Preface
Emergence of the adaptive immune system in vertebrates set the stage for evolution of an advanced symbiotic relationship with the intestinal microbiota. The defining features of specificity and memory that characterize adaptive immunity have afforded vertebrates mechanisms for efficiently tailoring immune responses to diverse types of microbes, whether to promote mutualism or host defense. These same attributes carry risk for immune-mediated diseases that are increasingly linked to the intestinal microbiota. Understanding how the adaptive immune system copes with the remarkable number and diversity of microbes that colonize the digestive tract, and how it integrates with more primitive innate immune mechanisms to maintain immune homeostasis, holds considerable promise for new approaches to modulate immune networks in order to treat and prevent disease.
Introduction
Each of us enters this world germ-free, devoid of microbial colonization by virtue of the sterile environment of the womb. This germ-free existence is short-lived, as the process of birth itself exposes the newborn to the microbiota of the mother, setting in motion the colonization of barrier tissues — the mucosal tissues (digestive, respiratory and urogenital tracts) and the skin — by a diverse microbiota with which we coexist throughout life. The complex, dynamic interaction between the microbiota and its human host is the culmination of nearly half a billion years of coevolution in vertebrates that have reciprocally shaped the repertoires of the microbiota and the immune system, such that our microbiota is normally restrained and well tolerated. This is particularly evident in the intestinal tract, where the greatest diversity and abundance of microbes reside. Estimated at ~100 trillion organisms, the overwhelming majority of which are Eubacteria (although the other two domains of life — Archeae and Eukarya are also represented), the microbiota numbers ~10 times the total cells in the human body, with the greatest density populating the distal ileum and colon 1. The collective genome, or metagenome, of the enteric microbiota contains over 100 times the number of genes in the human genome, with about 15% of the full diversity of the more than 1000 enteric bacteria represented in each of us 2 — reflecting substantial variability in the composition of the microbiota between individuals. Thus, there are ~10-fold more genes in each of our microbiomes than in each of us, encoding the greatest source of potential antigens with which the immune system must cope — substantially exceeding those of self and pathogen-derived antigens.
The relationship that has been forged between the intestinal microbiota and its human host provides mutual benefits. At homeostasis, the microbiota benefits from the warm, nutrient-rich environment of the gut to establish a relatively stable ecosystem. We, in turn, benefit from a highly adaptive metabolic engine, or bioreactor, that in addition to providing essential non-nutrient factors (eg, vitamins) also substantially extends our ability to harvest nutrients from food. This is primarily through the microbiota’s complementation of the limited diversity of enzymes encoded in the human genome capable of metabolizing complex carbohydrates. Further, by establishing robust, interlinked metabolic/nutrient networks and biofilms amongst its constituents, the microbiota limits resources available to potential pathogens that must outcompete well-adapted and entrenched resident microbes for metabolic and physical niche space. Resident microbes thereby establish a microbial buffer that limits access by those not part of the consortium.
However, the microbiota is not innocuous, and under conditions that compromise the host’s ability to limit its entry from the intestinal lumen, certain of its constituents can invade and threaten the host. Further, shifts in the composition of the microbiota, whether induced by dietary changes, antibiotic treatment or invasive pathogens, can disturb the balance of organisms in the microbiota so as to alter the metabolic network of the collective and favor outgrowth of potentially pathogenic constituents, referred to as dysbiosis. This can perturb immune regulatory networks that normally restrain intestinal inflammation, and may contribute to immune-mediated disease directed against antigens of the microbiota. Dysbiosis is most often associated with inflammatory bowel disease (IBD), including Crohn’s disease and ulcerative colitis, and necrotizing enterocolitis in premature infants, although it is increasingly linked to a number of extraintestinal immune-mediated diseases, including rheumatoid arthritis, multiple sclerosis, diabetes, atopic dermatitis and asthma, but also obesity and metabolic syndrome, all of which could well have their pathogenic origins in untoward reactivity of the immune system to the microbiota. Whether dysbiosis is a cause or effect of these disorders remains to be determined, although understanding the factors that lead to alterations in composition of the microbiota in these conditions promise to be informative, irrespective of their basis.
Here, we highlight studies that have advanced our understanding of immune mechanisms by which the dynamic interplay of the intestinal microbiota and its host normally favors a homeostatic, mutualistic relationship, as a basis for understanding causes of breakdowns in this relationship that lead to disease.
Coevolution of microbiota and adaptive immunity
Vertebrate evolution coincided with the emergence of the adaptive immune system, the principal components of which are T and B lymphocytes. The acquisition of new genetic recombinatorial mechanisms for generating diverse, anticipatory antigen recognition receptors on these cells (T-cell and B-cell receptors) enabled specific responses to a virtually limitless diversity of antigens and long-lived immune memory. While evolutionary pressures that have shaped immune strategies are often viewed in the context of host defense, it has been proposed that the invention of adaptive immunity could well have been driven as a means to foster, rather than limit, microbial colonization 3. In this view, a substantial survival advantage would have been afforded organisms that could harness the extended and more flexible metabolic capacity derived from a permanent, diverse intestinal microbiota — assuming that the attendant infectious risk could be adequately mitigated. The rise of adaptive immunity might well have enabled this.
By adding new layers to existing innate barrier defenses of (e.g., secretory immunoglobulin A, or sIgA), early vertebrates armed with adaptive immune mechanisms could begin to promote colonization of their alimentary tracts by exerting selective pressures that favored resident microbes which were relatively innocuous, yet metabolically useful. The survival benefits that accrued to derivation of nutrients from a broader range of foods could well have been a major factor in the evolutionary success of vertebrates 3. Thus, the emergence of adaptive immunity provided a means to recognize, and remember, both beneficial and detrimental members of the enteric microbiota to foster maintenance of the former at the expense of the latter. In so doing, the emerging adaptive immune system would have had to acquire mechanisms for tempering innate immune responses geared only for clearance of microbes. The development of a broad repertoire of immune cells that could suppress, as well as promote, innate inflammatory mechanisms contingent upon the threat level of the specific microbe recognized would become invaluable.
Support for this hypothesis comes from studies of the diversity of the microbiota in invertebrates, which appears to be far less complex than that of vertebrates 3. Although data on comparative metagenomics are limited at present, the density of bacteria found in the human colon are the highest measured to date, whether in other organisms or in the environment 1. And the complexity of the intestinal microbiota across vertebrate species as divergent as rodents and zebrafish share similar complexity to that of humans, and, as in humans, colonize the gastrointestinal tract soon after birth 4. Further, recent studies in comparative immunology have identified a homolog of sIgA in bony fish 5, extending to all vertebrate taxa except cartilaginous fish this unique class of immunoglobulin that is adapted for interactions with the commensal microbiota and mucosal pathogens. It is tempting to speculate that the invention of multimeric mucosal IgA and adaptations that effect its efficient transport across the mucosal epithelium, as well as the compartmentalization of the gut-associated lymphoid tissues (GALT) from the peripheral immune system, were essential adaptations that allowed vertebrates to harness a complex intestinal microbiota. While much work remains to be done to establish a comparative map of the microbiota in the digestive tracts of more vertebrate and non-vertebrate species, it is anticipated that the merger of comparative studies in metagenomics and immunology will yield important insights into the adaptive mechansims that have allowed vertebrates to become such an evolutionary success. It may yet prove that we owe it to the bugs that have become ‘us.’
Microbiota and the developing immune system
The mammalian immune system represents perhaps the most elaborate realization of the complex symbiotic relationship resulting from the coevolution of vertebrates and their microbiota. Unlike other vertebrates, placental mammals (eutherians) give birth to live young that are carried to term in utero and nurse their offspring with milk that is rich in maternal antibodies that provide passive transfer of immunity from mother to infant. This has important implications for the developing immune system of the infant and its microbiota.
Maternal effects on the neonatal microbiome
The greatest initial contribution to the composite human/microbiota ‘superorganism’ is the vertical transmission of components of the mother’s microbiota at birth. Normally, intestinal colonization of neonates is dominated by transmission of bacteria of the maternal vaginal flora, which is less diverse than that of the lower intestinal tract 6. The ‘pioneer’ species received from the mother appear to be important, as infants born by Cesarean section and initially colonized by bacterial species of epidermal, rather than vaginal, origin are predisposed to development of allergies and asthma later in life 7. Thus, first contact could be deterministic. Because as postpartum maternal-child transmission shifts to skin and oral bacterial ecologies, neonates delivered transvaginally are subsequently exposed to similar species as those delivered via Cesarean section. Indeed, while the microbiota of Cesarean-born infants lag in their acquisition of the two bacterial divisions dominant in adults (Firmicutes and Bacteroidetes), they do ‘catch up.’ Although unclear at present, this suggests that pioneer bacterial species might have substantial, lasting effects on the immune response, irrespective of the composition of the mature microbiota.
The neonatal microbiota varies erratically until approximately one year of age, when it stabilizes, establishing a consortium that resembles that of adults 8. During this period, the neonatal immune system is rapidly maturing under the influence of the microbiota. Whereas multiple environmental factors — including diet, exposure to new microbes, xenobiotics, bacteriophages and intestinal infections — play important roles in shaping the composition of the microbiota during this maturational window, the role of the neonatal immune system is less clear. What is clear is the initial and on-going maternal influence transmitted through breastfeeding. In addition to a unique mix of nutrients and antimicrobial proteins that influence the ecology of the neonatal microbiota, breast milk provides abundant sIgA, the specificities of which have been shaped by the maternal microbiota. The mucosal immune memory of the mother is thereby transmitted to her offspring. Thus, in breastfed infants delivered transvaginally, the intestinal microbiota is not only seeded by maternal bacterial species, its composition may be reinforced and shaped by the maternal sIgA repertoire that is influenced, in turn, by the maternal microbiota (see also Text Box 1).
Text Box 1. Conditioning of perinatal microbiota-host mutualism by maternal sIgA.
Because maturation of the infant mucosal immune system takes months, the presence of passive transfer of maternal sIgA plays a major protective role against potential pathogens that could perturb the ecological trajectory of the infant’s intestinal microbiota. Maternal sIgA also shields the neonatal immune system from its own microbiota, perhaps so as not to overwhelm the neonate’s defenses before they fully develop. Insofar as microbial antigens bound by sIgA are handled by the innate immune system in a ‘tolerogenic’ mode, due in part to IgA’s poor fixation of complement, transfer of maternal sIgA would appear to favor the establishment of regulatory immune networks in the infant that promote a mutualistic relationship with the microbiota. Collectively, then, in addition to direct effects on the microbiota of the infant, maternal sIgA also contributes immunomodulatory effects on the developing infant’s immune repertoire so as to indirectly influence its microbiota. In this way, the maternal-neonate metagenomic bond is extended postnatally, providing one mechanism by which microbial ecologies tend to cluster in family members.
Microbiome effects on developing host barrier defenses
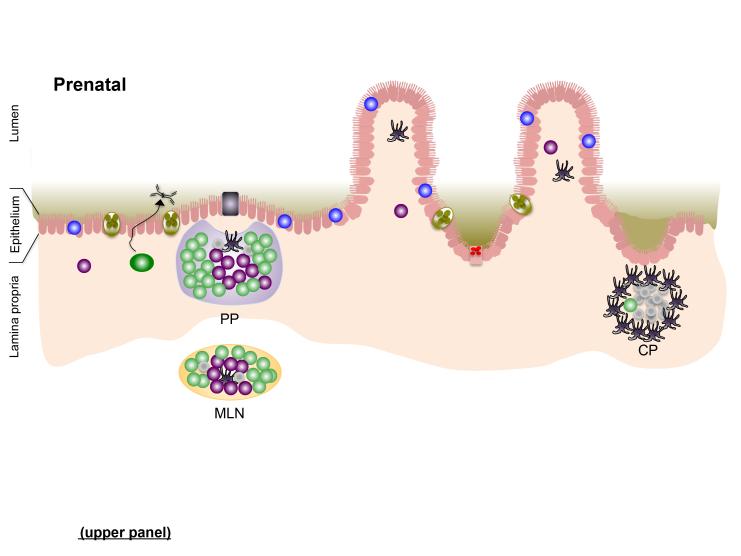
Maturation of the intestinal mucosa and its gut-associated lymphoid tissues (GALT) — Peyer’s patches (PPs) of the distal ileum, isolated lymphoid follicles (ILFs), and mesenteric lymph nodes (MLNs) — is initiated by, and contingent upon, intestinal colonization. Whereas PPs and MLNs develop prenatally, ILFs develop postnatally 9. However, each requires signals derived from sensing of the intestinal microbiota for complete development and/or recruitment of a mature complement of immune cells. Similarly, non-lymphoid structures of the intestinal mucosa that contribute to the establishment of host-microbiota mutualism are driven by colonization of the neonate.
Preparation for the adaptive immune response to neonatal colonization requires the prenatal actions of a subset innate lymphoid cells (ILCs), termed lymphoid tissue inducer (LTi) cells (Fig. 1). As their name implies, LTi cells are instrumental in organizing the development of lymphoid tissues, including each of the components of the GALT. LTi cells develop in the fetal liver from a common lymphoid precursor that gives rise to all lymphoid cells. During fetal development, LTi cells disseminate to MLN and PP anlagen, where they stimulate the development of these structures, as well as the recruitment and partitioning of B and T cells into B cell follicles and T cell zones that characterize secondary lymphoid tissues 9,10.
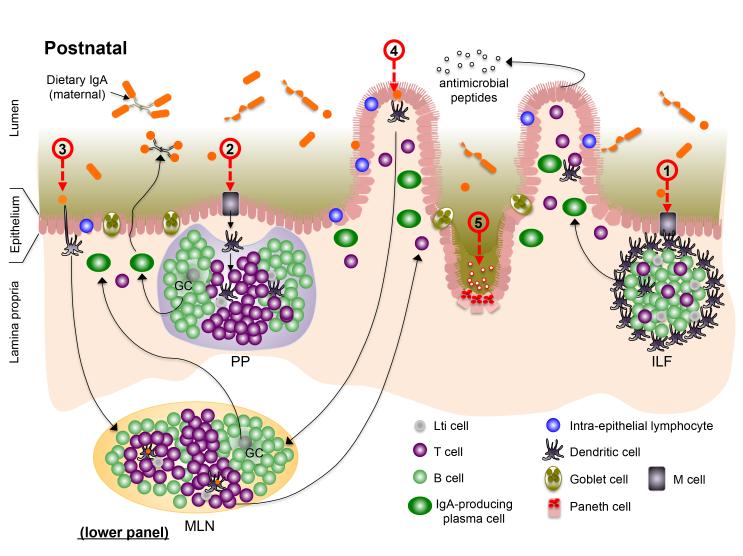
Figure 1. The gut-associated lymphoid tissues (GALT); establishing perinatal host-microbiota mutualism in the intestine.
In utero (prenatal; upper panel), secondary lymphoid tissues [Peyer’s patches (PP) and mesenteric lymph nodes (MLN)], as well as cryptopatches (CP), develop through spatiotemporal recruitment of lymphoid tissue inducer (LTi) cells to sites in the developing intestine and supporting neurovascular structures. The intestinal epithelium is populated by intra-epithelial lymphocytes (IELs) before birth. Bacteria that colonize the neonatal intestine immediately following birth initiate mutiple events that impact development or functional maturation of the mucosa and GALT. Microbe-assoaciated molecular patterns (MAMPs) sensed by patter recognition receptors (PRRs) on intestinal epithelial cells (IECs) and dendritic cells (DCs) adjacent to to cryptopatched (CP) stimultate recruitment of B cells and subsequent maturation of isolated lymphoid follicles (ILFs) (1), which can produce IgA plasma cells via T-dependent and –independent interactions. Increased transport of microbes and their products across the epithelium enter the PP (and ILFs) via M cells (2) before being endocytosed by dendritic cells (DCs) in the subepithelial dome. Antigen-loaded DCs in the PP interact with local lymphocyte subsets to induce T cell differentiation and T-dependent B cell maturation to induce development of IgA-producing plasma cells that home to the lamina propria where they release dimeric IgA for transport into the intestinal lumen. DC-mediated luminal sampling (3) or transcytosis of bacteria across the epithelium (4) results in antigen loading of lamina propria DCs, which migrate via the afferent lymphatics to a draining MLN where they induce differentiation of effector T cells that traffic to the lamina propria. Finally, sensing of MAMPs stimulates the proliferation of IECs in crypts, resulting in their increased depth and, in the small intestine, increased density of Paneth cells and their arming for release of AMPs.
The development of ILFs is similarly dependent on LTi cells, but is initiated after birth when these cells cluster in the lamina propria subjacent to the intestinal crypts to form so-called cryptopatches 11. ILFs develop from cryptopatches only following colonization by the intestinal microbiota. Central to the sensing of the colonizers of the intestinal tract is the display of a diversity of germline-encoded pattern recognition receptors (PRRs) by intestinal epithelial cells (IECs) and immune cells resident in the gut. This includes transmembrane toll-like receptors (TLRs) and C-type lectin receptors (CLRs) that reside on the cell surface or in endosomes, and cytosolic nucleotide binding, oligomerization domain (NOD)-like receptors (NLRs) by which cells of the intestinal mucosa detect microbe-associated molecular patterns, or MAMPS, expressed by constituents of the microbiota or pathogens. Through mechanisms not well understood, recognition of MAMPS by cells in the neonatal gut stimulates ILF development to generate a lymphoid structure capable of supporting the maturation of B cells that produce sIgA. At least one microbial product that induces this process is peptidoglycan derived from Gram-negative bacteria, recognition of which by NOD1 in IECs elicits production of CCL20 and β-defensin 3 that direct recruitment of B cells to LTi-DC clusters in cryptopatches to induce expression of sIgA 12.
Unlike MLNs and PP, wherein class switch recombination to produce IgA is T cell-dependent, IgA class switching in ILFs can occur via both T-dependent and – independent pathways 13. And while MLNs and PPs are unique to mammals, ILFs are found in non-mammalian vertebrates, suggesting that they represent evolutionary forerunners of the secondary lymphoid tissues of the GALT 14 and might well have evolved to accommodate, and promote, an increasingly complex intestinal microbiota in advance of specialized secondary lymphoid tissues. The importance of ILFs in regulating the microbiota is evident in mice devoid of these structures, in which Gram-negative bacteria are overrepresented 12. Similar to PPs, ILFs develop in the intestinal mucosa in intimate association with the overlying intestinal epithelium (Fig. 1), both structures strategically deployed to sample the microbiota via specialized microfold, or M, cells, which transport intact microbes or their products across the epithelial barrier. In this way, the innate and adaptive immune systems constantly monitor the microbiota and remain ‘informed’ of the dominant bacterial species that comprise members of the consortium that reside in close proximity to the epithelium.
Like the GALT, the epithelium of the fetal and newborn intestine is incompletely adapted to dense bacterial colonization and responds rapidly to perinatal colonization to accommodate to the microbiota. Expression of, and signaling by, TLR4, which is the receptor for the abundant product of Gram-negative bacteria, lipopolysaccharide (LPS), are significantly increased antecedent to intestinal colonization at birth 15. Within hours of exposure to the microbiota, the response of IECs to LPS is markedly attenuated via down-regulation of TLR4 and components of its signaling apparatus. This would appear to represent a mechanism whereby the fetal epithelium is primed to transmit transient inflammatory signals upon sensing the microbiota as a means to promote an early immune response, yet becomes rapidly desensitized as an adaptation to the mounting bacterial load.
An additional immune mechanism that prepares the neonate for commensal colonization is the seeding of the intestinal epithelium with specialized intraepithelial lymphocytes (IELs) before birth. IELs intercalate between IECs where they exist in an activated state and can rapidly mount responses to microbial encroachment. Throughout life, intestinal IELs help to maintain the epithelial cell barrier integrity, limit bacterial translocation, and facilitate epithelium repair following injury, via the secretion of antimicrobial peptides (AMPs) and other soluble mediators 16.
While the host has evolved elaborate developmental strategies to prepare for postnatal colonization, the microbiota reciprocates by inducing maturation of the host immune system. Thus, compared to conventionally-raised mice, germ-free (GF) or ‘germ-reduced’ mice display reduced size and cellularity of secondary lymphoid tissues as well as altered numbers, frequencies, and/or diversity of immune cell populations at mucosal sites, and therefore mount abnormal responses to infection and injury. In addition, recent studies point to a postnatal window of time during which regulatory T cells 17 and invariant natural killer (iNK) T cells 18 that receive their ‘primary’ education in the thymus, receive a ‘secondary’ education, courtesy of the microbiota, that establishes a durable immune repertoire critical to the prevention of inflammatory diseases into adulthood.
Innate immune pathways by which the host senses and restrains the microbiota
The intestinal tract is the single largest barrier tissue in the human body. With a surface area of ~300 m3 in adults, it is the body’s most extensive portal for entry of microbes, whether commensals or pathogens. Highly specialized barrier defenses have evolved to confine the microbiota and resist pathogens while maintaining the major function of nutrient uptake. The strategy is one of a layered defense that integrates a stratified mucous layer, a relatively impenetrable but highly responsive epithelium, and a lamina propria populated by innate and adaptive immune cells that actively participate in homeostatic responses that restrain the microbiota without undo inflammation, yet are poised for the induction of anti-microbial clearance responses and tissue repair should barrier breaches occur. Here we consider the deployment of components of the innate immune system and their function in promoting mutualism in the microbiota-replete host.
Reciprocity between the intestinal epithelium and the microbiota
Central to orchestration of intestinal defenses is the epithelium. Not simply a passive barrier to microbial translocation, the intestinal epithelium is an active sensor of, and conduit for, the dialogue between host and microbiota. Each of the major cell types of which the epithelium is comprised — absorptive enterocytes, goblet cells, Paneth cells, M cells and enteroendocrine cells — develops from a common stem cell found near the base of intestinal crypts and plays a specialized, integral role in intestinal homeostasis that is responsive to, and conditioned by, the microbiota (Fig. 1).
Critical to the function of the epithelial barrier is its maintenance of a polarized structure. By virtue of tight junctions that seal the interfaces of adjacent IECs, the epithelium is segregated into an apical, lumen-exposed surface and a basolateral surface anchored to the basement membrane. Depending on the localization of membrane-associated PRRs, whether arrayed on the apical or basolateral surface of IECs, and on which side of the epithelial barrier MAMPs are detected, cells of the epithelium initiate responses that promote release of protective factors that are directed luminally (eg, secreted mucins and AMPs), where they directly restrain the microbiota, or internally (eg, cytokines and chemokines), where they either promote immune quiescence at homeostasis or activate inflammatory immune responses when the epithelial barrier has been breached.
As a mucosal tissue, the intestinal epithelium continuously produces and is invested by a layer of mucus that serves as a first line of defense against microbes (Fig. 2). Mucus is produced by goblet cells. It is composed of heavily glycosylated mucin proteins, as well as other protective molecules such as trefoil factor that contribute to epithelial restitution and repair. Production of intestinal mucus is regulated by products of the microbiota. Thus, in GF mice the mucous layer in the colon is highly attenuated, despite normal numbers of mucin-laden goblet cells. Addition of the MAMPs LPS or peptidoglycan stimulates release of mucin by goblets cells and rapid reconstitution of the colonic inner mucous layer 19. Butyrate produced by benign constituents of the microbiota has also been shown to promote increased mucin, providing a positive feedback loop for maintenance of the mucous barrier and its colonization by butyrate-producing commensals. The importance of the mucus layer is evident in mice deficient for principal intestinal mucin, Muc2, which show greatly increased translocation of both commensal and pathogenic bacteria 20, and spontaneous development of colitis 21. Components of the normal microbiota therefore directly contribute to the barrier function of the intestine through their induction of mucin production and secretion by goblet cells.
Figure 2. The barrier function of the intestinal epithelium.
Distinct subpopulations of intestinal epithelial cells (IECs) are integreated into a continuous, single cell layer that is divided into apical and basolateral regions by tight juctions. Although apically expressed toll-like receptors (TLRs) are adapted to limit pro-inflammatory responses at homeostasis, there is on-going sensing of the microbiota to induce the production of anti-microbial peptides (AMPs), both by enterocytes and colonocytes of the the small and large intestine, and specialized Paneth cells in the bases of small intestinal crypts. Goblet cells produce mucin, that is organized into a dense, more highly cross-linked inner proteoglycan gel that forms an IEC-adherent inner mucous layer, and a less densely cross-linked outer mucous layer that is the result of proteolytic cleavage of the predominant intestinal mucin, MUC2. The outer layer is highly colonized by constitutents of the microbiota that attach to and degrade the complex O-linked glycans, providing short-chain fatty acids that are: toxic to potential pathogens; used by IECs as an energy source; and suppress epithelium-induced inflammation. The inner mucous layer is largely impervious to bacterial colonization or penetration, due to its high concentration of bactericidal AMPs, as well as commensal-specific secretory IgA (sIgA), which is ferried across IECs from their basolatral surface, where it is bound by the polymeric immunoglobulin receptor (pIgR), to the inner mucous layer where it is released by proteolytic cleavage of pIgR, apportion of which remins bound to sIgA. Innate lymphoid cells (ILCs), including RORγt- and AhR-expressing LTis and NK-22 cells, respond to microbiota to produce IL-22, an important stimulator of IEC AMP production and epithelial barrier integrity.
The thickness and continuity of intestinal mucus differs regionally; it is thinner and discontinuous in the proximal SI and becomes thicker and continuous in the distal SI and LI 22, correlating somewhat with the local bacterial load (103-105 organisms per gram of luminal contents in duodenum and jejunum; ~108 organisms per gram in ileum; 1010-1012 organisms per gram in colon). It is stratified into two functionally distinct layers: a compact, firmly adherent inner layer that is sparsely populated by bacteria and a more loosely structured, non-adherent outer layer that is at least 10-fold more densely populated by the microbiota 22.
Although both mucous layers have a similar composition dominated by Muc2, proteolytic cleavage of its polypeptide backbone in the outer layer results in its expanded volume and accessibility to colonization by components of the microbiota 22. Indeed, the outer mucous layer serves as an anchor for attachment of bacteria of the microbiota that can establish biofilms that exclude pathogens. Further, in addition to dietary glycans, mucin glycans serve as nutrients for certain species of the microbiota, such as Bifidobacteria and Bacteroides, thereby promoting their retention in the collective. These bacteria ferment complex O-linked mucin glycans to produce short-chain fatty acids derived from mucin catabolism (eg, acetate and lactate) that are toxic to certain pathogens 23, and they produce other metabolites (eg, proprionate and butyrate) that are the major nutrient source for colonic IECs. The latter also signal through G protein-coupled receptors on IECs (eg, GPR43) to downregulate host inflammatory responses 24. Thus, colonization of the outer mucous layer by commensals represents an important adaptation that supports a stable host-microbiota relationship.
In contrast to the outer mucous layer, the inner mucous layer provides a relatively impermeable barrier against the microbiota. This is due both to its compact physiochemical structure and its function as a reservoir for microbicidal products of the epithelium: a variety of AMPs specialized for killing different classes of microbes, and sIgA, which is retained in the mucous layer after being shuttled across the epithelium by the polymeric immunoglobulin receptor (pIgR) (Fig. 2). Effectively, the inner mucous layer is a ‘killing field’ that few pathogens or commensals have evolved strategies to penetrate. This microbe-sparse zone is enforced, in part, by Reg-IIIγ 25. Because Reg-IIIγ is selectively bactericidal for Gram-positive bacteria, it is likely that AMPs specific for Gram-negative bacteria are also contributory. A similar spatial segregation of bacteria from the epithelium has been identified in the colon, where a thicker inner mucous layer excludes most bacteria despite the substantially higher bacterial load 22.
It is notable that the region of the intestinal tract that has the poorest coverage by the protective mucous layer — the proximal SI 22 — also has the greatest exposed epithelial surface area due to its prominent villous structure. While this favors digestion and absorption of nutrients it would also appear to make the epithelium in this region particularly vulnerable to entry of the microbiota and pathogens alike. Nevertheless, the bacterial loads here are the lowest along the length of the intestine, with over a million-fold fewer bacteria per unit of luminal contents than the LI. This is in part due to the more vigorous peristaltic motility of the upper SI, which rapidly clears material, including microbes, from the lumen. It is also because the lumen of the proximal SI, as the site for introduction of contents of the gall bladder and stomach, contains high levels of bile salts and acid that have anti-microbial effects.
In addition, however, the base of the crypts of the SI are home to large numbers of Paneth cells, IECs that are arrayed with a range of PRRs and are specialized for production and release of abundant AMPs, including α-defensins, which are small, highly cationic microbicides unique to this subset of IECs. In contrast to other AMPs such as Reg-IIIγ, the synthesis of which requires signals from the microbiota 26, α-defensins are synthesized and stored in Paneth cell granules without requirement for sensing of MAMPs. However, release of Paneth cell granules is induced by MAMPs, and secretion of active α-defensins has been shown to control the composition of the microbiota 27. NOD2, which was the first susceptibility gene linked to Crohn’s disease 28,29, is a NLR that is important in sensing the microbiota to control release of AMPs by Paneth cells. Collectively, then, Paneth cells respond to the microbiota via PRRs to produce abundant AMPs that regulate the microbiota composition and density in the SI, and their deficiency contributes to impaired homeostasis and heightened susceptibility to intestinal inflammation.
Colonic IECs also regulate the composition of microbiota through PPR-dependent mechanisms. Mice deficient in the NLRP6 inflammasome in colonic IECs demonstrate an altered microbiota that confer increased susceptibility to colitis due to injury of the colonic epithelium 30. Notably, the increased susceptibility to colitis that results from NLRP6 deficiency is transmissible to WT mice, indicating that the dysbiotic flora is contributory. How deficiency of the NLRP6 inflammasome results in an altered microbiota is incompletely undefined, although reduced IL-18 levels in NLRP6-deficient mice suggests an important role for this cytokine.
Reciprocity between the intestinal epithelium and innate immune cells
Although the intestinal mucous layer largely insulates the intestinal epithelium from direct interactions with the microbiota, bacterial metabolites and components are able to permeate this zone and alter gene expression in IECs via microbial PRRs 31. In addition to the epithelial products that are secreted apically to restrict contact with the microbiota (eg, mucins and AMPs), the epithelium also produces chemokines and cytokines, among other factors, that are secreted basolaterally to signal immune cells that reside internal to the epithelium, particularly in the intestinal lamina propria (Fig. 3). Because activation of PRRs typically promotes pro-inflammatory innate responses, the intestinal epithelium has had to evolve strategies by which these responses are mitigated in the case of commensals such that, at homeostasis, cytokine signals transmitted to mucosal immune cells limit inflammation. Although this remains an area of active investigation, several mechanisms have emerged.
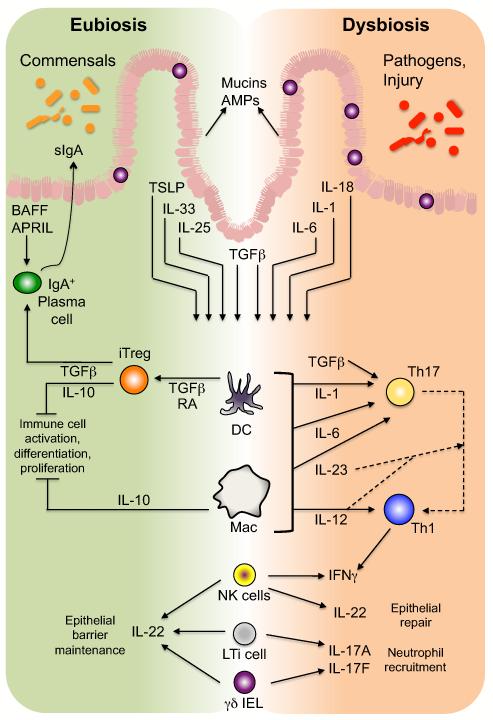
Figure 3. The epithelial-innate-adaptive continuum in the immune response to microbial antigens.
In response to the microbiota, IECs secrete mucins and AMPs that limit microbial interaction with epithelial cells. Also, under homeostatic, eubiotic conditions, epithelial stimulation by microbiota-derived antigens results in secretion of several cytokines (including TSLP, IL-33, IL-25, and TGFβ) that promote development of tolerogenic macrophages (MΦ) and DCs, which induce development of Treg cells via a TGFβ- and RA-dependent process. Intestinal Treg cells, through multiple mechanisms including secretion of TGFβ and IL-10, the latter of which is also produced by a subset of lamina propria macrophages, maintain an anti-inflammatory tone in the intestines by inhibiting or dampening potential effector responses. In addition, Treg cell-derived TGFβ promotes B cell antibody class-switching to IgA, which, coupled with the T-independent induction of IgA in isolated lymphoid follicles via epithelial-derived BAFF and APRIL, ensures an abundant supply of sIgA in the lumen, further limiting microbial interaction with the epithelium. Conversely, in the face of pathogen invasion or dysbiosis, intestinal DCs and macrophages are the targets of multiple IEC-derived pro-inflammatory cytokines and direct interactions with bacteria and/or their MAMPs that induce the development of effector CD4+ T cells, predominantly Th1 and Th17 cells, the latter of which can transition to the former. ILCs in the intestines, including NK-like cells and LTi cells, as well as γδ IELs, respond to cytokine signal derived from IECs and/or myeloid cells (DCs and macrophages) to upregulate cytokines similar to those of effector T cells: IFNγ, IL-17A, and IL-17F, as well as the epithelial barrier protectant, IL-22. Thus, innate cells of the intestinal immune system mediate the equilibrium between anti-inflammatory and pro-inflammatory signals induced by the microbiota under conditions of eubiosis versus dysbiosis, respectively.
The strategic distribution of TLRs and NLRs on and within IECs has considerable impact on whether bacterial MAMPs will be recognized, and if so, whether recognition promotes pro-inflammatory responses or represses them. Unlike pathogenic bacteria, which possess virulence factors that enable them to attach to and/or invade IECs, thereby introducing MAMPs into the IEC cytosol where they are recognized by NLRs, bacterial strains of the indigenous microbiota are non-invasive and therefore less potetnt activators of NLRs. Hence, in the intact epithelium, MAMPS derived from commensals are largely, albeit not absolutely, restricted to interactions with apically accessible TLRs, which appear to be functionally dampened by tonic exposure to the microbiota, whether through decreased expression, impaired signaling, or both.
In contrast, expression of TLRs on the basolateral aspects of IECs provides a means to signal a barrier breach by both commensals and pathogens. This is exemplified by TLR5, the receptor for flagellin, which is expressed on the basolateral aspects of the epithelium of the intestines where it can detect bacterial flagella, which are composed of repeated flagellin monomers 32. Detection of flagellin on the ‘host’ side of the epithelial barrier results in epithelial cell activation that initiates inflammatory responses by innate and adaptive immune cells aimed at eradicating the invading bacterium. The importance of flagellin as both an immune-activating MAMP and antigen targeted by adaptive immune cells is reflected in its high representation among antigens bound by serum antibodies from mice or patients with Crohn’s disease 33.
Apical expression of TLRs and restricted access to cytosolic NLRs represent adaptations to constrain interactions with luminal microbes, but some commensals have also evolved strategies that actively dampen TLR signaling in IECs should contact occur. Bacteroides and Lactobacillus spp. inhibit activation of the classical NF-κB pathway, which is central to induction of pro-inflammatory gene expression downstream of all PRRs. Active NF-κB dimers are sequestered in the cytosol by association with IκB complexes. Release of NF-κB for nuclear localization is contingent upon the phophorylation of IκB, by which it is targeted for ubiquitinylation and proteosomal degradation downstream of PRR signaling. Contact of IECs with these commensals inhibits IkB degradation, thereby blocking nuclear transport of NF-κB 34. Bacteroides spp. can further dampen NF-κB signaling by enhancing the nuclear export of NF-κB by inducing increased expression of the nuclear receptor family member, peroxisome-proliferation-activated receptor-γ (PPAR-γ). PPAR-γ binds NF-κB in the nucleus and shuttles it back to the cytoplasm, thereby terminating its transactivation of pro-inflammatory genes 35. Thus, in addition to host mechanisms that normally restrain direct contact between commensals and IECs, coadaptation of the microbiota and host have provided mechanisms that suppress pro-inflammatory engagement of PRRs in the intact epithelium.
An additional mechanism by which the epithelium supports tolerance of the microbiota is the conditioning of intestinal DCs through directional release of immunomodulatory cytokines into the lamina propria. At homeostasis, IECs secrete thymic stromal lymphopoietin (TSLP), IL-33, and IL-25, which promote tolerogenic activities of a subset of intestinal DCs defined by expression of integrin αEβ7 (CD103) 367 . Antigens presented by CD103+ DCs favor development of regulatory T cells and IgA+ plasma cells that home to the lamina propria where they repress inflammatory responses to the microbiota 37. IECs also produce abundant TGF-β, which suppresses NF-kB-dependent pro-inflammatory signaling in intestinal macrophages and DCs and promotes development and maintenance of regulatory T cells and IgA+ plasma cells at homeostasis. Finally, TLR-activated IECs can directly promote T-independent development of sIgA producing plasma cells in ILFs through release of B cell activating factor (BAFF) and B cell proliferation-inducing ligand (APRIL) 38. In addition to cytokines, IECs express other factors that limit pro-inflammatory responses of mucosal immune cells, including products of tryptophan metabolism and prostaglandins (eg, PGE2).
Like IECs, intestinal macrophages normally express lower levels of TLRs and are hyporesponsive to TLR signaling as a means to dampen inflammatory responses to the commensal microbiota, this despite their retention of highly active phagocytic and bactericidal activity towards commensals 39. As for IECs, this ‘inflammatory anergy’ is likely multifactorial, but also reflects the limited entry of commensal bacteria or their products into the cytosol where they can activate NLRs — despite their uptake and degradation in phagolysosomes. Interestingly, although phagocytosis of commensal bacteria induces pro-IL-1β in intestinal macrophages, its cleavage by caspase 1 to produce the active form requires actions of NLRP3- or NLRC4-containing inflammasomes, which are activated by MAMPs that are more effectively delivered into the cytosol by pathogenic, rather than commensal, bacteria 40. Therefore, phagocytes in the intestinal lamina propria are conditioned to mount inflammatory responses against pathogens that deliver MAMPs to cytosolic sensors, while efficiently clearing commensals that do not declare ‘danger.’
Innate lymphoid cells: rapid responders to epithelial barier breach
While LTi cells are indispensible for development of the GALT, they represent only one subset of a growing family of ILCs that have recently emerged as important contributors to intestinal homeostasis and barrier defense in the postnatal immune system (reviewed in refs. 41,42. It is now apparent that the diversity of ILC subsets parallels that of CD4+ T cells, with which they share many functional features. However, lacking antigen-specific receptors, ILCs are recruited to mucosal immune responses primarily by cytokines, which are produced by IECs or intestinal myeloid cells (DCs and macrophages).
ILCs are distributed in the intestinal lamina propria and GALT, where they are poised to respond rapidly to bacteria that penetrate epithelial defenses. Particularly important to barrier defense against bacterial incursions are LTi cells 43 and a subset of NK-like cells, referred to as NK-22, or ILC22 cells, which appear to share many features with LTi cells 44. Common to these cells, and to CD4+ T cells of the Th17 lineage discussed below, is their expression of the transcription factor, retinoic acid-related orphan receptor (ROR)γt, which is required for their development and function 9, and production of the cytokines IL-22 and/or IL-17A and IL-17F 45. IL-22 is a member of the IL-10 cytokine family that acts on epithelial cells of barrier tissues, including the intestine, to enhance anti-microbial defense and epithelial barrier integrity, whereas IL-17A and IL-17F target innate immune cells, stromal cells and epithelial cells to induce cytokines and chemokines (eg, G-CSF and CXCL8) that induce increased neutrophil production and recruitment, respectively.
RORγt+ ILCs appear to be the principal source of IL-22 at steady state and act in a continuous feedback loop with the microbiota, releasing IL-22 and/or IL-17 in response to microbiota-induced production of IL-23 by macrophages and DCs. These cytokines stimulate epithelial cell secretion of AMPs that inhibit or kill bacteria in the vicinity of the epithelial cell surface. Although ILCs can contribute to intestinal pathology under certain circumstances 46, they are critical in the early host response to enteropathogenic bacteria 41,47.
RORγt+ ILCs also express the aryl hydrocarbon receptor (AhR) by which they sense microbiota metabolites and xenobiotics 45,48; AhR signaling is required both for the maintenance and IL-22 production of these cells 48. Akin to its essential role in GALT development, ILC production of lymphotoxin provides an important amplifying loop for production of IL-22 by ILCs, stimulating dendritic cell production of IL-23 that, in turn, increases IL-22 production by the ILCs 49,50.
In addition to RORγt and AhR, another transcription factor shared between innate and adaptive immune cells that influences the composition of the microbiota is T-bet. A key transcription factor involved in Th1 cell development, T-bet is also expressed by innate immune cells. When maintained under pathogen-free conditions, recombinase-activating gene (RAG)-deficient mice, which lack B and T cells, are relatively resistant to inflammation driven by their microbiota despite absence of an adaptive immune system. This is due in large part to compensatory increases in ILCs. When deficient for T-bet, however, RAG-deficient mice develop spontaneous intestinal inflammation resembling ulcerative colitis (so-called, TRUC mice) 51. Disease results both from overexpression of TNFα by DCs and an absence of regulatory T cells. Remarkably, transfer of the microbiota from these mice to immunocompetent mice transfers disease, reflecting the outgrowth of two pathogenic bacterial strains: Klebsiella pneumoniae and Proteus mirabilis. Although details of the mechansims responsible for T-bet-dependent restraint of the microbiota remain to be defined, these results highlight the importance innate immune cells in regulating the composition of the microbiota. They also provide evidence that a dysbiotic flora can drive disease in otherwise normal hosts.
Adaptive immunity in homeostatic responses to the microbiota
The interplay between the microbiota, intestinal epithelium and innate and adaptive immune cells at homeostasis favors dominance of regulatory networks that prevent inflammation or immune-mediated disease. Remarkably, the innate defenses are sufficiently robust that much of the host response to the microbiota progresses without involvement of CD4+ T cell-dependent responses. Further, a portion of the sIgA response that contributes to partitioning of commensal organisms away from the epithelium is generated in ILFs without requirement for T cell help 13. And when the adaptive immune response is recruited, it is typically limited to the mucosal tissues such that systemic immunity is not generated. This is due to programming of adaptive immune cells to traffic back to the mucosae following their differentiation in PPs or MLNs. Finally, at steady state antigen presentation by intestinal dendritic cells favors the development of regulatory CD4+ T cells, which act to suppress the development of pro-inflammatroy innate and effector T cell responses to avert excessive inflammation.
Regulatory T cell responses to the microbiota
The function of CD4+ regulatory T cells is essential to maintain mutualism with the microbiota. The frequencies of these cells are considerably elevated in the intestine relative to other tissue sites 52, and microbiota-induced regulatory T cells impact the composition of the microbiota 53. The critical role of regulatory T cells in immune homeostasis to the microbiota is well documented by the consequences of their absence 54. Regulatory T cell deficiency, irrespective of the means by which it is induced, results in unopposed effector T cell responses and IBD driven by reactivity to antigens of the microbiota. Indeed, much of our understanding of the inflammatory potential of the commensal microbiota has been garnered through models in which regulatory pathways have been disrupted 54. The largely non-overlapping TCR specificities of regulatory and effector T cell subsets in the colonic lamina propria and associated lymphoid tissues suggest that distinct antigenic epitopes, whether derived from the same or distinct members of the microbiota, control these competing T cell fates 55.
Regulatory CD4+ T cells include subsets that are distinguished on the basis of their expression of the transcription factor Foxp3. Foxp3+ (Treg cells) or Foxp3− (T regulatory type 1, or Tr1 cells) are each characterized by their production of IL-10, a major immunoregulatory cytokine required for immune tolerance of the intestinal microbiota. The non-redundant role of IL-10 in intestinal immune homeostasis is well established, as IL-10-deficient mice develop spontaneous, unremitting colonic inflammation driven by the IL-23 and the Th17 pathway 56,57. The essential role of TLR sensing of the microbiota in this process is evidenced by absence of disease in germ-free IL-10-deficient mice and mice deficient for both IL-10 and MyD88 58,59. Although non-T cells produce IL-10, T cell-derived IL-10 is critical for intestinal immune homeostasis. Thus, IL-10 deficiency restricted to total CD4+ T cells results in spontaneous colitis that is comparable in severity to that in mice with global loss of IL-10 60. While deficiency of IL-10 limited to Foxp3+ Treg cells also induces colitis, its severity is reduced, indicating that Foxp3− CD4+ T cells contribute to the protective IL-10 response.
Myeloid cells of the innate immune system are primary targets for regulatory actions of IL-10 via 61. IL-10 signaling in CD4+ T cells also appears contributory, albeit less pronounced 62,63. Importantly, polyphormisms in the IL10 locus confer susceptibility to IBD in humans 64, and a subset of patients with early-onset IBD have been linked to mutations in IL-10 receptor components that impair signaling 65.
Remarkably, IL-10 producing T cells can be induced to develop in response to specific commensals or their products. The capsular polysaccharide A (PSA) moiety of the common commensal Bacteroides fragilis mediates interaction of B. fragilis with the colonic mucosa, enabling its colonization and activation of an anti-inflammatory cascade 66. PSA promotes expansion of colonic IL-10-expressing Foxp3+ Treg cells, acting via the TLR2/MyD88 pathway 66. The expression of TLR2 on CD4 T cells suggests the capacity for direct regulation by microbial products that favors PSA-dependent Treg cell over Th1 responses 67, although there are other known TLR2 ligands that do not promote induction of Foxp3 or IL-10 66.
A cocktail of Clostridia species, comprised primarily of clusters IV and XIVa, was shown to drive efficient expansion of Treg cells in the colons of germ-free mice, seemingly due to their superior ability to elicit TGFβ production in the intestinal mucosa 68. In contrast to induction of Treg cells by B. fragilis, Clostridium-dependent Treg expansion occurs independently of MyD88 via mechanisms yet to be defined 68. To date, studies utilizing otherwise ‘complete’ microbiota, that are devoid of B. fragilis or this specific collection of Clostridia spp. are lacking, leaving open the issue of the level of redundancy in this process. However, B. fragilis is absent from the microbiota of 20-30% of humans, indicating the existence of other pathways to Treg cell development. To this point, default induction/expansion of Treg cells can be achieved by colonization of germ-free mice with a defined altered Schaedler’s flora (ASF) that consists of eight species of the microbiota dominated by B. distasonis, but devoid of B. fragilis 69. Collectively, then, while there appears to be a proclivity of certain components of the microbiota to promote Treg cell development, this does not appear to a unique property of any one microbial component, reflecting substantial redundancy in the system to ensure that Treg cell dominance is established.
Effector T cell responses to the microbiota
Like other components of the adaptive immune system (ie, Treg cells and B cells), the largest deployment of effector CD4+ T cells in the normal immune system is in the intestines. While the intestines mount robust Th2 responses to infestations by helminthes, at least in industrialized countries, where these infections are less common, the major effector T cell subsets resident in the gut express cytokines characteristic of Th17 and Th1 cells. And like Treg cells and B cells, the development of these effector subsets is largely dependent on the microbiota.
Th17 cells in particular, thought to be the most ancient of effector T cell subsets 4, appear to have evolved to bolster mucosal barrier defenses as a means to promote mutualism with the microbiota. Th17 cells share with intestinal ILCs developmental requirements for RORγt and AhR 70, as well as a similar effector cytokine repertoire, including IL-17A, IL-17F and IL-22. Further, Th17 cells share developmental ties to Tregs through their mutual requirement for the abundant intestinal cytokine TGFβ 4. Finally, the Th17 developmental pathway is distinguished by considerable plasticity 71, enabling divergent functional programs contingent on local pro- or anti-inflammatory cues. Collectively, then, although a last line of defense, effector CD4+ T cells are perpetually engaged in the host-microbiota dialogue, favoring regulatory benefits at homeostasis, or pathologic consequences when dysregulated.
The contribution of the microbiota in intestinal Th17 cell development is highlighted by their virtual absence in germ-free mice 72. In addition to IL-6, which is required for Th17 differentiation and its deviation away from iTreg programming, multiple microbiota-dependent factors favor Th17 development in the intestines, including, in addition to TGFβ, IL-1β 73, IL-23 74 and even ATP derived from commensal bacteria 75. Similar to the ability of a limited quorum of commensal bacterial species to disproportionately influence iTreg cell development, minor constituents of the microbiota can amplify intestinal Th17 cell numbers. In mice, an unusually potent inducer of Th17 cells, albeit not unique 69, is the Clostridial spp., Candidatus arthromatus, or segmented filamentous bacteria (SFB) 72,76, which populates the ileum and cecum and has long been known to be a potent activator of intestinal immune responses 77. Induction of Th17 cells by SFB provides protection against gut pathogens 78, suggesting that general amplification of Th17 effector cells can be host-protective. However, Th17 induction by SFB is not entirely benign, as mono-association of mice with SFB induces Th17-mediated inflammatory arthritis 79 and multiple sclerosis (MS)-like symptoms in the experimental autoimmune encephalomyelitis (EAE) model 80. Remarkably, therefore, sensing of a single constituent of the intestinal microbiota can promote autoimmunity in extraintestinal tissues. At present, it is unclear whether SFB, or related organisms, exert similar effects in humans.
Whether at steady state, in response to infection, or in the context of chronic inflammation induced by IBD, the intestine contains in addition to effectors that express IL-17, T cells that express IFNγ, or both IL-17 and IFNγ. In this regard it is notable that Th17 cells retain a high capacity for divergent cytokine expression profiles and function, or plasticity, following their commitment to the Th17 pathway. Specifically, Th17 cells can give rise to IFNγ producers that resemble classical Th1 cells. This has been demonstrated for both human and mouse 71,81, and has raised the possibility that many of the ‘Th1’ cells found in the intestine arise from the Th17 pathway (Fig. 3). The relative contributions of Th17 and Th1 cells to immune protection or pathogenesis remain to be elucidated, whether derived from the Th17 pathway or not.
Balancing regulatory and effector responses to the microbiota
To balance accommodation of the microbiota with the necessity to mount host defenses against microbial invasion, the adaptive immune system has evolved to direct development of distinct CD4+ T cell fates by different APC subsets and the microbe-induced factors they produce. While Treg cell dominance is the default at homeostasis, the shared dependence of iTreg and Th17 cell development on TGFβ provides an elegant means to alternately divert programming of naïve CD4+ T cells from a homeostatic, microbe-tolerant response to an inflammatory, microbe-clearing response contingent on the co-factors integrated with TGFβ signaling 4 (Fig. 3).
At homeostasis, naïve CD4+ T cell recognition of antigens derived from the microbiota favors iTreg cell development by virtue of the production by intestinal DCs of the vitamin A metabolite, all-trans retinoic acid (RA) 82-84. Because vitamin A is not synthesized by the host, its influence on the regulatory tone of the mucosal immune response is predicated on adequate dietary intake. RA is derived from vitamin A by the sequential actions of the ubiquitous enzyme, alcohol dehydrogenases (ADH), and one of three retinal-specific dehydrogenases (RALDHs), the latter of which are more restricted in their tissue distribution, including CD103+ DCs and IECs in the intestine. The potency of RA as a co-factor for TGFβ-dependent development of iTreg cells — and suppression of Th17 development — and its constitutive production by a regulatory subset of intestinal DCs at steady state reflect the robust default pathway to T cell-mediated regulation. Under conditions where microbial antigens promote production of pro-inflammatory cytokines by DCs, RA is either repressed 85 or co-opted by pro-inflammatory pathways 86,87 to override Treg cell induction.
In common with Th17 cells, and perhaps reflecting an aspect of shared TGFβ-dependent programming that favors multipotency, iTreg cells display developmental plasticity that influences adaptive immunity to the microbiota. When acted on by pro-inflammatory cytokines, iTreg cells can down-regulate Foxp3 and up-regulate RORγt 88, and Treg cells can be converted into IL-17- or IFNγ-expressing effectors during infection 89. Notably, while Foxp3+ Treg cells may be diverted to effector responses, the reverse is less apparent; established effector T cells, including Th17 cells, appear resistant to reprogramming to become Foxp3+ regulatory cells. Accordingly, microbial antigens that are identified as products of a ‘pathogen’ are likely to be remembered as such through their imprinting of an effector T cell response. In contrast, microbial antigens that promote iTreg responses at homeostasis, may, in the context of an inflammatory response, reprogram iTregs to generate an effector T cell response that breaks ‘tolerance’ to that microbe.
In a further example of their developmental plasticity, recent studies report a role for Tregs in IgA class-switch recombination (CSR) in the gut, representing another arm of Treg cell function in promoting immune homeostasis to the microbiota. TGFβ has long been recognized as the principal switch factor for development of IgA-producing B cells. In a new twist on TGFβ’s role in this process, it was found that iTreg cells could down-modulate Foxp3 and acquire features of follicular helper T cells (Tfh) that promoted IgA production in PPs 90. Depletion of Treg cells also caused a rapid loss of IgA+ plasma cells and sIgA production by the intestine 91, indicating that Treg cells participate both in the development and maintenance of intestinal IgA+ B cells. While known to be important in host defense against infections, the primary role of sIgA is establishment of mutualism with the intestinal microbiota, as evidenced by its marked depletion in GF mice and systemic IgG responses to the microbiota in animals specifically lacking sIgA. Coupled with the fact that ~75% of sIgA reactive to the microbiota develops via the T cell-dependent pathway 91,92, this would suggest that sIgA is mainly regulated by intestinal Treg cells. In addition to providing evidence of further plasticity in the Treg developmental program, these findings identify an additional link between adaptive immune networks that cope with the commensal flora, and extend the ancient role of TGFβ in promoting host-microbiota mutualism 4.
Dysregulated immunity to the microbiota; IBD and beyond
Given their defined antigenic specificity and durable memory, effector CD4+ T cells are a liability when inappropriately directed against self-antigens or, in the case of the microbiota, the ‘extended’ self. In view of the enormous number of antigens expressed in the intestinal metgenome, it is remarkable that dysregulated effector responses against the microbiota are the exception. When they do occur, the result is inflammatory bowel disease, including Crohn’s disease (CD) or ulcerative colitis (UC), which have distinct clinical and histopathologic features, but a common requirement for the intestinal microbiota. As in autoimmunity directed against host self-antigens, dyregulated CD4+ T cell responses to antigens of the microbiota lead to chronic, typically relapsing and remitting disease that reflects the immune system’s inability to eliminate the antigens that drive the abnormal response.
That being said, inflammatory immune responses in the intestines result in readily detectable alterations of the resident microbiota, irrespective of causative agent 93. This presents a challenge in our attempts to understand how certain microbes positively or negatively affect the disease process in IBD since it is difficult to draw conclusions without the knowledge of the compostion of the microbiota pre-diagnosis. As such, our understanding of the antigenic components of the microbiota that are targets of immune attack in IBD are currently limited. However, studies from experimental mouse models and patients with CD reveal a remarkably shared reactivity to specific flagellin epitopes expressed by Clostridium spp., which appear to be immunodominant despite their minor representation within the commensal flora 33,94. What attributes of these commensals make them particularly common targets in a large subset of patients with CD — but not UC — are unknown. It is likely to reflect unique properties that place them in specific geographical niches in the microbiota and/or in intimate contact with the immune system where their expression of flagella, which is atypical for commensal bacteria, make them unusually proficient at inducing effector rather than regulatory responses. Irrespective of the basis, retention of these organisms in the collective despite the pathogenic risk they pose implies a benefit to host-microbiota mutualism that has been evolutionarily conserved and remains to be understood.
In view of its prominent role in adaptive immunity to the intestinal microbiota and propensity for generating inflammation promoted by both IL-17 and IFNγ, it is not surprising that the Th17 pathway has emerged as leading contributors to IBD pathogenesis. Whereas prior to the discovery of Th17, CD and UC were viewed in the context Th1- and Th2-centric mechanisms, respectively, increasingly data from genome-wide association studies (GWAS) implicate the contributions of the Th17 pathway in both disorders 95. Indeed, it is fitting that IL-23, discovery of which revolutionized views on the immunopathogenesis of autoimmunity and led to discovery of Th17 cells 96, was the first cytokine linked to IBD by GWAS, as variants of its receptor (IL-23R) have been found to confer both protection and susceptibility to CD and UC 95.
As the number of GWAS and next-gen sequencing studies has proliferated, so too has the number of genes linked to the Th17 pathway in IBD 95. This has been supplemented by genes of innate and adaptive immune pathways that are integrated with, and control, Th17 responses, spanning the gamut from epithelial barrier integrity maintenance and restitution to microbial sensing to immunomodulatory cytokines. Because individual susceptibility alleles confer minor risk, multiple genetic susceptibility alleles in individuals are required for disease. Defining this clustering of risk alleles is increasingly defining host genotypes that confer substantial risk, while also revealing interlaced susceptibility pathways that identify immunopathogenetic subsets of IBD and offering an explanation why early clinical trials targeting this pathway have met with mixed results. And just as these genotypes are being found to overlap with and reveal common links to extraintestinal immune-mediated diseases that share a Th17 pathogenesis, they also increasingly implicate a role for the microbiota in conferring disease risk beyond the confines of the gut. As we advance into the era of integrated metagenomics of the human-microbiota ‘superorganism,’ the opportunities for personalized medicine originally envisioned from sequencing of the human genome may well multiply in proportion to the increase in genes contributed by our microbiota, with attendant opportunities for more specifically targeted treatments and prevention of diseases that have their origins in the merger of vertebrates and their microbiomes millions of years ago.
Text Box 2. Modulation of the microbiota by intestinal blood group antigens.
An important aspect of the microbiota’s utilization of the outer mucus layer as an ecological niche is its decoration by blood group antigens. Analogous to blood group antigens on erythrocytes, the assembly of type A, B, or Lewis b glycans on intestinal mucins is contingent on generation of the core H glycan by the actions of fucosyltransferase 2. The gene encoding this glycosyltransferase, FUT2, is functional in most individuals (‘secretor’ genotype), but is non-functional in a significant minority (~20% of Caucasians, referred to as ‘non-secretors’) due to a missense mutation. Recent studies have found that the presence or absence of a functional FUT2 allele correlates strongly with composition of the microbiota 97. In particular, Bifidobacteria, which are a beneficial component of the microbiota, are dependent on the terminal blood group glycans for colonization of the intestinal mucus and are less abundant in non-secretor genotypes. Because the non-secretor phenotype is associated with necrotizing enterocolitis and gram-negative sepsis in premature infants 98 as well as Crohn’s disease 99, it would appear that the protective benefits of colonization by Bifidobacter spp. represent an mutually beneficial evolutionary adaptation of this commensal to its host. Given the predisposition to dybiosis and its deleterious health effects in non-secretors, it is likely that evolutionary pressure to retain the defective FUT2 allele in the population reflects the fact that mucin-linked blood group glycans also serve as sites of attachment for mucosal viruses (eg, norovirus), such that non-secretors are protected 100.
Acknowledgments
The authors thank David Randolph and Casey Morrow and for helpful discussions and/or critical review of this manuscript. CTW and COE are supported by grants from the National Institutes of Health, and CTW and CLM are supported by grants from the Crohn’s and Colitis Foundation of America. The authors extend their apologies to colleagues whose work could not be adequately acknowledged due to space limitations.
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 4.Weaver CT, Hatton RD. Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nature Reviews Immunology. 2009;9:883–889. doi: 10.1038/nri2660. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y-A, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nature Immunology. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clinical and Experimental Allergy. 2008;38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 8.Palmer C, Bik E, Digiulio D, Relman D, Brown P. Development of the Human Infant Intestinal Microbiota. PLoS Biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherrier M, Eberl G. The development of LTi cells. Current Opinion in Immunology. 2012;24:178–183. doi: 10.1016/j.coi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Mebius RE, Streeter PR, Michie S, Butcher EC, Weissman IL. A developmental switch in lymphocyte homing receptor and endothelial vascular addressin expression regulates lymphocyte homing and permits CD4+ CD3− cells to colonize lymph nodes. Proc Natl Acad Sci USA. 1996;93:11019–11024. doi: 10.1073/pnas.93.20.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanamori Y, et al. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+IL-7R+Thy1+ lympho-hemopoietic progenitors develop. J Exp Med. 1996;184:1449–1459. doi: 10.1084/jem.184.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 13.Tsuji M, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Lane PJL, et al. Lymphoid tissue inducer cells: bridges between the ancient innate and the modern adaptive immune systems. Mucosal Immunology. 2009;2:472–477. doi: 10.1038/mi.2009.111. [DOI] [PubMed] [Google Scholar]
- 15.Lotz M, et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismail AS, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci USA. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen CH, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7:e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012 doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersson J, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. American Journal of Physiology Gastrointestinal and Liver Physiology. 2011;300:G327–333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergstrom KSB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS pathogens. 2010;6 doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 24.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature Immunology. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 29.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411 doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 30.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Vijay-Kumar M, Aitken JD, Gewirtz AT. Toll like receptor-5: protecting the gut from enteric microbes. Seminars in Immunopathology. 2008;30:11–21. doi: 10.1007/s00281-007-0100-5. [DOI] [PubMed] [Google Scholar]
- 33.Lodes MJ, et al. Bacterial flagellin is a dominant antigen in Crohn disease. The J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neish AS, et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 35.Kelly D, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nature Immunology. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 36.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annual Review of Immunology. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rescigno M, Di Sabatino A. Dendritic cells in intestinal homeostasis and disease. J Clin Invest. 2009;119:2441–2450. doi: 10.1172/JCI39134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Smythies LE, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franchi L, et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nature Immunology. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nature Immunology. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 43.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. 9. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cella M, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takatori H, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nature Immunology. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 48.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ota N, et al. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nature Immunology. 2011;12 doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- 50.Tumanov AV, et al. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host and Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host and Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maynard CL, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 53.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annual Review of Immunology. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 55.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. 34. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 57.Yen D, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infection and immunity. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25:319–329. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Roers A, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–1297. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeda K, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 62.Chaudhry A, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huber S, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34:554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franke A, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nature Genetics. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 65.Glocker E-O, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. New England Journal of Medicine. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, et al. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J Exp Med. 2006;203:2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 70.Sawa S, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature Immunology. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 71.Lee YK, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanov, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host and Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaw MH, Kamada N, Kim Y-G, Núñez G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaph C, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atarashi K, et al. ATP drives lamina propria TH17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 76.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 77.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infection and Immunity. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu H-J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Annunziato F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204 doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 85.Maynard CL, et al. Contrasting roles for all-trans retinoic acid in TGF-β-mediated induction of Foxp3 and Il10 genes in developing regulatory T cells. J Exp Med. 2009;206:343–357. doi: 10.1084/jem.20080950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hall J, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DePaolo RW, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Current Opinion in Immunology. 2009;21:274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 89.Wohlfert E, Belkaid Y. Plasticity of Treg at infected sites. Mucosal Immunology. 2010;3 doi: 10.1038/mi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 91.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 93.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host and Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 94.Targan SR, et al. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn’s disease. Gastroenterology. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 95.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annual Review of Immunology. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 97.Wacklin P, et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morrow AL, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J Pediatr. 2011;158:745–751. doi: 10.1016/j.jpeds.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McGovern DPB, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Human Molecular Genetics. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rydell GE, Kindberg E, Larson G, Svensson L. Susceptibility to winter vomiting disease: a sweet matter. Reviews in Medical Virology. 2011;21:370–382. doi: 10.1002/rmv.704. [DOI] [PubMed] [Google Scholar]