Abstract
The reniform nematode, Rotylenchulus reniformis, is a sedentary semi-endoparasitic species with a host range that encompasses more than 77 plant families. Nematode effector proteins containing plant-ligand motifs similar to CLAVATA3/ESR (CLE) peptides have been identified in the Heterodera, Globodera, and Meloidogyne genera of sedentary endoparasites. Here, we describe the isolation, sequence analysis, and spatiotemporal expression of three R. reniformis genes encoding putative CLE motifs named Rr-cle-1, Rr-cle-2, and Rr-cle-3. The Rr-cle cDNAs showed >98% identity with each other and the predicted peptides were identical with the exception of a short stretch of residues at the carboxy(C)-terminus of the variable domain (VD). Each RrCLE peptide possessed an amino-terminal signal peptide for secretion and a single C-terminal CLE motif that was most similar to Heterodera CLE motifs. Aligning the Rr-cle cDNAs with their corresponding genomic sequences showed three exons with an intron separating the signal peptide from the VD and a second intron separating the VD from the CLE motif. An alignment of the RrCLE1 peptide with Heterodera glycines and Heterodera schachtii CLE proteins revealed a high level of homology within the VD region associated with regulating in planta trafficking of the processed CLE peptide. Quantitative RT-PCR (qRT-PCR) showed similar expression profiles for each Rr-cle transcript across the R. reniformis life-cycle with the greatest transcript abundance being in sedentary parasitic female nematodes. In situ hybridization showed specific Rr-cle expression within the dorsal esophageal gland cell of sedentary parasitic females.
Keywords: CLE, effector, host-parasite relationship, reniform nematode, Rotylenchulus
The reniform nematode (Rotylenchulus reniformis Linford and Oliviera) is a semi-endoparasitic species with a broad host range that includes more than 77 plant families (Robinson, 2007). In the United States, R. reniformis is a serious pest of multiple crops, including upland cotton, soybean, and pineapple. In fact, cotton yield losses due to R. reniformis infection can be >$100 million annually, making R. reniformis nearly as damaging to cotton production as the southern root-knot nematode (Blasingame and Patel, 2012). The R. reniformis life-cycle begins when eggs in the soil hatch, giving rise to second-stage juveniles (J2) with the initial molting event of J1 to J2 having occurred within the egg. Shortly after hatching, the J2 enter into a nonmotile state and continue their development through the J3 and J4 stages in the absence of feeding until vermiform female and male nematodes exsheath from the residual juvenile cuticles (Ganji et al., 2013). Host root infection is accomplished only by the female and can occur at any location along the root. Male nematodes do not infect but serve only to fertilize the mature sedentary females. The R. reniformis semi-endoparasitic nature is characterized by the embedding of the “head” region of the female nematode in the root whereas the posterior of the nematode remains outside and exposed to the soil (Robinson, 2007). As the sedentary female matures its body swells and takes on a kidney, i.e., reniform, shape. As a sedentary parasite, R. reniformis is dependent on the formation and maintenance of a permanent feeding site within the host root from which it can extract the nutrients required for reproduction. Feeding sites established by R. reniformis are similar to syncytia formed by Heterodera and Globodera cyst nematode species, with whom they share a number of characteristics such as increased metabolic activity, hypertrophied nuclei, and dense granular cytoplasm (Vovlas and Lamberti, 1990; Agudelo et al., 2005).
The current model of parasitism by sedentary plant-parasitic nematodes (PPN) posits that effector proteins, and possibly other signaling molecules, originating from the nematode modulate specific plant pathways so as to change an already differentiated root cell into a metabolically active feeding site (Davis et al., 2008; Mitchum et al., 2013). These effector proteins are encoded by “parasitism genes” that are expressed exclusively within the subventral and dorsal esophageal gland cells of the infective nematode life-stage and are injected into the host root cell through the nematode’s hollow stylet (Davis et al., 2008; Mitchum et al., 2013). Due to their worldwide importance in crop production, PPN belonging to the cyst (Heterodera and Globodera) and root-knot (Meloidogyne) genera have been the focus of effector discovery and characterization with a research history that spans almost three decades. The first effectors identified belonged to various groups of plant cell wall degrading enzymes, e.g., cellulase (reviewed by Haegeman et al., 2012). Effector discovery received a tremendous boost with the sequencing of cDNA libraries constructed from esophageal gland cell contents (Gao et al., 2001, 2003; Huang et al., 2004). The process of effector identification has continued to accelerate with the advent of next-generation sequencing platforms coupled with new means of isolating esophageal gland cell contents (Maier et al., 2013). With regards to R. reniformis, effector discovery and characterization are in the relatively early stages. An analysis of expressed sequence tags (EST) from sedentary parasitic females identified some putative effectors orthologous to those in cyst and root-knot nematodes (Wubben et al., 2010a). This EST dataset facilitated the cloning and characterization of a R. reniformis β-1,4-endoglucanse (Wubben et al., 2010b).
A major class of nematode effectors function as ligand mimics of plant CLE (CLAVATA3/ESR) peptides (Mitchum et al., 2012). CLEs act as secreted peptide hormones and are ubiquitous in dicot and monocot plant species where they help regulate the proliferation of the shoot and root apical meristems in addition to other physiological processes (Kiyohara and Sawa, 2012). All CLEs, be they of plant or nematode origin, possess an N-terminal signal peptide and a C-terminal 14-amino acid CLE motif that are separated by a variable domain (VD). The first nematode CLE gene identified was Hg-syv46, which was cloned as part of a yeast secretion signal peptide selection screen of cDNAs isolated specifically from H. glycines esophageal gland cells (Wang et al., 2001). In situ hybridization showed that Hg-syv46 was expressed exclusively within the dorsal esophageal glands of J2 through young adult female life-stages (Wang et al., 2001). The identity of Hg-syv46 as a potential CLE gene was determined serendipitously as part of a motif-based sequence database search (Olsen and Skriver, 2003). It was later shown that overexpression of Hg-syv46 in wild-type Arabidopsis plants phenocopied what had previously been observed in plants overexpressing plant CLEs (Wang et al., 2005). This study also demonstrated that overexpression of Hg-syv46 could rescue clv3-1 mutants (Wang et al., 2005). It has since been determined that H. glycines expresses two CLE genes (HgCLE1 and HgCLE2) (Wang et al., 2010a). CLEs also have been isolated from sugar beet cyst nematode (H. schachtii) (Wang et al., 2011) and a large amount of work has been performed characterizing CLE peptides from the potato cyst nematode (Globodera rostochiensis) (Lu et al., 2009; Guo et al., 2011). Most recently, so-called “Meloidogyne avirulence protein” (MAP) genes have been found to contain CLE-like motif sequences (Rutter et al., 2014).
RNA-interference (RNAi) experiments have demonstrated that nematode CLE gene expression is required for the full parasitic ability of Heterodera juveniles. Gene silencing by soaking H. glycines J2 in dsRNA solution that targeted HgCLE1/Hgsyv46, resulted in a decrease in the number of J2 that were able to establish feeding sites on soybean roots and shifted the sexual fate of established juveniles to male vs. female (Bakhetia et al., 2007). Furthermore, transgenic Arabidopsis plants expressing a hairpin RNAi construct specific for the H. schachtii orthologue of Hgsyv46 resulted in a 32% to 36% reduction in females that were able to develop (Patel et al., 2008). These experiments indicate that PPN CLEs, and the plant signaling pathways they target, may provide insight into the development of novel, transgenic-based resistance in PPN-susceptible hosts.
In this report, we describe the isolation of three R. reniformis cDNAs (Rr-cle-1, Rr-cle-2, and Rr-cle-3) that are predicted to encode peptides having a signal peptide for secretion and a single CLE-motif at their C-terminus. We also demonstrate that Rr-cle expression is restricted largely to the sedentary female life-stage and exclusively within the dorsal esophageal gland cell.
Materials and Methods
Polymerase chain reaction (PCR) and cloning:
To clone full-length Rr-cle sequences, PCR reactions were performed on cDNA and genomic DNA templates with the following forward and reverse primers, respectively: 5′-CCCAATCTTGAGGTCATAATTCAAA-3′ and 5′-CAATCATTCCCATTCCTAATCCAC-3′. PCR products were separated on agarose gels, purified using the MinElute™ Gel Purification Kit (Qiagen, Valencia, CA), and ligated into the pCR4.0 TOPO T/A cloning vector (Invitrogen, Carlsbad, CA) per the manufacturer’s instructions. Clone sequencing was performed by the USDA-ARS Mid-South Area Genomics Facility (Stoneville, MS). cDNA and genomic DNA sequence analyses and alignments were performed using Sequencher v4.10.01 (Gene Codes Corp., Ann Arbor, MI).
Nucleic acid isolation:
R. reniformis life-stages were isolated according to Ganji et al. (2013). Total RNA and genomic DNA extractions were performed as previously described (Ganji et al., 2014). For qRT-PCR, RNA was extracted from three separate samples, i.e., replicates, of each life-stage. For sedentary females, each sample was comprised of 200 to 500 individuals. Approximately 50,000 individuals comprised each egg and vermiform life-stage sample destined for RNA extraction.
Reverse-transcription and quantitative RT-PCR:
First-strand cDNA was synthesized from 100 ng of DNase-treated total RNA using the iScript™ Select cDNA Synthesis Kit with oligo dT primer per the manufacturer’s instructions (Bio-Rad, Hercules, CA). Reverse-transcription reactions used in qRT-PCR experiments also included 0.1 µM of a R. reniformis-specific 18S ribosomal RNA primer (5′-AACCAGGGCGCTCATTGAGTCTTA-3′). Reverse-transcription reactions were diluted 1:10 for use as template in qRT-PCR. Quantitative RT-PCR reactions were performed in triplicate in 96-well plates on a CFX96™ Real-Time System (Bio-Rad). Rr-cle-specific qRT-PCR forward primers were as follows: Rr-cle-1 (5′-GCAATCCAACAACAACAACAGCTCAA-3′), Rr-cle-2 (5′-GCAATCCAACAACAACAGCTCAATTC-3′), and Rr-cle-3 (5′-GCGACCCAACAACAACAGCTCAA-3′). Each forward primer was coupled with a universal Rr-cle qRT-PCR reverse primer (5′-AAAGGCTCAATGATGTTTAGGATCCG-3′). Slope and R2 values for each primer pair were determined using a 1:5 serial dilution series of cDNA template: Rr-cle-1 (−3.683, 0.997), Rr-cle-2 (−3.729, 0.998), and Rr-cle-3 (−3.711, 0.999). Rr-cle expression values were normalized by R. reniformis 18S ribosomal RNA levels as previously described (Ganji et al., 2014).
In situ hybridization of Rr-cle:
A 237-bp dioxigenin (DIG)-labeled sense and antisense probe was amplified from purified Rr-cle cDNA using asymmetric PCR and a dot blot of the DIG probe was performed to insure the incorporation of DIG molecules before the in situ hybridization assay. Primers used for probe generation were (sense) 5′-GGAAGTCTCGGAGGGATTGG-3′ and (antisense) 5′-GGATTCTCGTTTGGATTCATTGTA-3′. Sample fixation, permeablization, and probe hybridization were as previously described (Ganji et al., 2014).
Results
Identification of Rr-cle genes:
An R. reniformis EST dataset developed from sedentary parasitic females (Wubben et al., 2010a) was queried by tBLASTn using the 13-mer H. glycines CLE peptide motif “KRLSPSGPDPHHH” under low stringency conditions. This search identified three R. reniformis ESTs (GenBank accession no. GT736832, GT737621, and GT737367) as encoding putative CLE motifs. A cross comparison of the EST sequences revealed they were redundant; therefore, further analysis was restricted to the longest sequence GT736832 (498 nt). Translation of GT736832 in the reading frame producing the putative CLE motif revealed that the 5'-end of the cDNA was missing as no start codon (ATG) was present; however, a stop codon (TGA) was present immediately following the putative CLE motif. Attempts to acquire the cDNA ends by rapid amplification of cDNA ends using GT736832 as a starting point were unsuccessful. In an independent series of experiments, a transcriptome survey of R. reniformis sedentary females had been conducted using the Illumina MiSeq next-generation sequencing platform. Transcripts assembled from this survey were queried by BLASTn using the GT736832 sequence. Transcripts that overlapped the 5′-end of GT736832 were identified and assembled into a consensus full-length Rr-cle cDNA sequence. Forward and reverse primers designed from this consensus sequence were used to amplify putative full-length Rr-cle cDNA and genomic sequences from R. reniformis sedentary females.
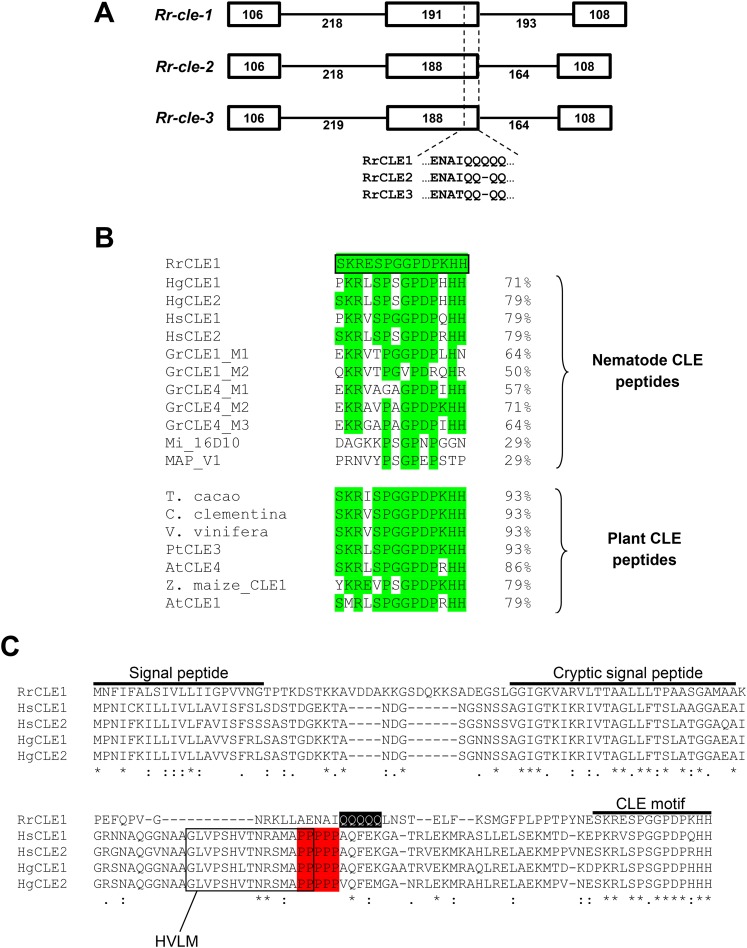
PCR using either cDNA (from sedentary female nematodes) or genomic DNA template produced single specific bands of approximately 450 and 800 nt, respectively. The PCR products were cloned and bi-directional sequencing of multiple clones identified three distinct full-length cDNA sequences containing the CLE motif. These cDNAs were subsequently named Rr-cle-1 (470 nt), Rr-cle-2 (467 nt), and Rr-cle-3 (467 nt) (GenBank accession no. KR011024, KR011025, and KR011026, respectively). The Rr-cle cDNAs shared >98% identity with one another with each cDNA possessing indentical 25 nt 5′-UTRs (untranslated region) and 40 nt 3′-UTRs. Sequencing of genomic clones identified three sequences that corresponded one-to-one with the individual cDNA sequences at 100% identity. An alignment of the cDNAs with their corresponding genomic sequences identified three exons and two introns for each Rr-cle gene (Fig. 1A). Differences in nucleotide sequence composition and length occurred primarily in exon 2 and intron 2 of the Rr-cle genes (Fig. 1A). Relative to the predicted peptides, exon/intron splice sites were situated shortly downstream of the protein signal peptide and a short distance upstream of the CLE motif (Fig. 1A). These two exon/intron junctions delimit a single VD within the RrCLE peptides.
Fig. 1.
DNA and amino acid sequence analysis of the Rotylenchulus reniformis CLE genes (Rr-cle-1, Rr-cle-2, and Rr-cle-3). A. Genetic structure of the Rr-cle genes. Exons (boxes) and introns (lines) are shown with corresponding nucleotide lengths. Amino acid sequence divergence between the three RrCLE predicted proteins is shown at the 3'-end of the second exon. B. Alignment of the RrCLE1 CLE motif (outlined in black box) with CLE motifs from other plant-parasitic nematodes and with selected plant CLE motifs. Regions of sequence identity are highlighted in green. Percent identity shared is shown on the right. C. Alignment of RrCLE1 peptide with CLE1/CLE2 peptides from Heterodera schachtii (Hs) and Heterodera glycines (Hg). Regions of amino acid identity are marked by an asterisk whereas regions of functional residue identity or similarity are marked by “:” and “.”, respectively. Sequence regions corresponding to the signal peptide, cryptic signal peptide, and CLE motif are noted above the alignment. The polyproline tract in the Hs and Hg CLEs is highlighted in red whereas the polyglutamine tract in RrCLE1 is highlighted in black. The Heterodera variable domain-like motif (HVLM), which is absent in RrCLE1, is shown within the black boxed region.
Translation of the Rr-cle cDNAs yielded peptides of 134 amino acids (aa) (Rr-cle-1) and 133 aa (Rr-cle-2 and Rr-cle-3) with predicted isoelectric points of 9.44. The RrCLE peptides shared 100% identity with each other with the exception of a short stretch of residues at the C-terminus of the VD (Fig. 1A). BLASTp of the full-length RrCLE1 peptide did not result in any hits with E-values < 0.005. Top hits resulting from BLASTp of RrCLE1 included putative CLE peptides from wheat, rice, and millet (E ≥ 0.005). Among nematodes, the greatest homology was shared with CLE peptides from H. schachtii and H. glycines. When analyzed using DELTA-BLAST, weak similarity (E = 0.02) was identified between the RrCLE1 VD and the bacterial Exosortase D, VPLPA-CTERM-specific domain (Cdd:TIGR04152). DELTA-BLAST analysis of HgCLE1&2, HsCLE1&2, and GrCLE1 peptides did not identify this domain. Analysis of the RrCLE peptides using Signal P v4.1 (Petersen et al., 2011) identified a signal peptide at the N-terminus from residues 1 to 20 (Fig. 1C).
Each RrCLE peptide contained a single, identical CLE motif that possessed the canonical well-conserved residues (R−5P−2G0P+1P+3H+5) as determined by Oelkers et al. (2008). The RrCLE1 peptide CLE motif was aligned with CLE motifs from other plant-parasitic nematode peptides and with plant CLE peptides (Fig. 1B). Among nematodes, homology was greatest with CLE motifs from Heterodera species and with motif 2 of GrCLE4 from Globodera. Only 29% identity was shared with CLE-like motif sequences from Meloidoyne spp. (Fig. 1B). In contrast to the nematode proteins, the R. reniformis CLE motif showed 93% identity with motifs from Theobroma cacao and Citrus clementina, whereas 79% identity was shared with the CLE1 motifs from Arabidopsis thaliana and Zea maize (Fig. 1B).
The full-length RrCLE1 peptide was aligned with the H. glycines and H. schachtii CLE peptides using Clustal Omega (Sievers et al., 2011). This alignment revealed a number of interesting similarities but also marked differences (Fig. 1C). Although each CLE peptide possessed an N-terminal signal peptide, only three amino acid residues were conserved across the proteins in this region. In contrast, amino acid sequence identity was highly conserved within the cryptic signal peptide of the RrCLE1 VD and VDI of HsCLE1/2 and HgCLE1/2. The presence of a cryptic signal peptide within the RrCLE1 VD was also predicted by Signal P v4.1 by systematically testing truncated RrCLE1 protein sequences similar to that described by Wang et al. (2010b). Heterodera CLE peptides possess a proline(P)-rich sequence upstream of the CLE motif at the start of VDII (Fig. 1C). This feature is also shared by Globodera CLE peptides (Lu et al., 2009). Strikingly, instead of a proline-rich sequence, the RrCLE peptides showed a glutamine(Q)-rich sequence of the same number of residues (Fig. 1C). While proline is classified as a nonpolar amino acid, glutamine is polar/uncharged. We also observed that RrCLE1 lacked the HVLM sequence (Heterodera Variable domain-Like Motif) (Rutter et al., 2014) that is shared by H. schachtii and H. glycines (Fig. 1C).
Quantitative RT-PCR of Rr-cle-1, Rr-cle-2, and Rr-cle-3:
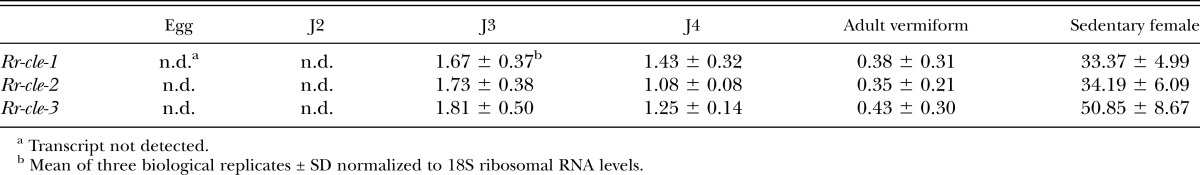
The relative transcript quantity of each Rr-cle cDNA was measured across all R. reniformis life-stages by qRT-PCR. We determined that Rr-cle transcript could not be reliably detected in total RNA collected from the egg or J2 life-stages. In contrast, the transcript of each Rr-cle gene was detected in replicate RNA samples collected from J3, J4, adult vermiform, and sedentary female nematodes (Table 1). Each Rr-cle gene showed transcript levels that were similar between J3 and J4 with a slight decrease observed in adult vermiform nematodes. Maximum Rr-cle expression was found in sedentary females. For example, compared to the J3 life-stage, transcript levels increased 19.9-, 19.7-, and 28.1-fold in sedentary females for Rr-cle-1, Rr-cle-2, and Rr-cle-3, respectively.
Table 1.
Relative measurement of Rr-cle transcript levels across Rotylenchulus reniformis life-stages by quantitative RT-PCR.
In situ hybridization:
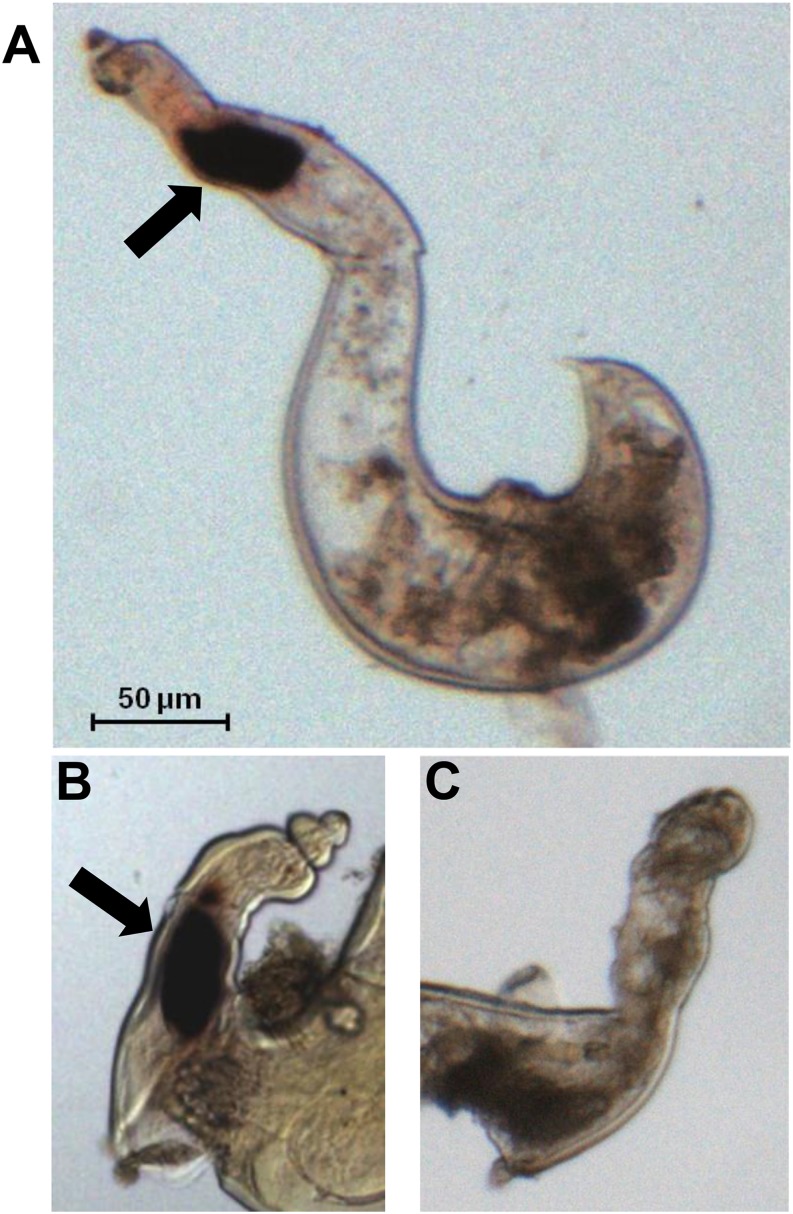
To determine the spatial expression pattern of the Rr-cle cDNAs, sense and antisense DIG-labeled ssDNA probes were synthesized and hybridized with fixed nematodes from vermiform and sedentary life-stages. Hybridization of the antisense probe with vermiform nematodes failed to detect Rr-cle transcript (data not shown), which reflects the relatively low levels of Rr-cle expression in this life-stage as measured by qRT-PCR (Table 1). In contrast, strong DIG staining was observed in the dorsal esophageal gland region of R. reniformis sedentary females (Fig. 2A,B). No DIG staining was observed in sedentary females hybridized with the sense DIG probes (Fig. 2C).
Fig. 2.
Spatial localization of Rr-cle transcript in sedentary parasitic female Rotylenchulus reniformis nematodes by in situ hybridization. A 237 bp sense and antisense DIG-labeled probe was hybridized with fixed sedentary females. A,B. DIG staining specific to the dorsal esophageal gland cell is shown (black arrow). C. Nematode samples hybridized with DIG-labeled sense probes did not show staining.
Discussion
The similarities in feeding site structure between cyst and reniform nematodes suggest these parasites implement a similar suite of effectors to bring about feeding site formation. Thus far, molecular studies on R. reniformis have supported this viewpoint. Multiple cyst nematode effector orthologs have been identified within R. reniformis cDNA sequencing datasets (Wubben et al., 2010a). The results presented in this report on the identification of the Rr-cle genes provide yet another example of commonality between the cyst and reniform nematodes. Our finding of peak Rr-cle gene expression during the sedentary female life-stage and specific Rr-cle expression within the dorsal esophageal glands of such females strongly indicate a role for the RrCLE peptides in facilitating syncytium formation as has been concluded for cyst nematode species (Mitchum et al., 2012); however, on closer inspection, a number of interesting disparities were also present between R. reniformis CLE peptides and their cyst nematode counterparts.
An alignment of the R. reniformis CLE motif with motifs from other nematodes clearly showed a high degree of identity was shared with CLEs from H. glycines and H. schachtii with lesser identity being shared with the various Globodera CLE motif sequences. This hierarchy of shared identity also extended over the entire length of the RrCLE protein; however, with regard to gene structure, i.e., exon/intron number and position, the Rr-cle genes were more akin to G. rostochiensis orthologs. In contrast to Heterodera CLE genes that possess an intron within the coding region of the VD, giving rise to VDI and VDII (Wang et al., 2010a, 2011), R. reniformis and Globodera CLE genes do not have this VD-splitting intron; consequently, these nematodes have a single VD (Lu et al., 2009). As R. reniformis, and other semi-endoparasitic species for that matter, are sometimes considered evolutionary intermediates between the migratory and fully endoparasitic life-styles, it is plausible that the Rr-cle genes would share characteristics of both Heterodera and Globodera. The high level of conservation shared between RrCLEs and Hg/Hs CLEs within the region of the cryptic signal peptide suggests that a similar in planta trafficking mechanism would shuttle processed RrCLE peptides to the plant apoplast, as has been shown for Heterodera and Globodera CLEs (Wang et al., 2010b, 2011; Guo et al., 2011).
Heterodera and Globodera CLE peptides all contain a proline-rich region within the VD (Lu et al., 2009; Wang et al. 2010a, 2011). The importance of this region in the processing or functionality of CLE peptides is not completely clear; however, the similarity of these regions to some proline recognition domains (e.g., Src homology 3 domain) raises the possibility of them being involved in intracellular signaling at some level (Lu et al., 2009). Structural modeling of HgCLE2 VDII identified a 24-amino-acid helix immediately following the poly-proline tract (Wang et al., 2010a). The same study strongly implicated the VD in mediating host-specificity of the nematode CLE protein (Wang et al., 2010a). We found that instead of nonpolar proline residues, R. reniformis CLE proteins possess a stretch of polar glutamine residues at the same relative position within the protein. Compared to Heterodera and Globodera cyst nematodes, R. reniformis has a much broader host range and is able to parasitize a number of plant species that are in addition to those infected by cyst nematodes. Therefore, it is tempting to speculate that perhaps this glutamine-rich tract provides more flexibility to R. reniformis in establishing feeding sites across a broader range of hosts.
The R. reniformis CLE motifs showed 93% identity with motifs from predicted CLE peptides of T. cacao and C. clementina. Rotylenchulus reniformis is a major pathogen of cotton (Gossypium hirsutum), which is in the same family as T. cacao (i.e., Malvaceae); however, T. cacao is not considered a suitable host for R. reniformis (Khan, 2005). This observation leads us to question how large of a role the CLE motif sequence plays in mediating host specificity or host range. Recent studies suggest that residues N-terminal to the CLE motif, within VDII of the Heterodera CLEs, strongly influence the host specificity of a particular CLE peptide (Wang et al., 2010a). The citrus nematode, Tylenchulus semipentrans, is in the Hoplolaimidae along with R. reniformis and also shows a sedentary parasitic life-style. An assessment of the CLE gene complement of T. semipenetrans in comparison to R. reniformis may shed more light on the role CLE peptides play in governing host range.
In a recent study of MAP encoding genes from different Meloidogyne species, 14 variations of CLE-like motifs were identified (Rutter et al., 2014). In more than one instance, these CLE-like motifs were tandemly repeated within the gene and interspersed with another motif the authors named HVLM (Heterodera variable domain-like motif). The HVLM consists of 15 amino acids within the VD of all Heterodera CLE peptides that was identified as a conserved motif in the majority of MAP studied (Rutter et al., 2014). Interestingly, this motif was completely absent from the R. reniformis CLE peptides. Although the functional significance of this observation remains obscure, it provides another example of a departure from the homology shared between R. reniformis and cyst nematode CLE proteins.
It has been demonstrated by a variety of means that nematode CLEs can mimic plant CLEs in their ability to modulate plant signaling pathways. For example, the A. thaliana receptor kinases CLAVATA2 (CLV2) and CORYNE (CRN) function in mediating the effects of CLE peptides secreted by H. glycines and H. schachtii (Replogle et al., 2011). This was demonstrated by observing that loss of function mutations in CLV2 and CRN abolish the effects of synthetic nematode CLE peptide treatment on root growth and the effects of nematode CLE overexpression on plant development (Replogle et al., 2011). The authors also observed decreased H. schachtii susceptibility and decreased feeding site size in the clv2-1 and crn-1 mutants compared to wild type (Replogle et al., 2011). The repertoire of A. thaliana receptors that recognize cyst nematode CLEs was later expanded to include CLV1 and RPK2 (Replogle et al., 2013). Rotylenchulus reniformis can parasitize Arabidopsis (Urwin et al., 2000); therefore, it would be possible to observe the effect of mutations in these receptors on the ability of R. reniformis to infect the plant.
Literature Cited
- Agudelo P, Robbins RT, Stewart JM, Bell A, Robinson AF. Histological observations of Rotylenchulus reniformis on Gossypium longicalyx and interspecific cotton hybrids. Journal of Nematology. 2005;37:444–447. [PMC free article] [PubMed] [Google Scholar]
- Bakhetia M, Urwin PE, Atkinson HJ. qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Molecular Plant-Microbe Interactions. 2007;20:306–312. doi: 10.1094/MPMI-20-3-0306. [DOI] [PubMed] [Google Scholar]
- Blasingame D, Patel MV. 2012. Cotton disease loss estimate committee report. Proceedings of the 2012 Beltwide Cotton Conferences, 3–6 January 2012, Orlando, FL:341–344.
- Davis EL, Hussey RS, Mitchum MG, Baum TJ. Parasitism proteins in nematode-plant interactions. Current Opinion in Plant Biology. 2008;11:360–366. doi: 10.1016/j.pbi.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Ganji S, Wubben MJ, Jenkins JN. Two simple methods for the collection of individual life-stages of reniform nematode, Rotylenchulus reniformis. Journal of Nematology. 2013;45:87–91. [PMC free article] [PubMed] [Google Scholar]
- Ganji S, Jenkins JN, Wubben MJ. Molecular characterization of the reniform nematode C-type lectin gene family reveals a likely role in mitigating environmental stresses during plant parasitism. Gene. 2014;537:269–278. doi: 10.1016/j.gene.2013.12.048. [DOI] [PubMed] [Google Scholar]
- Gao B, Allen R, Maier T, Davis EL, Baum TJ, Hussey RS. Identification of putative parasitism genes expressed in the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Molecular Plant-Microbe Interactions. 2001;14:1247–1254. doi: 10.1094/MPMI.2001.14.10.1247. [DOI] [PubMed] [Google Scholar]
- Gao B, Allen R, Maier T, Davis EL, Baum TJ, Hussey RS. The parasitome of the phytonematode Heterodera glycines. Molecular Plant-Microbe Interactions. 2003;16:720–726. doi: 10.1094/MPMI.2003.16.8.720. [DOI] [PubMed] [Google Scholar]
- Guo Y, Ni J, Denver R, Wang X, Clark SE. Mechanisms of molecular mimicry of plant CLE peptide ligands by the parasitic nematode Globodera rostochiensis. Plant Physiology. 2011;157:476–484. doi: 10.1104/pp.111.180554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Mantelin S, Jones JT, Gheysen G. Functional roles of effectors of plant-parasitic nematodes. Gene. 2012;492:19–31. doi: 10.1016/j.gene.2011.10.040. [DOI] [PubMed] [Google Scholar]
- Huang G, Dong R, Maier T, Allen R, Davis EL, Baum TJ, Hussey RS. Use of solid-phase subtractive hybridization for the identification of parasitism gene candidates from the root-knot nematode Meloidogyne incognita. Molecular Plant Pathology. 2004;5:217–222. doi: 10.1111/j.1364-3703.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- Khan MR. Hosts and non-hosts of reniform nematode, Rotylenchulus reniformis Linford & Oliveira, 1940 – A critical review. Environment and Ecology. 2005;23:124–140. [Google Scholar]
- Kiyohara S, Sawa S. CLE signaling systems during plant development and nematode infection. Plant and Cell Physiology. 2012;53:1989–1999. doi: 10.1093/pcp/pcs136. [DOI] [PubMed] [Google Scholar]
- Lu S-W, Chen S, Wang J, Yu H, Chronis D, Mitchum MG, Wang X. Structural and functional diversity of CLAVATA3/ESR (CLE)-like genes from the potato cyst nematode Globodera rostochiensis. Molecular Plant-Microbe Interactions. 2009;22:1128–1142. doi: 10.1094/MPMI-22-9-1128. [DOI] [PubMed] [Google Scholar]
- Maier TR, Hewezi T, Peng J, Baum TJ. Isolation of whole esophageal gland cells from plant-parasitic nematodes for transcriptomic analyses and effector identification. Molecular Plant-Microbe Interactions. 2013;26:31–35. doi: 10.1094/MPMI-05-12-0121-FI. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wang X, Wang J, Davis EL. Role of nematode peptides and other small molecules in plant parasitism. Annual Review of Phytopathology. 2012;50:175–195. doi: 10.1146/annurev-phyto-081211-173008. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, Wubben MJ, Davis EL. Nematode effector proteins: An emerging paradigm of parasitism. New Phytologist. 2013;199:879–894. doi: 10.1111/nph.12323. [DOI] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology. 2008;8:1. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Skriver K. Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends in Plant Science. 2003;8:55–57. doi: 10.1016/S1360-1385(03)00003-7. [DOI] [PubMed] [Google Scholar]
- Patel N, Hammouch N, Li C, Hussey R, Mitchum M, Baum T, Wang X, Davis EL. Similarity and functional analyses of expressed parasitism genes in Heterodera schachtii and Heterodera glycines. Journal of Nematology. 2008;40:299–310. [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Replogle A, Wang J, Bleckmann A, Hussey RS, Baum TJ, Sawa S, Davis EL, Wang X, Simon R, Mitchum MG. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant Journal. 2011;65:430–440. doi: 10.1111/j.1365-313X.2010.04433.x. [DOI] [PubMed] [Google Scholar]
- Replogle A, Wang J, Paolillo V, Smeda J, Kinoshita A, Durbak A, Tax FE, Wang X, Sawa S, Mitchum MG. Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Molecular Plant-Microbe Interactions. 2013;26:87–96. doi: 10.1094/MPMI-05-12-0118-FI. [DOI] [PubMed] [Google Scholar]
- Robinson AF. Reniform in U.S. cotton: When, where, why, and some remedies. Annual Review of Phytopathology. 2007;45:263–288. doi: 10.1146/annurev.phyto.45.011107.143949. [DOI] [PubMed] [Google Scholar]
- Rutter WB, Hewezi T, Maier TR, Mitchum MG, Davis EL, Hussey RS, Baum TJ. Members of the Meloidogyne avirulence protein family contain multiple plant ligand-like motifs. Phytopathology. 2014;104:879–885. doi: 10.1094/PHYTO-11-13-0326-R. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular and Systems Biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin PE, Levesley A, McPherson MJ, Atkinson HJ. Transgenic resistance to the nematode Rotylenchulus reniformis conferred by Arabidopsis thaliana plants expressing proteinase inhibitors. Molecular Breeding. 2000;6:257–264. [Google Scholar]
- Vovlas N, Lamberti F. Histological alterations induced by Rotylenchulus reniformis on Coffea arabica roots. Nematologia Mediterranea. 1990;18:77–81. [Google Scholar]
- Wang J, Joshi S, Korkin D, Mitchum MG. Variable domain I of nematode CLEs directs post-translational targeting of CLE peptides to the extracellular space. Plant Signaling & Behavior. 2010a;5(12):1633–1635. doi: 10.4161/psb.5.12.13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lee C, Replogle A, Joshi S, Korkin D, Hussey R, Baum TJ, Davis EL, Wang X, Mitchum MG. Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytologist. 2010b;187:1003–1017. doi: 10.1111/j.1469-8137.2010.03300.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Replogle A, Hussey R, Baum T, Wang X, Davis EL, Mitchum MG. Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Molecular Plant Pathology. 2011;12:177–186. doi: 10.1111/j.1364-3703.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen R, Ding X, Goellner M, Maier T, de Boer JM, Baum TJ, Hussey RS, Davis EL. Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Molecular Plant-Microbe Interactions. 2001;14:536–544. doi: 10.1094/MPMI.2001.14.4.536. [DOI] [PubMed] [Google Scholar]
- Wang X, Mitchum MG, Gao B, Li C, Diab H, Baum TJ, Hussey RS, Davis EL. A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Molecular Plant Pathology. 2005;6:187–191. doi: 10.1111/j.1364-3703.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Wubben MJ, Callahan FE, Scheffler BS. Transcript analysis of parasitic females of the sendentary semi-endoparasitic nematode Rotylenchulus reniformis. Molecular and Biochemical Parasitology. 2010a;172:31–40. doi: 10.1016/j.molbiopara.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Wubben MJ, Ganji S, Callahan FE. Identification and molecular characterization of a β-1,4-endoglucanase gene (Rr-eng-1) from Rotylenchulus reniformis. Journal of Nematology. 2010b;42:342–351. [PMC free article] [PubMed] [Google Scholar]