Abstract
The 5′-leader of the HIV-1 genome contains conserved elements that direct selective packaging of the unspliced, dimeric viral RNA into assembling particles. Using a 2H-edited NMR approach, we determined the structure of a 155-nucleotide region of the leader that is independently capable of directing packaging (Core Encapsidation Signal; ΨCES). The RNA adopts an unexpected tandem three-way junction structure, in which residues of the major splice donor and translation initiation sites are sequestered by long-range base pairing, and guanosines essential for both packaging and high-affinity binding to the cognate Gag protein are exposed in helical junctions. The structure reveals how translation is attenuated, Gag binding promoted, and unspliced dimeric genomes selected, by the RNA conformer that directs packaging.
Assembly of HIV-1 particles is initiated by the cytoplasmic trafficking of two copies of the viral genome and a small number of viral Gag proteins to assembly sites on the plasma membrane (1–6). Unspliced, dimeric genomes are efficiently selected for packaging from a cellular milieu that includes a substantial excess of non-viral mRNAs and more than 40 spliced viral mRNAs (7, 8). RNA signals that direct packaging are located primarily within the 5′-leader of the genome and are recognized by the nucleocapsid (NC) domains of Gag (4). Transcriptional activation, splicing, and translation initiation are also dependent on elements within the 5′-leader, the most conserved region of the genome (9), and there is evidence that these and other activities are temporally modulated by dimerization-dependent exposure of functional signals (6, 10–13).
Understanding the RNA structures and mechanisms that regulate HIV-1 5′-leader function has been based on phylogenetic, biochemical, nucleotide reactivity, and mutagenesis studies (4). The dimeric leader selected for packaging appears to adopt a highly branched secondary structure, in which there are structurally discrete hairpins and helices that promote transcriptional activation (TAR), tRNA primer binding (PBS), packaging (ψ), dimer initiation (DIS), splicing (SD), and dimer stability (U5:AUG) (4, 14) (Fig. 1). Although NMR signals diagnostic of TAR, PBS, ψ, DIS, U5:AUG, and Poly(A) helices have been observed in spectra obtained for the full-length dimeric leader (13, 15) (Fig. 1A), signals diagnostic of a putative SD hairpin have not been detected (colored magenta in Fig. 1A) (15), and there is little agreement among more than 20 different structure predictions for residues adjacent to the helices (4). For example, predictions vary for stretches of residues shown by in vivo nucleotide reactivity (16) and crosslinking with immunoprecipitation (CLIP) (17) to reside at or near sites of Gag binding (4). The TAR, Poly(A), and PBS hairpins of the HIV-1 leader are not required for efficient encapsidation (15), and a minimal HIV-1 packaging element, the Core Encapsidation Signal (ΨCES), exhibits NC binding properties and NMR spectral features similar to those of the intact 5′-leader and is independently capable of directing vector RNAs into virus-like particles (15). To gain insights into the mechanism of HIV-1 genome selection, we determined the structure of ΨCES by NMR.
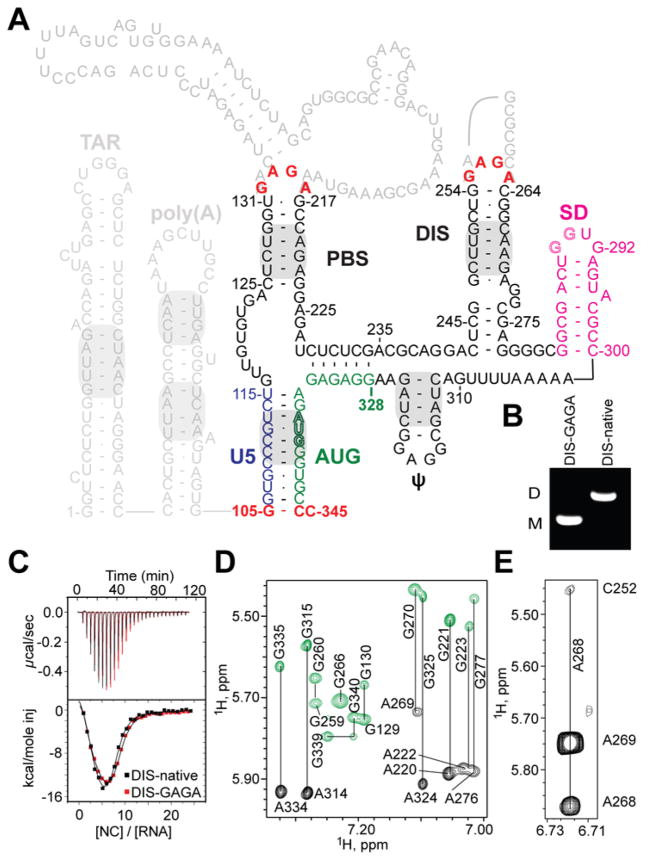
Figure 1.
HIV-1NL4-3 5′-leader and ΨCES RNA construct. (A) Predicted secondary structure of the HIV-1 5′-leader (16); gray shading denotes elements detected in the intact leader by NMR (13, 15); dark letters denote ΨCES (non-native residues colored red; see text). (B and C) Substitution of the native DIS loop residues (DIS-native) by GAGA (DIS-GAGA) prevents dimerization (B) but does not affect NC binding (C). (D) Representative NOESY spectra for G8A-ΨCES (black) and G-ΨCES (green); lines connect H8 (vertical labels) and H1′ (horizontal labels) signals. (E) Representative very long-range NOE (A268-H2 to C252-H1′; ~ 7 Å separation) obtained for A2rCr-ΨCES.
Contributions of slow molecular rotational motion to NMR relaxation were minimized by substituting the dimer promoting GC-rich loop of the ΨCES DIS hairpin by a GAGA tetraloop (Fig. 1A). This prevented dimerization (Fig. 1B) but did not affect NC binding (Fig. 1C) or NOESY NMR spectral patterns (18), indicating that the modified RNA retains the structure of the native dimer. Non-exchangeable aromatic and ribose H1′, H2′ and H3′ 1H NMR signals were assigned for nucleotides of the U5:AUG, lower-PBS, DIS, and ψ helices by sequential residue analysis of 2D NOESY spectra obtained for nucleotide-specific 2H-labeled samples (18–20) (Fig. 1D). Very long-range A-H2 NOEs (1H-1H distances up to ~7 Å) were detected in spectra of highly deuterated samples (Fig. 1E) (as observed for proteins (21)), facilitating assignments.
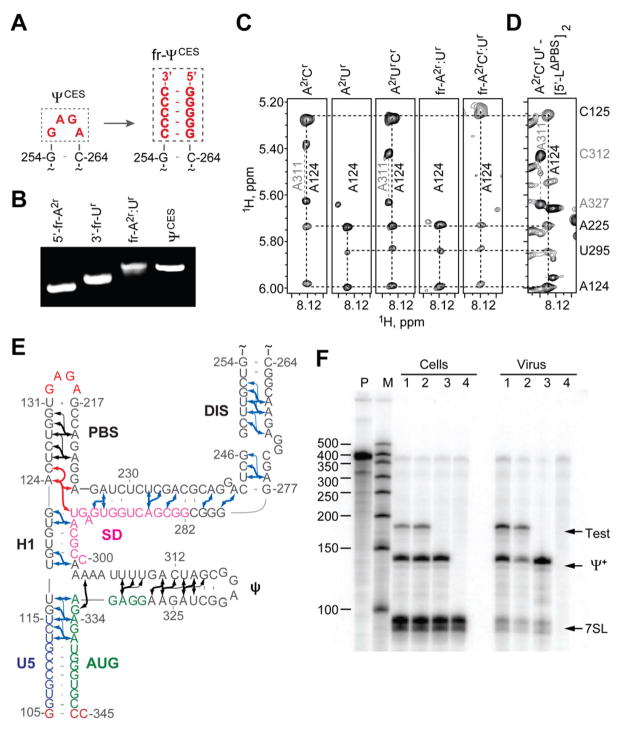
NMR signals that could not be assigned by nucleotide-specific labeling were identified by a fragmentation-based segmental 2H-labeling approach we developed, in which differentially labeled 5′- and 3′-fragments of ΨCES were prepared separately and non-covalently annealed (Fig. 2, A and B, and fig S1). The dimer-promoting loop of the DIS hairpin served as the fragmentation site, and was substituted by a short stretch of intermolecular G:C base pairs (Fig. 2A). Differential 2H labeling afforded the following fragment-annealed RNAs (fr-ΨCES; denoted 5′-fragment:3′-fragment-ΨCES; D = perdeuterated fragment; superscripts denote sites of protonation, all other sites deuterated; e.g., G = fully protonated guanosines, A2r = adenosines protonated at C2 and ribose carbons): A2r:Ur-ΨCES, A2rCr:Ur-ΨCES Gr:A2rCr-ΨCES, D:A2rCr-ΨCES, A:D-ΨCES, and D:A-ΨCES (fig. S1). Except for residues at the sites of substitution, the NMR spectra of the fr-ΨCES RNAs were consistent with those of the parent, non-fragmented RNA. For example, NOEs that correlate A124-H2 with cytosine and uridine H1′ protons in 2D NOESY spectra obtained for non-fragmented A2rCr-ΨCES, A2rUr-ΨCES, and A2rCrUr-ΨCES samples were also detected in spectra obtained for fragment-annealed A2r:Ur-ΨCES and A2rCr:Ur-ΨCES constructs, indicating that A124 resides near a cytosine (C125) in the 5′-fragment and a uridine (U295) in the 3′-fragment (Fig. 2C). More than 80 long-range and sequential A-H2 NOEs were identified using the 2H-edited NMR approach (Fig. 2E). 1H NMR assignments were validated by NOE cross peak pattern redundancy and database chemical shift analyses (18, 22) (fig. S2).
Figure 2.
Fragmentation-based 2H-edited NMR approach and observed ΨCES secondary structure. (A) The DIS loop of ΨCES served as the fragmentation site and was substituted by a stretch of intermolecular G-C base pairs. (B) Fragment annealing efficiency as measured by native polyacrylamide gel electrophoresis. (C) 2D NOESY spectra of uniformly labeled A2rCr-, A2rUr-, and A2rCrUr-ΨCES and segmentally-labeled fr-A2r:Ur- and fr-A2rCr:Ur-ΨCES samples used to make long-range NOE assignments. (D) Similarities in NOESY spectra obtained for A2rCrUr-labeled [5′-LΔPBS]2 and ΨCES confirm that the tandem three-way junction structure is present in both constructs. (E) NMR-derived secondary structure of ΨCES. Black and blue arrows denote A-H2 NOEs observable in ΨCES and fr-ΨCES samples, respectively; red arrows highlight NOEs shown in panel (C and D); thin arrows denote very long-range NOEs. (F) Packaging of native HIV-1NL4-3 5′-L and 5′-LΔPBS RNAs under competition conditions assayed by means of ribonuclease protection. P, undigested probe; M, RNA sizes marker. Lanes 1 and 2: native HIV-1NL4-3 helper versus test vectors containing 5′-LΔPBS (1) or native HIV-1NL4-3 (2). Lane 3: HIV-1NL4-3 helper expressed without test RNA. Lane 4: mock transfected-cells. Samples obtained from transfected cells (Cells) or viral containing media (Virus) are indicated. Bands corresponding to host 7SL RNA, HIV-1NL4-3 helper RNA (Ψ+) and co-packaged test RNAs (Test) are labeled.
The NMR data indicate that residues proximal to the major splice donor site do not form a hairpin, but instead participate in long-range base pairing within an extended DIS stem and a short helical segment, H1 (Fig. 2E). To determine if this secondary structure is also adopted by the native 5′-leader, we obtained NOESY data for dimeric, 2H-labeled 5′-leader constructs. Adenosine-H2 signals diagnostic of the U5:AUG, DIS, PBS, and ψ helices were observed in spectra obtained for the native leader ([5′-L]2), as expected (15). However, signals diagnostic of H1 were only detectable upon removal of the upper PBS loop (substituted by a GAGA tetraloop; [5′-LΔPBS]2), which eliminated broad upper PBS signals that overlapped with the A124-H2 signal of H1 (Fig. 2D). This construct exhibits dimerization, NC binding, and NMR properties similar to those of the intact leader (15), and directs both non-competitive (15) and competitive RNA packaging with near-wild type efficiency (94 ± 4 % and 93 ± 18%, respectively) (Fig. 2F). Thus, the secondary structure observed for ΨCES, including the H1 helix, is also adopted by the 5′-leader.
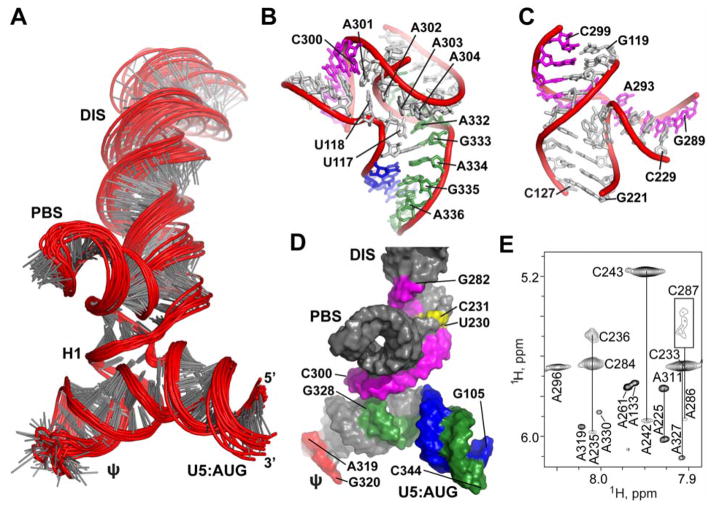
NOE-restrained structure calculations (18) reveal that ΨCES adopts a tandem three-way junction structure (Fig. 3, A-C and fig. S3). The overall shape is quasi-tetrahedral, with the U5:AUG, H1, and ψ-helices forming a plane that is nearly perpendicular to the plane formed by the H1, PBS, and DIS helices (Fig. 3A). Splice site residues G289 and G290 are base paired with C229 and U228, respectively, and adjacent residues are base paired within or near the H1-PBS-DIS (three-way-2) junction (Fig. 3, B-D), and residues of AUG are base paired within the U5:AUG-H1-ψ (three-way-1) junction (Fig. 3B and D). A227-U291 forms an extended DIS hairpin with two internally stacked but non-paired guanosines (G272 and G273) and a G240(syn):G278(anti)-G241(anti) base triple. Sequentially stacked pyrimidines (U230*U288 and C231*C287) exhibit broad line widths indicative of millisecond timescale conformational exchange (Fig 3E). These residues appear to function as a flexible hinge that connects the extended DIS hairpin with the tandem three-way junction (Fig. 3D). U307-G330 forms an extended ψ-hairpin structure that contains three non-canonical base pairs [G310(anti)*A327(anti), G328*U309, G329*U308] and a stacked A-A bulge [A311(anti)-A326(anti)] (Fig. 2E), and a flexible GAGG loop (Fig. 3D). Adenosines A302-A305 exhibit pseudo A-form stacking but are not base paired (Fig. 3B), which supports proposals that genomic adenosine enrichment occurs primarily at non-base paired sites (23). A302 and A303 also make A-minor contacts with the U5:AUG helix (Fig. 3B).
Figure 3.
Three-dimensional structure of ΨCES. (A) Ensemble of 20 refined structures (residues 105–344 shown). (B and C) Expanded views of the (B) three-way-1 and (C) three-way-2 junctions. (D) Surface representation of ΨCES highlighting U5 (blue):AUG (green) base pairing and the integral participation of SD residues (pink) in the tandem three-way junction structure. (E) Severe line broadening indicative of slow (millisecond) conformational averaging was observed for stacked, mismatched pyrimidines in the extended DIS stem (yellow in (D); broadened C287-H1′ signal boxed in (E)). NOE patterns and sharp NMR signals also indicate that the ψ hairpin loop is unstructured (red in (D)).
To determine if the tandem three-way junction is evolutionarily conserved, we analyzed published HIV-1 leader sequences that contained full coverage of the 5′-UTR (278 total sequences). Representatives from B, C, and F1 subtypes were included in the analysis (18). Of the 48 base paired nucleotides at or near the three-way junction, 31 were either strictly (16 sites) or very highly (>99%, 15 sites) conserved, and 13 displayed high (90.2% – 98.9%) identity (Table S2). Only 11 of 126 substitutions resulted in loss of base pairing. The remaining 4 sites – 227A, G279, A286 and U288 – exhibited significant variation, ranging from 12% (U288) to 50.3% (227A). Most changes mapped to terminal branches of the ΨCES phylogeny. Thus the tandem three-way junction structure is highly conserved, and the rare variations that disrupt base pairing are due to transient polymorphisms.
The PBS, DIS, and ψ helices of ΨCES are consistent with models derived from nucleotide reactivity experiments (4), but the SD structure differs significantly. Recent in-gel chemical probing of resolved monomeric and dimeric leader RNAs (24), and probing studies under solution conditions that favor either the monomeric or dimeric species (25), showed that SD loop residues are relatively unreactive in the dimeric RNA, consistent with the ΨCES structure. Pseudo-free energy calculations indicate that the in-gel reactivity data for the dimeric leader (24) are in better agreement with the ΨCES NMR structure than the proposed model (~25% lower experimental pseudo-free energy (18); fig. S4). These findings support proposals that variations in structure predictions are at least partly due to site-specific structural heterogeneity associated with the monomer-dimer equilibrium (13, 24).
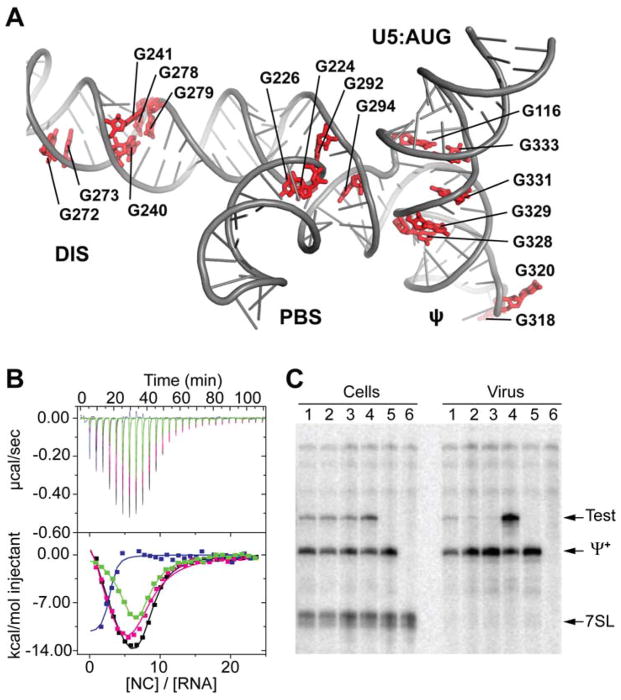
HIV-1 NC binds with high affinity to oligonucleotides that contain exposed guanosines (4, 26, 27). ΨCES contains five unpaired Gs (excluding the non-native GAGA tetraloops), a GGG base triple in the DIS stem, and five additional guanosine mismatches clustered at or near the two three-way junctions (G*U, G*A, or G*G) that could serve as NC binding sites (Fig. 4A). Potential contributions of these “junction guanosines” to NC binding were tested by isothermal titration calorimetric (ITC) studies of G-to-A substituted ΨCES RNAs. Free energy calculations indicate that these substitutions, which include three G*U to A-U substitutions, should not alter the secondary structure of the RNA (18). Replacement of the ψ GGAG loop by GAAA eliminated one NC binding site, as expected (27), and substitution of the three-way-1 junction guanosines by adenosines (G116A/G333A/G328A/G329A/G331A) eliminated three additional NC sites (Fig. 4B). Mutation of the unpaired (G226, G292 and G294) and mismatched (G224) three-way-2 junction guanosines to adenosines eliminated one NC binding site (Fig. 4B). The influence of these guanosines on RNA encapsidation was evaluated using a competitive in situ RNA packaging assay. Human embryonic kidney 293T cells were co-transfected with plasmids that produce vector RNAs containing the wild type (Ψ+, which also encodes for viral proteins) and mutant (Test) leader sequences (18). When co-expressed at similar levels, Ψ+ and Test vector RNAs with native leader sequences were packaged into HIV-1 virus-like particles with similar efficiencies (Fig. 4C). In contrast, significant packaging defects were observed upon G-to-A mutation of the three-way-2 junction guanosines (17% ± 2%), the ψ-loop and three-way-1 junction guanosines (10% ± 2%), or all junction and -loop guanosines (5%±1%) (Fig. 4C). Our findings indicate that the tandem three-way junction serves as a scaffold for exposing clusters of unpaired or weakly paired junction guanosines, thereby enabling their binding to the zinc knuckle domains of NC.
Figure 4.
Junction guanosines mediate NC binding and packaging. (A) ΨCES contains 17 unpaired or weakly paired guanosines (red) that serve as potential NC binding sites. (B) Mutation of the three-way-2 (green) or ψ (magenta) guanosines to adenosines modestly reduces NC binding (N = 7.0 ± 0.3 and 7.0 ± 0.5, respectively) relative to WT ΨCES (black; N = 8.0 ± 0.3). Mutation of ψ and three-way-1 guanosines to adenosines (blue) severely inhibits high affinity NC binding (N = 2.0 ± 0.1). (C) Competitive packaging of HIV-1NL4-3 vectors containing native and mutant 5′-leader sequences, assayed by means of ribonuclease protection. Lanes 1 to 4: native HIV-1NL4-3 versus test vectors containing 5′-L3way2-G/A (1), 5′-L3way1-G/A (2), 5′-L3way1,2-G/A (3), and 5′-L (4). Lane 5: HIV-1NL4-3 helper expressed without test RNA. Lane 6: mock transfected-cells. Samples obtained from transfected cells (Cells) or viral containing media (Virus) are indicated. Bands corresponding to host 7SL RNA, HIV-1NL4-3 helper RNA (Ψ+) and co-packaged test RNAs (Test) are labeled.
The ΨCES structure explains biochemical, nucleotide reactivity, and phylogenetic results and suggests a mechanism by which the 5′-leader structure regulates translation and splicing (4). In vitro translational activity and chemical reactivity of the AUG residues are suppressed upon dimerization (28), and this can be attributed to sequestration of the 5′ end of the gag open reading frame within the three-way-1 junction (Fig. 3D). Enhanced in vitro translational activity caused by mutations immediately downstream of the major splice donor site (ΔA296/A301U and A293C/U295C/ΔG298) can be explained by destabilization of the H1 helix and, for ΔA296/A301U, stabilization of the SD hairpin (29), both of which should favor the monomer. Mutations in AUG that inhibit genome dimerization and suppress packaging (30, 31) are expected to disrupt base pairing in the U5:AUG helix and ψ-hairpin stem, thereby destabilizing the tandem three-way junction structure required for Gag binding. In vitro splicing activity is also attenuated by dimerization (12, 32), and this can be attributed to sequestration of the major splice site recognition sequence within the three-way-2 junction. Antisense oligonucleotides with complementarity to the SD loop inhibit dimerization, and this is likely due to their ability to competitively block formation of the tandem three-way junction (25).
The ΨCES structure also explains the exquisite selectivity of HIV-1 to package its unspliced genome (1, 2). Residues immediately downstream of the major splice site are base paired within the H1 helix and are thus integral to the formation of the tandem three-way junction structure. Although unspliced and spliced HIV-1 mRNAs contain identical 5′ sequences (G1-G289), differences in spliced mRNA sequences derived from 3′-exons would preclude formation of the packaging competent junction structure. Similarly, because SD appears to exist as a hairpin in the monomeric, unspliced 5′-leader (12), it is likely that monomeric genomes are also ignored during virus assembly because they do not adopt the tandem three-way junction structure.
Compared to the proteins of HIV-1, structural information for the viral nucleic acids is sparse. RNAs in general are vastly underrepresented in the structural databanks (99,000 proteins versus 2,700 RNA structures), due in part to NMR technical challenges and difficulties obtaining suitable crystals for X-ray diffraction (19, 20). The fr-RNA 2H-edited NMR approach enables efficient segmental labeling without requiring enzymatic ligation. Given the ubiquity of hairpin elements that can serve as fragmentation/annealing sites, this method should be generally applicable to modest-sized RNAs (~ 160 nucleotides).
Supplementary Material
One Sentence Summary.
An NMR structure of a region of the HIV-1 5′-leader gives insight into how the viral genome is selected for packaging.
Acknowledgments
This research was supported by grants from the National Institute of General Medical Sciences (NIGMS, R01 GM42561 to MFS and AT, P50 GM 103297 to MS, BJ and DAC). SB, NCB, and SM were supported by a NIGMS grant for enhancing minority access to research careers (MARC U*STAR 2T34 GM008663), and SB, JS, NCB, and SM were supported by an HHMI undergraduate education grant. We thank the HHMI staff at UMBC for technical assistance, and Brittany Rife (University of Florida) for advice regarding the phylogenetic analysis. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3 from Dr. Malcolm Martin. Atomic coordinates have been deposited into the Protein Data Bank with accession code 2N1Q. NMR chemical shifts and restraints have been deposited into the Biological Magnetic Resonance Bank with accession code 25571.
Footnotes
REFERENCES AND NOTES
- 1.Coffin JM, Hughes SH, Varmus HE. Cold Spring Harbor Laboratory Press; Plainview, N.Y: 1997. [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff SP. Curr Top Microbiol Immun. 1996;214:177. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, et al. Proc Natl Acad Sci USA. 2009 Aug 11;106:13535. [Google Scholar]
- 4.Lu K, Heng X, Summers MF. J Mol Biol. 2011;410:609. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jouvenet N, Simon SM, Bieniasz PD. J Mol Biol. 2011 Jul 22;410:501. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzembayeva M, Dilley K, Sardo L, Hu WS. Virology. 2014 Apr;454–455:362. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S, Felber BK, Benko DM, Fenyo EM, Pavlakis GN. J Virol. 1990 Jun;64:2519. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolaitchik OA, et al. PLoS Pathog. 2013 Mar;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lever AM. Adv Pharmacol. 2007;55:1. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 10.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Nature Revs Microbiol. 2004;2:461. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 11.Greatorex J. Retrovirology. 2004;1 doi: 10.1186/1742-4690-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbink TEM, Berkhout B. J Virol. 2008;82:3090. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu K, et al. Science. 2011;344:242. [Google Scholar]
- 14.Abbink TEM, Berkhout B. J Biol Chem. 2003;278:11601. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 15.Heng X, et al. J Mol Biol. 2012;417:224. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson KA, et al. PLoS Biology. 2008;6:883. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutluay SB, et al. Cell. 2014 Nov 20;159:1096. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Information on materials and methods is available at the Science website.
- 19.Lu K, Miyazaki Y, Summers MF. J Biomol NMR. 2009;46:113. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duss O, Lukavsky PJ, Allain FH. Adv Exp Med Biol. 2012;992:121. doi: 10.1007/978-94-007-4954-2_7. [DOI] [PubMed] [Google Scholar]
- 21.Koharudin LM, Bonvin AM, Kaptein R, Boelens R. J Magn Reson. 2003 Aug;163:228. doi: 10.1016/s1090-7807(03)00149-6. [DOI] [PubMed] [Google Scholar]
- 22.Barton S, Heng X, Johnson BA, Summers MF. J Biomol NMR. 2012 Nov 23; doi: 10.1007/s10858-012-9683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Hemert FJ, van der Kuyl AC, Berkhout B. RNA Biol. 2013 Feb;10:211. doi: 10.4161/rna.22896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenyon JC, Prestwood LJ, Le Grice SF, Lever AM. Nucleic Acids Res. 2013 Oct;41:e174. doi: 10.1093/nar/gkt690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deforges J, Chamond N, Sargueil B. Biochimie. 2012 Jul;94:1481. doi: 10.1016/j.biochi.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 26.South TL, Summers MF. Protein Sci. 1993;2:3. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Guzman RN, et al. Science. 1998;279:384. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 28.Baudin F, et al. J Mol Biol. 1993;229:382. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 29.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. Biochemistry. 2005 Jun 28;44:9058. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 30.Poon DT, Chertova EN, Ott DE. Virology. 2002 Feb 15;293:368. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- 31.Nikolaitchik O, Rhodes TD, Ott D, Hu WS. J Virol. 2006;80:4691. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jablonski JA, Buratti E, Stuani C, Caputi M. J Virol. 2008 Aug;82:8038. doi: 10.1128/JVI.00721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.