Abstract
The human epidermal growth factor receptor (HER) tyrosine kinases homo- and hetero-dimerize to activate downstream signaling pathways. HER3 is a catalytically impaired member of the HER family that contributes to the development of several human malignancies and is mutated in a subset of cancers. HER3 signaling depends on heterodimerization with a catalytically active partner, in particular EGFR (the founding family member, also known as HER1) or HER2. The activity of homodimeric complexes of catalytically active HER family members depends on allosteric activation between the two kinase domains. To determine the structural basis for HER3 signaling through heterodimerization with a catalytically active HER receptor, we solved the crystal structure of the heterodimeric complex formed by the isolated kinase domains of EGFR and HER3. The structure visualized HER3 as an allosteric activator of EGFR and revealed a conserved role of the allosteric mechanism in activation of HER family members through heterodimerization. To understand the effects of cancer-associated HER3 mutations at the molecular level, we solved the structures of two HER3 kinase mutants, each in a heterodimeric complex with the kinase domain of EGFR. These structures, combined with biochemical analysis and molecular dynamics simulations, indicated that the cancer-associated HER3 mutations enhanced the allosteric potential of HER3 by redesigning local interactions at the dimerization interface.
Introduction
Receptor tyrosine kinases play a major role in the process of converting diverse extracellular cues into the activation of distinct intracellular signaling networks. The activity of the human epidermal growth factor receptors (HERs) controls indispensable signaling processes in both the developing and adult organism through the stimulation of pathways that regulate cellular proliferation, survival, and motility (1). The human genome contains four HER genes, encoding the founding member EGFR (also known as HER1 or ERBB1), HER2 (also known as ERBB2), HER3 (also known as ERBB3), and HER4 (also known as ERBB4). Activation of the HER family of receptors occurs upon binding of extracellular ligands that promote receptor oligomerization and catalytic activation of the intracellular kinase domains. Subsequent phosphorylation of intracellular regions of the receptors triggers recruitment and activation of downstream components, initiating signaling cascades. A feature that enables the HER family of receptors to control diverse signaling outputs is their ability to utilize their phosphorylation sites in a combinatorial manner through receptor heterodimerization. The combined outputs from specific pairs of HER family members are important for many fundamental biological processes, including heart function (2, 3), the proliferation of Schwann cells (4), and neurogenesis (5).
The formation of heterodimeric complexes is particularly relevant for signaling by HER3. The kinase domain of HER3 contains several inactivating substitutions that result in an almost complete loss of catalytic activity, and HER3 is thus denoted as a pseudokinase receptor. Consequently, HER3 has not been demonstrated to signal as a homodimer, but rather to efficiently dimerize with and become phosphorylated by its heterodimerization partners, primarily EGFR and HER2 (6-8). Signaling pathways activated by heterodimeric complexes containing HER3 are essential for proper embryogenesis and development (1, 9). Heterodimerization of HER3 with EGFR or HER2 also plays a role in oncogenic signaling by the HER family (10-16), and contributes to cellular mechanisms that cause resistance to cancer therapeutics targeting EGFR and HER2 (13, 17, 18). The design of next-generation inhibitors that could overcome this developed resistance is now focused on directly targeting HER3 or HER3-containing heterodimers (15).
Cancer genomics studies have identified the first HER3 mutations in primary human tumors (19-21). Interestingly, the missense mutations located in the catalytically inactive HER3 kinase domain are the strongest gain-of-function substitutions (19). However, signaling by these HER3 mutants still requires the presence of a catalytically active HER partner, suggesting that these mutations enhance signaling only when HER3 forms heterodimeric complexes. The molecular mechanism behind the activating effect of HER3 mutations has not been described, but the identification of cancer mutations in this critically important receptor underscores the need to understand how HER3 forms active complexes with other members of the HER family under both normal and oncogenic conditions.
Current structural understanding of the molecular mechanism underlying catalytic activation of HER family of receptors is informed by studies of homodimeric complexes, formed by the catalytically active HERs, primarily EGFR and HER4 (22, 23). Catalytic activation of these complexes relies on the formation of an asymmetric head-to-tail dimer between the two kinase domains (22). In this dimer, the kinase domain of one receptor (the “activator” kinase) allosterically activates the kinase domain of the dimerization partner (the “receiver” kinase). This mechanism effectively yields one activated kinase molecule per receptor dimer (the receiver), which is thought to phosphorylate tyrosine residues in both receptor subunits’ cytoplasmic tails. In this arrangement, the allosteric activator kinase does not require catalytic activity. Biochemical studies indicate that HER3 functions as the allosteric activator kinase in dimers with other HERs (24) and is therefore capable of functionally participating in the active heterodimeric complex as both upstream activator and downstream substrate.
Here, we solved the crystal structure of an active complex formed by the isolated kinase domains of HER3 and EGFR. This structure illustrated how HER family of receptors utilizes conserved binding interfaces to enable combinatorial signaling in response to extracellular cues. We also report crystal structures of two heterodimeric complexes formed by the kinase domain of EGFR and mutant kinase domains of HER3 identified in cancer patients. We leveraged these structural insights into HER3-containing heterodimers to mechanistically explain how HER3 cancer mutations increase HER3-dependent signaling. These studies revealed a mechanism for the aberrant activation of members of the HER family in cancer through the enhancement of their noncatalytic allosteric “activator” function. These results emphasize the utility of specific inhibition of the allosteric function of HERs as an approach to expand the available pool of HER-targeted therapeutics.
Results
The structure of the EGFR/HER3 kinase domain heterodimer visualizes HER3 as an allosteric activator of EGFR
To elucidate the structural basis for the heterodimeric interactions that underlie signaling by receptor complexes containing HER3, we pursued a crystal structure of the heterodimeric complex of the kinase domains of EGFR and HER3. Both of these isolated kinase domains are well-behaved monomers and each has been well characterized previously through X-ray crystallography (22, 24). To promote formation of EGFR/HER3 heterodimers, we utilized the EGFR-V924R mutant which prevents EGFR from functioning as an allosteric activator kinase, thereby limiting its function to that of the receiver kinase within the active asymmetric dimer (22). We anticipated that this property would promote specific interaction of the EGFR kinase domain with the HER3 kinase domain, which is predicted to preferentially occupy the activator position in heterodimeric complexes (22). In addition, we designed the EGFR and HER3 constructs to include different fragments of the juxtamembrane (JM) segment, a region immediately N-terminal to the kinase domain that increases the dimerization affinity between EGFR monomers (25, 26). On the basis of sequence conservation within this region, we hypothesized that JM segment-mediated interactions should also promote heterodimerization between EGFR and HER3.
Initial screening, performed with equimolar concentrations of EGFR and HER3 constructs, yielded several conditions that produced crystals of the EGFR-V924R construct alone. In these crystal lattices, EGFR-V924R formed an inactive, symmetric dimer mediated by the C-terminal tail fragments present in the crystallization construct that encompassed residues 960-998. This dimer was previously reported as an autoinhibited EGFR complex (fig. S1A) (26). We hypothesized that the presence of EGFR-V924R nucleation centers arising from this autoinhibited symmetric dimer might have prevented the growth of crystals of the active asymmetric complex between the EGFR and HER3 kinase domains. To favor the specific interaction between EGFR and HER3, we introduced a double mutation into the C-terminal tail region of the EGFR constructs (F973A/L977A) which is predicted to disrupt a dimerization interface that stabilizes this autoinhibited, symmetric dimer. The EGFR-V924R/F973A/L977A variant retained full function as a receiver kinase when reconstituted with HER3 in an in vitro kinase assay (fig. S1B). Combining the EGFR construct containing these substitutions and the C-terminal half of the JM segment (residues 658-998) with a HER3 construct limited to the pseudokinase domain (residues 674-1001) yielded crystals of the heterodimeric complex (fig. S1C). We biochemically confirmed that the crystals contained both EGFR and HER3 subunits by silver staining an SDS-PAGE sample of an isolated crystal (fig. S1D). These crystals diffracted X-rays to 3.1-Å resolution and contained an active asymmetric dimer formed between the EGFR and HER3 kinases (Fig. 1A, Table 1).
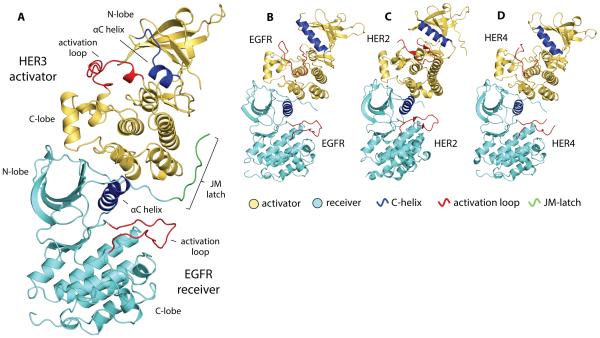
Figure 1.
Crystal structure of the EGFR receiver/HER3 activator kinase domain heterodimer compared to homodimeric complexes of HER kinase domains. (A) EGFR/HER3 heterodimer (PDB ID: 4RIW), (B) EGFR/EGFR homodimer (PDB ID: 2GS6), (C) HER2/HER2 homodimer (PDB ID: 3PP0), and (D) HER4/HER4 homodimer (PDB ID: 3BCE). All structures are aligned on the N-lobe of the receiver kinase domain.
Table 1.
Summary of crystallographic data and refinement. No., number
| Data collection | EGFR/HER3 | EGFR/HER3-Q790R | EGFR/HER3-E909G |
|---|---|---|---|
| Space group | P 1 21 1 | P 1 21 1 | P 1 21 1 |
| Unit cell dimensions | |||
| a, b, c (Å) | a = 65.67 b = 154.52 c = 86.98 |
a = 64.65 b = 155.05 c = 86.86 |
a = 65.33 b = 155.14 c = 87.30 |
| α, β, γ (°) | A = 90.00 β = 110.94 γ = 90.00 |
A = 90.00 β = 111.09 γ = 90.00 |
A = 90.00 β = 110.93 γ = 90.00 |
| Resolution, Å | 60.48-3.10 (3.27- 3.10) |
59.77-3.10 (3.27- 3.10) |
60.00-3.00 (3.21- 3.00) |
| Rmerge | 0.191 (0.602) | 0.173 (0.587) | 0.176 (0.573) |
| I/σ(I) | 5.2 (2.0) | 6.9 (2.4) | 6.5 (2.7) |
| Completeness (%) | 97.6 (99.4) | 95.4 (96.8) | 99.5 (99.9) |
| Multiplicity | 2.7 (2.7) | 1.7 (1.7) | 3.8 (3.9) |
| Refinement | |||
| Resolution, Å | 60.48-3.10 | 59.77-3.10 | 56.78-3.00 |
| No. unique reflections |
28,729 | 27,509 | 32,340 |
| Rwork/Rfree | 22.7/28.7 | 20.7/25.8 | 21.6/26.7 |
| No. atoms | |||
| Protein | 9,023 | 9,027 | 9,002 |
| Ligand | 120 | 120 | 120 |
| B-factors, mean (Å2) | |||
| Protein | 41.2 | 60.0 | 45.8 |
| Ligand | 33.4 | 85.2 | 58.9 |
| RMS deviations | |||
| Bond lengths (Å) | 0.005 | 0.006 | 0.005 |
| Bond angles (°) | 0.982 | 1.206 | 1.093 |
The overall organization of the dimer formed between the EGFR and HER3 kinase domains (Fig. 1A) is similar to those observed in the EGFR/EGFR (Fig. 1B) (22), HER2/HER2 (Fig. 1C) (27), and HER4/HER4 (Fig. 1D) (23) homodimer structures . In all previously solved structures of active HER homodimers, the packing of molecules in the crystal lattice results in a daisy chain of asymmetric dimers in which every receiver kinase simultaneously functions as the allosteric activator for the next molecule in the chain. These interactions stabilize an active conformation in all kinase molecules, eliminating an unbiased insight into the conformation of the activator kinase within the asymmetric dimer. In the structure of the EGFR/HER3 complex, daisy chaining is prevented due to the inherent inability of the catalytically impaired HER3 to take the receiver position (24). In the EGFR/HER3 heterodimer, the HER3 pseudokinase domain is in an inactive conformation known as the “Src/CDK-like” inactive state (28, 29) (Fig. 1A). In this conformation, the αC helix is swung away from the active site and the N-terminal portion of the activation loop is folded into a single helical turn. In addition, as observed in previous structures of the isolated HER3 pseudokinase domain (24, 30), the αC helix in HER3 is markedly shortened in comparison to that of EGFR.
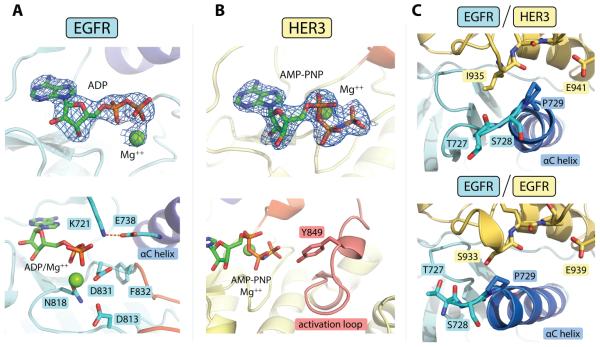
Despite large structural differences in the conformation of the N-lobe of HER3 in comparison to the conformation of the activator kinases in previously solved asymmetric homodimer structures (fig. S2A, B), the C-lobe of HER3 forms contacts with the EGFR N-lobe that are analogous to the contacts visualized in homodimeric structures. The C-lobe of the HER3 kinase domain rests on the N-lobe of the EGFR kinase domain, stabilizing a canonical active conformation of EGFR in which the catalytically important αC helix is swung toward the active site, allowing the conserved glutamate (Glu738) to form a salt bridge with the catalytic lysine (Lys721) at a favorable distance of 2.9 Å (Fig. 2A). We found that the chemically stabilized ATP analog (AMP-PNP), which we included in our crystallization mix, had been hydrolyzed to its diphosphate form within the EGFR active site (Fig. 2A). Hydrolysis of AMP-PNP has been observed in previous crystal structures of active ATPases (31, 32), and its occurrence in the crystals of the EGFR/HER3 complex further emphasizes the active catalytic state of EGFR. In contrast to EGFR, the intact AMP-PNP molecule was observed in the HER3 nucleotide-binding site (Fig. 2B).
Figure 2.
Detailed structural features of the EGFR/HER3 heterodimer. (A) Top: electron density for the ADP/Mg++ complex observed in the EGFR active site (at 3.0 σ above the mean value). The difference electron density maps shown in all panels were calculated using a model of the protein at a refinement stage before the inclusion of the nucleotide. Bottom: detailed view of the EGFR active site, showing an optimal catalytic center. (B) Top: Electron density for the AMP-PNP/Mg++ complex bound to the HER3 nucleotide-binding site (3.0 σ). Bottom: Stable conformation of the HER3 activation loop enables visualization of Tyr849 in one of the HER3 subunits in the asymmetric unit. (C) Conformational variability observed at the N-terminus of the αC helix in the EGFR/HER3 (top) and EGFR/EGFR (bottom, PDB ID: 2GS6) asymmetric dimers. In all panels, carbon atoms contained within the αC helix are blue and activation loop sequences are red.
Although the overall architecture of the HER3 pseudokinase domain in the EGFR/HER3 heterodimer does not display large conformational changes in comparison to previously solved monomeric HER3 structures (24, 30) (fig. S3), the HER3 activation loop, which has consistently been predominantly disordered in monomeric structures, is fully ordered in one of the heterodimers in the asymmetric unit (Fig. 2B). In this dimer, the HER3 activation loop adopts a folded conformation in which the activation loop tyrosine (Tyr849) points directly toward the nucleotide bound in HER3, with a closest distance of 3.4 Å separating the tyrosine hydroxyl and the triphosphate linkage of AMP-PNP. The activation loop tyrosine is conserved throughout tyrosine kinases and plays a central regulatory role in the activation of many of these enzymes (33). Although this closed conformation appears to be stabilized by a nonspecific lattice contact present only in this HER3 monomer, it is in agreement with the canonical organization of the Src/CDK-like inactive conformation. Therefore, the observed closed conformation of the HER3 activation loop may represent a native state in its conformational equilibrium. By analogy to other protein kinases, phosphorylation of HER3 Tyr849 is expected to destabilize the Src/CDK-like inactive conformation (34). Although it is currently unclear whether this conformation has specific functional consequences for HER3-dependent signaling processes, the HER3 activation loop is well conserved throughout evolution (fig. S4), suggesting it may be involved in regulation of HER3.
The conformation of the EGFR kinase domain in the heterodimeric EGFR/HER3 complex is largely unchanged compared to structures of active homodimeric EGFR complexes, with one notable exception. The N-terminal end of the EGFR αC helix shifts ~4Å outward from the active site in the EGFR/HER3 heterodimer in comparison to its position the EGFR/EGFR homodimer. This allows the αC helix to tuck into a shallow pocket in the C-lobe of the HER3 kinase domain (Fig. 2C). This structural shift likely occurs due to a difference in the identity of the C-lobe residues in this region of the activator interface, which contains an isoleucine in HER3 (Ile935) and a serine in EGFR (Ser933). This region has been implicated as a determinant in activator/receiver pairing specificity (35), and our heterodimeric structure highlights the conformational plasticity of the EGFR kinase domain, which allows it to engage in allosteric interaction with either HER3 or EGFR activator subunits.
Juxtamembrane segment interactions enhance EGFR/HER3 heterodimerization
Previous studies on the activation mechanism of EGFR have identified an essential role of the intracellular JM segment in the stabilization of the catalytically active kinase domain dimer (25, 26). The sequence of the JM segment can be separated into two functionally distinct regions (Fig. 3A). The N-terminal region, known as JM-A, is thought to directly dimerize with the JM-A sequence of the other receptor subunit in the active receptor dimer through the formation of a short antiparallel coiled-coil motif (26, 36, 37). The C-terminal region of the JM segment, known as the JM-latch (or JM-B), is immediately adjacent to the kinase domain, and, in contrast to JM-A, does not have a defined secondary structure. In the homodimeric crystal structures of EGFR and HER4 asymmetric dimers, the JM-latch of the receiver kinase runs along the surface of the C-lobe of the activator kinase, creating a narrow interaction surface ~20Å in length that directly extends the central receiver/activator interface between the two kinase domains in the dimer (25, 26, 38).
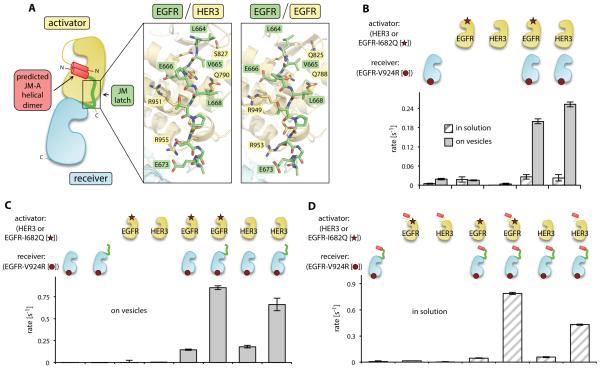
Figure 3.
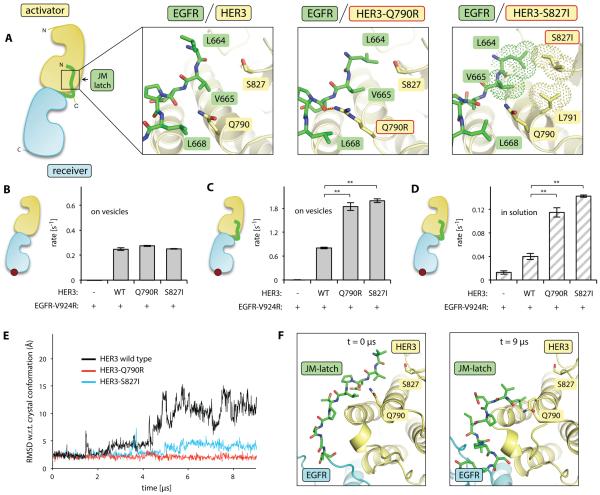
Contribution of the JM segment to EGFR/HER3 heterodimerization and EGFR activation. (A) Cartoon diagram showing the architecture of the asymmetric kinase domain dimer formed by HER family proteins. Insets show a detailed view of the JM-latch interfaces observed in the EGFR/HER3 heterodimer and the EGFR/EGFR homodimer (PDB ID: 3GOP). (B) The activity of the receiver kinase domain EGFR-V924R in the presence of indicated activator kinase domains in the vesicle-based in vitro kinase assay. (C) Activity of the kinase domain EGFR-V924R construct containing the JM-latch (green) in the presence of indicated activator kinase domains in the vesicle-based in vitro kinase assay. (D) Activity of the kinase domain EGFR-V924R construct containing the full JM segment (red and green) measured in solution in the presence of the indicated activator kinase domains with or without the JM-A segment (red). Error bars in all figures represent the standard deviation of three independent measurements.
The contribution of JM segment-mediated interactions to heterodimerization of HERs is not known. We obtained crystals of the EGFR/HER3 heterodimer only when the EGFR construct contained the JM-latch (Fig. 1A), suggesting that this region likely supports heterodimerization events. We found that the interaction between the JM-latch of EGFR and the C-lobe of HER3 involved two backbone hydrogen bonds, two charge-charge interactions (HER3 Arg951/EGFR Glu666 and HER3 Arg955/EGFR Asp673), and a region of alternating polar and nonpolar residues (EGFR Leu664/Val665/Leu668 and HER3 Gln790/Ser827), which form nearly identical contacts as those observed in the EGFR/EGFR and HER4/HER4 homodimers (25)(38)(Fig. 3A).
To test the functional relevance of the JM segment-mediated interactions observed in the crystal structure of the EGFR/HER3 heterodimer, we measured the catalytic activation of the EGFR kinase domain in the presence or absence of the JM segments. To specifically detect the ability of the pseudokinase domain of HER3 to allosterically activate EGFR, we utilized the EGFR-V924R mutant, which cannot function as an allosteric activator but retains catalytic activity as a receiver kinase. We also measured the allosteric activator function of the kinase domain of EGFR using constructs with a mutation that allows function only as an activator, EGFR-I682Q (22). Robust kinase activity occurred only when EGFR-V924R was mixed with the pseudokinase domain of HER3 or with the kinase domain of EGFR-I682Q (Fig. 3B). These results indicated that, although the N-terminal end of the EGFR αC helix is shifted in the EGFR/HER3 heterodimer in comparison to its position in the EGFR/EGFR homodimer as shown in Fig. 2C, pairing of the isolated kinase domains in EGFR/HER3 or EGFR/EGFR complexes resulted in approximately equal kinase activation..
The dimerization affinity of isolated kinase domains of HERs without the presence of the JM segments is quite low (22). As previously observed (22, 24), detection of the dimerization-driven kinase activity of the isolated kinase domain fragments required the concentration of His-tagged constructs on the surface of Ni-NTA lipid vesicles (Fig. 3B). Using the vesicle-based assay, we found that the presence of the EGFR JM-latch sequence potentiated HER3-dependent activation of the kinase domain of EGFR to a similar extent as observed for the homodimeric EGFR/EGFR interaction (Fig. 3C), indicating that the interaction between the JM-latch of EGFR and the C-lobe of the HER3 pseudokinase domain contributes to kinase activation.
We also investigated the importance of the JM-A dimer in EGFR/HER3 activation. Inclusion of the complete JM segments on both activator and receiver EGFR constructs produces in vitro kinase activity in the absence of vesicles due to increased dimerization affinity (26). Comparison of EGFR-I682Q and HER3 kinase domain activator subunits containing the full JM-A regions to those lacking these JM-A regions showed that the presence of the JM-A sequences potentiated the activity of the EGFR/HER3 kinase domain heterodimer in solution (Fig. 3D). Overall, this analysis indicated that JM segment-mediated interactions are both structurally and functionally conserved among HERs, and contribute to the activation of kinase activity by different dimerization partners.
Cancer-associated mutations in the HER3 kinase domain map to the active dimer interface
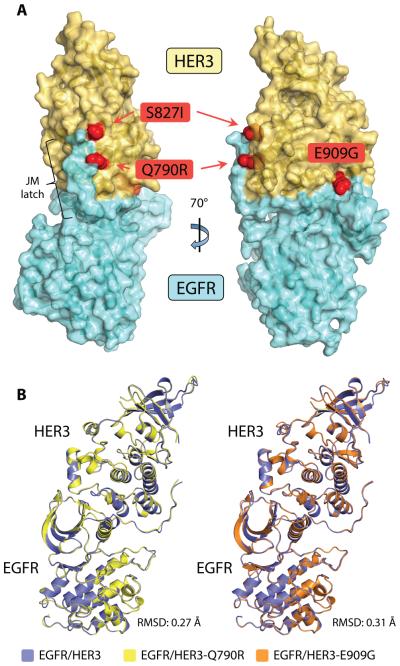
Several HER3 mutations have been identified in primary human tumors (19-21). These mutations confer increased signaling potential to the mutant HER3 when it is coexpressed with a catalytically active HER family member (19). Three strongly activating cancer-associated HER3 mutations -- Q790R, S827I, and E909G -- localize to the pseudokinase domain of HER3. In agreement with previous studies (19), we found that the kinase domain mutations do not increase the catalytic activity of the HER3 pseudokinase (fig. S5). However, mapping the pseudokinase domain mutations onto the structure of the EGFR/HER3 heterodimer unveiled a common theme in their distribution. Although the mutations are not concentrated in a single region of the HER3 pseudokinase domain, each mutation sits at the periphery of the EGFR/HER3 dimerization interface (Fig. 4A). Thus, we hypothesized that cancer mutations in the HER3 pseudokinase domain modulate the ability of HER3 to function as an allosteric activator.
Figure 4.
Cancer-associated mutations in the HER3 pseudokinase domain. (A) The gain-of-function HER3 mutations Q790R, S827I, and E909G mapped onto a surface representation of the EGFR/HER3 kinase heterodimer crystal structure. (B) Crystal structures of EGFR/HER3 heterodimers carrying HER3 cancer mutations. Left: Ribbon diagram overlay of EGFR/HER3 heterodimeric complexes containing either the wild-type or Q790R HER3 kinase domain. Right: Ribbon diagram overlay of EGFR/HER3 heterodimeric complexes containing either the wild-type or E909G HER3 kinase domain.
To gain structural insight into the functional role of the HER3 mutations, we solved the structures of the EGFR/HER3-Q790R and EGFR/HER3-E909G complexes (Table 1). As in the crystal structure of EGFR with the wild-type HER3 pseudokinase domain, the EGFR kinase domain carried the V924R/F973A/L977A mutations and contained the JM-latch fragment. In each complex, EGFR was bound to ADP, a result of AMP-PNP hydrolysis in the active site of EGFR, and the HER3 subunit was bound to AMP-PNP (Fig. S6). Both EGFR/mutant HER3 complexes had an almost identical overall structure as that of the EGFR/wild-type HER3 heterodimer (Fig. 4B). These results indicated that the HER3 Q790R and E909G mutations are compatible with the architecture of the asymmetric dimer that supports allosteric activation of EGFR by HER3. Thus, we hypothesized that the cancer-associated mutations may enhance HER3 signaling by stabilizing the asymmetric dimer interface. Although we could grow crystals of the EGFR/HER3-S827I heterodimer under the same crystallization conditions, we were unable to obtain diffraction-quality crystals of this heterodimer. However, we predict that this complex adopts an overall structure similar to that observed in the EGFR/HER3-Q790R and EGFR/HER3-E909G dimers.
A subset of HER3 cancer-associated mutations promote the interaction between the kinase domain and the juxtamembrane latch
The HER3 mutations Q790R and S827I are located in the region of the HER3 kinase domain that interacts with the EGFR JM-latch (Fig. 4A, 5A). Consequently, we reasoned that these mutations might enable increased HER3-dependent activation of EGFR through the stabilization of the JM-latch interface. We measured the ability of the HER3-Q790R and HER3-S827I mutant kinase domains to allosterically activate the EGFR-V924R kinase domain construct, which lacks the JM-latch, in the vesicle-based in vitro kinase assay. In this context, the wild-type HER3 kinase domain and the two mutants allosterically activated EGFR similarly (Fig. 5B). However, in the vesicle-based assay with the EGFR-V924R kinase containing the JM-latch, both the Q790R and S827I HER3 mutants were more effective at activating EGFR kinase activity than was the wild-type HER3 kinase domain (Fig. 5C). This enhancement was even more pronounced in solution-based experiments, in which the role of JM segment-mediated interactions can be assessed independently from the vesicle-mediated increase in local protein concentration. In solution, the wild-type HER3 kinase domain produced a modest activation of the EGFR construct containing the JM-latch, whereas the Q790R and S827I HER3 mutant kinase domains exhibited substantially increased allosteric activator function in stimulating EGFR activity (Fig. 5D). Thus, the Q790R and S827I mutations improved the allosteric activator function of HER3 by enhancing interaction with the EGFR JM-latch sequence.
Figure 5.
Effect of HER3 cancer-associated mutations in the JM-latch binding site on allosteric activator function. (A) Cartoon diagram highlighting the interaction of the JM-latch of EGFR (green) with its binding site on the HER3 pseudokinase domain (yellow). Insets show the side chain interaction networks observed in the wild-type EGFR/HER3 dimer (crystal structure), the EGFR/HER3-Q790R dimer (crystal structure), and the EGFR/HER3-S827I dimer (after 9 μs of simulation). The hydrogen bond unique to the EGFR/HER3-Q790R dimer is shown in orange dashes, while the hydrophobic cluster unique to the EGFR/HER3-S827I dimer is shown in dot representation. (B) Activity of the EGFR-V924R kinase domain in the presence of the indicated HER3 kinase domain in the vesicle-based assay. (C) Activity of the EGFR-V924R kinase domain containing the JM-latch (green) in the presence of the indicated HER3 kinase domain in the vesicle-based assay. (D) Activity of the EGFR-V924R kinase domain containing the JM-latch (green) in the presence of the indicated HER3 kinase domain in solution. In B-D, the red circle in the cartoons of the dimers represents the EGFR-V924R mutation; WT, wild-type. Error bars are plotted as the standard deviation of three independent measurements, **p<0.001. (E) Root-mean-square deviation (RMSD) of the alpha carbons of the EGFR JM-latch sequence observed in molecular dynamics simulations of the EGFR/HER3 heterodimer. RMSD calculations were completed after alignment of each trajectory snapshot on the HER3 C-lobe. (F) Molecular dynamics simulations of the wild-type EGFR/HER3 heterodimer. Left: The native conformation of the JM-latch sequence observed at the start of the simulation. Right: Conformation of the JM-latch sequence observed after 9 μs of simulation. This conformation is not predicted to be stable, as the backbone hydrogen bonds and charge-charge interactions that secure the JM-latch interface in the EGFR/HER3 crystal structure are lost.
Inspection of the region of the JM-latch interface containing HER3 Gln790 and Ser827 revealed a nonoptimal interaction motif: Three hydrophobic residues from the EGFR subunit (Leu664, Val665, and Leu668) alternate with two polar residues from the HER3 C-lobe (Gln790 and Ser827) (Fig. 5A). This structural configuration suggested that the Q790R and S827I substitutions might functionally redesign this interface through the introduction of stabilizing interactions. Indeed, the crystal structure of the EGFR/HER3-Q790R heterodimer showed that the Q790R substitution resulted in a hydrogen bond between the arginine guanidinium headgroup and the backbone carbonyl of EGFR Glu666 (Fig. 5A). This hydrogen bond is inaccessible to the shorter side chain of the glutamine residue in wild-type HER3.
To further investigate the hypothesis that the Q790R mutant enhances the interaction between the EGFR and HER3 kinases, we performed molecular dynamics simulations starting from the crystal structure of the EGFR/wild-type HER3 heterodimer. In this heterodimer the EGFR JM-latch lost contact with the HER3 C-lobe over time (Fig. 5E, F). The simulation data indicated that, although the presence of the JM-latch contributed to EGFR/HER3 heterodimer formation, this interaction was relatively weak, at least in the context of the EGFR fragment containing only the kinase domain and JM-latch. In contrast, inclusion of the Q790R mutation in the HER3 kinase domain resulted in a stable JM-latch interface that persisted over the 9-μs simulation (Fig. 5E). The additional hydrogen bond observed in the EGFR/HER3-Q790R crystal structure was readily apparent in the molecular dynamics simulations.
Since we were not able to solve a crystal structure of the EGFR/HER3-S827I complex, we used molecular dynamics simulations to gain structural insight into the role of this mutation in increasing the allosteric potential of HER3. Similarly to the Q790R mutation, simulation of the EGFR/HER3-S827I heterodimer also showed a stabilized JM-latch interaction (Fig. 5E). Additionally, these simulations suggested the presence of a molecular interaction unique to the EGFR/HER3-S827I complex. The HER3 S827I mutation appeared to drive the formation of a stabilizing hydrophobic cluster comprised of EGFR Leu664/Val665, and HER3 Leu791/Ile827 (Fig. 5A). This hydrophobic grouping is disrupted in the wild-type EGFR/HER3 heterodimer by the polar nature of the wild-type HER3 Ser827 side chain.
A previous study characterized an activating cancer-associated mutation on the opposite side of this interface, the EGFR V665M substitution in the JM-latch sequence (25). EGFR V665 is located directly across from the Gln790 and Ser827 residues in HER3 (Fig. 5A). The EGFR V665M substitution was proposed to facilitate formation of a small hydrophobic cluster, similar to the HER3 S827I mutation (25). Thus, activating cancer-associated mutations on either side of the JM-latch binding site appear to redesign the interaction network connecting these five residues, leading to a stabilized interface between the activator and receiver kinase domains.
The HER3-E909G cancer-associated mutation disrupts an electrostatic network at the activator/receiver interface
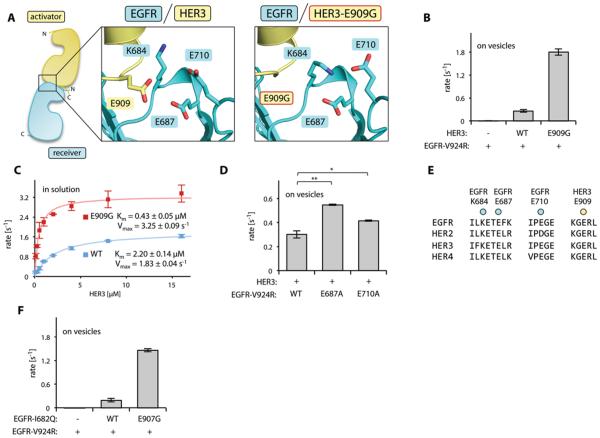
Another cancer-associated mutation, HER3 E909G, lies in the section of the activator/receiver interface formed by the kinase domains of HER3 and EGFR (Fig. 4A, 6A). In agreement with a previous study (39), we found that the E909G mutation enhanced the allosteric activating potential of the HER3 pseudokinase domain in vitro (Fig. 6B). We did not observe any major alteration in the dimerization interface in the structure of the EGFR/HER3-E909G heterodimer (Fig. 4B, 6A). This result suggested that the E909G mutation might primarily enhance EGFR/HER3 dimerization affinity rather than, as was previously postulated (39), increasing the catalytic rate of the activated EGFR.
Figure 6.
Effect of the E909G mutation on the allosteric activator function of HER3. (A) Diagram showing the cluster of charged residues in the vicinity of HER3 Glu909 at the activator/receiver interface of the EGFR/HER3 dimer (left) and in the EGFR/HER3-E909G dimer (right). (B) Activity of the EGFR-V924R kinase domain in the presence of the indicated HER3 kinase domain constructs in the vesicle-based assay. (C) Activity of the EGFR-V924R kinase domain upon titration with the wild-type (WT, blue) or E909G (red) HER3 kinase domain. Both EGFR and HER3 contained the kinase domains and the full JM segments. (D) The activity of the EGFR-V924R kinase domain without additional mutation (WT) or with the indicated mutation in the presence of the wild-type HER3 kinase domain in the vesicle-based assay. (E) Sequence conservation of amino acid residues in the acidic cluster surrounding Glu909. (F) Activity of the EGFR-V924R kinase domain in the presence of the HERG1-I682Q kinase domain containing either no additional mutations (WT) or the E907G mutation in the vesicle-based assay. In panels B, C, D, and F, data are plotted as the mean and standard deviation of three independent experiments, *p<0.005, **p<0.001.
We compared the dimerization affinity of the EGFR kinase domain with wild-type and E909G mutant HER3 by measuring the in vitro kinase activity of EGFR-V924R under a titration with either wild-type or E909G HER3 constructs. Both the EGFR-V924R and HER3 constructs contained the full JM segments (the JM-A regions and the JM-latch), allowing the observation of robust kinase activity in solution-based experiments. In the presence of increasing concentrations of either wild-type HER3 or the E909G mutant, we were able to achieve maximal EGFR activity, an indication of saturated binding (Fig. 6C). Comparison of the Km values from these titrations showed that the HER3-E909G mutant kinase domain had ~5-fold increased affinity for EGFR-V924R as compared to the wild-type HER3 kinase domain. In addition, the HER3-E909G mutant elicited a ~2-fold enhanced catalytic rate of the EGFR kinase at HER3 concentrations that saturated the EGFR/HER3 dimer. These results indicated that the HER3-E909G mutant enhanced both kinetic parameters of HER3 allosteric activator function, but that the majority of the in vitro activating effect of the E909G substitution derived from enhanced EGFR/HER3 dimerization affinity.
Although the structures of the wild-type and HER3-E909G heterodimeric complexes with EGFR did not show large conformational changes in this region, we noted the presence of a cluster of charged residues that surround Glu909 (Fig. 6A). HER3 Glu909 lies across the EGFR/HER3 interface from the EGFR Lys684, Glu687, and Glu710 positions. The presence of three closely spaced negatively charged residues adjacent to only one positively charged side chain creates an unfavorable electrostatic network that is likely inhibitory to EGFR dimerization with the wild-type HER3. Thus, we predicted that neutralization of the repulsive charge across the interface, due to the HER3 E909G substitution, increased the dimerization affinity between the EGFR and HER3 kinase domains.
Furthermore, we predicted that modulation of the acidic electrostatic network (Fig. 6A) through the mutation of other glutamates, EGFR Glu687 and Glu710, should phenocopy the activating effect of the HER3-E909G mutant. We measured the HER3-mediated activation of EGFR-V924R kinase domains with the E687A or E710A mutation and observed that the EGFR E687A mutation increased HER3-mediated EGFR activity (Fig. 6D). The E710A mutation had a less pronounced effect, likely because it is a less stable component of the interaction network due to its position on a flexible loop (fig. S7). Although neither mutation fully replicated the highly activating effect of the HER3-E909G kinase domain, we note that substitutions in the N-lobe of the EGFR kinase domain are likely to have pleiotropic effects due to the direct mutation of a regulatory subdomain of the kinase, which may partially obscure a gain-of-function phenotype. We were unable to produce a sample of the E687A/E710A double mutant due to the instability of this EGFR construct.
All of the residues involved in the acidic cluster surrounding HER3 Glu909 are highly conserved in the HER family (Fig. 6E), suggesting that this interaction network might play a role in regulating the activation of other HER family members. We therefore, investigated whether a mutation in EGFR analogous to the HER3-E909G substitution (EGFR-E907G, introduced in the context of the I682Q mutant) enhanced the allosteric activator function of the EGFR kinase domain toward EGFR-V924R receiver kinase domain using the vesicle-based in vitro kinase assay. EGFR-E907G exhibited an ~8-fold enhancement in activator activity compared with the kinase domain of EGFR-I682Q, which was similar to the ~7-fold enhancement provided by the HER3 E909G substitution (Fig. 6B, F). Thus, the activator-enhancing effect of this mutation was not HER3-specific, but rather represented a general effect on the function of the activator kinase domain.65
Discussion
One of the defining features of the EGFR family is their ability to engage in heterodimeric signaling complexes, enabling connections to distinct biological outputs. Although many studies have improved our understanding of the molecular architecture of active homodimeric HER complexes (22, 23, 25, 26, 37, 40-46), no direct structural insights into the interaction between subunits of heterodimer HER complexes have been made. Studies of HER homodimers have revealed that the molecular mechanisms underlying receptor activation, which involve both the extracellular and intracellular domains, are highly conserved between different homodimeric complexes. Here, we reported a structural visualization of an active complex between two different HER family members, EGFR and HER3. The heterodimeric EGFR/HER3 structure demonstrated that this pairing of HERs also utilizes the highly conserved set of interactions located within the kinase domains and JM segments that characterize the interactions between HER homodimers. This mechanistic conservation highlights that conserved binding interfaces have evolved to enable combinatorial receptor pairing, thereby providing diverse and tailored signaling output from differential growth factor stimulation.
During the course of our analysis, we identified two nonoptimal interaction motifs in the EGFR/HER3 allosteric dimerization interface (Fig. 5A, 6A). Although the protein-protein interfaces containing these regions are essential for receptor activation, biochemical analysis and molecular dynamics simulation indicated that these interactions are relatively weak and transient in nature. We posit that these nonoptimal interactions exist for the precise purpose of controlling receptor activation. By weakening the individual domain interactions in the allosterically activated state of the dimeric HER complex, these inhibitory motifs help to suppress the possibility of aberrant receptor activation caused by the extensive dimer interface. Although our structure-function analysis focused on the EGFR/HER3 heterodimer, the principles derived in our experiments are likely applicable to other HER complexes, including the disease-relevant HER2/HER3 heterodimer. Residues involved in the JM-latch interaction (26) and the electrostatic cluster surrounding HER3 Glu909 (Fig. 6E) are conserved in all HERs.
The EGFR/HER3 heterodimer structure also provided insight into the disease-relevant HER3 pseudokinase in its role as an allosteric activator of catalytically active HER family members. HERs are often aberrantly expressed or mutated in human cancers. These mutations generally facilitate oncogenic signaling by increasing catalytic activity of the receptor (47). Unless oncogenic mutations in HER3 restore catalytically required residues, enhanced function as a catalytically active kinase is not an activating mechanism applicable to HER3. Our structural and biochemical analyses of cancer-associated mutations in the HER3 pseudokinase domain revealed an alternative mechanism by which HER complexes can undergo aberrant activation. The crystal structures of active heterodimeric EGFR/HER3 complexes showed that several gain-of-function cancer-associated mutations in the HER3 pseudokinase domain localize to the periphery of the EGFR/HER3 interaction interface. These missense mutations alter the protein-protein interface used by the HER3 pseudokinase to allosterically activate the EGFR kinase domain. Thus, these mutations act by increasing the allosteric activator function of a HER family member, underscoring the importance of the allosteric activation mechanism used by this set of receptors.
A conceptually similar mechanism for enhancing HER3 signaling has been described in breast cancer cells treated with an inhibitor targeting the active site of HER2 (13). In these cells, increased allosteric potential of HER3 is achieved through increased abundance of HER3, driving an amplified amount of HER2/HER3 signaling which buffers against HER2 inhibition. Whether catalytically active HERs are also deregulated in human disease through the direct enhancement of their allosteric function remains to be investigated. We searched the literature for evidence of the EGFR-E907G substitution, which increased the allosteric activator function of the EGFR kinase domain in vitro. Cell-based experiments have identified this sequence alteration as the top hit in a screen for EGFR mutants with resistance to an ATP-competitive inhibitor (48). It is tempting to speculate that the EGFR-E907G substitution may be found in future studies as a bona fide oncogenic mutant or as a variant driving drug resistance.
Our finding that the identified gain-of-function mutations in the HER3 pseudokinase domain enhance the allosteric activator potential of HER3 supports the development of next-generation inhibitors that target this allosteric function. Furthermore, our analysis of the EGFR/HER3 heterodimer provides a structural framework for this drug development effort by visualizing HER3 in an active complex with one of its catalytically active EGFR dimerization partners.
Materials and Methods
Cloning and protein purification
The human HER3 kinase domain fragment, residues 674-1001 (numbering without the 19-aa signal peptide) and the EGFR kinase domain, residues 672-998 (numbering without the 24-aa signal peptide), were expressed in SF9 cells using the Bac-to-Bac expression system (Invitrogen) and purified as previously described (22, 24). Mutations were introduced by QuikChange site-directed mutagenesis (Stratagene) and confirmed by DNA sequencing. Constructs including the full JM segments contained HER3 residues 648-1001 and EGFR residues 645-998. The EGFR construct including only the JM-latch extension contained residues 658-998.
Crystallization and data collection
To form the EGFR/HER3 asymmetric dimer, an equimolar amount of HER3 (residues 674-1001) and EGFR-V924R/F973A/L977A (residues 658-998) were combined in crystallization buffer (10 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT, 1 mM TCEP, 2 mM AMP-PNP, 5 mM MgCl2) to a final concentration of 7 mg/mL total protein. This crystallization mix was combined with an equal volume of mother liquor (100 mM HEPES pH 7.5, 14% PEG-6K, 200 mM MgCl2) and incubated in the hanging drop format to produce crystals. After incubation at room temperature for ~15 days, individual crystals were cryoprotected by soaking in mother liquor plus 20% glycerol and flash frozen in liquid nitrogen. Diffraction data was collected on beamline 8.3.1 of the Advanced Light Source at the Lawrence Berkeley National Laboratory in Berkeley, CA. Mosflm10 and Scala11 were used to index, integrate, and scale the diffraction data. Data from two nonoverlapping sweep angles were collected from opposite ends of the same crystal and merged to obtain a highly complete dataset. Crystals of the EGFR/HER3 heterodimers carrying cancer-associated HER3 mutations were grown using a mother liquor composed of 100 mM HEPES pH 7.5, 15% PEG-5K MME, 5 mM MgCl2, and 500 mM ammonium acetate, at a total protein concentration of 10 mg/mL.
Phasing and structure determination
We solved the phase problem by molecular replacement with the HER3 and EGFR kinase domains (PDB IDs 3KEX (24) and 2GS6 (22), respectively) after the removal of all ligands. Following a determination of the predicted number of kinase domains in the asymmetric unit, we utilized Phaser (49) to place HER3 and EGFR kinase domains in an unbiased fashion. Manual model building in COOT (50) and automated refinement in Phenix (51) were used alternately to refine the structure to completion. Detailed statistics for data collection and refinement can be found in Table 1.
In vitro kinase assay
Kinase activity was measured using a continuous enzyme-coupled reaction system as previously described (22). The reaction buffer contained 20 mM Tris pH 7.5, 10 mM MgCl2 and 500 μM ATP. Poly 4Glu:Tyr peptide (Sigma) was used as the phosphorylation substrate at a concentration of 1 mg/mL. Small unilamellar vesicles were produced by extrusion through a membrane containing 100 nm pores (Whatman) using a mix of 90% DOPC and 10% Ni-NTA-DGS lipids (Avanti Polar Lipids). The kinetic analysis described in Figure 6 was performed using GraphPad Prism software.
Molecular dynamics
Simulations were initiated from the crystal structure of the EGFR/HER3 kinase domain heterodimer and included residues 664–960 of EGFR and 680–960 of HER3. The heterodimer was placed in the middle of a cubic simulation box (with periodic boundary conditions) of 128 Å per side. Explicitly represented water molecules were added to fill the system, and Na+ and Cl− ions were included to maintain physiological salinity (150 mM) and to obtain a neutral total charge for the system. Each simulation contained approximately 200,000 atoms in total. The systems were parameterized using the TIP3P water model (52) and the Amber99SB*-ILDN force field (53-55), which was developed based on Amber99 (56) and Amber99SB (57), with Amber03 charges used to model the side-chains of Arg, Lys, Glu, and Asp. The simulation systems were equilibrated in the NPT ensemble at 1 bar and 300 K for 4 ps. Equilibrium molecular dynamics simulations were performed on the special-purpose machine Anton (58) in the NPT ensemble at 310 K and 1 bar, using the Nose-Hoover thermostat and barostat (59) and Martyna-Tobias-Klein barostat (60) with relaxation times of 1.0 ps and 10.0 ps, respectively, and a time step of 2.5 fs. Bond lengths to hydrogen atoms were constrained using a recently developed implementation (61) of M-SHAKE (62). Lennard-Jones and Coulomb interactions were calculated using a force-shifted cutoff of 9.5 Å (63).
Supplementary Material
Acknowledgements
We thank F. Findeisen for valuable help in analyzing and processing diffraction datasets and C. Kimberlin for advice during the refinement process. We thank M. Grabe, M.M. Moasser, and K. Shokat for critical reading of the manuscript and insightful discussions. We also thank the staff at the Advanced Light Source at the Lawrence Berkeley National Laboratory. This work was supported in part by a grant from the National Institute of General Medical Sciences to N.J. (R01 RGM109176A) and by American Heart Association Beginning-in-Aid Grant 11BGIA7440051.
Footnotes
Author Contributions
P.L. performed all experiments. L.L assisted with crystallization and data collection for the EGFR/HER3 heterodimers containing the Q790R and E909G substitutions. V.M. and Y.S. designed, performed, and analyzed the simulations. D.E.S. participated in overseeing aspects of the computational research. P.L. and N.J. conceived the study and P.L. and N.J. analyzed the data and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
Data and materials availability. Coordinates and structure factors for wild-type and mutant EGFR/HER3 complexes have been deposited in the Protein Data Bank under PDB IDs 4RIW, 4RIX, and 4RIY.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature reviews. Molecular cell biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Soria P, Camenisch TD. ErbB signaling in cardiac development and disease. Semin Cell Dev Biol. 2010;21:929–935. doi: 10.1016/j.semcdb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Li X, Zhou M. Neuregulin-1/ErbB signaling: a druggable target for treating heart failure. Current opinion in pharmacology. 2009;9:214–219. doi: 10.1016/j.coph.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Levi AD, Bunge RP, Lofgren JA, Meima L, Hefti F, Nikolics K, Sliwkowski MX. The influence of heregulins on human Schwann cell proliferation. J Neurosci. 1995;15:1329–1340. doi: 10.1523/JNEUROSCI.15-02-01329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chausovsky A, Waterman H, Elbaum M, Yarden Y, Geiger B, Bershadsky AD. Molecular requirements for the effect of neuregulin on cell spreading, motility and colony organization. Oncogene. 2000;19:878–888. doi: 10.1038/sj.onc.1203410. [DOI] [PubMed] [Google Scholar]
- 6.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-Dependent Regulation of Her2/Neu Oncogenic Signaling by Heterodimerization with Her3. Embo J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzahar E, Waterman H, Chen XM, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger MB, Mendrola JM, Lemmon MA. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. Febs Lett. 2004;569:332–336. doi: 10.1016/j.febslet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Erickson SL, O'Shea KS, Ghaboosi N, Loverro L, Frantz G, Bauer M, Lu LH, Moore MW. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 10.Frolov A, Schuller K, Tzeng CWD, Cannon EE, Ku BC, Howard JH, Vickers SM, Heslin MJ, Buchsbaum DJ, Arnoletti JP. ErbB3 expression and dimerization with EGFR influence pancreatic cancer cell sensitivity to erlotinib. Cancer Biol Ther. 2007;6:548–554. doi: 10.4161/cbt.6.4.3849. [DOI] [PubMed] [Google Scholar]
- 11.Koutras AK, Fountzilas G, Kalogeras KT, Starakis I, Iconomou G, Kalofonos HP. The upgraded role of HER3 and HER4 receptors in breast cancer. Crit Rev Oncol Hematol. 2010;74:73–78. doi: 10.1016/j.critrevonc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Liles JS, Arnoletti JP, Tzeng CW, Howard JH, Kossenkov AV, Kulesza P, Heslin MJ, Frolov A. ErbB3 expression promotes tumorigenesis in pancreatic adenocarcinoma. Cancer Biol Ther. 2010;10:555–563. doi: 10.4161/cbt.10.6.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007 doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell MR, Amin D, Moasser MM. HER3 comes of age: new insights into its functions and role in signaling, tumor biology, and cancer therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:1373–1383. doi: 10.1158/1078-0432.CCR-09-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nature Reviews Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 16.Amin DN, Campbell MR, Moasser MM. The role of HER3, the unpretentious member of the HER family, in cancer biology and cancer therapeutics. Semin Cell Dev Biol. 2010;21:944–950. doi: 10.1016/j.semcdb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, Manning HC, Chang J, Arteaga CL. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, Janakiraman V, Scholz RP, Bowman KK, Lorenzo M, Li H, Wu JS, Yuan WL, Peters BA, Kan ZY, Stinson J, Mak M, Modrusan Z, Eigenbrot C, Firestein R, Stern HM, Rajalingam K, Schaefer G, Merchant MA, Sliwkowski MX, de Sauvage FJ, Seshagiri S. Oncogenic ERBB3 Mutations in Human Cancers. Cancer Cell. 2013;23:603–617. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L, Mardis ER, Wilson RK, Ally A, Balasundaram M, Butterfield YSN, Carlsen R, Carter C, Chu A, Chuah E, Chun HJE, Coope RJN, Dhalla N, Guin R, Hirst C, Hirst M, Holt RA, Lee D, Li HYI, Mayo M, Moore RA, Mungall AJ, Pleasance E, Robertson AG, Schein JE, Shafiei A, Sipahimalani P, Slobodan JR, Stoll D, Tam A, Thiessen N, Varhol RJ, Wye N, Zeng T, Zhao YJ, Birol I, Jones SJM, Marra MA, Cherniack AD, Saksena G, Onofrio RC, Pho NH, Carter SL, Schumacher SE, Tabak B, Hernandez B, Gentry J, Nguyen H, Crenshaw A, Ardlie K, Beroukhim R, Winckler W, Getz G, Gabriel SB, Meyerson M, Chin L, Park PJ, Kucherlapati R, Hoadley KA, Auman JT, Fan C, Turman YJ, Shi Y, Li L, Topal MD, He XP, Chao HH, Prat A, Silva GO, Iglesia MD, Zhao W, Usary J, Berg JS, Adams M, Booker J, Wu JY, Gulabani A, Bodenheimer T, Hoyle AP, Simons JV, Soloway MG, Mose LE, Jefferys SR, Balu S, Parker JS, Hayes DN, Perou CM, Malik S, Mahurkar S, Shen H, Weisenberger DJ, Triche T, Lai PH, Bootwalla MS, Maglinte DT, Berman BP, Van den Berg DJ, Baylin SB, Laird PW, Creighton CJ, Donehower LA, Getz G, Noble M, Voet D, Saksena G, Gehlenborg N, DiCara D, Zhang JH, Zhang HL, Wu CJ, Liu SY, Lawrence MS, Zou LH, Sivachenko A, Lin P, Stojanov P, Jing R, Cho J, Sinha R, Park RW, Nazaire MD, Robinson J, Thorvaldsdottir H, Mesirov J, Park PJ, Chin L, Reynolds S, Kreisberg RB, Bernard B, Bressler R, Erkkila T, Lin J, Thorsson V, Zhang W, Shmulevich I, Ciriello G, Weinhold N, Schultz N, Gao JJ, Cerami E, Gross B, Jacobsen A, Sinha R, Aksoy BA, Antipin Y, Reva B, Shen RL, Taylor BS, Ladanyi M, Sander C, Anur P, Spellman PT, Lu YL, Liu WB, Verhaak RRG, Mills GB, Akbani R, Zhang NX, Broom BM, Casasent TD, Wakefield C, Unruh AK, Baggerly K, Coombes K, Weinstein JN, Haussler D, Benz CC, Stuart JM, Benz SC, Zhu JC, Szeto CC, Scott GK, Yau C, Paul EO, Carlin D, Wong C, Sokolov A, Thusberg J, Mooney S, Ng S, Goldstein TC, Ellrott K, Grifford M, Wilks C, Ma S, Craft B, Yan CH, Hu Y, Meerzaman D, Gastier-Foster JM, Bowen J, Ramirez NC, Black AD, Pyatt RE, White P, Zmuda EJ, Frick J, Lichtenberg T, Brookens R, George MM, Gerken MA, Harper HA, Leraas KM, Wise LJ, Tabler TR, McAllister C, Barr T, Hart-Kothari M, Tarvin K, Saller C, Sandusky G, Mitchell C, Iacocca MV, Brown J, Rabeno B, Czerwinski C, Petrelli N, Dolzhansky O, Abramov M, Voronina O, Potapova O, Marks JR, Suchorska WM, Murawa D, Kycler W, Ibbs M, Korski K, Spychala A, Murawa P, Brzezinski JJ, Perz H, Lazniak R, Teresiak M, Tatka H, Leporowska E, Bogusz-Czerniewicz M, Malicki J, Mackiewicz A, Wiznerowicz M, V. Le X, Kohl B, Tien NV, Thorp R, Bang NV, Sussman H, Phu BD, Hajek R, Hung NP, Tran VTP, Thang HQ, Khan KZ, Penny R, Mallery D, Curley E, Shelton C, Yena P, Ingle JN, Couch FJ, Lingle WL, King TA, Gonzalez-Angulo AM, Mills GB, Dyer MD, Liu SY, Meng XL, Patangan M, Waldman F, Stoppler H, Rathmell WK, Thorne L, Huang M, Boice L, Hill A, Morrison C, Gaudioso C, Bshara W, Daily K, Egea SC, Pegram MD, Gomez-Fernandez C, Dhir R, Bhargava R, Brufsky A, Shriver CD, Hooke JA, Campbell JL, Mural RJ, Hu H, Somiari S, Larson C, Deyarmin B, Kvecher L, Kovatich AJ, Ellis MJ, King TA, Hu H, Couch FJ, Mural RJ, Stricker T, White K, Olopade O, Ingle JN, Luo CQ, Chen YQ, Marks JR, Waldman F, Wiznerowicz M, Bose R, Chang LW, Beck AH, Gonzalez-Angulo AM, Pihl T, Jensen M, Sfeir R, Kahn A, Chu A, Kothiyal P, Wang ZN, Snyder E, Pontius J, Ayala B, Backus M, Walton J, Baboud J, Berton D, Nicholls M, Srinivasan D, Raman R, Girshik S, Kigonya P, Alonso S, Sanbhadti R, Barletta S, Pot D, Sheth M, Demchok JA, Shaw KRM, Yang LM, Eley G, Ferguson ML, Tarnuzzer RW, Zhang JS, Dillon LAL, Buetow K, Fielding P, Ozenberger BA, Guyer MS, Sofia HJ, Palchik JD, Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Reid JG, Santibanez J, Shinbrot E, Trevino LR, Wu YQ, Wang M, Gunaratne P, Donehower LA, Creighton CJ, Wheeler DA, Gibbs RA, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, Stojanov P, McKenna A, Lander ES, Gabriel S, Getz G, Ding L, Fulton RS, Koboldt DC, Wylie T, Walker J, Dooling DJ, Fulton L, Delehaunty KD, Fronick CC, Demeter R, Mardis ER, Wilson RK, Chu A, Chun HJE, Mungall AJ, Pleasance E, Robertson AG, Stoll D, Balasundaram M, Birol I, Butterfield YSN, Chuah E, Coope RJN, Dhalla N, Guin R, Hirst C, Hirst M, Holt RA, Lee D, Li HI, Mayo M, Moore RA, Schein JE, Slobodan JR, Tam A, Thiessen N, Varhol R, Zeng T, Zhao Y, Jones SJM, Marra MA, Bass AJ, Ramos AH, Saksena G, Cherniack AD, Schumacher SE, Tabak B, Carter SL, Pho NH, Nguyen H, Onofrio RC, Crenshaw A, Ardlie K, Beroukhim R, Winckler W, Getz G, Meyerson M, Protopopov A, Zhang J, Hadjipanayis A, Lee E, Xi R, Yang L, Ren X, Zhang H, Sathiamoorthy N, Shukla S, Chen PC, Haseley P, Xiao Y, Lee S, Seidman J, Chin L, Park PJ, Kucherlapati R, Auman JT, Hoadley KA, Du Y, Wilkerson MD, Shi Y, Liquori C, Meng S, Li L, Turman YJ, Topal MD, Tan D, Waring S, Buda E, Walsh J, Jones CD, Mieczkowski PA, Singh D, Wu J, Gulabani A, Dolina P, Bodenheimer T, Hoyle AP, Simons JV, Soloway M, Mose LE, Jefferys SR, Balu S, O'Connor BD, Prins JF, Chiang DY, Hayes DN, Perou CM, Hinoue T, Weisenberger DJ, Maglinte DT, Pan F, Berman BP, Van den Berg DJ, Shen H, Jr TT, Baylin SB, Laird PW, Getz G, Noble M, Voet D, Saksena G, Gehlenborg N, DiCara D, Zhang J, Zhang H, Wu CJ, Liu SY, Shukla S, Lawrence MS, Zhou L, Sivachenko A, Lin P, Stojanov P, Jing R, Park RW, Nazaire MD, Robinson J, Thorvaldsdottir H, Mesirov J, Park PJ, Chin L, Thorsson V, Reynolds SM, Bernard B, Kreisberg R, Lin J, Iype L, Bressler R, Erkkila T, Gundapuneni M, Liu Y, Norberg A, Robinson T, Yang D, Zhang W, Shmulevich I, De Ronde JJ, Schultz N, Cerami E, Ciriello G, Goldberg AP, Gross B, Jacobsen A, Gao J, Kaczkowski B, Sinha R, Aksoy BA, Antipin Y, Reva B, Shen R, Taylor BS, Chan TA, Ladanyi M, Sander C, Akbani R, Zhang N, Broom BM, Casasent T, Unruh A, Wakefield C, Hamilton SR, Cason RC, Baggerly KA, Weinstein JN, Haussler D, Benz CC, Stuart JM, Benz SC, Sanborn JZ, Vaske CJ, Zhu J, Szeto C, Scott GK, Yau C, Ng S, Goldstein T, Ellrott K, Collisson E, Cozen AE, Zerbino D, Wilks C, Craft B, Spellman P, Penny R, Shelton T, Hatfield M, Morris S, Yena P, Shelton C, Sherman M, Paulauskis J, Gastier-Foster JM, Bowen J, Ramirez NC, Black A, Pyatt R, Wise L, White P, Bertagnolli M, Brown J, Chan TA, Chu GC, Czerwinski C, Denstman F, Dhir R, Doerner A, Fuchs CS, Guillem JG, Iacocca M, Juhl H, Kaufman A, Kohl B, Van Le X, Mariano MC, Medina EN, Meyers M, Nash GM, Paty PB, Petrelli N, Rabeno B, Richards WG, Solit D, Swanson P, Temple L, Tepper JE, Thorp R, Vakiani E, Weiser MR, Willis JE, Witkin G, Zeng Z, Zinner MJ, Zornig C, Jensen MA, Sfeir R, Kahn AB, Chu AL, Kothiyal P, Wang Z, Snyder EE, Pontius J, Pihl TD, Ayala B, Backus M, Walton J, Whitmore J, Baboud J, Berton DL, Nicholls MC, Srinivasan D, Raman R, Girshik S, Kigonya PA, Alonso S, Sanbhadti RN, Barletta SP, Greene JM, Pot DA, Shaw KRM, Dillon LAL, Buetow K, Davidsen T, Demchok JA, Eley G, Ferguson M, Fielding P, Schaefer C, Sheth M, Yang L, Guyer MS, Ozenberger BA, Palchik JD, Peterson J, Sofia HJ, Thomson E. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal A, Bornmann WG, Lemmon MA, Cole PA, Leahy DJ. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci U S A. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Molecular cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aertgeerts K, Skene R, Yano J, Sang BC, Zou H, Snell G, Jennings A, Iwamoto K, Habuka N, Hirokawa A, Ishikawa T, Tanaka T, Miki H, Ohta Y, Sogabe S. Structural Analysis of the Mechanism of Inhibition and Allosteric Activation of the Kinase Domain of HER2 Protein. Journal of Biological Chemistry. 2011;286:18756–18765. doi: 10.1074/jbc.M110.206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Molecular cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 29.Xu WQ, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Molecular cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 30.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci U S A. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olesen C, Picard M, Winther AML, Gyrup C, Morth JP, Oxvig C, Moller JV, Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–U1035. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson AD, Sheth PR, Basso AD, Paliwal S, Gray K, Fischmann TO, Le HV. Structural basis of CX-4945 binding to human protein kinase CK2. Febs Lett. 2011;585:104–110. doi: 10.1016/j.febslet.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 33.Adams JA. Activation loop phosphorylation and catalysis in protein kinases: Is there functional evidence for the autoinhibitor model? Biochemistry. 2003;42:601–607. doi: 10.1021/bi020617o. [DOI] [PubMed] [Google Scholar]
- 34.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Molecular cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monsey J, Shen W, Schlesinger P, Bose R. Her4 and Her2/neu Tyrosine Kinase Domains Dimerize and Activate in a Reconstituted in Vitro System. Journal of Biological Chemistry. 2010;285:7035–7044. doi: 10.1074/jbc.M109.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arkhipov A, Shan YB, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and Membrane Interactions of the EGF Receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood ER, Shewchuk LM, Ellis B, Brignola P, Brashear RL, Caferro TR, Dickerson SH, Dickson HD, Donaldson KH, Gaul M, Griffin RJ, Hassell AM, Keith B, Mullin R, Petrov KG, Reno MJ, Rusnak DW, Tadepalli SM, Ulrich JC, Wagner CD, Vanderwall DE, Waterson AG, Williams JD, White WL, Uehling DE. 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc Natl Acad Sci U S A. 2008;105:2773–2778. doi: 10.1073/pnas.0708281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collier TS, Diraviyam K, Monsey J, Shen W, Sept D, Bose R. Carboxyl Group Footprinting Mass Spectrometry and Molecular Dynamics Identify Key Interactions in the HER2-HER3 Receptor Tyrosine Kinase Interface. Journal of Biological Chemistry. 2013;288:25254–25264. doi: 10.1074/jbc.M113.474882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi LZ, Lu CF, Li ZL, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18:984–U1502. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 42.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Molecular cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 43.Alvarado D, Klein DE, Lemmon MA. Structural Basis for Negative Cooperativity in Growth Factor Binding to an EGF Receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouyain S, Longo PA, Li SQ, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. P Natl Acad Sci USA. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrett TPJ, McKern NM, Lou MZ, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 46.Garrett TPJ, McKern NM, Lou MZ, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, Ward CW. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Molecular cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 47.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Z, Boggon TJ, Kobayashi S, Jin C, Ma PC, Dowlati A, Kern JA, Tenen DG, Halmos B. Resistance to an irreversible epidermal growth factor receptor (EGFR) inhibitor in EGFR-Mutant lung cancer reveals novel treatment strategies. Cancer research. 2007;67:10417–10427. doi: 10.1158/0008-5472.CAN-07-1248. [DOI] [PubMed] [Google Scholar]
- 49.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams P, Afonine P, Bunkoczi G, Chen V, Davis I. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 53.Best RB, Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. The journal of physical chemistry. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joung IS, Cheatham TE., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. The journal of physical chemistry. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 57.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins-Structure Function and Bioinformatics. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, Chow E, Eastwood MP, Ierardi DJ, Klepeis JL, Kuskin JS, Larson RH, Lindorff-Larsen K, Maragakis P, Moraes MA, Piana S, Shan YB, Towles B. Millisecond-Scale Molecular Dynamics Simulations on Anton. Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis. 2009 [Google Scholar]
- 59.Hoover WG. Canonical Dynamics - Equilibrium Phase-Space Distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 60.Martyna GJ, Tobias DJ, Klein ML. Constant-Pressure Molecular-Dynamics Algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 61.Lippert RA, Bowers KJ, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Shaw DE. A common, avoidable source of error in molecular dynamics integrators. The Journal of chemical physics. 2007;126:046101. doi: 10.1063/1.2431176. [DOI] [PubMed] [Google Scholar]
- 62.Krautler V, Van Gunsteren WF, Hunenberger PH. A fast SHAKE: Algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22:501–508. [Google Scholar]
- 63.Beck DAC, Armen RS, Daggett V. Cutoff size need not strongly influence molecular dynamics results for solvated polypeptides. Biochemistry-Us. 2005;44:609–616. doi: 10.1021/bi0486381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.