Abstract
Despite the growing use of nanofiber scaffolds for tissue engineering applications, there is not a validated, readily available, free solution for rapid, automated analysis of nanofiber diameter from scanning electron microscope (SEM) micrographs. Thus, the goal of this study was to create a user friendly ImageJ/FIJI plugin that would analyze SEM micrographs of nanofibers to determine nanofiber diameter on a desktop computer within 60 seconds. Additional design goals included 1) compatibility with a variety of existing segmentation algorithms, and 2) an open source code to enable further improvement of the plugin. Using existing algorithms for centerline determination, Euclidean distance transforms and a novel pixel transformation technique, a plugin called “DiameterJ” was created for ImageJ/FIJI. The plugin was validated using 1) digital synthetic images of white lines on a black background and 2) SEM images of nominally monodispersed steel wires of known diameters. DiameterJ analyzed SEM micrographs in 20 seconds, produced diameters not statistically different from known values, was over 10-times closer to known diameter values than other open source software, provided hundreds of times the sampling of manual measurement, and was hundreds of times faster than manual assessment of nanofiber diameter. DiameterJ enables users to rapidly and thoroughly determine the structural features of nanofiber scaffolds and could potentially allow new insights to be formed into fiber diameter distribution and cell response.
Introduction
Electrospun polymeric nanofiber scaffolds have wide applicability across a variety of fields including tissue engineering [1,2], filtration [3–6], catalysis [7–9] and in biosensors [10–12]. In all of these fields a strong correlation has been found between nanofiber mat morphology and performance. Nanofiber morphology features such as fiber diameter, orientation, and mesh hole size (pore size) have been shown to correlate strongly with mechanical properties [13–15], surface area to volume ratios (influencing catalytic efficiency [16,17], signal to noise ratios in biosensors [18,19], and filtration efficiency [20]), and in tissue engineering to determine cell morphology [21–23], phenotype [24,25], and differentiation [26,27]. Thus, there is a need for characterization tools of the scaffolds that can quickly and accurately assess these fiber properties.[28] Currently, a variety of manual assessment techniques are commonly used to obtain distributions of fiber diameters. [28–30] Due to the size of the fibers being spun (< 1000 nm) a widely used tool for the assessment of nanofiber scaffolds is the scanning electron microscope (SEM). [31,32] Therefore, image analysis tools that can analyze SEM images, the most prevalent in the field, would have the largest user-base if developed.
The gold standard for assessment of nanofiber morphology has historically been manual measurement in SEM images using line tools in image analysis programs such as ImageJ/FIJI (National Institute of Health, MD) [33–35]. In addition to being low-throughput, manual measurements may be biased. Operators may avoid “non-representative” fibers, such as blebs or very thin or thick fibers as well as introduce small systemic biases in measurement. Also, because a distribution of fiber diameters is present in electrospun nanofiber preparations, obtaining enough manual measurements to reduce the standard deviation to a level where meaningful statistical comparisons can be performed is time-consuming.[37,38] To help expedite this analysis and reduce bias, several laboratories have developed tools to assess nanofiber orientation [36–39], mesh hole size [34], and nanofiber diameter [39–41] from SEM images of nanofiber mats. Several of the tools to assess nanofiber orientation have been validated [36–39] and are available to the community for free on ImageJ/FIJI. Similarly, mesh hole analysis tools have been developed and validated and are available for ImageJ/FIJI [42–45]. Currently, only one open source tool can be used to assess nanofiber diameter; which uses Dougherty et al.’s method, and is available in the BoneJ plugin for ImageJ [46–48]. Though BoneJ has been used to measure nanofiber diameters, [47,49], it was not created to assess these types of structures and has not yet been validated for them. Several other labs have developed tools to assess nanofiber diameter using edge detection algorithms [40], Radon Transforms [39], or principal component analysis [41] however, these tools were never released to the community and were therefore never validated.
Additionally, there are two commercially available pieces of software that have been compared in other studies, but both require tens to hundreds of thousands of dollars worth of investment in order for researchers to use these pieces of software.[38] Also, the algorithms in these commercial software packages are not open source and cannot be viewed or modified by the community. Therefore, there is currently not a validated, readily available (commercial or open source) tool for rapid, automated analysis of nanofiber diameter in scanning electron micrographs.
For these reasons we have developed an algorithm named “DiameterJ” for determining nanofiber diameter. Our goal was to create an automated, accessible, open-access, and simple-to-use image analysis tool to enable automatic and efficient measurement of nanofiber diameters in scanning electron micrographs. The algorithm was implemented as an ImageJ/FIJI plugin because these programs are free, easy to use, and widely used by the biological and biomaterials research communities. [46,48] Additionally, although image segmentation has a significant role in image analysis, previously existing segmentation algorithms were used in this work and therefore segmentation is not a contribution of the proposed technique. The proposed plugin was validated by using a series of 1) digital synthetic image files made with black and white lines of known pixel diameters and 2) scanning electron micrographs of stainless steel wires of nominally monodispersed diameter.
Materials and Methods
Generating Digital Synthetic Images
In order to validate the proposed technique, 118 digital synthetic images of white lines of known pixel diameter on a black background were generated using Inkscape graphics package (www.inkscape.org) and converted to Tiff images using ImageJ (see Figure 1 and Figure 2 in reference [53] for Tiff images). In parallel to the current manuscript, a “Data-in-Brief” article [53] has been published which contains an extended description of the Materials and Methods, the image files that were analyzed and all of the raw data to support the figures in this manuscript. White line diameters were chosen to span 3 px to 250 px in diameter. Images were also created that contained lines with different diameters within the same image, in order to mimic the distribution of fiber diameters that are observed in electrospun nanofiber scaffolds. A detailed description of how digital synthetic images were generated can be found in Section 2.1 of reference [53]. Briefly, five types of image sets were created: sets with straight lines of a single diameter (Ordered-1D); sets of disordered, curved lines of a single diameter (Disordered- 1D); sets of straight lines of 3 diameters (Ordered-3D); sets of disordered, curved lines of three diameters (Disordered-3D); and sets of disordered, curved lines of 2, 3, 4 or 6 diameters (Multi-Dia.).
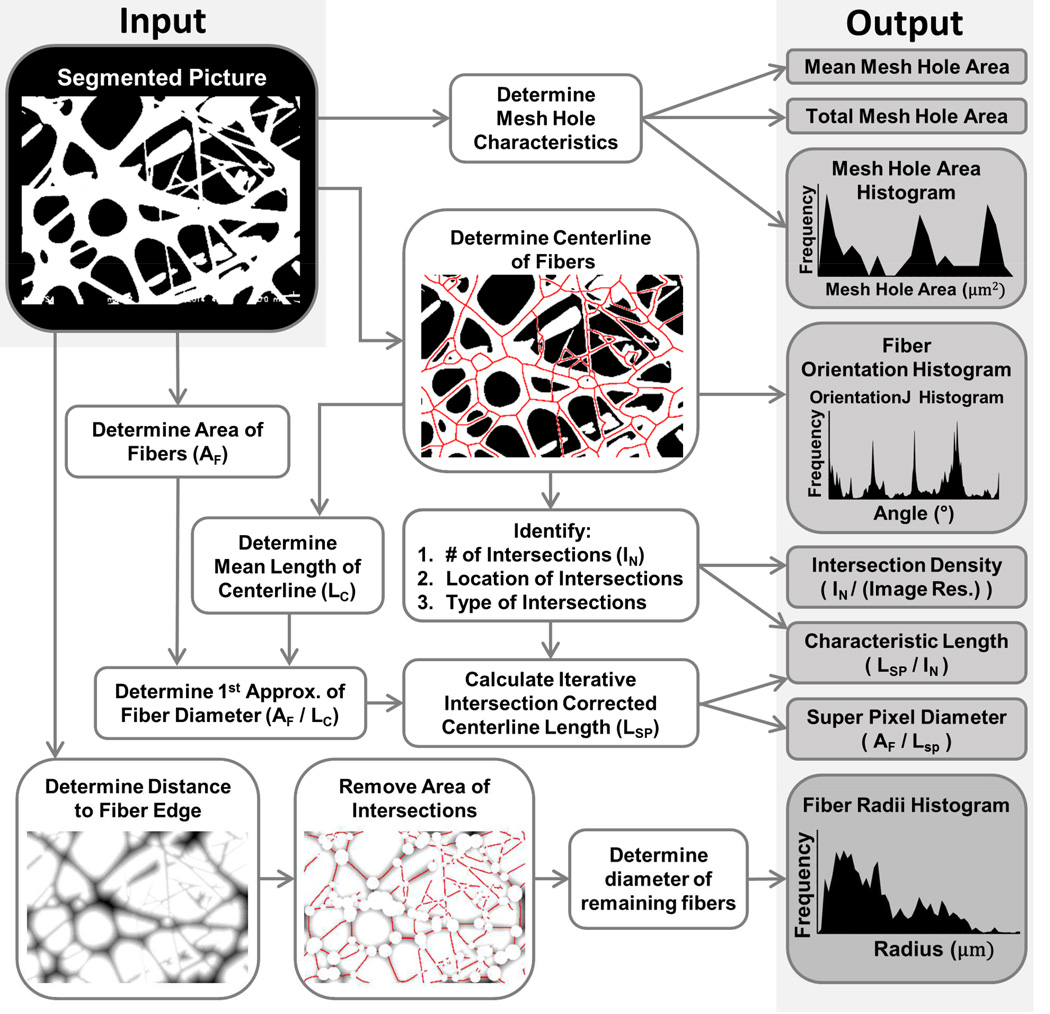
Figure 1.
Diagram of fiber analysis algorithm. The starting input point is the black box labeled “Segmented Picture”. Grey Boxes are outputs of DiameterJ. Algorithms were not able to effectively determine fiber diameter at fiber intersections requiring subroutines to remove fiber intersections from the analysis leading to interconnected outputs within the code as indicated.
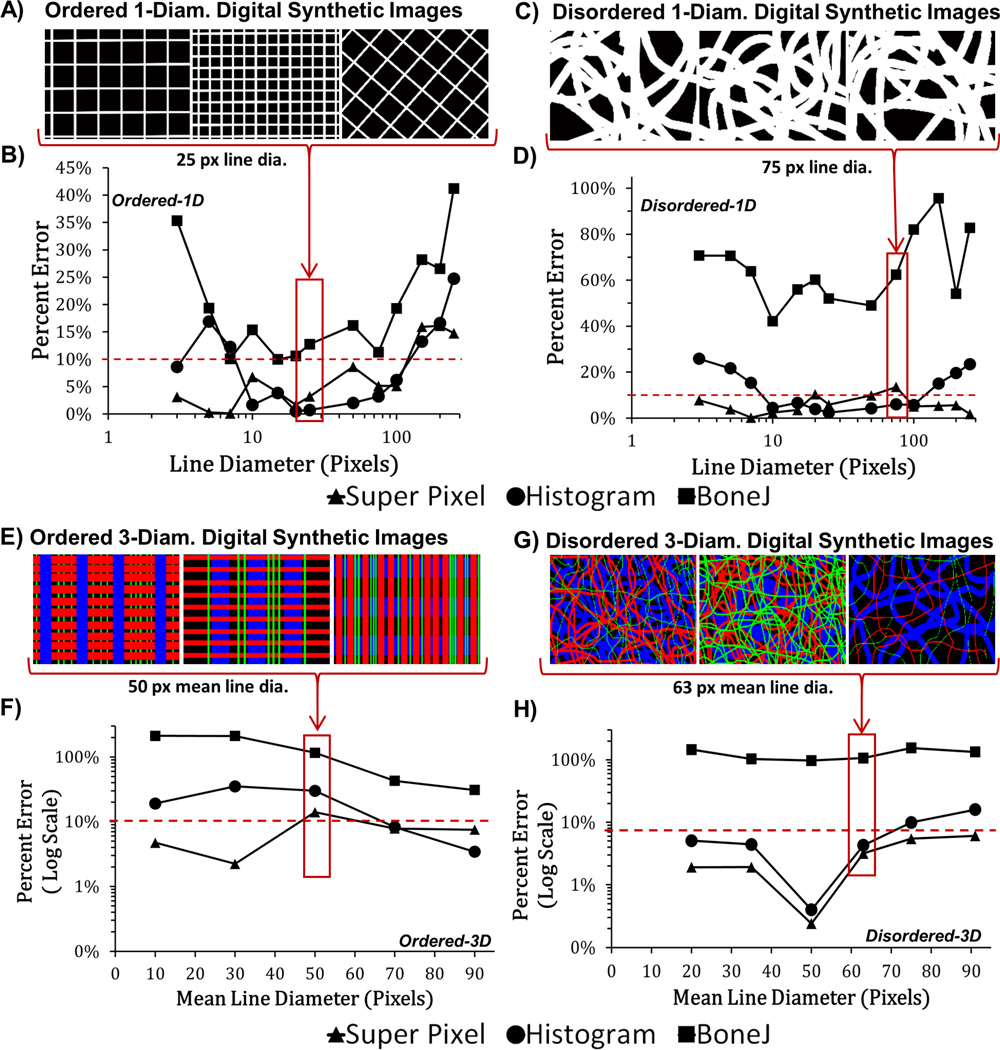
Figure 2.
Analysis of calibration images to determine the mean line dia. Digital synthetic images were analyzed by DiameterJ (Super Pixel, Histogram) or BoneJ. Lines in (E) and (G), are colored by size (Blue>Red>Green) for ease of visual inspection but actual analysis was performed on images with white lines. The plot of the percent error between the diameter produced by the programs and the ground truth from the calibration images can be seen in Figures (B), (D), (F), and (H). The red dashed line represents 10 % error. N = 3 images for each data point in all graphs. Error bars were omitted for clarity.
Determining Average Line Diameter in Digital Synthetic Images
The average diameter, calculated independently of outputs from DiameterJ, for the single diameter white line digital synthetic images was simply taken as the diameter of the white lines in the image, as defined by Inkscape. The average diameter for the Ordered-3D and Disordered-3D images was calculated via number averaging of each of the line diameters and does not account for line length. The number of lines at a given diameter in an image was defined by continuous line length within the image that did not leave the image border. The average line diameter was calculated as follows: , where Fi is the frequency of a fiber in an image and Di is the diameter at that frequency. For example an image with 3 fibers that were 10px in diameter, 3 fibers at 20px in diameter, and 3 fibers with 30 px in diameter would have an average diameter of 20px [(3*10+3*20+3*30)/(3+3+3)] regardless of the length of each line within the image.
Steel Wire Samples
Three gauges (48 ga., 50 ga. and 53 ga.) of steel wire (316 stainless steel wire, HSM Wire, Inc.) of a nominally monodispersed diameter were used as test samples to provide a base for measuring fiber diameters in scanning electron micrographs. Mean wire diameter was verified using white-light transmitted microscopy (Nikon Eclipse TE300 microscope) and a NIST-traceable stage micrometer (Klarmann Rulings, Inc.). Ten images of linear wire (no overlaps) were captured randomly over a meter of each wire and 10 wire diameter measurements were made in each image using the Line tool in ImageJ (100 total measurements for each ga.). Taking images of linear wire removed the complication of having wire overlaps that are present in images of entangled fibers, making the wire measurements more accurate. The mean diameter was found to be 31.1 ± 0.1 µm, 25.6 ± 0.1 µm, 16.7 ± 0.1 µm for the 48, 50 and 53 ga. wires respectively. The manufacturer also provided mean wire diameters from electric resistivity and digital caliper measurements (31.0 µm, 25.5 µm, and 16.75 µm respectively) and these values fell within the standard deviation of our optical image measurements. All micrographs of the steel wire that were taken and analyzed can be found in Figure 3 of reference [53].
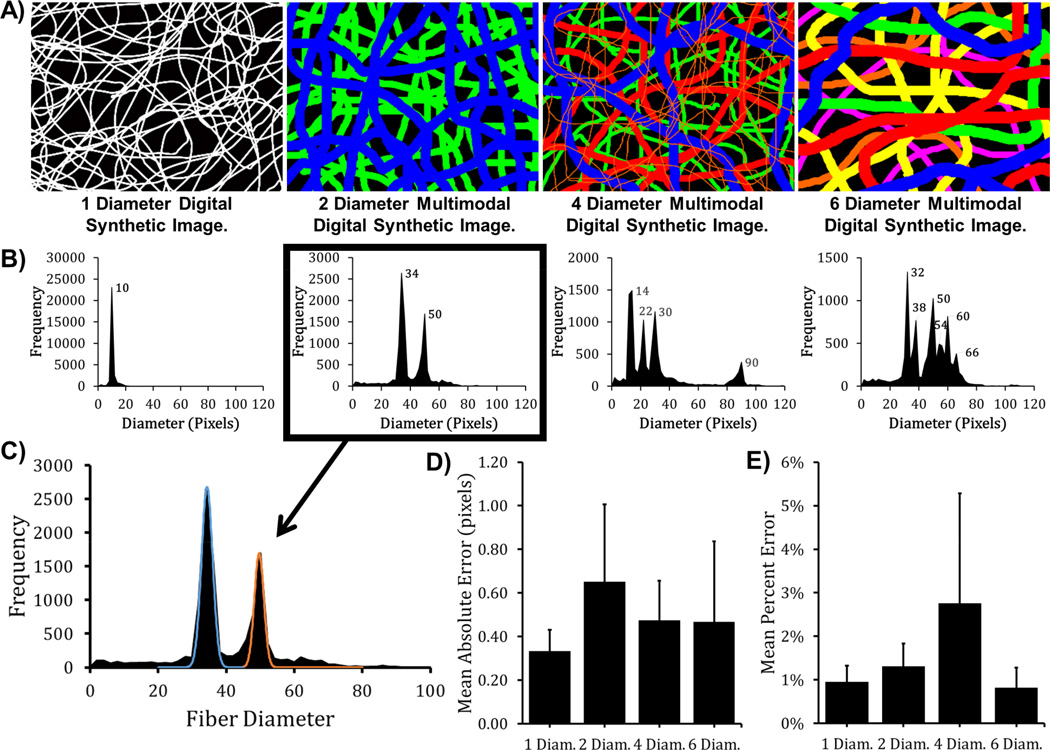
Figure 3.
DiameterJ Histogram algorithm peak fitting analysis to determine multi-modal line diameter distributions in Multi-Dia. calibration images. Lines in the digital synthetic images (A) are colored by size (Blue > Red > Green > Orange > Yellow > Purple) for visualization but analysis was performed on images with white fibers. (B) DiameterJ’s Histogram algorithm’s analysis of images in (A). Peak modes are indicated on histograms. (C) Gaussian peak fits for the “2 dia.” histogram in (B). The mean absolute error and percent error of the DiameterJ Histogram algorithm when Gaussian peaks are fit to each histogram in (D) and (E), respectively. Error bars represent standard error for N = 3 images.
In order to prepare samples for scanning electron microscopy (SEM), a meter of wire was cut, folded in half and rolled between two latex gloved-hands in opposing circular motions until the wire was fully entangled. Samples were prepared for each of the three gauges individually (48 ga., 50 ga. and 53 ga.). In addition, a “mixed-gauge” sample was prepared where a half meter of each wire ga. was entangled as described above. To remove dust, the entangled samples were washed with successive immersion (5 mL) and vortexing (5 min) in each of the following solvents: dichloromethane, dimethyl sulfoxide, tetrahydrofuran, and hexafluoroisopropanol (HFIP) (Sigma-Aldrich). Prior to SEM, representative images of the entangled wire were taken with a Leica MZ16 (Leica) stereo-optical microscope.
Electrospun Polymeric Nanofibers
Poly(lactic-co-glycolic acid) (PLGA 50:50 molar ratio of L to G, relative molecular mass 35 000 g/mol, Akina Inc., Polyscitech) was dissolved at 25 % by mass in hexafluoroisopropanol and electrospun (25 ga. steel needle, 0.3 mL/h, tip to collector distance of 15 cm, 1 h spin time, aluminum foil target) at 12.5 kV (high voltage generator, ES30P-5W, Gamma High Voltage Research) to yield monodisperse PLGA nanofibers. The PLGA nanofiber mat was removed from the foil and cut into 5 mm × 5 mm squares for SEM.
Nanofiber samples with two layers of different diameter (bimodal dia.) were commercially available (Stellenbosch Nanofiber Company; Cape Town, South Africa). The bimodal samples were composed of PLGA 50:50 (PLGA 50:50 molar ratio of L to G, inherent viscosity 0.4 – 1.0 dl/g). The manufacturer reported values for fiber diameter in the two layers were 580 nm ± 226 nm and 1800 nm ± 775 nm. All micrographs of either the monodispersed PLGA nanofibers or the bimodal PLGA fibers that were taken and analyzed can be found in Figure 4 of reference [53].
Figure 4.
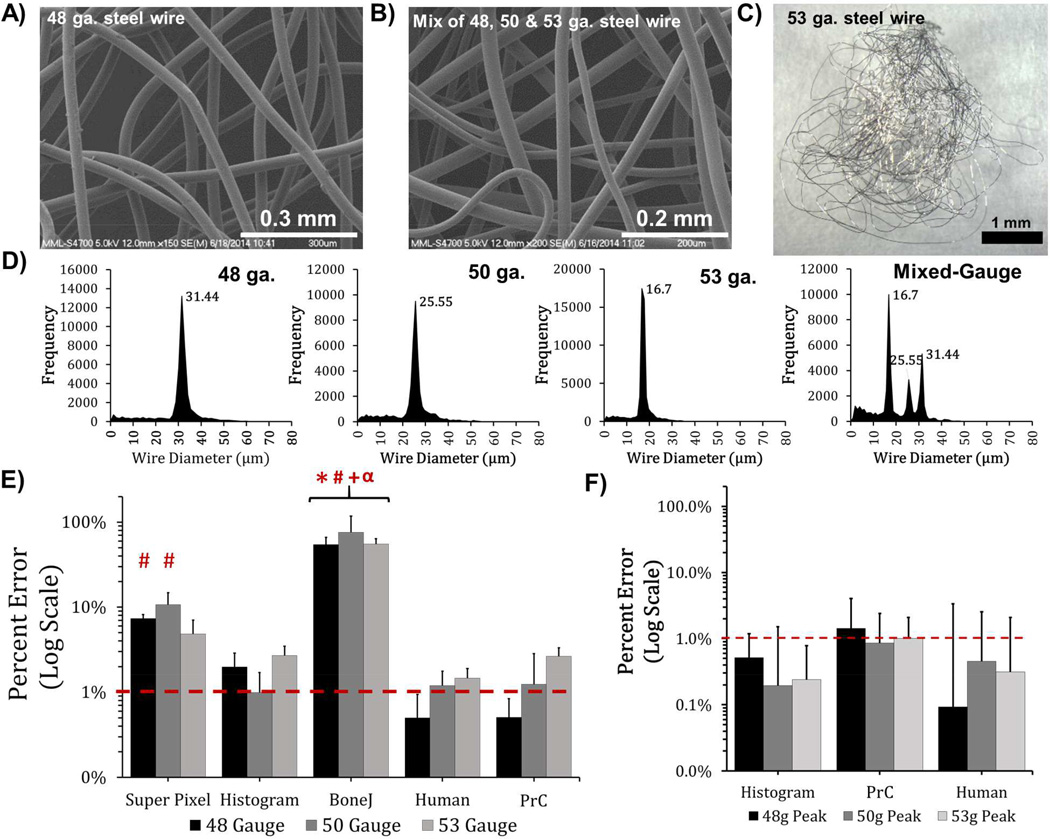
Analysis of SEM micrographs of 316 stainless steel reference wire. (A, B) SEM micrographs of steel wire. (C) Optical image of 53 ga. steel wire. (D) DiameterJ’s Histogram algorithm output. Error in mean wire diameter of the single diameter (E), or mixed three diameter (F) steel wire micrographs for 5 different methodologies/algorithms. N= 6 images for all analyses. Error bars represent the standard deviation. Asterisks (*) indicate statistically different from DiameterJ’s Histogram algorithm; number signs (#) indicate statistical difference from human measure; plus signs (+) indicate statistical difference from DiameterJ’s Super Pixel algorithm; and an alpha sign (α) indicates statistical difference from PrC (P < 0.05).
Scanning Electron Microscopy
Steel wire samples (each ga. individually and mixed-ga. samples), PLGA monodisperse nanofibers (removed from the aluminum foil target) and PLGA bimodal nanofibers were mounted onto SEM stubs using carbon tape (Ted Pella, Inc.), placed under house vacuum overnight with desiccant and gold sputter-coated (120 s at 75 mA, Desk V HP, Denton Vacuum, approximately 10 nm of gold deposition). Six scanning electron micrographs were captured of each sample (Hitachi S4700 SEM, 5kV, 10mA, ≈13 mm working distance). Wire with a gauge of 48 was imaged at 150X, and 50 ga., 53 ga., and mixed-gauge samples were imaged at 200X. Monodisperse nanofibers were imaged at 5000X and bimodal nanofibers were imaged at 2500X. In total 36 SEM images were analyzed (24 steel wire and 12 PLGA nanofiber).
Manual Measurement
In order to validate DiameterJ, fiber diameter for the wire samples and nanofibers were determined manually from SEM images using the Line tool in ImageJ. Two people experienced in this practice each measured fiber diameter at 25 random locations in each micrograph. Each person analyzed six micrographs for 6 samples: wire samples (48 ga., 50 ga., 53, ga., mixed ga.), PLGA monodisperse nanofibers and PLGA bimodal nanofibers, for a total 36 images and 150 data points per sample per person. The 300 data points for each sample (150 data points from 2 people) were plotted on a histogram and peak-fitting (described below) was used to assess the results. This manual measurement was not seen as a “ground truth” for the images and the protocol was chosen by using a method that is indicative of that which is used in the literature.[54] All manual human measures and their histograms can be seen in Figure 3 and Figure 4 of reference [53].
DiameterJ Algorithm
A design requirement for the DiameterJ algorithm was to be able to analyze an 8-bit SEM image of any resolution using a desktop computer in less than 60 seconds. DiameterJ was tested on a Dell Optiplex 9010 (Windows 8, 8 GB of RAM, Intel 3rd generation i7-3770) and functionality was also verified on an older system (Windows 8.1, 4 GB RAM, AMD Phenom II 2.5 Ghz processor, built in 2009). The DiameterJ algorithm has been published to the ImageJ repository and is submitted to be incorporated into the Fiji base release. The DiameterJ plugin can be downloaded from the ImageJ Wiki, http://imagej.net/DiameterJand installed into ImageJ 1.48 or newer or the latest release of Fiji. See Fig. 1 for an overview of how the DiameterJ algorithm analyzes fiber diameter and other scaffold properties.
Segmentation
The first step to image analysis is segmenting the image into foreground and background. SEM micrographs were segmented using eight different thresholding techniques available as plugins within ImageJ/FIJI. Section 2.2 of reference [53] contains an in-depth description of the segmentation algorithms used and the post-processing of segmented images. Generally, images were segmented with either global, local, machine learning or edge detection methods and then noise was removed and line smoothing was performed on resulting images.
Super Pixel Diameter Determination
The Super Pixel approach yields a single fiber diameter for each image and does not give a histogram. The Super Pixel algorithm divides the area of all the fibers in an image by the total length of all the fibers’ centerlines. This transforms the unit measurement size from a single pixel to a new value called the “Super Pixel” diameter which is equal to the mean fiber diameter. To obtain this calculation, white pixels from either digital synthetic or segmented images were summed for total fiber area in each image. Additionally, two different centerlines were calculated for nanofibers in each image. The first centerline was determined using a thinning algorithm developed by Zhang and Suen[55] which is sensitive to changes in the fiber surface and results in branches that are not necessarily fibers (sensitive centerline (SC) determination). A second centerline was determined using a Voronoi spatial tessellation[56] which was built based on connected clusters of black (background) pixels and is insensitive to fiber surface morphology (insensitive centerline (IC) determination). The length of the centerlines was calculated and the total area of fibers was divided by either the SC or IC length. This calculation gave the first two approximations of the Super Pixel diameter (DSP): . The Super Pixel name was chosen because the fiber area (in pixels) was divided by the centerline lengths (in pixels, producing a unit-less value that is equivalent to a transformed (larger) pixel unit, equivalent to the mean fiber diameter. This method for calculation of global mean fiber diameter is based on the assumption that fibers are long and rectangular when segmented in 2D images (fiber length × diameter = fiber area).
Using uncorrected centerline lengths to determine fiber diameter gave an overly large diameter value, and thus an intersection correction step was introduced which removed all intersections from the image. Intersection correction takes the length of either the SC or IC and subtracts a radius value (obtained from first approximation of the diameter as determined without intersection correction) for each three-point intersection or a diameter value (obtained from first approximation) for each four-point intersection of the fibers. Intersections of each centerline were found using the algorithm developed by Arganda-Carreras et al.[57]. A new diameter was calculated using the new intersection-subtracted length and the total fiber area. This processes was repeated until the diameter converged 1/1000th of a pixel. A thousandth of a pixel was chosen to drop the error to below 0.01 percent on images where fibers were 10 px in diameter. The results from each method (SC or IC) were averaged to produce the final Super Pixel diameter.
Additionally, the number of intersections and their location was saved and the intersection density (ID) was calculated for a 100 × 100 pixel area by dividing the total number of intersections by the total image area (in pixels) and multiplying by 104: . This value can be converted to intersections per unit area by the user. The intersections for the SC method were used for this calculation because the IC method was found to frequently miss branches of fibers if they did not segment mesh holes completely. The characteristic fiber length (CFL) was defined as the mean length of fiber between intersections and was calculated by dividing the total centerline length (LengthFiber) by the number of intersections: . The centerline length (LengthFiber) was determined by taking the average of the SC and IC fiber lengths.
Fiber Diameter Histogram
DiameterJ includes a second algorithm for determining fiber diameter (referred to as “Histogram”) which determines the fiber diameter at every pixel along the fiber lengths to produce a histogram of fiber diameters. To obtain the distribution of fiber diameters, the segmented image was transformed with a Euclidian distance transformation algorithm[58]. This algorithm takes a fiber pixel and finds the distance to the nearest mesh hole using the ‘square root of the sum of the square of the vertical and horizontal distances to the hole’ and then transforms the fiber pixel to a grey scale value equal to that distance. The resulting image is a greyscale image rather than black and white. The SC (sensitive centerline determination) was overlaid on top of the distance transformed image. At each intersection of the centerline, the greyscale value was found and radius values within that range were subtracted out from the centerline (this step removes intersections). The greyscale values under the remaining centerline were obtained and multiplied by 2 to get the value of all diameters not in an intersection area. The subsequent histogram of greyscale values was found and placed in a ".csv” file along with the overall average, standard deviation, median and mode of all diameter values. The SC was used because it gave the more correct answer when used to analyze digital synthetic images with known intersection densities. It was hypothesized that using the SC produced an answer that was closer to known values than when using IC because SC removes more intersections than IC and because SC has a higher likelihood of eliminating pixels that have a higher value than the true radius.
Mesh Hole Analysis
As a convenience to the user, mesh hole analysis was included in the plugin using a previously developed algorithm [35]. Black pixels in segmented images (void space) were analyzed in ImageJ using the Analyze Particles command, which counts the number of pixels in discrete clusters of black pixels (“Pixel units” was selected, “Circularity” set to range of 0.00 to 1.00, “Exclude clusters that touch the edge” was checked). The particle analysis was used to generate mesh hole histograms, mean mesh hole area (produced by averaging all cluster areas), and percent mesh hole (produced by taking the total number of black pixels and dividing it by the total image resolution).
Fiber Orientation
As a convenience to the user, fiber orientation analysis was also included in DiameterJ using OrientationJ, an established algorithm for ImageJ. [36] To determine fiber orientation, the SC was used and enlarged by 2 px (using the Enlarge command in ImageJ/FIJI) to ensure accurate measure of the line. The Fourier gradient was used with a Gaussian window equal in size to the mean fiber diameter as determined by the DiameterJ Super Pixel algorithm. The subsequent frequency histogram of fiber orientation was reported. In addition, a metric found in the literature to define the orientation of a scaffold, [43] the normalized orientation index (NOI), was calculated as follows: NOI = (90-x)/90 × 100, where x is the number of degrees you must expand from the prevalent orientation angle in both directions encompass 50% of the total orientations. In this way, an NOI close to 0% or 100% indicates highly oriented fibers while an NOI near 50% indicates random orientation.
Image Analysis
Image analysis was performed using BoneJ an open source ImageJ plugin, FibraQuant (proprietary code, PrC) (NanoScaffold Technologies) a commercial tool that is able to produce histograms of fiber diameter, and using DiameterJ. Explanations of how absolute and percent errors were calculated are described can be found in section 2.3 of reference [53].
Validating Particle Analysis, OrientationJ and AnalyzeSkeleton
DiameterJ incorporates the Particle Analysis algorithms within ImageJ for mesh hole analysis, the OrientationJ Plugin for fiber orientation [36], and uses the AnalyzeSkeleton plugin[57] from FIJI. These algorithms were validated using the digital synthetic images discussed above. The output from these algorithms was used to calculate normalized orientation index (NOI), mesh hole size, porosity, intersection density and characteristic fiber length (CFL).
Statistical Analysis
All comparisons between groups were performed via a repeated measures 2-Way ANOVA with Tukey’s post-test using an α < 0.05 for significance using R[59] unless otherwise indicated.
Results
DiameterJ Performance on Digital Synthetic Images
DiameterJ Super Pixel was validated with a set of 118 digital synthetic images (Ordered-1D, Disordered-1D, Ordered-3D, Disordered-3D, see Figure 1 in reference [53] for Tiff images). Examples of digital synthetic image sets can be seen in Fig. 2 A, C, E and G. An analysis was performed on the digital synthetic images for the DiameterJ Super Pixel algorithm where centerlines determined by the IC and SC approaches were compared (Table 1). Generally, the IC derived diameter was smaller than the known diameter and the SC derived diameter was larger. The average of the IC and SC error when averaged across all 118 digital synthetic images was 31% lower than that of either the IC or SC derived diameters. Thus, the remainder of the analyses were performed using the average of the IC and SC diameter which was designated the Super Pixel diameter.
Table 1.
Representative Digital Synthetic Image Analyses Using DiameterJ Super Pixel: Comparison of Results from the IC and SC Centerline Determinations*
| Image Type | Actual Diameter (px) |
Insensitive - IC (px) |
Sensitive - SC (px) |
Average of IC & SC (px) |
Insensitive - IC (% Error) |
Sensitive - SC (% Error) |
Average of IC & SC (% Error) |
|---|---|---|---|---|---|---|---|
| Ordered-1D | 50 | 50.7 | 53.5 | 52.1 | 1.4% | 7.1% | 4.3% |
| Disordered-1D | 15 | 12.7 | 16.0 | 14.4 | 15.1% | 6.6% | 4.3% |
| Ordered-3D | 70 | 70.4 | 77.9 | 74.1 | 0.5% | 11.3% | 5.9% |
| Disordered-3D | 35 | 31.7 | 36.9 | 34.3 | 9.4% | 5.34% | 2.0% |
Next, the 112 digital synthetic images were used to compare the performance of DiameterJ Super Pixel, DiameterJ Histogram and BoneJ (Fig. 2). Both Super Pixel and the mean Histogram diameter had less than 10% error for digital synthetic images with single line diameters that were greater than 10 px and less than 125 px. The average Super Pixel error was lower than both mean Histogram and BoneJ when analyzing images with three diameters (Ordered-3D and Disordered-3D). For either the Super Pixel or Histogram algorithms, when analyzing single diameter images (Ordered-1D, Disordered-1D), the percent error had a high initial value at smaller diameters, then dropped below 10% at a diameter of 10 px and remained below 10% until diameters rose above 125 px. For Ordered-3D and Disordered-3D images, which had three line diameters, percent error generally increased with increasing mean line diameter and the Super Pixel algorithm performed better than did the Histogram or BoneJ. In almost every digital synthetic image measured, BoneJ had a higher mean percent error than did Super Pixel or Histogram. Table 2 shows the mean error of the algorithms across all digital synthetic images with line diameters between 10px and 125px. DiameterJ’s Histogram algorithm had the lowest percent error for single diameter images (Ordered-1D, Disordered-1D) while Super Pixel had the lowest percent error for Ordered-3D and Disordered-3D images.
Table 2.
Percent Error of the Super Pixel, Histogram, and BoneJ Algorithms for Digital Synthetic Images with Line Diameters between 10 px and 125 px
| Ordered-1D (% Error) |
Disordered-1D (% Error) |
Ordered-3D (% Error) |
Disordered-3D (% Error) |
|
|---|---|---|---|---|
| Super Pixel | 4.9 ± 2.2 | 7.2 ± 4.0 | 7.2 ± 4.4 | 7.8 ± 4.3 |
| Histogram | 2.5 ± 1.9 | 4.7 ± 1.4 | 19.1 ± 13.5 | 16 ± 10.7 |
| BoneJ | 13.6 ± 3.4 | 57.6 ± 12.7 | 122.6 ± 87.6 | 158.9 ± 23.7 |
An advantage of DiameterJ Histogram is that many fiber diameter measurements are made on each image enabling histograms of fiber diameter distributions to be generated and analyzed via peak-fitting (Fig. 3). Gaussian curves were fit to histograms of Multi-Dia. digital synthetic images to find the mean diameter of each peak (Fig. 3C). The data and fits for the histograms as well as the original digital synthetic images can be seen in Figure 2 of reference [53]. For images with multiple line diameters that were close together multimodal peak-fitting became more difficult due to overlap between the peaks (see six line diameter image in Fig. S1). To measure the deviation of the peak-fits, the absolute (Fig. 3D) and percent (Fig. 3E) error between the known diameters in digital synthetic images and the peak locations were calculated. The mean absolute error of the peak-fits was less than 0.6 px from the known value in digital synthetic images with a maximum error of 1.5 px. This absolute error translated to a mean percent error of less than 3 % for all Mult-Dia digital synthetic images (Fig. 3E). In Fig. 3E, the larger percent error for the 4-diameter images was caused by the fact that these images had a high percentage of lines less than 10 px in diameter, leading to an absolute error of 0.5 px to 1.0 px. Measurements on 10-diameter Multi-Dia. digital synthetic images were not successful since the peaks were too close to one another to resolve via peak-fitting. Thus, peaks should be separated by at least 3 px in order to enable accurate peak-fitting. These results validate that Super Pixel and Histogram were able to effectively determine line diameters in digital synthetic images. FibreQuant was not designed to analyze segmented images ((black and white images) and, thus, could not be validated on digital synthetic images.
DiameterJ Validation on Wire Micrographs
After validating DiameterJ with digital synthetic images, the program was tested on SEM images of wires with “known” diameters. Obtaining polymer nanofibers with a “known” diameter is not possible because there is a distribution of fiber diameters in electrospun nanofiber mats. Thus, small gauge (48, 50, or 53 ga.) 316 stainless steel wire with nominally monodispersed diameter was used to provide a measurement base for DiameterJ (Fig. 4, Fig. S2, Fig. S3). Manual measurement of wires produced an error less than 1% for both the single wire and three wire micrographs (Fig. 4, Fig. S3). The two analysts’ measurements were shown to produce statistically not different results for the wire diameter (Fig. S3). The modes of the DiameterJ Histogram results shown in Fig. 4D (before peak-fitting) differed by less than 1.2% from known diameter of the wires. The histograms showed a tight dispersion of measured wire diameters with none of the Gaussian peak-fits having a standard deviation greater than 0.64 ± 0.1 µm. The data and fits for the histograms as well as the original SEM images and their segments can be seen in Figure 3 of reference [53].
Analysis of wire micrographs with different measurement tools (Super Pixel, Histogram, BoneJ, Manual, PrC) revealed that each tool produced a similar amount of error when comparing measurements between the three sizes of single ga. micrographs (Fig. 4E–F). When comparing percent error between different tools, DiameterJ’s Histogram was not statistically different from Manual or PrC in either single ga. or mixed-ga. micrographs (Fig. 4E–F). The Super Pixel algorithm had a statistically higher error than the Manual measurement when measuring the 48 ga. and 50 ga. wire. BoneJ had a statistically higher error when compared to any other tool across all three gauges. BoneJ included all fiber intersections in its analysis which most likely led to inflated average diameters as shown Fig. S3. For mixed-ga. image analysis in Fig. 4F, all methods tested produced similar levels of mean percent error (less than 1.5 %) and were statistically different from one another. Note that only DiameterJ Histogram, Manual measurement, and PrC were included in Fig. 4F (and Fig. 5F) because they provide fiber diameter histograms capable of providing multiple fiber diameters (BoneJ and Super Pixel only provide a global average fiber dia). Taken together, these results indicate that Super Pixel and Histogram produced fiber diameter measurements from SEMs of steel wire that were equivalent to the measures produced using other measuring methodologies.
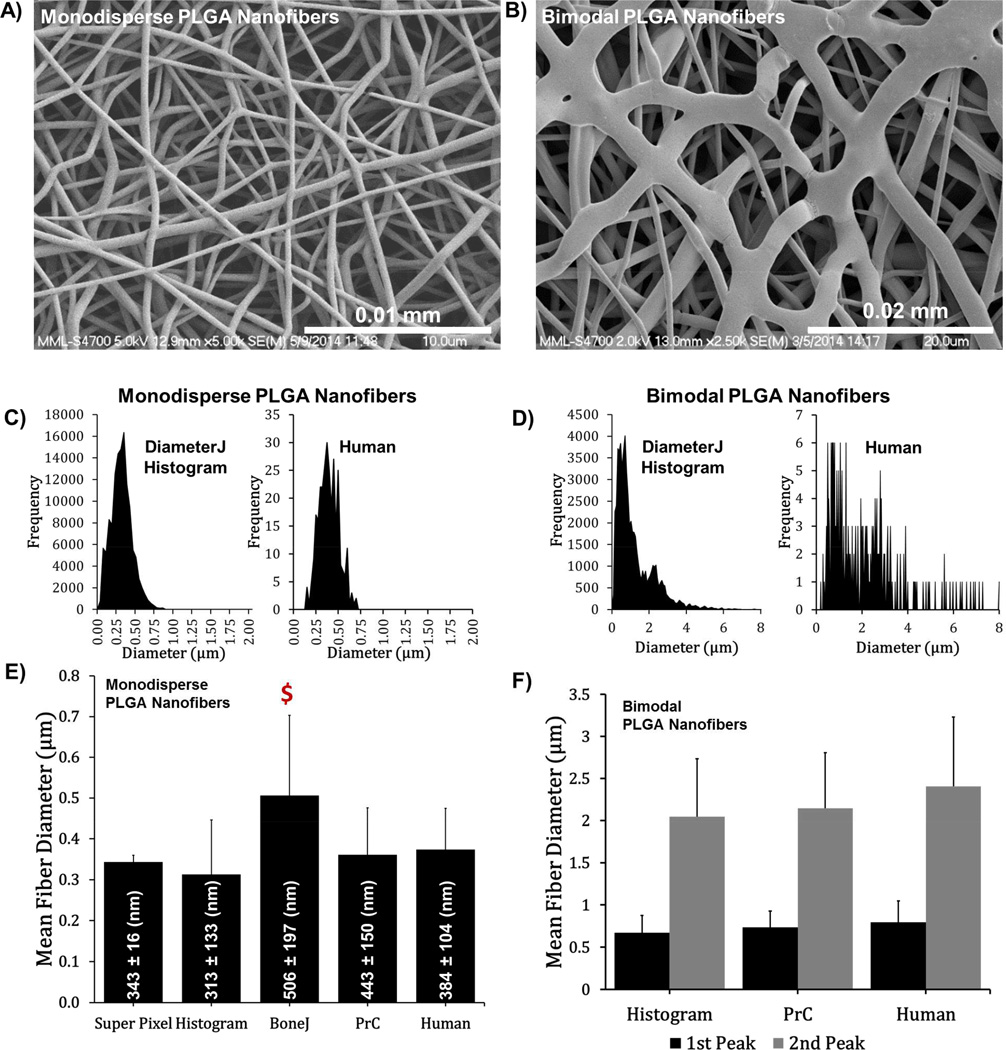
Figure 5.
Analysis of SEM micrographs of PLGA nanofibers. (A, B) SEM micrographs of PLGA nanofibers. (C, D) Cumulative fiber diameter histograms generated by DiameterJ Histogram (left) and by human measure (right). (E, F) Mean fiber diameter for different methodologies/algorithms. The mean and standard deviation in nm are given on the bars in (E). N = 6 images for all histograms and graphs. Error bars are standard deviation. Money sign ($) represents statistically different (P<0.1) from DiameterJ’s Histogram algorithm.
PLGA Nanofiber Measurements
DiameterJ was used to analyze SEM micrographs of monodisperse and bimodal PLGA nanofibers (Fig. 5). DiameterJ Histogram generated between 3000 and 12000 fiber diameter counts per image, depending on the image properties (number of fibers present, diameter of fibers, etc.). Thus, the signal to noise of the histograms generated by DiameterJ Histogram is higher and more robust peak-fitting is possible. Manual measurements of PLGA nanofiber diameters were not statistically different from Super Pixel, Histogram or PrC (Fig. 5). BoneJ calculated a diameter that was statistically higher than DiameterJ Histogram for monodispersed nanofibers. For the bimodal PLGA nanofibers (Fig. 5F), all techniques were found to be statistically equal further confirming the results seen in Figure 4. The data and fits for the histograms as well as the original digital synthetic images can be seen in Figure 4 of reference [53].
Of note was the time taken to analyze images. For manual measurement each analyst spent approximately 8 minutes per image leading to a total time of analysis of approximately 5 hours per analyst to generate 300 data points for each of the six samples (48 ga., 50 ga., 53 ga., monodispersed PLGA fibers, and bimodal PLGA fibers). Compared to DiameterJ (with included automated segmentation algorithms) taking less than 40 minutes to analyze all six samples while generating tens of thousands of measurements per sample. In summary, these results demonstrate that DiameterJ algorithms agreed with manual measures of PLGA fiber diameter, but were 10s of times faster while providing 100s of times the data for peak fitting.
Segmentation and Fiber Diameter Analysis
In order to enable flexibility, DiameterJ does not prescribe a segmentation algorithm and analyzes images after segmentation. SEM image properties vary widely between instruments, instrument settings, users, sample preparations and fiber compositions. The best results can be obtained if the user selects the segmentation algorithm that works best for their image sets. For convenience, several segmentation tools have been packaged with DiameterJ to help users obtain an adequate segmentation for fiber analysis. These segmentation tools use 16 common algorithms to segment an SEM image and provides them to the user so that the best approach may be selected for fiber diameter analysis via DiameterJ. In addition, Fiji has more than 25 global and local segmentation algorithms, as well as several machine learning and edge detection algorithms built into its latest release. Further, a tool has also been packaged with DiameterJ, for convenience, which allows the user to identify where in the image DiameterJ is obtaining any given fiber diameter measure.
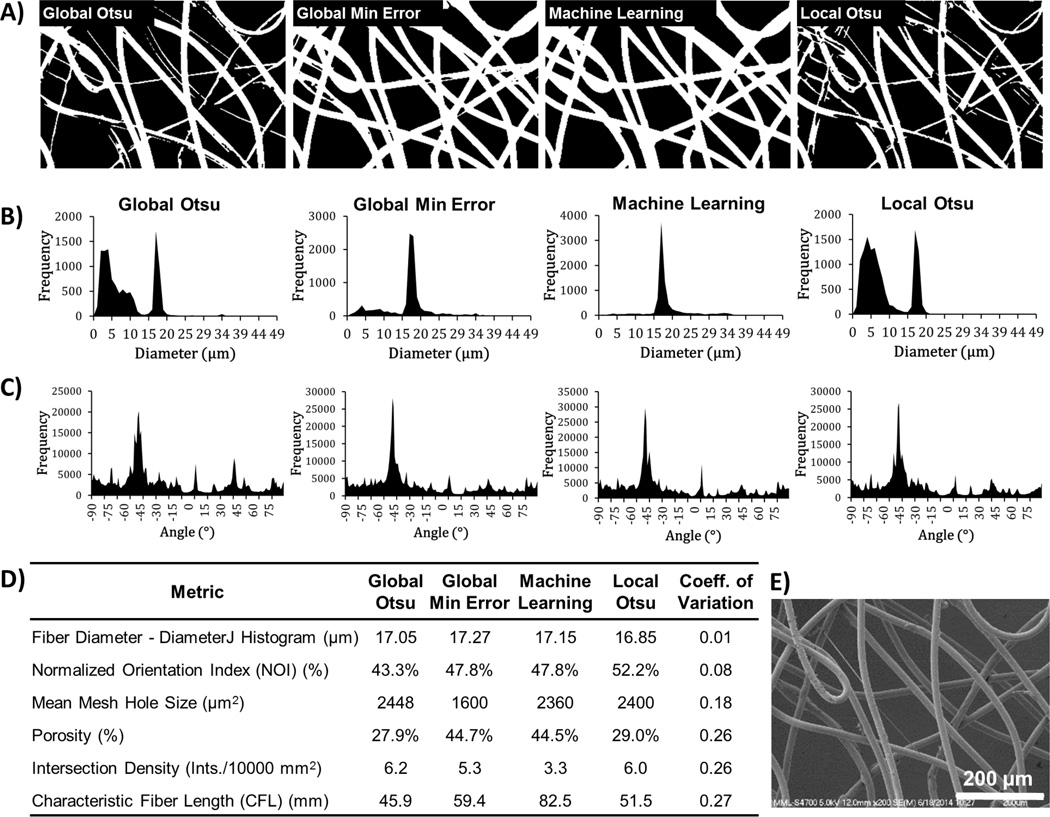
Fig. 6A shows 4 different segmentations of a steel wire SEM image and visual inspection reveals that two had little to no partial fiber segmentations (Global – Min Error and Machine Learning) and two segmented many fibers partially (Global – Otsu and Local Otsu). The Histogram fiber diameter distributions are dependent on the segmentation algorithm and thus, errant peaks appear between 0 µm and 10 µm for the segmentation algorithms that partially segmented fibers (Fig. 6B). Fig. S4 shows further confirmation of the heterogeneity of segmentation algorithms when analyzing other SEM images. However, despite the differences in the segmentation, peak fitting of DiameterJ Histogram results for the 4 segmentations were consistent (0.01 coefficient of variation). These results suggest that DiameterJ Histogram fiber diameter measurement and analysis were relatively insensitive to the choice of segmentation algorithm.
Figure 6.
Segmentation algorithms and analysis of fiber dia., fiber orientation, mesh holes, porosity, intersection density and fiber length. (A) An SEM micrograph of 53 gauge wire (dia. 16.7 mm) was segmented with the indicated algorithms. Diameter (B) and Orientation (C) histograms produced by DiameterJ’s Histogram algorithm and OrientationJ given below their corresponding segmented images in (A). (D) Summary table of DiameterJ analysis of the segmented images. (E) Original SEM image of 53 ga. steel wire.
Mesh Hole Size, Fiber Orientation, Intersection Density, Porosity and Fiber Length
DiameterJ incorporates several previously published plugins including mesh hole size, fiber orientation [36], and fiber intersection density[57]. Prior to incorporation into DiameterJ, the Particle Analyzer algorithm and OrientationJ were validated using digital synthetic images (Fig. S5). Ordered-1D digital synthetic images were analyzed because their mesh hole size and fiber orientations were known. The mesh hole size, porosity and fiber orientation results produced by the Analyze Particle algorithm and OrientationJ differed by less than 1 % from the known values for the digital synthetic images. Intersections were counted manually in all digital synthetic images to determine the accuracy of the algorithm and DiameterJ was found to be > 95 % consistent with manual measurements for intersection density calculations. Finally, the method used for image segmentation induced greater variability on these additional fiber metrics than it had on fiber diameter (Fig. 6). While the fiber diameter measurements had a coeff. of variation of only 1 % when a wire image was segmented with four different algorithms, NOI, mean mesh hole size, porosity, intersection density and CFL had a coeff. of variation almost an order of magnitude higher as can be seen in Fig. 6D (8.0% to 27%). The data that was analyzed for this analysis can be seen in Figure 5 of reference [53].
Discussion
Two fundamentally distinct algorithms for determining fiber diameter, Super Pixel and Histogram, were included in DiameterJ so that the program could analyze a larger range of image types. Super Pixel yields a single mean fiber diameter value from each image analyzed. In contrast, Histogram yields a fiber diameter measurement at every pixel along the length of every fiber, yielding thousands of data points for each image. These large data sets enable fiber diameter distributions to be plotted and analyzed via peak-fitting; providing better confidence in fiber diameter measurements. When comparing data from Tables 1 and 2 as well as the information from Figure 2 it can be seen that the Super Pixel algorithm has lower error when reporting a global mean diameter in images with multiple diameters and Histogram has less error when analyzing images with monodispersed fiber diameters. However, because Histogram can show distributions of fiber diameter and because it can show multiple modes in a given image the Authors recommend using the Histogram algorithm with peak fitting for most use cases unless the User truly only needs a global mean of multi-modal fibers. Additionally, when comparing this work to the results found by Stanger et al. DiameterJ appears to produce results that are not statistically different from that of all the commercially reviewed pieces of software in that study. For a more in-depth discussion of the underlying algorithmic reasons for the error trends shown in these Tables and Figures see the Supplemental Discussion of this report.
A primary reason that DiameterJ placed a priority on measurement of fiber diameter was because two-dimensional (2D) images of tubular structures, such as fibers, can be used to determine the diameter of a tube. The relationship between the measured fiber orientation, mesh holes, percent porosity, intersection density and fiber length and the true structure of a nanofiber scaffold is unclear and an ongoing area of research. For a more in-depth discussion of the relationship between these metrics and weighted fiber diameter estimates produced from the Histogram algorithm see the Supplemental Discussion of this report.
The results yielded specifications for using DiameterJ effectively. First, fibers in images should be at least 10 px in diameter. Second, fibers should not be greater than 10% of the smallest dimension of the image. Measurements on fiber images outside of this range yielded percent errors over 10 %. For example, to stay within this range SEM imaging of 500 nm fibers should be conducted at a magnification between 1500X and 10000X for a 1280 px by 960 px image capture. Third, samples with multi-modal fiber diameter distributions must have modes that are separated by greater than 3 px in order for DiameterJ Histogram to be reliable. All three of these specifications can be met by choosing the appropriate magnification and image resolution during SEM image capture. Fourth, after obtaining the histogram data from DiameterJ a “common sense” check between the histogram and the image is needed. Fibers running along the edge of an image, poor segmentation, or images where many fibers overlap or become entwined can yield errant peaks in the histogram (as shown in Fig. 6B). A quick review of the original image can confirm that these measurements are in error and should not be included in the analysis. If there are questions about the source of a fiber diameter peak in a histogram, then a locator tool, included with DiameterJ, can be used to visualize which lines in an analyzed image produced fiber diameters for a given range.
DiameterJ is an open source program and thus its value lies not just in the fact that it is free but that it can be modified by others to adapt to their needs. For example, natural matrices, such as collagen or extracellular matrix, were not assessed in this study, but, due to their fibrous nature, they could be analyzed with DiameterJ. However, if the density of fibers was too high for DiameterJ to obtain enough measurements for a reliable histogram, a researcher could modify DiameterJ’s intersection correction algorithms to better represent their particular type of sample. Further, due to DiameterJ’s separation of segmentation and analysis, if a technique other than SEM is used, such as transmission electron microscopy, fluorescence microscopy, brightfield microscopy or X-ray micro-computed tomography, then DiameterJ would be able to analyze these images as well, as long as they were segmented effectively. Thus, DiameterJ is the only tool that can be used independent of imaging modality.
Conclusions
DiameterJ is a rapid, validated, open-source tool for analyzing fiber diameter in scanning electron micrographs. DiameterJ was ten times faster and obtained at least 2 orders of magnitude more fiber diameter measurements (3000 to 12000 data points in 10 s) than manual measurements using software Line tools (25 data points in ~100 s). The large data sets afforded by DiameterJ enabled fiber diameter distributions to be plotted and analyzed via peak-fitting, providing better confidence in fiber diameter measurements. DiameterJ was validated on a wide range of digital synthetic images that mimic segmented fiber images (white lines on a black background) and with SEM images of steel wires of known diameters. Further, when analyzing fiber diameter in wire SEM images, DiameterJ results were comparable to manual operator measurements, demonstrating that throughput was not purchased at the expense of accuracy. Most importantly, DiameterJ provides an accessible, rapid, validated, flexible, free and easy-to-use tool that engineers, biologists, and other scientists can use to efficiently analyze fiber diameter in nanofiber scaffolds.
Supplementary Material
Acknowledgements
We thank Daniel Krogstad for help with Igor Pro peak-fitting software, Desu Chen for help with image processing and Antonio D’Amore for help with SEM image analysis. The “standard deviation” (S.D.) is the same as the “combined standard uncertainty of the mean” for the purposes of this work. This article, a contribution of NIST, is not subject to US copyright. Certain equipment and instruments or materials are identified in the paper to adequately specify the experimental details. Such identification does not imply recommendation by NIST, nor does it imply the materials are necessarily the best available for the purpose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mo XM, Xu CY, Kotaki M, Ramakrishna S. Electrospun P(LLA-CL) nanofiber: a biomimetic extracellular matrix for smooth muscle cell and endothelial cell proliferation. Biomaterials. 2004;25:1883–1890. doi: 10.1016/j.biomaterials.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 2.Yoshimoto H, Shin YM, Terai H, Vacanti JP. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials. 2003;24:2077–2082. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 3.Gopal R, Kaur S, Ma Z, Chan C, Ramakrishna S, Matsuura T. Electrospun nanofibrous filtration membrane. J Membr Sci. 2006;281:581–586. [Google Scholar]
- 4.Qin X-H, Wang S-Y. Filtration properties of electrospinning nanofibers. J Appl Polym Sci. 2006;102:1285–1290. [Google Scholar]
- 5.Zhang Q, Welch J, Park H, Wu C-Y, Sigmund W, Marijnissen JCM. Improvement in nanofiber filtration by multiple thin layers of nanofiber mats. J Aerosol Sci. 2010;41:230–236. [Google Scholar]
- 6.Barhate RS, Ramakrishna S. Nanofibrous filtering media: Filtration problems and solutions from tiny materials. J Membr Sci. 2007;296:1–8. [Google Scholar]
- 7.Formo E, Lee E, Campbell D, Xia Y. Functionalization of Electrospun TiO2 Nanofibers with Pt Nanoparticles and Nanowires for Catalytic Applications. Nano Lett. 2008;8:668–672. doi: 10.1021/nl073163v. [DOI] [PubMed] [Google Scholar]
- 8.Chronakis IS. Novel nanocomposites and nanoceramics based on polymer nanofibers using electrospinning process—A review. J Mater Process Technol. 2005;167:283–293. [Google Scholar]
- 9.Demir MM, Gulgun MA, Menceloglu YZ, Erman B, Abramchuk SS, Makhaeva EE, et al. Palladium Nanoparticles by Electrospinning from Poly(acrylonitrile-co-acrylic acid)−PdCl2 Solutions. Relations between Preparation Conditions, Particle Size, and Catalytic Activity. Macromolecules. 2004;37:1787–1792. [Google Scholar]
- 10.Choi S-W, Park JY, Kim SS. Synthesis of SnO2–ZnO core–shell nanofibers via a novel two-step process and their gas sensing properties. Nanotechnology. 2009;20:465603. doi: 10.1088/0957-4484/20/46/465603. [DOI] [PubMed] [Google Scholar]
- 11.Shin YJ, Wang M, Kameoka J. Electrospun Nanofiber Biosensor for Measuring Glucose Concentration. J Photopolym Sci Technol. 2009;22:235–237. [Google Scholar]
- 12.Li D, Frey MW, Baeumner AJ. Electrospun polylactic acid nanofiber membranes as substrates for biosensor assemblies. J Membr Sci. 2006;279:354–363. [Google Scholar]
- 13.Keun Kwon I, Kidoaki S, Matsuda T. Electrospun nano- to microfiber fabrics made of biodegradable copolyesters: structural characteristics, mechanical properties and cell adhesion potential. Biomaterials. 2005;26:3929–3939. doi: 10.1016/j.biomaterials.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Shin HJ, Lee CH, Cho IH, Kim Y-J, Lee Y-J, Kim IA, et al. Electrospun PLGA nanofiber scaffolds for articular cartilage reconstruction: mechanical stability, degradation and cellular responses under mechanical stimulation in vitro. J Biomater Sci Polym Ed. 2006;17:103–119. doi: 10.1163/156856206774879126. [DOI] [PubMed] [Google Scholar]
- 15.Lee SJ, Oh SH, Liu J, Soker S, Atala A, Yoo JJ. The use of thermal treatments to enhance the mechanical properties of electrospun poly(ε-caprolactone) scaffolds. Biomaterials. 2008;29:1422–1430. doi: 10.1016/j.biomaterials.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Ma H, Huang Y, Shen M, Hu D, Yang H, Zhu M, et al. Enhanced decoloration efficacy of electrospun polymer nanofibers immobilized with Fe/Ni bimetallic nanoparticles. RSC Adv. 2013;3:6455–6465. [Google Scholar]
- 17.Prastomo N, Ayad M, Kawamura G, Matsuda A. Synthesis and characterization of polyaniline nanofiber/TiO2 nanoparticles hybrids. J Ceram Soc Jpn. 2011;119:342–345. [Google Scholar]
- 18.Ren G, Xu X, Liu Q, Cheng J, Yuan X, Wu L, et al. Electrospun poly(vinyl alcohol)/glucose oxidase biocomposite membranes for biosensor applications. React Funct Polym. 2006;66:1559–1564. [Google Scholar]
- 19.Patel AC, Li S, Yuan J-M, Wei Y. In Situ Encapsulation of Horseradish Peroxidase in Electrospun Porous Silica Fibers for Potential Biosensor Applications. Nano Lett. 2006;6:1042–1046. doi: 10.1021/nl0604560. [DOI] [PubMed] [Google Scholar]
- 20.Qin X-H, Wang S-Y. Filtration properties of electrospinning nanofibers. J Appl Polym Sci. 2006;102:1285–1290. [Google Scholar]
- 21.Zhu Y, Leong MF, Ong WF, Chan-Park MB, Chian KS. Esophageal epithelium regeneration on fibronectin grafted poly(l-lactide-co-caprolactone) (PLLC) nanofiber scaffold. Biomaterials. 2007;28:861–868. doi: 10.1016/j.biomaterials.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 22.Lee CH, Shin HJ, Cho IH, Kang Y-M, Kim IA, Park K-D, et al. Nanofiber alignment and direction of mechanical strain affect the ECM production of human ACL fibroblast. Biomaterials. 2005;26:1261–1270. doi: 10.1016/j.biomaterials.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Ku SH, Park CB. Human endothelial cell growth on mussel-inspired nanofiber scaffold for vascular tissue engineering. Biomaterials. 2010;31:9431–9437. doi: 10.1016/j.biomaterials.2010.08.071. [DOI] [PubMed] [Google Scholar]
- 24.Bartneck M, Heffels K-H, Bovi M, Groll J, Zwadlo-Klarwasser G. The role of substrate morphology for the cytokine release profile of immature human primary macrophages. Mater Sci Eng C. 2013;33:5109–5114. doi: 10.1016/j.msec.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zhang C, Wu Y, Han Y, Cui W, Jia L, et al. Interleukin-12p35 Deletion Promotes CD4 T-Cell–Dependent Macrophage Differentiation and Enhances Angiotensin II–Induced Cardiac Fibrosis. Arterioscler Thromb Vasc Biol. 2012;32:1662–1674. doi: 10.1161/ATVBAHA.112.249706. [DOI] [PubMed] [Google Scholar]
- 26.Xin X, Hussain M, Mao JJ. Continuing differentiation of human mesenchymal stem cells and induced chondrogenic and osteogenic lineages in electrospun PLGA nanofiber scaffold. Biomaterials. 2007;28:316–325. doi: 10.1016/j.biomaterials.2006.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, et al. Selective Differentiation of Neural Progenitor Cells by High-Epitope Density Nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 28.Simon CG, Yaszemski MJ, Ratcliffe A, Tomlins P, Luginbuehl R, Tesk JA. ASTM international workshop on standards and measurements for tissue engineering scaffolds. J Biomed Mater Res B Appl Biomater. 2014 doi: 10.1002/jbm.b.33286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar G, Tison CK, Chatterjee K, Pine PS, McDaniel JH, Salit ML, et al. The determination of stem cell fate by 3D scaffold structures through the control of cell shape. Biomaterials. 2011;32:9188–9196. doi: 10.1016/j.biomaterials.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziabari M, Mottaghitalab V, Haghi AK. Simulated image of electrospun nonwoven web of PVA and corresponding nanofiber diameter distribution. Korean J Chem Eng. 2008;25:919–922. [Google Scholar]
- 31.Lin K, Chua K-N, Christopherson GT, Lim S, Mao H-Q. Reducing electrospun nanofiber diameter and variability using cationic amphiphiles. Polymer. 2007;48:6384–6394. [Google Scholar]
- 32.Warnke PH, Alamein M, Skabo S, Stephens S, Bourke R, Heiner P, et al. Primordium of an artificial Bruch’s membrane made of nanofibers for engineering of retinal pigment epithelium cell monolayers. Acta Biomater. 2013;9:9414–9422. doi: 10.1016/j.actbio.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 33.You Y, Min B-M, Lee SJ, Lee TS, Park WH. In vitro degradation behavior of electrospun polyglycolide, polylactide, and poly(lactide-co-glycolide) J Appl Polym Sci. 2005;95:193–200. [Google Scholar]
- 34. [accessed July 28, 2014];Fiji Is Just ImageJ n.d. http://fiji.sc/wiki/index.php/Fiji.
- 35.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. [accessed July 28, 2014];ImageJ n.d. http://imagej.nih.gov/ij/index.html.
- 37.Gitman IM, Askes H, Sluys LJ. Representative volume: Existence and size determination. Eng Fract Mech. 2007;74:2518–2534. [Google Scholar]
- 38.Stanger JJ, Tucker N, Buunk N, Truong YB. A comparison of automated and manual techniques for measurement of electrospun fibre diameter. Polym Test. 2014;40:4–12. [Google Scholar]
- 39.Rezakhaniha R, Agianniotis A, Schrauwen JTC, Griffa A, Sage D, Bouten CVC, et al. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol. 2012;11:461–473. doi: 10.1007/s10237-011-0325-z. [DOI] [PubMed] [Google Scholar]
- 40.Woolley AJ, Desai HA, Steckbeck MA, Patel NK, Otto KJ. In situ characterization of the brain–microdevice interface using Device Capture Histology. J Neurosci Methods. 2011;201:67–77. doi: 10.1016/j.jneumeth.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu ZQ. Scale space approach to directional analysis of images. Appl Opt. 1991;30:1369–1373. doi: 10.1364/AO.30.001369. [DOI] [PubMed] [Google Scholar]
- 42.Schaub NJ, Kirkpatrick SJ, Gilbert RJ. Automated Methods to Determine Electrospun Fiber Alignment and Diameter Using the Radon Transform. BioNanoScience. 2013;3:329–342. [Google Scholar]
- 43.D’Amore A, Stella JA, Wagner WR, Sacks MS. Characterization of the complete fiber network topology of planar fibrous tissues and scaffolds. Biomaterials. 2010;31:5345–5354. doi: 10.1016/j.biomaterials.2010.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomba E, Facco P, Roso M, Modesti M, Bezzo F, Barolo M. Artificial Vision System for the Automatic Measurement of Interfiber Pore Characteristics and Fiber Diameter Distribution in Nanofiber Assemblies. Ind Eng Chem Res. 2010;49:2957–2968. [Google Scholar]
- 45.Igathinathane C, Pordesimo LO, Batchelor WD. Major orthogonal dimensions measurement of food grains by machine vision using ImageJ. Food Res Int. 2009;42:76–84. [Google Scholar]
- 46.Igathinathane C, Pordesimo LO, Columbus EP, Batchelor WD, Methuku SR. Shape identification and particles size distribution from basic shape parameters using ImageJ. Comput Electron Agric. 2008;63:168–182. [Google Scholar]
- 47.Papadopulos F, Spinelli M, Valente S, Foroni L, Orrico C, Alviano F, et al. Common Tasks in Microscopic and Ultrastructural Image Analysis Using ImageJ. Ultrastruct Pathol. 2007;31:401–407. doi: 10.1080/01913120701719189. [DOI] [PubMed] [Google Scholar]
- 48.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doube M, Klosowski MM, Arganda-Carreras I, Cordelieres FP, Dougherty RP, Jackson JS, et al. BoneJ: free and extensible bone image analysis in ImageJ. Bone. 2010;47:1076–1079. doi: 10.1016/j.bone.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Thomopoulos S, Chen C, Birman V, Buehler MJ, Genin GM. Modelling the mechanics of partially mineralized collagen fibrils, fibres and tissue. J R Soc Interface. 2014;11:20130835. doi: 10.1098/rsif.2013.0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert P, Dougherty K-HK. Computing Local Thickness of 3D Structures with ImageJ. Microsc Microanal - MICROSC MICROANAL. 2007:13. [Google Scholar]
- 52.Lyu S, Huang C, Yang H, Zhang X. Electrospun Fibers as a Scaffolding Platform for Bone Tissue Repair. J Orthop Res Off Publ Orthop Res Soc. 2013;31:1382–1389. doi: 10.1002/jor.22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hotaling NA, Bharti K, Kriel H, Simon Jr. CG. Dataset for the Validation and use of DiameterJ an Open Source Nanofiber Diameter Measurement Tool. Data Brief. doi: 10.1016/j.dib.2015.07.012. n.d.:Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tutak W, Sarkar S, Lin-Gibson S, Farooque TM, Jyotsnendu G, Wang D, et al. The support of bone marrow stromal cell differentiation by airbrushed nanofiber scaffolds. Biomaterials. doi: 10.1016/j.biomaterials.2012.12.020. n.d. [DOI] [PubMed] [Google Scholar]
- 55.Zhang TY, Suen CY. A Fast Parallel Algorithm for Thinning Digital Patterns. Commun ACM. 1984;27:236–239. [Google Scholar]
- 56.Okabe A. Spatial Tessellations: Concepts and Applications of Voronoi Diagrams. 2 edition. Chichester; New York: Wiley; 2000. [Google Scholar]
- 57.Arganda-Carreras I, Fernández-González R, Muñoz-Barrutia A, Ortiz-De-Solorzano C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc Res Tech. 2010;73:1019–1029. doi: 10.1002/jemt.20829. [DOI] [PubMed] [Google Scholar]
- 58.Leymarie F, Levine MD. Fast raster scan distance propagation on the discrete rectangular lattice. CVGIP Image Underst. 1992;55:84–94. [Google Scholar]
- 59.R Core Team. R: A language and Enviroonment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 60.Otsu N. A threshold selection method from gray-level histograms. Automatica. 1975;11:23–27. [Google Scholar]
- 61.Huang L-K, Wang M-JJ. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 1995;28:41–51. [Google Scholar]
- 62.Kittler J, Illingworth J. Minimum error thresholding. Pattern Recognit. 1986;19:41–47. [Google Scholar]
- 63.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA Data Mining Software: An Update. SIGKDD Explor Newsl. 2009;11:10–18. [Google Scholar]
- 64.Deriche R. Using Canny’s criteria to derive a recursively implemented optimal edge detector. Int J Comput Vis. 1987;1:167–187. [Google Scholar]
- 65.Gonzalez RC, Eddins SL. Digital Image Processing Using MATLAB, 2nd ed. 2nd edition. S.I.: Gatesmark Publishing; 2001. [Google Scholar]
- 66.Lam L, Lee S-W, Suen CY. Thinning Methodologies-A Comprehensive Survey. IEEE Trans Pattern Anal Mach Intell. 1992;14:869–885. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.