Figure S3).
Utilizing the same nanocoating, localized regions of the material can be selectively dissolved to release single cells from the substrate at room temperature. This second mechanism of release relies on the mechano-responsive behavior of thixotropic hydrogels formed by particle aggregates that are bonded to each other through non-covalent interactions (e.g., hydrogen bonds, electrostatic interactions). Thixotropic hydrogels will undergo flow (i.e., shear-thinning) when inertial forces are applied.[22] We theorized that the gelatin nanocoating can be disrupted locally by applying a normal force through a frequency-controlled 80-μm microtip (Figure 1g-h). The mechanism of release would likely be a combination of three effects: fluidic shear stresses generated by the oscillatory motion, the momentum of the cell, and the disruption of the nanocoating under such stresses. We explored the theoretical contribution of each effect using a finite element model (see Supporting Information).
Based on our theoretical model, an experimental prototype of the frequency-controlled microtip system was built (Figure S4) and used to release localized areas of the nanocoating. The microtip produced a controlled vibration at the surface of the microfluidic device, producing a normal inertial force that could selectively remove a single cell. The surface area of the film removed (and thus the number of cells released) depended on the frequency of vibration, resulting in the ability to tune a release radius for individual CTC recovery. From these results, the optimal release radius was determined to be between 145 and 215 μm, corresponding to vibration frequencies between 15 and 30 Hz (Figure 1i).
Initial experiments to evaluate the performance of our nanocoating revealed that our gelatin-based material alone was not as efficient in capturing CTCs in comparison to our traditional surface chemistry, as evidenced by a 70% decline in capture efficiency (Figure S5). To increase target cell capture, we incorporated streptavidin-coated polystyrene nanoparticles onto the top surface of the gelatin nanocoating which increased the local concentration of antibodies available for target cell binding (Figure S6). The mean size of our nanoparticles was determined to be 166 nm ± 56 nm at a concentration of 7.2×108 particles / ml. Using atomic force microscopy to evaluate the surface of our material, we calculated an average of 241 particles per 2.5 x 2.5 um2, with the nanoparticles covering 22% of the surface (Figure S7). In the past, nanostructure materials such as nanopillars, graphene sheets, and nanoparticles[23] have been previously shown to enhance sensitive cell or biomolecule recognition.[24] The addition of the nanoparticles onto our gelatin coating dramatically increased capture efficency for cancer cells with high EpCAM expression levels (Figure S5), resulting in statistically similar capture rates in comparison to our control (p = 0.11).
To quantify the performance of our material, the inner surface of our microfluidic HBCTC-Chip[7] was functionalized with our gelatin nanocoating and exposed to whole blood spiked with cancer cell lines. The optimal nanoparticle concentration for the capture of prostate cancer cells (PC3) from whole blood was determined to be 0.11 mg/ml, producing a capture efficiency of 95.9 % ± 1.5 %. Higher concentrations of nanoparticles did not improve efficiency (Figure 2a). The specificity of antibody capture was demonstrated by fitting the x-y coordinates of the captured cells to an exponential equation (Figure 2b). A decay pattern in captured cells was observed along the length of the devices, indicative of specific cell capture (Figure S8). For high-EpCAM-expression cancer cell lines[5b] (e.g., PC3, H1650, SKBR3), the nanocoating achieved capture efficiencies between 75.0 % ± 6.5 % to 95.7 % ± 4.0 % (Figure 2c), which were comparable to other cell capture technologies.[7, 15, 25] Of particular interest, the nanocoating achieved a five-fold increase in capture efficiency for a low-EpCAM-expression cancer cell line, MDA-MD-231,[8] relative to our previous surface chemistry (p < 0.001, n = 4). Moreover, when a cocktail of cell capture antibodies was used to facilitate capture of a heterogeneous population of CTCs[6] (i.e., anti-EpCAM, anti-EGFR, anti-HER2), the capture of MDA-MD-231 cells increased to 94.0 % ± 2.3 % (Supplementary Table 2). The functionalized nanocoating also demonstrated a higher specificity of capture, isolating fewer contaminating white blood cells (WBCs) in comparison to devices functionalized with our standard surface chemistry (Figure 2d).
Figure 2. Validation of nanocoating performance.
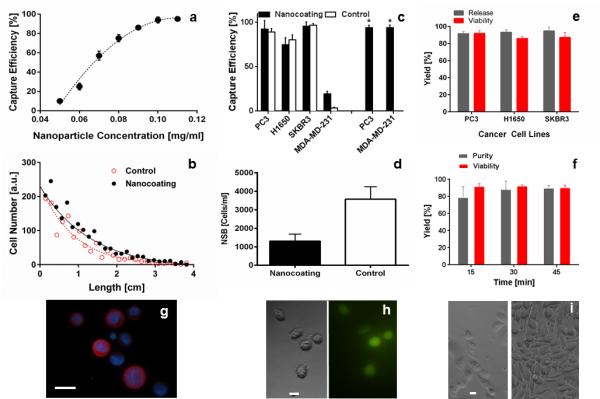
(a) Effect of nanoparticle concentration on cell capture. Prostate cancer cells (PC3s) were spiked into whole blood at 1000 cells/ml to determine the capture efficiency (b) Plotting the location of target cell capture on the microfluidic device reveals a distinct decay pattern, indicating the specificity of capture (Figure S8). (c) Cell capture efficiency for different cancer cell lines. PC3 and MDA-MD-231 cells were captured on the nanocoating functionalized with anti-EpCAM (black bars = nanocoating, white bars = control chemistry), or our antibody cocktail (black bars with *). (d) Quantification of non-specific binding (NSB) of contaminating leukocytes on the nanocoating. (e) Comparison of release efficiency and viability for different cancer cell lines using the temperature degradation mechanism of the nanocoating. (f) Quantification of viability and purity of PC3 cells released from the nanocoating using the selective, single cell release mechanism. 15 cells were released at each time point in a sequential manner (n = 3). (g) Immunofluorescence microscopy image of released PC3 cells on a glass slide (see Supporting Information for a list of antibodies, scale bar 20 μm). (h) Bright field and fluorescent micrographs of released cells, cultured for 6 hours post-release (scale bar 10μm). (i) Micrographs of released cells, 3 days (left) and 7 days (right) post-release (scale bar 10 μm).
We measured the release and subsequent recovery of viable cells using both release mechanisms. For the thermally driven, bulk-population release mode (Supplementary Movie 1 and Movie 2), the nanocoating allowed an average cell recovery and viability of 93.2 % and 88.3 %, respectively (Supplementary Table 2). A novel feature of the nanocoating is the ability to collect cells in selective release mode (Supplementary Movie 3 and Movie 4). Individual target cells were dislodged from the material within a tuned release radius between 145 and 215 μm (Figure 2f). Captured cells were released at three different time points: 15, 30, and 45 min, and the mean viability was 91.5 % with no significant differences between time points (p > 0.05). This constitutes a major improvement compared to enzymatic release strategies, which show a detrimental effect towards cell viability over time (Figure S9). Recovered cells were either stained for CTC markers (Figure 2g) or placed into cell culture, adhering shortly after selective release (Figure 2h) and ultimately, forming confluent colonies after seven days of cell culture (Figure 2e-i).
Our antibody cocktail was used with the nanocoating to facilitate the capture of CTCs from the blood of cancer patients.[4] An average of 3.5 ml of blood was processed from 16 metastatic cancer patients at different stages of treatment (n = 8 for breast cancer, and n = 8 for lung cancer). Blood from healthy individuals were also processed as controls (n = 6). CTCs were identified using immunofluorescence staining, and were isolated in 14 of 16 cancer patients (87.5 %, 7 out of 8 each for breast and lung cancer). Cells were identified as CTCs when stained positive for DNA (DAPI), positive for tumor markers (wide spectrum cytokeratin, MET, SOX2, and EGFR) and negative for leukocyte markers (CD45). Samples were defined as being positive for CTCs when more than 2 CTCs/ 3.5 ml were detected, with this threshold defined by the number of events detected in healthy donor controls (median = 0.5 CTCs/3.5 ml, mean = 1 ± 0.44 CTCs/3.5 ml, n = 6). CTC counts were obtained (Figure 3a) for breast cancer patients (0 to 159 CTCs/3.5 ml, medianBreast = 13 CTCs/3.5 ml, meanBreast = 29 ± 18.7 CTCs/3.5 ml), and lung cancer patients (0 to 18 CTCs/3.5 ml, medianLung = 13 CTCs/3.5 ml, meanLung = 12.5 ± 2.6 CTCs/3.5 ml). Using image processing techniques, the size distribution of CTCs (mean diameter of 11.4 ± 1.2 μm (breast) and 13.5 ± 1.3 μm (lung)), and contaminating WBCs (mean diameter of 10.1 ± 2.3 μm) were quantified (Figure 3b).
Figure 3. Characterization of patient CTCs.
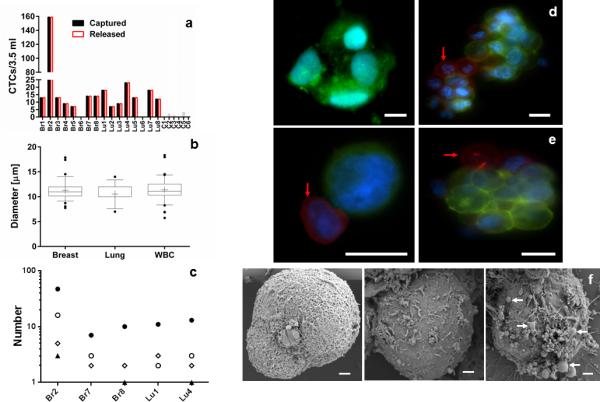
(a) Enumeration of patient CTCs captured (black bars) and released (white bars) from the nanocoating. Breast (Br#) and Lung (Lu#) cancer patients were analyzed along with healthy donors (C#). (b) Size comparison between single CTCs and white blood cells (WBC) from patient samples. Cell diameter was calculated from the area values obtained from stained cells. (c) Frequency distribution for single, doublet, triplet and CTC cluster capture for five patients. The population of isolated CTCs were grouped and quantified in four categories: Single (•), double (ο), triple (▲), and cluster (◊) of CTCs. (d) Micrographs of clusters of CTCs captured on the nanocoating from lung (left) and breast (right) cancer patients. (e) Micrographs of CTCs from a breast cancer patient released from the nanocoating. Red arrows indicate WBCs within single or cluster of CTCs (scale bar 10 μm). (f) Scanning electron microscopy image of breast cancer CTCs of the same patient. The images revealed a heterogeneous surface morphology with the presence of membrane ruffles and in some cases extracellular vesicles (white arrows).
Clusters of CTCs have been isolated from the blood of metastatic cancer patients, and are of clinical interest.[7, 10, 26] When using the nanocoating, clusters of CTCs were found at a frequency of 37.5% (breast) and 25% (lung) in the patient samples analyzed for this study. The frequency of CTC cluster isolation from this small cohort of patients is higher than what we previously observed with our traditional microfluidic chemistry.[7] We defined a cluster of CTCs as an aggregate of cells that contained four or more tumor cells. Using this definition, a number-based distribution was calculated for the frequency of single, double, triple or cluster of CTCs for different patients. We found that single CTCs are more common than clusters of CTCs within breast and lung cancer patients (Figure 3c). Clusters of lung CTCs (Figure 3d, left) showed less defined intercellular boundaries when compared to clusters of breast CTCs (Figure 3d, right). We also observed the presence of WBCs attached to single or clusters of CTCs (Figure 3e).
Comparisons of the membrane morphology of different CTCs captured on the surface of the nanocoating revealed their heterogeneity (Figure 3f), with some CTCs having extracellular vesicles attached to their surface. Although the presence of such extracellular vesicles may suggest signs of apoptotic CTCs; it is also possible that the vesicles serve as delivery vehicles of tumorigenic cargo (e.g., mRNA) to different regions of the body.[27] Additionally, cell-independent micrometer-size vesicles and vesicle clusters were also found on the nanocoating surface (Figure S10), suggesting that the coating may also serve to isolate smaller cancer-derived vesicles.
For genotyping analysis, we selected three breast and two lung cancer patients with metastatic disease that were previously characterized to contain hotspot mutations in the PIK3CA and EGFR oncogenes, respectively. At the time of diagnosis, fine needle aspirates (FNAs) from the primary tumor site were used to biopsy the tissues (Figure 4a), and the mutational profile of each sample was obtained using SNaPshot genotyping at a MGH genomics facility. Specifically, three breast cancer patients resulted positive for the 3140A/G (H1047R) heterozygous mutation in the PIK3CA oncogene. For lung cancer patients, tumors were positive for the exon 19 deletion and the 2573T/G (L858R) point mutation in the EGFR oncogene. For this cohort of patients, CTCs were isolated with the nanocoating, and individual CTCs (identified with a vital fluorescent stain) were recovered using the frequency-controlled microtip. For each single CTC isolated, its release was observed under the microscope (Figure 4b, Supplementary Movie 5 and Figure S11), and the recovered cells were placed into a tube containing lysis buffer. Targeted PCR was performed on genomic DNA extracted from the CTC to amplify bands associated with specific mutations prior to sequencing with a fluorescently-labeled dideoxy-nucleotide chain termination method (Figure 4c). For all single CTCs isolated from the breast cancer patient samples, we identified the 3140A/G (H1047R) mutation in the PIK3CA gene (Figure 4d). For the lung patient samples, we identified the presence of the exon 19 deletion, and the 2573T/G (L858R) mutations in the EGFR gene in each patient, but not in every CTC isolated from the individual draw (2 cells did not successfully sequence out of 5).
Figure 4. Single cell genomics of CTCs from patients.
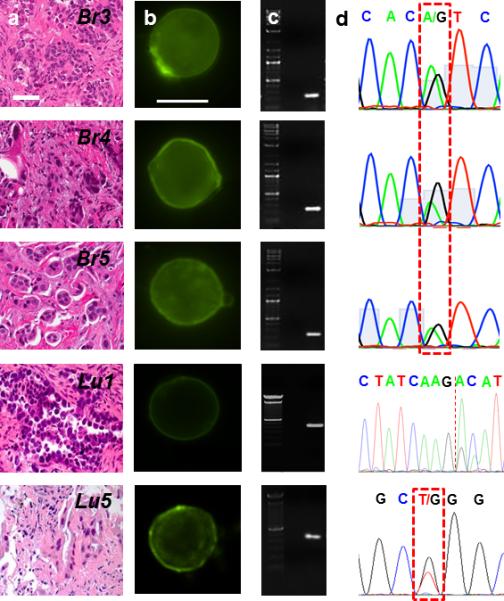
(a) H & E staining of the primary tumor of metastatic breast and lung cancer patients. Tissue biopsies were used to determine the presence of DNA mutations on the oncogene PIK3CA and EGFR. (b) Panel of CTCs from the same metastatic breast and lung cancer patients in (a). Micrographs of the CTCs identified and subsequently released for molecular analysis using our selective release mechanism (scale bar 10 μm). (c) Micrographs of amplified DNA of the single CTCs shown in (b). (d) Sequencing of the amplified DNA from the single CTCs shown in (b). The 3140A/G (H1047R) point mutation in the PIK3CA oncogene as well as the exon 19 deletion and the 2573T/G (L858R) point mutation in the EGFR oncogene were detected at the single cell level.
In summary, we have developed a nanocoating that was synergistically incorporated into microfluidic devices using a LbL process. The unique dual-mode release mechanism of the nanocoating enables efficient release of viable CTCs (in bulk or individually) from peripheral blood and opens the possibility for different biomedical applications. The uniform, conformal nature of the nanocoating facilitates its use on complex three-dimensional structures, and the room-temperature operating conditions and standard refrigeration temperature storage (4°C) make the nanocoating conducive to large-scale production and clinical operation. Our system was used to perform population counts and size-based analysis of CTCs as well as molecular assays for which different levels of purity and cellular integrity are required. The results demonstrate genotyping of single CTCs for the detection of somatic mutations (e.g., PIK3CA, EGFR) from a blood sample and can be applied to other cancer types. This technique could be broadly used to efficiently and safely recover other rare cells, exosomes, proteins, and DNA from biological specimens.
Supplementary Material
Acknowledgements
The authors acknowledge all our patients who participated in this study and the healthy volunteers who kindly donate their blood. Blood specimens for CTC isolation were obtained after informed patient consent according to institutional review board (IRB) protocol (05-300), at the Massachusetts General Hospital. This work was supported by grants from Stand Up to Cancer (D.A.H., M.T., S.M.), Howard Hughes Medical Institute (D.A.H.), NIH CA129933 (D.A.H.), National Institute for Biomedical Imaging and bioengineering (NIBIB) EB008047 (M.T., D.A.H.), NIH P41 EB002503-11 (M.T.). Thanks to Laura Libby and Octavio Hurtado for expert technical support. Thanks to Bavand Keshavarz for helping with microtip design, Matthew Phillips for the CTC enumeration scans, Thomas Carey for the Atomic Force Microscopy analysis, and Charles P. Lai for the nanoparticle size characterization.
Footnotes
Supporting Information
Experimental section and additional figures are available from the Wiley Online Library or from the author.
Contributor Information
Eduardo Reátegui, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129; Shriners Hospital for Children, Harvard Medical School 51 Blossom Street, Boston, MA 02114; Department of Surgery, Massachusetts General Hospital, Harvard Medical School 55 55 Fruit Street, Boston, MA 02114.
Nicola Aceto, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston 55 Fruit Street, Boston, MA 02114.
Eugene J. Lim, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129 Department of Electrical Engineering, Massachusetts Institute of Technology; 77 Massachusetts Ave, Cambridge, MA 02139.
James P. Sullivan, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston 55 Fruit Street, Boston, MA 02114
Anne E. Jensen, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129
Mahnaz Zeinali, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129.
Joseph M. Martel, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129 Shriners Hospital for Children, Harvard Medical School 51 Blossom Street, Boston, MA 02114; Department of Surgery, Massachusetts General Hospital, Harvard Medical School 55 55 Fruit Street, Boston, MA 02114.
Alexander. J. Aranyosi, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129
Wei Li, Department of Chemical Engineering, Massachusetts Institute of Technology; 77 Massachusetts Ave, Cambridge, MA 02139.
Steven Castleberry, Department of Chemical Engineering, Massachusetts Institute of Technology; 77 Massachusetts Ave, Cambridge, MA 02139.
Aditya Bardia, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston 55 Fruit Street, Boston, MA 02114.
Lecia V. Sequist, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston 55 Fruit Street, Boston, MA 02114
Daniel A. Haber, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston 55 Fruit Street, Boston, MA 02114 Howard Hughes Medical Institute 4000 Jones Bridge Road, Chevy Chase, MD 20815.
Shyamala Maheswaran, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston 55 Fruit Street, Boston, MA 02114.
Paula T. Hammond, Department of Chemical Engineering, Massachusetts Institute of Technology; 77 Massachusetts Ave, Cambridge, MA 02139
Mehmet Toner, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129; Shriners Hospital for Children, Harvard Medical School 51 Blossom Street, Boston, MA 02114; Department of Surgery, Massachusetts General Hospital, Harvard Medical School 55 55 Fruit Street, Boston, MA 02114.
Shannon L. Stott, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School Building 114 16th Street, Charlestown, MA 02129; Shriners Hospital for Children, Harvard Medical School 51 Blossom Street, Boston, MA 02114; Department of Medicine, Massachusetts General Hospital, Harvard Medical School 55 Fruit Street, Boston, MA 02114.
References
- 1.Yu M, Stott S, Toner M, Maheswaran S, Haber DA, Cell Biol J. 2011;192:373. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. New Engl. J. Med. 2004;351:781. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 3.Cristofanilli M, Mendelsohn J. Proc. Natl. Acad. Sci. 2006;103:17073. doi: 10.1073/pnas.0608651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Science. 2013;339:580. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Jeffrey SS. Plos One. 2012;7:33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P.-i., Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Sci. Transl. Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. New Engl. J. Med. 2008;359:366. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Mitra D, Sullivan Ryan J., Wittner Ben S., Kimura Anya M., Pan S, Hoang Mai P., Brannigan Brian W., Lawrence Donald P., Flaherty Keith T., Sequist Lecia V., McMahon M, Bosenberg Marcus W., Stott Shannon L., Ting David T., Ramaswamy S, Toner M, Fisher David E., Maheswaran S, Haber Daniel A. Cell Rep. 2014;7:645. doi: 10.1016/j.celrep.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA. Nature. 2012;487:510. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aceto N, Bardia A, Miyamoto David T., Donaldson Maria C., Wittner Ben S., Spencer Joel A., Yu M, Pely A, Engstrom A, Zhu H, Brannigan Brian W., Kapur R, Stott Shannon L., Shioda T, Ramaswamy S, Ting David T., Lin Charles P., Toner M, Haber Daniel A., Maheswaran S. Cell. 2014;158:1110. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, OuYang W-H, Xu X, Zhang Z, Song M, Lee T, Garcia MA, Xiong B, Hou S, Tseng H-R, Fang X. Adv. Mater. 2013;25:2368. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo S-C, Tseng H-R, Yu H.-h. Adv. Mater. 2013;25:1547. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie M, Lu N-N, Cheng S-B, Wang X-Y, Wang M, Guo S, Wen C-Y, Hu J, Pang D-W, Huang W-H. Anal. Chem. 2014;86:4618. doi: 10.1021/ac500820p. [DOI] [PubMed] [Google Scholar]
- 15.Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Anal. Chem. 2012;84:3682. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proc. Natl. Acad. Sci. 2012;109:19626. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee Y-K, Chung LWK, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng H-R. Angew. Chem. Int. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S. Adv. Mater. 2011;23:4376. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 18.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang W-H, He R, Zhao X-Z, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng H-R. Angew. Chem. Int. 2013;52:3379. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L, Lu Y-T, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang W-H, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LWK, Rettig M, Tseng H-R, Posadas EM. Adv. Mater. 2013;25:2897. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran R, Li L, Wang M, Wang S, Zheng Z, Lin PP. Anal. Bioanal. Chem. 2013;405:7377. doi: 10.1007/s00216-013-7156-y. [DOI] [PubMed] [Google Scholar]
- 21.Westwood M, Gunning AP, Parker R. Macromolecules. 2010;43:10582. [Google Scholar]
- 22.Pek YS, Wan ACA, Sherakan A, Zhuo L, Ying JY. Nat. Nanotech. 2008;3:671. doi: 10.1038/nnano.2008.270. [DOI] [PubMed] [Google Scholar]
- 23.Yoon HJ, Kozminsky M, Nagrath S. ACS Nano. 2014;8:1995. doi: 10.1021/nn5004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobrovolskaia M. a., McNeil SE. Nat. Nanotech. 2007;2:469. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 25.a Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Nat. Nanotech. 2013;8:735. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, Ko AH, Korn WM, Kolatkar A, Bethel K, Kuhn P. Phys. Biol. 2012;9:16001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Glaves D, Huben RP, Weiss L. Br. J. Cancer. 1988;57:32. doi: 10.1038/bjc.1988.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.a Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, Holowka DA, Cerione RA. Proc. Natl. Acad. Sci. 2011 doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Vizio DD, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, Mulholland D, Rotinen M, Hager MH, Insabato L, Moses MA, Demichelis F, Lisanti MP, Wu H, Klagsbrun M, Bhowmick NA, Rubin MA, D'Souza-Schorey C, Freeman MR. Am. J. Pathol. 2012;181:1573. doi: 10.1016/j.ajpath.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


