Abstract
Fungus isolated from soil has been evaluated for its antimicrobial activity which showed broad spectrum antimicrobial activity against all the pathogenic microorganisms used. Optimization was done by response surface methodology (RSM) to further optimize the medium which could further enhance the antimicrobial activity by 1.1–1.9 folds. Column chromatography was used to isolate the active compound which was characterized to be by various spectroscopic techniques such NMR, IR and LCMS and it was found to be apparently novel compound 7-methoxy-2,2-dimethyl-4-octa-4′,6′-dienyl-2H-napthalene-1-one. MIC of the active compound ranged from (0.5–15 µg/mL which was found to be comparable with the standard antibiotics. Viable cell count studies of the active compound showed it to be bactericidal in nature. Further, the compound when tested for its biosafety was found neither to be cytotoxic nor mutagenic. Cytotoxicity studies of the compound on cancer cell lines showed a valuable cytotoxic potential against all tested human cancer cell lines. Further, the compound induces apoptosis in lung cancer (A549) cells reveled by increase the distribution of nuclear DNA in Sub G1 phase as observed in flow cytometry. The study demonstrated that an apparently novel compound isolated from Penicillium sp. seems to be a stable and potent antimicrobial.
Electronic supplementary material
The online version of this article (doi:10.1186/s13568-015-0120-9) contains supplementary material, which is available to authorized users.
Keywords: Antimicobial, Fungi, Penicillium, Purification, Cytotoxicity
Introduction
Microorganisms have been used as a source for the production of variety of bioactive metabolites. These include bacteria, algae, fungi, actinomycetes etc. Numbers of bioactive metabolites have been isolated from these natural sources. The discovery and development of antibiotics was one of the most significant advances in the medicine in the 20th century. Till now many antibiotics have been commercialized to fight against various diseases. The bacterial resistance is spreading throughout the world, revealing the steadily decreasing potencies of relevant antibiotics (Gould 2008) thus necessitating the discovery of novel compounds or modification of already existing antimicrobial stock. Now days, multiple drug-resistant microorganisms such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant S. aureus (VRSA), vancomycin-resistant Enterococci (VRE), have increased around the world (Gould 2008). Extended spectrum β-lactamases (ESBLs) producing Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa, are becoming resistant to almost every available antibiotic (Livermore 2009). Soil holds an enormous biodiversity that can be screened for antibiotic production. Because of huge expenditure on synthetic molecules with effective antimicrobial properties, natural products are still worth promise (Newman and Cragg 2007).
Filamentous fungi are widely distributed in the environment. The wonder drug directed the interest of the researchers towards natural resources having different biological activities. Since then, a number of compounds with antibiotic activity have been isolated from different fungi such as, Penicillium janczewskii, Penicilliumcanescens (Kozlovskii et al. 1997), Penicillium sclerotiorium, Penicillium janthinellum, Penicillium citrinum (Takahashi et al. 2008), Myrothecium cinctum (Kobayashi et al. 2004) and Aspergillus fumigatus (Furtado et al. 2005), etc. Fungi are historically important sources of secondary metabolites and they continue to provide new chemical entities with novel biological activities (Baker and Alvi 2004). They provide a rich source of compounds for therapeutic applications including antibacterial (Takahashi et al. 2008), antifungal (Nicoletti et al. 2007), antiviral (Nishihara et al. 2000), immunosuppressant and cholesterol-lowering agents (Kwon et al. 2002). Number of fungi showing differ rent biological activities have been listed in the literature; still a lot remains untapped from diverse soil habitats. Screening of novel strains is bringing about microorganisms, not yet assayed for their antibacterial activity that can yield innovative molecules or useful templates for development of new antibiotics (Takahashi et al. 2008). There is a wide variety of fungi used in the production of these compounds including Penicillium spp. which are also exploited to produce bioactive molecules such as antibiotics, enzymes, and organic acids. A large number of fungal extracts and or extracellular products have been found to possess antimicrobial activity, mainly from the filamentous fungi Penicillium (Petit et al. 2009). Many Penicillium spp. have been screened for bioprocessing since the discovery of penicillin, still many of the species and strains remain untapped which may be useful for various pharmaceutical purposes. Taking cognizance of wide biological importance of Penicillium spp., the present study was designed to isolate and screen the fungi from soil collected from different areas of Punjab (30° 4′ N, 75° 5′ E), India. One such promising isolate, Penicillium sp. showing antimicrobial activity was selected for isolation, purification and characterization of the compound.
Materials and methods
Test organisms
The reference strains of bacteria and yeasts were obtained from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India and the clinical isolate methicillin resistant Staphylococcus aureus (MRSA) was obtained from Post graduate Institute of Medical Education and Research (PGIMER), Chandigarh, India. Reference strains included Gram positive bacteria: Enterococcus faecalis (MTCC-439), S. aureus (MTCC-740), Staphylococcusepidermidis (MTCC-435), Gram negative bacteria: E. coli (MTCC-119), K. pneumoniae 1 (MTCC-109), K. pneumoniae 2 (MTCC-530), P. aeruginosa (MTCC-741), Salmonella typhimurium 1 (MTCC-98), S. typhimurium 2 (MTCC-1251), Shigella flexneri (MTCC-1457) and two yeast strains: Candida albicans (MTCC-227), and Candida tropicalis (MTCC-230).
Fungal isolation and extract preparation
Fungus was isolated as described earlier (Kaur et al. 2014) using yeast extract glucose agar (YGA) medium. Fifty millilitre YPDS medium (g/L) with pH-6, taken in 250 mL flasks, was inoculated with four fungal discs (8 mm) of 6–7 days old culture grown on YGA. After 7 days of incubation at 25°C as stationary culture, the contents were filtered through Whatman filter paper no. 1 and the filtrate obtained was used for assaying its antimicrobial potential.
Antimicrobial screening
Inoculum preparation
A loopful of bacterial and yeast colonies were respectively inoculated into 5 mL of nutrient broth and yeast malt medium and incubated at 37 and 25°C, respectively for 4 h. The turbidity of actively growing microbial suspension was adjusted to match the turbidity standard of 0.5 Mc Farland units (Kaur and Arora 2009). The inoculum thus prepared was used to assay their sensitivity to fungal extract by agar well diffusion assay (Bauer et al. 1996).
Statistical optimization of the medium
On the basis of our previous results (Arora and Kaur 2011) obtained from screening of different carbon and nitrogen sources through one-factor-at-a-time classical method; starch, dextrose and yeast extract were taken as independent variables for further optimization by response surface methodology (RSM) using Box–Behnken design (Arora and Sharma 2011; Box and Behnken 1960). Each variable was studied at three levels (−1, 0, +1); for starch and dextrose these were 0.5, 1.25 and 2%, while for yeast extract it was 0.4, 1.2 and 2%. The experimental design included 17 flasks with five replicates having all the three variables at their central coded values. The mathematical relationship of response G (for each parameter) and independent variable X (X1, dextrose; X2, Starch; X3, Yeast extract) was calculated by the following quadratic model equation.
where, G is the predicted response; β0 is intercepted; β1, β2 and β3 are linear coefficients; β11, β22 and β33 are squared coefficients and β12, β13 and β23 are interaction coefficients. MINITAΒ statistical software was used to obtain optimal working conditions and generate response surface graphs.
Isolation and purification of bioactive compound
For the extraction and purification of active group/component from Pencillium sp., 3 L of the culture broth was extracted with equal volume of butanol (1:1). The organic layer was separated and treated with Na2SO4 and then evaporated to dryness in vacuum and the resulting solids 4.5 g were subjected to column chromatography using silica gel (100–200 mesh size, column 18 mm × 300 mm; Hi-media) packed and pre-equilibrated with hexane. The column was first eluted with equilibration solvent i.e. hexane (two bed volumes) followed by linear gradients of hexane: chloroform (100:0, 90:10, 80:20, 70:30, 60:40, 50:50, 40:60, 30:70, 20:80, 10:90, 0:100) at a flow rate of 1 mL/min followed by linear gradients of hexane: ethyl acetate (100:0, 90:10, 80:20, 70:30, 60:40, 50:50, 40:60, 30:70, 20:80, 10:90, 0:100). Different fractions of 20 mL each were collected and after concentration were subjected to agar disc diffusion assay and thin layer chromatography. Hexane: chloroform (8:2) was used as screening system to develop the chromatograms which were observed under UV light (254 and 365 nm) and in iodine chamber. Fractions which showed similar TLC pattern were pooled, concentrated and again loaded to silica gel (100–200 mesh size), column (10 mm × 300 mm) and pre-equilibrated with hexane (two bed volumes). The column after elution with hexane was eluted with linear gradients of Hexane: Chloroform (as above) and fraction size reduced to 5 mL. The collected fractions were tested for antimicrobial activity and thin layer chromatography. The single bands with similar Rf value were pooled and again checked for antimicrobial activity against S. aureus and C. albicans with disc diffusion assay. These pooled single bands were then loaded to HPLC and were further subjected to various spectroscopic analyses.
HPLC analysis
The purified active fractions as obtained above were further subjected to high pressure liquid chromatography (HPLC) using Dionex P680 HPLC. Acetonitrile (75% aqueous solution) was used as mobile phase at a flow rate 0.3 mL/min and injection volume was 20 µL at a column temperature of 25°C. The detections were monitored at different wavelengths (225, 250, 275 and 300 nm).
Spectroscopic analysis
The purified fraction from column chromatography was further subjected to various spectroscopic analyses for the characterization of the compound. Bruker AC-400 FT (400 MHz) spectrometer was used to record 1H and 13C nuclear magnetic resonance (NMR) spectra. Chemical shifts (δ) are reported as downfield displacements from trimethylsilane (TMS) used as internal standard and coupling constants are reported in Hz. Infrared (IR) spectra was recorded on Varian 660-IR Fourier Transform Infra-Red Spectrometer in KBr pellets coated with chloroform film. A mass spectrum by EI and ESI methods was recorded on Bruker microTOF using ESI-TOF method.
Minimum inhibitory concentration (MIC)
Minimum inhibitory concentration of the butanolic extract as well as for the purified compound was worked out by agar dilution method (Kaur and Arora 2009). A stock solution of butanolic extract (25 mg/mL) and for purified compound (10 mg/mL) concentration was prepared and incorporated into Mueller–Hinton agar medium for bacteria and yeast malt extract medium for yeast. The final concentrations of the compound in the medium containing plates ranged from (0.1–20 mg/mL) for butanolic extract and (0.5–15 µg/mL) for pure compound. The experiment was performed in duplicate along with standard drugs (gentamicin 0.059 mg/mL and amphotericin B 0.75 mg/mL).
Time kill assay
A stock solution 25 mg/mL of butanolic extract of the fungus and for the purified compound stock solution of (10 mg/mL) was prepared and the time kill assay for butanolic extract and the purified compound was performed by viable cell count method (VCC) (Kaur and Arora 2009).
Post antibiotic studies
The PAE of butanolic extract and the compound was studied as described by Raja et al. (2011). The extract was mixed with each test organism (in equal volume) containing approximately 1 × 105 cfu/mL suspended in suitable broth medium. After 2 h exposure the drug activity was stopped by placing 1:1,000 dilution of microbial suspension in a broth drug free pre-warmed suitable double strength broth. CFU sampling was done at an interval of 1 h until visual cloudiness was noted. The PAE was calculated as follows PAE = T − C, where T represents the time required for the count in the test culture to increase 1log 10 cfu/mL above the count observed immediately after drug removal and C represents the time required for the count of the untreated control tubes to increase by 1log 10 cfu/mL.
Biosafety evaluation
Ames mutagenicity testing
Butanolic extract and the purified compound were subjected to both spot and plate incorporation methods of Ames test to evaluate their mutagenicity (Maron and Ames 1983). This Salmonella reverse mutation test is based on histidine dependence and mutations in S. typhimurium (MTCC-1251, IMTECH, Chandigarh, India). In case of plate method, 0.1 mL of the bacterial inoculum was added which was adjusted according to its MIC value and 0.1 mL of butanolic extract (25 mg/mL) as well as purified compound (10 mg/mL) to 5 mL of top agar containing 0.25 mL of 0.5 mM histidine–biotin mixture (1:1 ratio). The contents were mixed and poured onto glucose minimal agar plates immediately. Similarly, the same protocol was followed for spot method except that butanolic extract and the purified compound were impregnated on to disc which was placed in the centre of the plate. Sodium azide (5 μL of 17.2 mg/mL) was used as a positive control while DMSO was used as negative control. The plates were prepared in duplicate and incubated at 37°C for 48 h. The number of visible revertant colonies was counted. The mutagenic potential of the extracts was determined on the basis of number of colonies as compared to positive control.
Cellular toxicity testing using MTT assay
The cellular toxicity of butanolic extract and the purified compound was determined by MTT (3-[(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl] tetrazolium bromide) assay (Ciapetti et al. 1993). Ten millilitre sheep blood was taken into injection syringe containing 3 mL Alsever’s solution (anticoagulant) and transferred to sterile centrifuge tubes. The blood was centrifuged at 1,600×g at room temperature (25 ± 3°C) for 20 min to separate the plasma from the cells. The supernatant was discarded and centrifuged again adding 6 mL phosphate buffer saline (PBS). The blood cells were washed thrice with PBS by centrifugation and the pellet was resuspended in 6 mL of PBS. Various dilutions of these cells using PBS were prepared and counted with the help of a haemocytometer under a light microscope so as to obtain cells equivalent to 1 × 105 cells. The following formula was used to determine the required number of cells: number of cells/mL = number of cells counted in 25 squares × dilution factor × 104. The cell suspension thus prepared was dispensed into Elisa plates (100 µL/well) and incubated at 37°C for overnight. The supernatant was removed carefully and 200 µL of the extract and the purified compound was added and incubated further for 24 h. Supernatant was removed again and added 20 µL MTT solutions (5 mg/mL) to each well and incubated further for 3 h at 37°C on orbital shaker at 60 rpm. After incubation, the supernatant was removed without disturbing the cells and 50 µL DMSO was added to each well to dissolve the formazan crystals. The absorbance was measured at 590 nm using an automated microplate reader (Biorad 680-XR, Japan). The wells with untreated cells served as control.
Cytotoxic activity
The sulforhodamine B (SRB) assay is a colorimetric assay used for cytotoxic screening to assess cell growth inhibition (Skehan et al. 1990). Cells are cultured in a 96 well tissue culture plate and the cell growth which depends upon the rate of multiplication is measured indirectly by the intensity of the color of the dye which is directly proportional to the number of cells present. The human cancer cell lines (procured from N.C.I., Frederick, USA) were grown in tissue culture flasks in complete growth medium (RPMI-1640 with 2 mM glutamine, pH 7.4, supplemented with 10% fetal calf serum, 100 µg/mL streptomycin and 100 units/mL penicillin) in a carbon dioxide incubator (37°C, 5% CO2, 90% RH). The cells at sub-confluent stage were harvested from the flask by treatment with trypsin [0.05% in PBS (pH 7.4) containing 0.02% EDTA]. Cells with viability of more than 98% as determined by trypan blue exclusion were used. For determination of cytotoxicity the cell suspension of 1 × 105 cells/mL was prepared in complete growth medium. Stock solution of the purified compound was prepared in DMSO and it was serially diluted with complete growth medium containing 50 µg/mL of gentamicin to obtain working test solutions of required concentrations. In vitro cytotoxicity against five human cancer cell lines of different tissues was determined using 96-well tissue culture plates. The 100 µL of cell suspension was added to each well of the 96-well tissue culture plate. The cells were allowed to grow in carbon dioxide incubator (37°C, 5% CO2, 90% RH) for 24 h. Test materials in complete growth medium (100 µL) were added after 24 h of incubation to the wells containing cell suspension. The plates were further incubated for 48 h in a carbon dioxide incubator. The cell growth was stopped by gently layering trichloroacetic acid (50%, 50 µL) on top of the medium in all the wells. The plates were incubated at 4°C for 1 h to fix the cells attached to the bottom of the wells. The liquid of all the wells was gently pipette out and discarded. The plates were washed five times with distilled water to remove trichloroacetic acid, growth medium, low molecular weight metabolites, serum proteins etc. The plates were stained with sulforhodamine B dye [0.4% (w/v) in 1% acetic acid (v/v), 100 µL] for 30 min. The plates were washed five times with 1% acetic acid and then air-dried (Skehan et al. 1990). The adsorbed dye was dissolved in Tris–HCl buffer (100 µL, 0.01 M, pH 10.4) and plates were gently stirred for 10 min on a mechanical stirrer. The optical density (OD) was recorded on ELISA reader at 540 nm. The cell growth was determined by subtracting mean OD value of respective blank from the mean OD value of experimental set. Considering the growth in the absence of any test material as 100%, percent growth inhibition was calculated. IC50 values were determined by non-linear regression analysis using Graph Pad Software (2236 Avenida de l Playa La Jolla, CA 92037, USA).
Flow cytometry analysis of nuclear DNA
Analysis of nuclear DNA by flow cytometry is interesting in fundamental research and has broadly contributed to improved knowledge on cell DNA content and their distribution in the various phases of cell cycle. DNA amount in cells is often the single parameter measured for cell cycle studies by flow cytometry. In order to obtain a linear relationship between cellular fluorescence intensity and DNA amount, analyses are performed with fluorescent molecules that bind specifically and stoichiometrically to DNA. Some dyes possess an intercalative binding mode such as propidium iodide etc., whereas others such as Hoechst 33342, DAPI etc. possess an affinity for DNA A-T rich region. To analyze the nuclear DNA content, lung cancer cells (2 × 106 cells/mL/well) were treated with purified compound for 24 h at concentrations 10 and 20 µM, and washed twice with ice-cold PBS, harvested, fixed in cold 70% ethanol in PBS and stored at −20°C for 30 min. After fixation, the cells were incubated with RNase A (0.1 mg/mL) at 37°C for 30 min, stained with propidium iodide (50 µg/mL) for 30 min and then measured for nuclear DNA content using BD-LSR Flowcytometer (Becton–Dickinson, USA) equipped with electronic doublet discrimination capability using blue (488 nm) excitation from argon laser.
Statistical analysis
The values have been presented as mean of three independent experiments. Single factor analysis of variance (ANOVA) was used to calculate the significant difference.
Results
Antimicrobial screening
The isolated fungus was identified as Penicillium sp. close to P. citrinum group by National Fungal Culture Collection of India (NFCCI), Agharkar Research Institute, Pune, India (Accession No. NFCCI 2555) Extracellular culture broth of the selected fungus. It showed antimicrobial activity against two Gram positive bacteria: S. aureus (34 mm) S. epidermidis (36 mm) and MRSA (17 mm), one Gram negative bacteria: K. pneumoniae 1 (17 mm) and one yeast C. albicans (18 mm),while rest of the organisms were found to be resistant to extracellular culture broth. Further, to enhance the antimicrobial activity, RSM experiment was designed using the carbon and nitrogen sources.
Box–Behnken design for statistical optimization of carbon and nitrogen sources for antimicrobial activity of Penicillium sp.
On the basis of results obtained from screening of different carbon and nitrogen sources through one-factor-at-a-time classical method; these were optimized by RSM using Box–Behnken design of experiments
- S. aureus (G1)
Linear effect of starch and squared effect of dextrose was highly significant with P value ≤0.001. Similarly linear effect of yeast extract and squared effect of starch was significant with P value ≤0.05 and R2 value of 94.4%. The response surface graph showed the highest activity at 2% starch, 1–1.5% dextrose, with yeast extract 0.2% (Figure 1a).
- S. epidermidis (G2)
Figure 1.
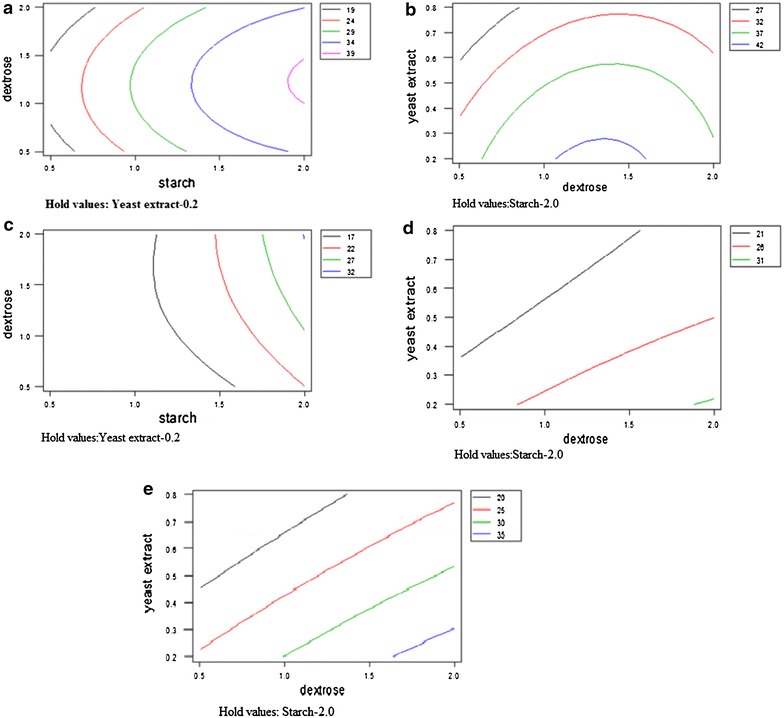
a Contour plot of Staphylococcus aureus. b Contour plot of Staphylococcus epidermidis. c Contour plot of Klebsiella pneumoniae 1. d Contour plot of MRSA. e Contour plot of C. albicans.
Linear effect of starch and squared effect of dextrose was highly significant with P value ≤0.001. Similarly linear effect of yeast extract was found to be significant with P value ≤0.05. Interactive effect of starch and yeast extract was also found to be significant P value ≤0.05 and R2 value observed was 92.7%. The response surface graph showed the highest activity at 1–1.5% dextrose, starch 2% and yeast extract 0.2% (Figure 1b).
- K.pneumoniae 1 (G3)
Linear effect of starch was highly significant with P value ≤0.001. Linear effect of dextrose and squared effect of starch was also significant with P value ≤0.05. Interactive effect of starch and yeast extract, starch and dextrose was significant with P value ≤0.05. R2 value observed was 91.3%. The response surface graph showed the highest activity at starch 2%, dextrose 1.5–2% and yeast extract 0.2% (Figure 1c).
- MRSA (G5)
Linear effect of starch was highly significant with P value ≤0.005. Squared effect of starch and interactive effect starch and yeast extract was significant with P value ≤0.05 and R2 value of 87.1%. The response surface graph showed the highest antimicrobial activity at dextrose 1.5–2%, starch 2% and yeast extract 0.2% (Figure 1d).
- C. albicans
Linear effect of starch was highly significant with P value ≤0.005. Linear effect of dextrose and squared effect of starch was significant with P value ≤0.05. Interactive effect of starch and dextrose; starch and yeast extract was significant with P value ≤0.05 and R2 value of 90.8%. The response surface graph showed the highest antimicrobial activity at dextrose 1.5–2%, starch 2% and yeast extract 0.2% (Figure 1e).
Validation of results
Thus from the overall assessment 2% starch, dextrose 1–2%, yeast extract 0.2% and 1% peptone in YPDS medium may be regarded as the optimized conditions for antimicrobial activity. The F value and R2 value showed that the model correlated well with measured data and was statistically significant. To confirm the adequacy of the model the verification experiments using optimum medium composition as described above were carried out in triplicates. The antimicrobial activity (Table 1) was enhanced by 1.1 folds in S. aureus and S. epidermidis. However, an increase of 1.8 fold was observed for K. pneumoniae1 and MRSA while in C. albicans, it increased by 1.9 folds.
Table 1.
Result of Box–Benhken design experiment for antimicrobial activity of Penicillium sp
| Std. | Dextrose | Starch | Yeast extract | S. aureus | S. epidermidis | K. pneumoniae | C. albicans | MRSA |
|---|---|---|---|---|---|---|---|---|
| (g/100 mL) | Zone of inhibition (mm) | |||||||
| 1 | 0.5 | 0.5 | 0.5 | 15 | 17 | 14 | 17 | 15 |
| 2 | 0.5 | 2 | 0.5 | 27 | 28 | 16 | 17 | 18 |
| 3 | 2 | 0.5 | 0.5 | 20 | 17 | 14 | 17 | 17 |
| 4 | 2 | 2 | 0.5 | 34 | 36 | 28 | 32 | 27 |
| 5 | 1.25 | 0.5 | 0.2 | 20 | 20 | 15 | 16 | 17 |
| 6 | 1.25 | 2 | 0.2 | 40 | 41 | 30 | 34 | 30 |
| 7 | 1.25 | 0.5 | 0.8 | 21 | 25 | 16 | 18 | 18 |
| 8 | 1.25 | 2 | 0.8 | 30 | 32 | 18 | 18 | 18 |
| 9 | 0.5 | 1.25 | 0.2 | 30 | 30 | 14 | 17 | 15 |
| 10 | 2 | 1.25 | 0.2 | 25 | 26 | 16 | 18 | 17 |
| 11 | 0.5 | 1.25 | 0.8 | 22 | 20 | 15 | 16 | 17 |
| 12 | 2 | 1.25 | 0.8 | 25 | 20 | 16 | 17 | 18 |
| 13 | 1.25 | 1.25 | 0.5 | 33 | 34 | 16 | 17 | 16 |
| 14 | 1.25 | 1.25 | 0.5 | 30 | 28 | 17 | 17 | 17 |
| 15 | 1.25 | 1.25 | 0.5 | 32 | 33 | 16 | 18 | 17 |
| 16 | 1.25 | 1.25 | 0.5 | 35 | 33 | 16 | 18 | 16 |
| 17 | 1.25 | 1.25 | 0.5 | 32 | 33 | 16 | 17 | 16 |
Isolation and purification of bioactive compound
For the extraction and purification of active group/component from Penicillium sp., three liters of the culture broth was extracted with equal volume of butanol (1:1). All the fractions (120) obtained from column chromatography were pooled according to similar pattern of chromatogram on TLC plates. Three sets of fractions (A, B and C) were obtained from column chromatography having similar Rf value and antimicrobial activity against various pathogenic bacteria and yeast strains such as S. aureus, S. epidermidis, K. pneumoniae 1, MRSA, C. albicans, S. typhimurium 1, S. typhimurium 2, Sh. flexneri and E. coli. The second set (B) of pooled fraction showed a single spot on TLC with Rf value (0.65 cm). It was further subjected to HPLC analysis to determine the purity of active compound which showed single peak at retention time 8.643 min. However, Set A and C also showed antimicrobial activity but had some impurities. First set (A) showed antimicrobial activity with zone of inhibition ranging from 20 to 23 mm followed by second set (B) which showed zone of inhibition ranging from 17 to 20 mm against various microbial strains. As the second set (B) showed single spot on TLC, it was pursued for further spectroscopic analysis. The compound responsible for antimicrobial activity was characterized by various spectroscopic techniques such as IR, 1H & 13C NMR and mass. Colour of compound: Yellowish brown: IR (KBr, CHCl3): λmax = 2,940, 2,915, 2,876, 1,666, 1,651, 1,535, 1,454, 1,373, 1,284, 1,161, 1,107, 1,072, 968, 756 cm−1; 1H NMR (400 MHz, CDCl3): δ 9.35 (s, 1H, C8-H), 8.26–8.12 (d, 1H, J = 8.0 Hz C5-H), 7.61 (d, 1H, J = 8.0 Hz, C6-H), 7.39–7.20 (m, 2H, C4′-H & C6′-H), 6.91–6.84 (m, 3H, C5′-H, C7′-H & C3-H), 4.37 (s, 3H, OCH3), 2.04 (t, 2H, J = 5.2 Hz, C1′-H), 1.92–1.87 (m, 2H, C2′-H), 1.40–1.24 (m, 2H, C3′-H), 1.11 (s, 6H, 2 × CH3); 13C NMR (100 MHz, CDCl3): δ 170.2 (C=O), 169.1 (olefinic CH), 166.4 (arom. q), 163.7 (olefinic CH), 160.1 (arom. q), 147.9 (C4), 125.6 (olefinic CH), 124.5 (olefinic CH), 121.9 (olefinic CH), 118.8 (arom. CH), 116.2 (arom. CH), 67.0 (OCH3), 57.2 (q), 45.8 (CH2), 44.5 (CH2), 27.8 (CH2), 21.9 (CH3), 19.1 (CH3); HR-MS (TOF, ESI): m/z: calculated for C21H26O2 + [Na]: 333.1803; found: 333.1766 [M + Na]+ (Additional file 1: Figure S1).
Minimum inhibitory concentration (MIC)
Equal volume of the culture broth from Penicillium sp. was extracted with equal volume of butanol in separating funnel. The organic phase was separated and the solvent was then evaporated to dryness under vacuum and the residue obtained was reconstituted in dimethyl sulfoxide (DMSO). Minimum inhibitory concentration was worked out for butanolic extract of the fungus as well as for the purified compound by agar dilution method (Table 2). Butanolic extract showed significant antimicrobial activity with MIC 0.1 mg/mL against S. aureus, and K. pneumoniae 1, followed by MIC 0.2 mg/mL for S. epidermidis and C. albicans, against MRSA, 0.7 mg/mL, S. typhimurium 2 and P. aeruginosa 1 mg/mL. The butanolic extract also showed good inhibitory activity against Sh. flexneri and E. faecalis with MIC 5 mg/mL, followed by MIC value of 10 mg/mL and 20 mg/mL against S. typhimurium 1 and K. pneumoniae 2, respectively. MIC of purified compound against C. albicans and K. pneumoniae 1 with 1 µg/mL as compared to standard antibiotics, amphotericin B (99 µg/mL) and gentamicin (1 µg/mL). It was followed by MIC 2 µg/mL against S. aureus and S. epidermidis whereas against MRSA 5 µg/mL and S. typhimurium showed highest MIC (10 µg/mL).
Table 2.
Comparison of MIC of butanolic extract and pure compound with two standard antibiotics
| Microorganisms | MIC butanolic extract | MIC of compound | MIC of gentamicin | MIC of amphotericin B |
|---|---|---|---|---|
| S. aureus | 0.1c | 2b | 1a | ND |
| S. epidermidis | 0.2c | 2b | 1a | ND |
| K. penumoniae 1 | 0.1c | 1b | 0.19a | ND |
| C. albicans | 0.2b | 1a | ND | ND |
| MRSA | 0.5c | 5b | 0.19a | 99d |
| P. aeruginosa | 1b | ND | 10a | ND |
| S. typhimurium 2 | 0.7c | 5b | 2a | ND |
| Sh. flexneri | 5b | ND | 2a | ND |
| E. faecalis | 5b | ND | 10a | ND |
| E. coli | 10b | ND | 1a | ND |
| S. typhimurium 1 | 15b | ND | 1a | ND |
| K. penumoniae 2 | 20b | ND | 1a | ND |
The values represents mean of three values, different lower case letters (a–c) show statistical significant (P ≤ 0.05) difference between columns.
Time kill assay
Time kill assay of butanolic extract and purified compound of Penicillium sp.
On the basis of 1× MIC, viable cell count studies were carried out. A stock solution 25 mg/mL of butanolic extract of Penicillium sp. and 10 mg/mL for purified compound respectively was prepared. Complete killing of E. coli was observed at 0 h of incubation. K. pneumoniae 1 and C. albicans got completely killed at 2 h. E. faecalis took 4 h for complete killing, MRSA 6 h, S. epidermidis and P. aeruginosa got completely killed at 8 h whereas S. aureus and Sh.flexneri were killed at 10 h of incubation. S. typhimurium 1 took 14 h for complete inhibition and K. pneumoniae2 took the longest time of 24 h for complete killing (Figure 2).
Figure 2.

Time kill studies of butanolic extract of Penicillium sp.
Time kill assay using purified compound was also studied by viable cell count method. Among the tested organism, complete killing of E. coli and K. pneumoniae 1 was observed at 0 h. C. albicans got killed at 2 h of incubation, whereas MRSA and S. typhimurium 2 took 4 h whereas S. epidermidis took 6 h of incubation. S. aureus took the longest time and killed at 10 h of incubation (Figure 3).
Figure 3.
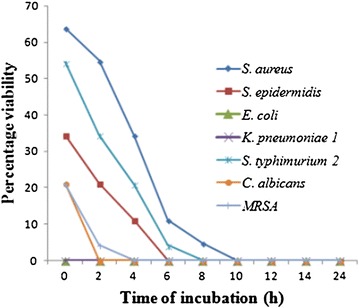
Time kill studies of purified compound.
Post antibiotic effect (PAE)
PAE is persistent suppression of bacterial growth after their brief exposure (1 or 2 h) to an antimicrobial agent even in the absence of host defense mechanisms. The concentrations used in kill time assay were applied in PAE studies. Butanolic extract of Penicillium sp. and the purified compound induced a varied PAE amongst test organisms and was concentration dependent. Butanolic extract of Penicillium sp. induce PAE ranging from 2 to 20 h in the microorganisms tested. Sh. flexneri 2 h, S. aureus 4 h, S. epidermidis 4 h, MRSA 6 h, K. pneumoniae 1 6 h, S. typhimurium 2 6 h, C. albicans 8 h and E. coli 20 h, Similarly, purified compound induced PAE ranging from 4 h to 22 h. S. epidermidis 4 h, S. typhimurium 2 6 h, S. aureus 8 h, MRSA 10 h, C. albicans 12 h, K. pneumoniae 1 20 h, and E. coli 22 h (Figure 4).
Figure 4.
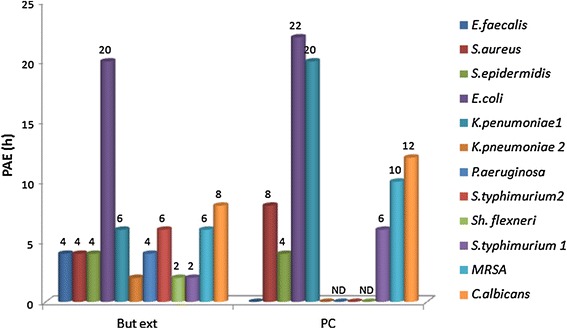
Post antibiotic effect of the butanolic extract and pure compound.
Toxicity testing
To work out the biosafety of the butanolic extract and purified compound they were subjected to Ames mutagenicity test and MTT cytotoxicity assay. While performing the Ames test the number of revertant colonies in the positive control was numerous, whereas the bacteria incubated with the butanolic extract and the purified compound did not show any revertant colonies. The glucose minimal agar media plates layered with top agar containing excess of biotin and no histidine also served as control as no colonies were observed.
In MTT assay, since the reduction of 3-[(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl] tetrazolium bromide (MTT) can only occur in metabolically active cells, as the mitochondrial succinate dehydrogenase enzymes in living cells reduce the yellow colored water soluble substrate MTT into an insoluble, formazan crystals which are then dissolved in DMSO and the absorbance of purple colored solutions directly represents the viability of the cells. In the present studies, 90 and 92% of the viable cells were observed in the butanolic extract and the purified compound respectively, showing both to be noncytotoxic.
Cytotoxic activity against human cancer cell lines
In the present study, purified compound was tested for its cytotoxicity against human cancer cell lines. The compound was found to be endowed with valuable cytotoxic potential against all tested human cancer cell lines (Table 3). The purified compound was found to display promising cytotoxicity with IC50 = 28 against leukemia cancer cell line (THP-1). Similarly, against prostate cancer cell line (PC-3), considerable inhibitory activity with (IC50 = 31) of the compound was observed. While for breast cancer cell line (MCF-7) the IC50 observed was 35 and for lung cancer cell line (A549), +the compound displayed IC50 > 50.
Table 3.
Percentage growth inhibition against different cancer cell lines
| Compounds | Conc (µg/mL) | A549 | THP-1 | PC-3 | Colo-205 |
|---|---|---|---|---|---|
| Lung | Leukemia | Prostate | Colon | ||
| KB14 | 1 | 25 | 0 | 10 | 0 |
| 5 | 28 | 10 | 27 | 20 | |
| 10 | 31 | 22 | 38 | 39 | |
| 30 | 55 | 39 | 45 | 50 | |
| 50 | 75 | 65 | 70 | 72 | |
| IC50 | 28 | >50 | 31 | 35 | |
| 5-FU | 1 | 72 | 74 | – | – |
| Adriamycin | 1 | 67 | 70 | – | 80 |
| Mitomycin-c | 1 | – | 67 | 71 | – |
Flow cytometery analysis of nuclear DNA
Morphological change is a primary indication of cell apoptosis. One of the specific characteristics of cell apoptosis is DNA fragmentation, which leads to the appearance of hypodiploid cells. The cell cycle is a key factor in unregulated cell proliferation leading to cancer and differential sensitivity to apoptosis is linked to the distinct phases of cell cycle (Pucci et al. 2000). The hypo diploid sub-G1 DNA fraction (<2n DNA) was found to increase from 37 to 58% for compound in a concentration dependent manner (Figures 5, 6). In contrast to camptothecin (positive control) showed 48% hypo diploid DNA content at a concentration of 2 µM.
Figure 5.
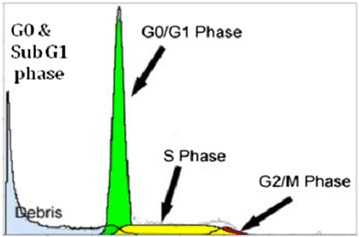
Overview of flowcytometric analysis of nuclear DNA.
Figure 6.
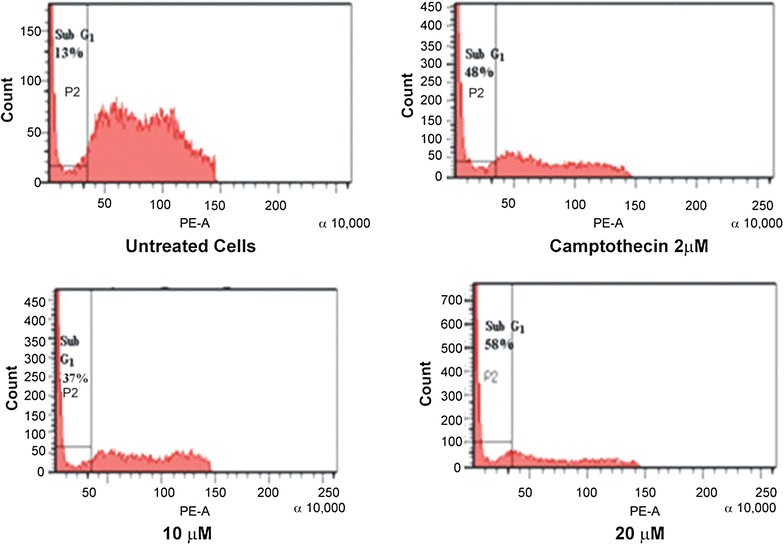
Flow cytometric analysis of nuclear DNA treated compound and the standard.
These results indicate the capability of the purified compound to induce apoptosis in lung cancer (A549) cells.
Discussion
The emerging problem of increasing resistance led the researchers to look for the novel sources for antimicrobial agents or novel compounds from the existing sources. Penicillium spp. have been well known for their bioactive compounds with wide variety of biological activities. A number of Penicillium spp. have been studied but a lot more still remain untapped. Penicillium sp. in this study has been evaluated for its antimicrobial potential and further the active compound has been purified. Even small changes in the culture medium may not only impact the quantity of certain compounds but also the general metabolic profile of microorganisms. The application of statistical experimental design techniques in the fermentation process development can thus result in improvement of product yield and reduce process variability.
The statistical optimization of various nutritional parameters by RSM resulted in 1.1–1.9 folds enhancement in antimicrobial activity. A good agreement between the predicted and experimental results verified the validity of the model and the improvement of antimicrobial activity indicated the RSM to be a powerful tool for determining the exact optimal values of the individual factors and the maximum response value. Our results corroborate well with other studies where the activity was also enhanced by 1.1–1.8 folds (Wang et al. 2011; Zhengyan et al. 2012).
The purified compound is apparently novel compound with broad spectrum antimicrobial activity. The structure is elucidated on the basis of spectroscopic techniques. Proton NMR of the compound showed a signal of C8-H appeared as singlet at δ 9.35 downfield due to anisotropic effect and hydrogen bonding with nearing carbonyl group. The aliphatic multiplet was also observed in the region of δ 1.92–1.87 and δ 1.40–1.24. The 13C NMR spectrum showed aromatic and olefinic carbon resonances in the region of δ 169.1–166.4. The signals of aliphatic carbons appeared at the region of δ 57.2–15.1 and signal of carbonyl group appeared at δ 170.2, which further analyzed by stretching appeared in IR spectrum at 1,666 cm−1. The structure of compound was further corroborated by mass spectrum which showed a molecular ion peak at m/z 333.1766 [M + Na] +. On the basis of these observations the purified compound is proposed to be 7-methoxy-2,2-dimethyl-4-octa-4′,6′-dienyl-2H-napthalene-1-one (Figure 7).
Figure 7.
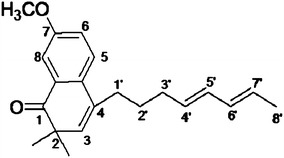
7-Methoxy-2,2-dimethyl-4-octa-4′,6′-dienyl-2H-napthalene-1-one.
MIC of butanolic extract of the fungi ranged from (0.1–20 mg/mL) which supported the data obtained by agar well diffusion assay. The importance of the study was further highlighted where the purified compound showed better or equal activity when compared with standard antibiotics. MIC of amphotericin B against C. albicans was found to be 99 µg/mL whereas, purified compound was effective at much lower MIC of 1 µg/mL. The study showed its further importance when MRSA showed sensitivity to the purified compound with MIC of 5 µg/mL Similarly, compound showed significant inhibitory potential against K. pneumoniae 1 with MIC 1 µg/mL which is comparable with the gentamicin having MIC of 1 µg/mL. S. aureus also showed MIC of 2 µg/mL, S. epidermidis, 2 µg/mL. The kill kinetics provides the more accurate description of antimicrobial agents than does the MIC (Mandal et al. 2009). Time kill studies not only give the information about the nature of the antimicrobial agent whether the particular agent is bactericidal or bacteriostatic but also the time taken by the antimicrobial agent for complete killing of the microorganism. Viable cell count studies which showed the effectiveness of the compound with complete killing of E. coli, and K. pneumoniae 1 at 0 h and our observations are better than previous viable cell count studies with medicinal plants (Kaur and Arora 2009). The viable cell count studies thus endorse that the compound is having potent bactericidal activity.
The design of antimicrobial dosing regimens is mainly based on the susceptibility of pathogen and pharmacokinetic parameters such as drug concentration in serum and tissue. PAE is being applied increasingly to allow antimicrobial dosing regimens to be developed in a more scientific manner. Different antimicrobials induce varied duration of PAE against different types of microbes. The purified compound was found to be effective even after removing it and showed the PAE ranging from 4 to 22 h. More prolonged intermittent dosing regimens would apply primarily to antimicrobials that exhibit a prolonged PAE. On the other hand more continuous dosing would be necessary for antimicrobials that exhibit a shorter PAE or lack of PAE (Lorian 1996).
Further the isolated compound did not show any cytotoxicity as done by MTT assay and mutagenicity by Ames test indicating its suitability to proceed for further pharmacological use.
Many compounds showing biological activities have also been listed in the literature from Penicillium sp. having no mutagenicity and cytotoxicity (Kaur et al. 2014; Arora and Chandra 2011). Cancer is a multifaceted and multimechanistic disease, which is one of the leading causes of death. Beside surgical and other interventions, chemotherapy remains the mainstay of treatment of cancer (Harrison et al. 2009). The importance of this study was further highlighted when the compound was found to be endowed with valuable cytotoxic potential against all tested human cancer cell lines. Further the compound induces apoptosis in lung cancer (A549) cells as observed by flow cytometry.
This study conclude that the antimicrobial activity of the purified compound was found to be comparable with standard drugs. Moreover the compound was found to be noncytotoxic which will facilitate the further studies to gain better understanding of the production of bioactive metabolites from fungi. Further, the compound also showed promising anticancerous activity which could be further use for various pharmaceutical purposes.
Authors’ contributions
DSA and HK: Substantial contributions to conception and design, or acquisition of data, or analysis and interpretation of data. JGO: Designing of cytotoxicity test. VS: Designing of purification experiments and interpretation of NMR, IR and Mass. DSA: Final approval of the version to be published. All authors read and approved the final manuscript.
Acknowledgements
Harpreet Kaur is thankful to University Grants Commission for Maulana Azad National Fellowship [F.40-50(M/S)/2009(SA-III/MANF)].
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional file
Figure S1. Mass spectrum of the purified compound.
Contributor Information
Harpreet Kaur, Email: harpreet40@gmail.com.
Jemimah Gesare Onsare, Email: jemimah_g@yahoo.com.
Vishal Sharma, Email: vishalmedchem@hotmail.com.
Daljit Singh Arora, Email: daljit_02@yahoo.co.in.
References
- Arora DS, Chandra P. Antioxidant activity of Aspergillus fumigatus. ISRN Pharmacol. 2011 doi: 10.5402/2011/619395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora DS, Kaur H. Study of Penicillium expansum and Penicillium citrinum for their antibacterial potential and optimizing their activity. Res J Biotech. 2011;6:49–56. [Google Scholar]
- Arora DS, Sharma RK. Effect of different supplements or bioprocessing of wheat straw by Phlebia brevispora: changes in its chemical composition, in vitro digestibility and nutritional properties. Bioresour Technol. 2011;102:8085–8091. doi: 10.1016/j.biortech.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Baker DD, Alvi KA. Small molecule natural products: new structures, new activities. Curr Opin Biotechnol. 2004;15:576–583. doi: 10.1016/j.copbio.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1996;43:493–496. [PubMed] [Google Scholar]
- Box GEP, Behnken DW. Some new three level design for study of quantitative variables. Technomet. 1960;2:455–475. doi: 10.1080/00401706.1960.10489912. [DOI] [Google Scholar]
- Ciapetti G, Cenni E, Pratelli L, Pizzoferrato A. In vitro evaluation of cell/biomaterial interaction by MTT assay. Biomaterials. 1993;14:359–364. doi: 10.1016/0142-9612(93)90055-7. [DOI] [PubMed] [Google Scholar]
- Furtado NAJC, Fonseca MJV, Bastos JK. The potential of an Aspergillus fumigatus brazillian strain to produce antimicrobial secondary metabolites. Braz J Microbiol. 2005;36:357–362. doi: 10.1590/S1517-83822005000400010. [DOI] [Google Scholar]
- Gould IM. The epidemiology of antibiotic resistance. Int J Antimicrob Ag. 2008;32:S2–S9. doi: 10.1016/j.ijantimicag.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Harrison M, Holen K, Liu G. A review of novel agents that target mitotic tubulin and microtubules, kinases, and kinesins. Clin Adv Hematol Oncol. 2009;7:54–64. [PMC free article] [PubMed] [Google Scholar]
- Kaur GJ, Arora DS. Antibacterial and phytochemical screening of Antheum graveolens, Foeniculum vulgare and Trychyspermum ammi. BMC Complement Altern Med. 2009 doi: 10.1186/1472-6882-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Arora DS, Sharma V. Isolation, purification and characterization of antimicrobial compound 6-[1, 2-dimethyl-6-(2-methyl-allyloxy)-hexyl]-3-(2-methoxy-phenyl)-chromen-4-one from Penicillium sp. HT 28. Appl Biochem Biotechnol. 2014 doi: 10.1007/s12010-014-0979-y. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Kanasaki R, Ezaki M, Sakamoto K, Takase S, Fujie A, et al. FR227244, a novel antifungal antibiotic from Myrothecium cinctum no. 002. J Antibiot. 2004;57:780–787. doi: 10.7164/antibiotics.57.780. [DOI] [PubMed] [Google Scholar]
- Kozlovskii AG, Vinokurova NG, Zhelifonova VP, Adanin VM. Secondary metabolites of fungi belonging to the species Penicillium janczewskii. Appl Biochem Microbiol. 1997;33:70–74. [Google Scholar]
- Kwon OE, Rho MC, Song HY, Lee SW, Chung MY. Phenyl propene A and B, new inhibitors of acyl-CoA: cholesterol acyl transferase produced by Penicillium griseofulvum f 1959. J Antibiot. 2002;55:1004–1008. doi: 10.7164/antibiotics.55.1004. [DOI] [PubMed] [Google Scholar]
- Livermore DM. Has the era of untreatable infections arrived? Antimicrob Chemother. 2009;64:29–36. doi: 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- Lorian V, editor. Antibiotic in laboratory medicine. Baltimore: William and Wilkins; 1996. pp. 296–329. [Google Scholar]
- Mandal S, Pal NK, Chowdhry IH, Mandal M. Antibacterial activity of ciprofloxacin and trimethoprim, alone and in combination against Vibrio cholerae O1 biotype E1 T or serotype Ogawa isolates. Pol J Microbiol. 2009;58:57–60. [PubMed] [Google Scholar]
- Maron D, Ames B. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Nicoletti R, Lopez-Gresa MP, Manzo E, Carella A, Ciavatta ML. Production and fungitoxic activity of Sch 642305, a secondary metabolite of Penicillium canescens. Mycopathol. 2007;163:295–301. doi: 10.1007/s11046-007-9015-x. [DOI] [PubMed] [Google Scholar]
- Nishihara YE, Tsujii S, Takase Y, Tsurumi T, Kino M, Hino M, et al. FR191512, a novel anti-influenza agent isolated from fungus strain no. 17415. J Antibiot. 2000;53:1333–1340. doi: 10.7164/antibiotics.53.1333. [DOI] [PubMed] [Google Scholar]
- Petit P, Lucas EMF, Breu LMA, Flenning LHP, Takahashi JA. Novel antimicrobial secondary metabolites from a Penicillium sp. isolated from Brazilian cerrado soil. Electron J Biotechnol. 2009;12:1–9. [Google Scholar]
- Pucci B, Katsten M, Giordano A. Cell cycle and apoptosis. Neoplasia. 2000;29:1–9. doi: 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja AF, Ali F, Khan IA, Shawl AS, Arora DS. Antistaphylococcal and biofilm inhibitory activities of acetyl-11-keto-β-boswellic acid from boswellic acid from Boswellia serrata. BMC Microbiol. 2011;11:54. doi: 10.1186/1471-2180-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;13:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Takahashi JA, Monteiro de Castro MC, Souza GG, Lucas EMF, Bracarense Abreu LM, Marriel IE. Isolation and screening of fungal species isolated from Brazilian cerrado soil for antibacterial activity against Escherichia coli, Staphylococcus aureus, Salmonella typhimurium,Streptococcus pyogenes and Listeria monocytogenes. J Med Mycol. 2008;18:198–204. doi: 10.1016/j.mycmed.2008.08.001. [DOI] [Google Scholar]
- Wang Y, Fang X, An F, Wang G, Zhang X. Improvement of antibiotic activity of Xenorhabdus bovienii by medium optimization using response surface methodology. Microb Cell Fact. 2011;10:98. doi: 10.1186/1475-2859-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhengyan G, Ling S, Zhiqin J, Wenjun W. Enhanced production of a novel cyclic hexapeptide antibiotic (NW-G01) by Streptomyces alboflavus 313 using response surface methodology. Int J Mol Sci. 2012;13:5230–5241. doi: 10.3390/ijms13045230. [DOI] [PMC free article] [PubMed] [Google Scholar]


