Abstract
Interpreting variants of uncertain significance (VUS) is a central challenge in medical genetics. One approach is to experimentally measure the functional consequences of VUS, but to date this approach has been post hoc and low throughput. Here we use massively parallel assays to measure the effects of nearly 2000 missense substitutions in the RING domain of BRCA1 on its E3 ubiquitin ligase activity and its binding to the BARD1 RING domain. From the resulting scores, we generate a model to predict the capacities of full-length BRCA1 variants to support homology-directed DNA repair, the essential role of BRCA1 in tumor suppression, and show that it outperforms widely used biological-effect prediction algorithms. We envision that massively parallel functional assays may facilitate the prospective interpretation of variants observed in clinical sequencing.
Keywords: deep mutational scanning, BRCA1, variants of uncertain significance, human genetic variation, protein function
IN an era of increasingly widespread genetic testing, DNA sequencing identifies many missense substitutions with unknown effects on protein function and disease risk. In the absence of genetic evidence, experimental measurement is the most reliable way to determine the functional impact of a variant of uncertain significance (VUS). However, initiating an experiment for each new variant observed in a gene is often impractical. When experiments are done, they are nearly always performed in a retrospective manner (Bouwman et al. 2013), such that the resulting data are not useful for the patient in whom the VUS was observed.
By prospectively measuring, in a high-throughput fashion, the consequences of all possible missense mutations on a gene’s function, we can generate a look-up table for interpreting newly observed VUS. Although functional analysis at this scale is made possible by deep mutational scanning (Fowler and Fields 2014), a central challenge is that any single assay may not recapitulate all the activities of a given protein in human disease. To address this challenge, we hypothesized that integrating the results of assays of multiple biochemical functions would strengthen estimates of the effects of mutations on disease risk (strategy outlined in Figure 1A). As a proof-of-concept, we initiated massively parallel functional analysis of BRCA1, a protein for which there are multiple biochemical functions as well as known pathogenic and benign missense substitutions to benchmark results.
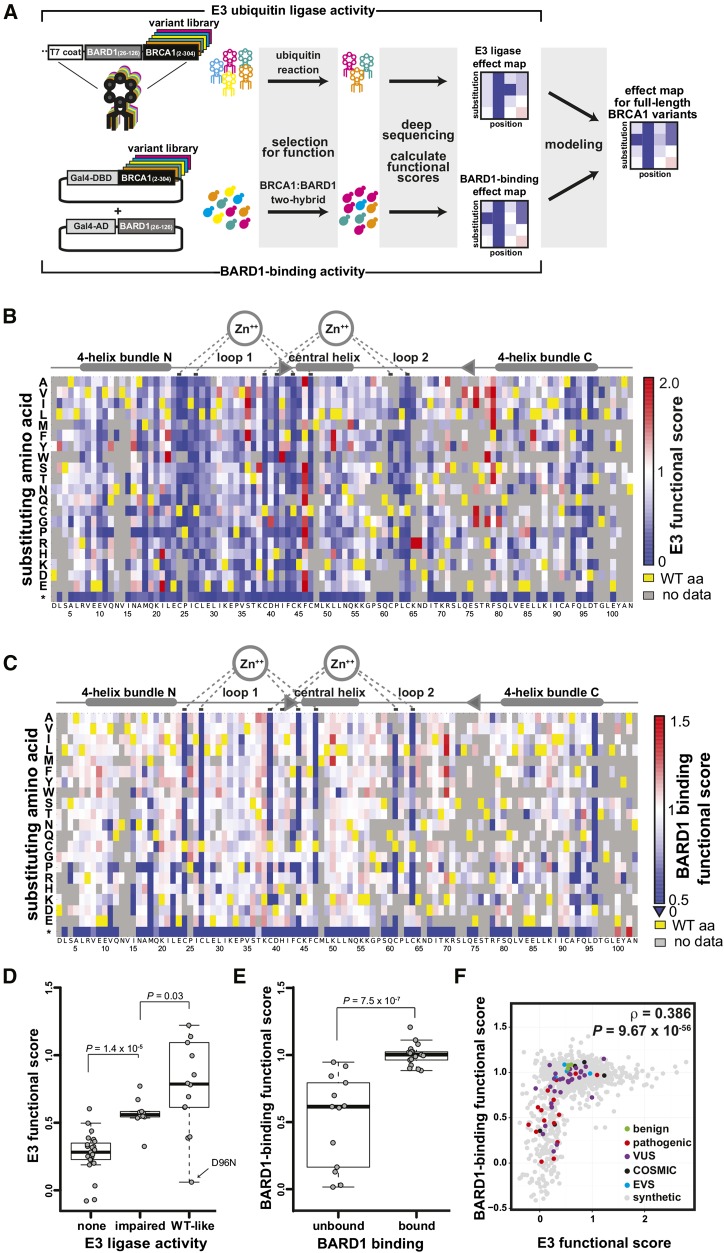
Figure 1.
(A) Scheme for leveraging scores from parallelized assays for BRCA1 RING function into predictions for the function of the full-length BRCA1 protein in homology-directed DNA repair. (B-F) Scoring the E3 ligase and BARD1-binding activities of BRCA1 RING domain variants. (B) A sequence-function map of the effect of missense mutations in the BRCA1 RING domain on E3 ligase function. The functional score for each variant is the slope of the fit curve, normalized by setting stop codons to a score of 0 and the wild-type to a score of 1. Each position in BRCA1(2-103) is arranged along the x-axes, structural features of the RING domain are diagrammed above. The amino acid substitutions, grouped by side-chain properties, are on the y-axes. The E3 ligase scores range from improved activity versus wild-type (red), equivalent to wild-type (white), to less than wild-type (blue). Yellow represents the wild-type residue and gray missing or low confidence data. (C) A sequence-function map of the effect of missense mutations in the BRCA1 RING domain on BARD1-RING binding. Coloring as in panel B. (D) Comparison of the variant scores from the deep mutational scan for E3 ligase activity versus literature-reported E3 ligase activities for the same BRCA1 variants (Brzovic et al., 2003; Morris et al., 2006). The Wilcoxon rank sum test (WRST) was used to test for significant differences between the categories. The biggest outlier in the wild type-like category, D96N, not only performed poorly as an E3 ligase score but also failed to bind to BARD1 and to support homology-directed repair in cells (Table S2). (E) Comparison of BARD1-binding scores from the two-hybrid experiment versus literature-reported BARD1 binding by the same BRCA1 variants (Brzovic et al., 2003; Ransburgh et al., 2010). The WRST was used to test for significant differences between categories. (F) The relationship between the quality-filtered E3 ligase functional scores and the BARD1-binding scores. Colors indicate the clinical classification or database of origin for each variant.
BRCA1 has been subject to intense study since its implication in hereditary, early onset breast and ovarian cancer (Miki et al. 1994). All missense substitutions in BRCA1 that are known to be pathogenic occur in either the amino-terminal RING domain or the carboxy-terminal BRCT repeat (http://brca.iarc.fr/LOVD/home.php?select_db=BRCA1). Although the RING domain represents only 5% of the BRCA1 protein, 58% of the pathogenic missense substitutions occur within this domain. Sixty-two missense substitutions in the RING domain have been observed in patients, the general population, or tumor samples. Of these, only 22 have been classified—19 as pathogenic and 3 as benign (Supporting Information, Table S1)—by multifactorial models based on information from personal history, family history, and pathological profile and by A-GVGD (Tavtigian et al. 2006), a conservation-based, biological-effect prediction algorithm (reviewed in Lindor et al. 2012).
Although BRCA1 has multiple roles in the cell, its activity in homology-directed DNA repair (HDR) is most closely associated with cancer risk (Moynahan et al. 1999; Towler et al. 2013). Cell-based HDR rescue assays on the full-length BRCA1 protein have been performed for a small number of variants (Ransburgh et al. 2010; Towler et al. 2013). However, those assays are too laborious to be applied to each possible BRCA1 variant. We therefore sought to implement alternative BRCA1 functional assays that are more amenable to multiplexing.
The BRCA1 RING domain heterodimerizes with the RING domain of BARD1 to comprise an E3 ubiquitin ligase (Hashizume et al. 2001). The structural stability of the heterodimer is required for the stability of full-length BRCA1 (Wu et al. 2010). BRCA1 variants that cannot dimerize result in defects in HDR and loss-of-tumor suppression (Drost et al. 2011; Ransburgh et al. 2010). Assays for both BRCA1 E3 ligase activity and interaction with BARD1 are sensitive to amino acid substitutions that destabilize the structure of the heterodimer (Brzovic et al. 2003; Morris et al. 2006; Ransburgh et al. 2010). We therefore developed massively parallel assays (Fowler et al. 2010) to measure the impact of thousands of missense substitutions on these two functions.
To assay E3 ligase activity, we subjected an allelic series (Kitzman et al. 2015) (Figure S1) of the BRCA1 N terminus amino acids (2–304) to a phage display assay (Starita et al. 2013) that selects for protein variants capable of autoubiquitination (Christensen et al. 2007). We expressed BRCA1(2–304) variants on the surface of phage and selected for BRCA1 ubiquitination activity over five sequential rounds of selection in the presence of an E1, an E2, and Flag–ubiquitin by capturing phage with anti-Flag beads (Figure S2). Phages that encode active BRCA1 RING variants increase in abundance and those that encode inactive variants decrease in abundance over the multiple rounds of selection. We used deep sequencing to count each allele in the input phage population and after each round. We calculated E3 ligase scores by tracking the changes in the relative abundance of each allele during the selection (Araya et al. 2012). The scores were normalized such that the wild type had a score of one and the mean score for variants with premature termination codons had a score of zero. We obtained scores for 5154 of the 5757 possible single-amino-acid substitutions (Table S2). Using an input frequency threshold (Figure S3A), we filtered these to a high-confidence set corresponding to 3170 amino acid substitutions, with the six replicates having Spearman’s rank correlation values between 0.76 and 0.83 (Figure S3B).
E3 ligase activity for variants with missense substitutions ranged from completely nonfunctional (scores of zero) to nearly three times higher than wild type. Scores for residues in the RING domain (2–103) are shown in Figure 1B and for residues 104–304 in Figure S4; all scores are reported in Table S2. As expected, substitutions in the residues that coordinate zinc ions and the residues in loop 1 and the central helix that contact the E2 enzyme (Brzovic et al. 2003) were the most intolerant to mutation (Figure 1B; Wilcoxon rank sum test (WRST), P = 0.0008), with the exception of Phe46, where most substitutions were hyperactivating. We compared the E3 ligase scores to previous work by splitting the published activities of BRCA1 RING domain variants in in vitro ubiquitination assays (Brzovic et al. 2003; Morris et al. 2006) into three categories: completely nonfunctional, impaired, or wild-type like. E3 ligase scores corresponding to variants in the nonfunctional category were lower than those in the impaired category (WRST, P = 1.4 × 10–5), which were in turn lower than those in the wild-type-like category (WRST, P = 0.03, Figure 1D).
In separate experiments, we used a multiplexed yeast two-hybrid assay to select for the ability of BRCA1 RING domain (2–103) (Brzovic et al. 2001) variants to interact with the RING domain of BARD1. The DNA-binding domain of the yeast transcription factor Gal4 was fused to the BRCA1(2–304) allelic series and the Gal4 activation domain was fused to BARD1(26–126) (Figure S5). Here, BRCA1 binding to BARD1 drives the expression of a selectable reporter gene such that yeast expressing BRCA1 variants that bind to BARD1 increase in abundance during the selection and those expressing nonfunctional variants decrease. We used deep sequencing to quantify the relative abundance of alleles after transformation into the yeast and at multiple time points during the selection (Materials and Methods and Table S2). We calculated a BARD1-binding score for 1855 of 1938 possible amino acid substitutions, excluding the carboxy-terminal 201 amino acids, which were required only for the autoubiquitination assay but not the BARD1-binding assay (Brzovic et al. 2001). Using an input frequency threshold, we filtered these to a high-confidence subset corresponding to 1529 substitutions, whose scores were highly reproducible (ρ = 0.82–0.95, Figure S6 and Table S2).
Overall, BARD1-binding scores agreed with what is known about the RING–RING interaction. The residues that coordinate the zinc ions were the most intolerant to substitution with the exception of H41 (Brzovic et al. 2001) (Figure 1C). The effect size for most other substitutions was small, which was expected given the large interface between the two RING domains (Brzovic et al. 2001). We compared our results with those published for co-immunoprecipitation of BRCA1 RING domain variants with BARD1 (Brzovic et al. 2003; Ransburgh et al. 2010). While the scores from the yeast two-hybrid BARD1-binding assay were lower for BRCA1 variants reported not to bind to BARD1 (WRST, P = 7.5 × 10–7), these scores spanned the entire range from zero to one (Figure 1E). Intermediate BARD1-binding scores for BRCA1 variants with weak or no BARD1 binding in co-immunoprecipitation assays may derive from differences in variant thermostability between the yeast assay (carried out at 30°) and the mammalian cell culture assay (carried out at 37°), and the in vivo transcriptional readout of the two-hybrid assay being more sensitive than co-immunoprecipitation.
We compared the E3 ligase scores to the BARD1-binding scores and observed that neither assay was sufficient alone to accurately discriminate BRCA1 variants with respect to their pathogenicity (Figure 1F, colored points). Because BARD1-binding is required for E3 ligase function, the scores from both assays were modestly correlated (ρ = 0.386; P = 9.67 × 10–56), but many more positions were intolerant to substitutions in the E3 ligase assay (Figure 1F). Although the E3 ligase activity of BRCA1 may not be required for HDR and therefore tumor suppression (Reid et al. 2008; Shakya et al. 2011), the E3 ligase and BARD1-binding activities likely reflect the structural stability of the RING domain. Indeed both assays had some power to discriminate BRCA1 variants with respect to their pathogenicity (Figure 1F, colored points). We hypothesized that these two rich mutational data sets could be combined to accurately identify deleterious substitutions in the BRCA1 RING domain.
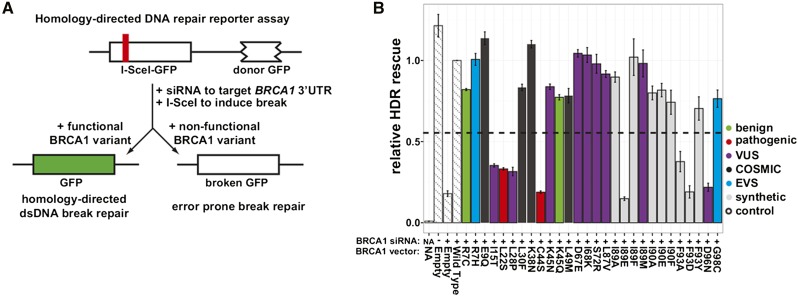
A test of whether the results from these high-throughput biochemical assays can be used to discriminate disease risk alleles needs “gold standards” as benchmarks. Since only 22 mutations in the BRCA1 RING domain have been classified for pathogenicity, we required a larger set of BRCA1 variants with established, disease-relevant functional consequences. Therefore, we tested additional full-length BRCA1 variants in the assay that best correlates with tumor suppression: rescue of HDR at an induced double-strand break by expression of a BRCA1 variant following siRNA knockdown of endogenous BRCA1 (Figure 2A). We curated results from this assay (Ransburgh et al. 2010; Towler et al. 2013) for 17 missense substitutions in the BRCA1 RING domain and tested an additional 28 (Figure 2B) for a total of 45. Of the 19 known pathogenic mutants, 8 have now been tested for HDR rescue. As expected, after excluding R71G, a variant that affects BRCA1 splicing (Vega et al. 2001), these pathogenic mutants all had low HDR rescue scores (mean = 0.19, max = 0.33) that separate them from the three known benign variants, which have much higher scores (mean = 0.88, min = 0.77; Figure 2B and Table S2). We defined a BRCA1 HDR rescue score of 0.53—the value midway between the average HDR rescue score for known pathogenic BRCA1 variants and the average score for known benign variants—as the inflection point for discriminating between functional and nonfunctional variants, as was done for BRCA2 (Guidugli et al. 2014). With this inflection point, the HDR rescue assay has 100% sensitivity and 100% specificity.
Figure 2.
Testing BRCA1 variants for their ability to rescue homology-directed DNA repair. (A) Integrated into the genome of the HDR reporter strain is one copy of the GFP gene containing an I-SceI homing endonuclease site that introduces an in-frame stop codon, along with another copy of the GFP gene lacking both its start and stop codons that functions as a donor for DNA repair (Pierce et al. 2001). The cell line is depleted for BRCA1 by transfection with an siRNA that targets the 3′-UTR of the endogenous gene. I-SceI and a full-length variant of BRCA1 are then transfected into the cells. After 3 days, the GFP+ population is assessed by flow cytometry. (B) The wild-type normalized percentage of cells that were GFP+ in the BRCA1 HDR rescue assay is shown. Experiments were performed in triplicate and error bars represent the standard error. siRNA, BRCA1 variant, and clinical classification or database of origin is indicated by color. Dashed horizontal line represents the midpoint between the average HDR scores for known pathogenic and benign variants.
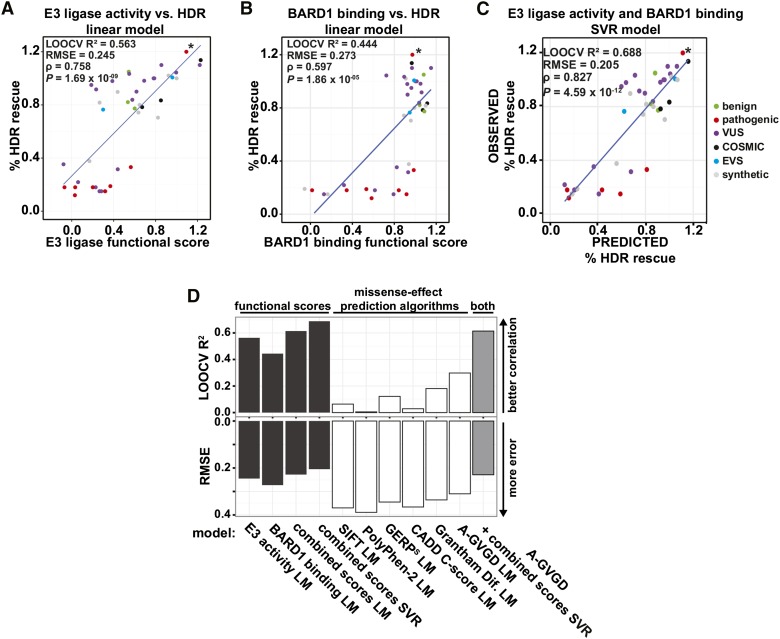
We then asked whether models trained on the E3 ligase and BARD1-binding scores can predict the effects on HDR rescue of substitutions in the full-length protein. We evaluated the accuracy of several models using leave-one-out cross-validation (LOOCV), wherein we serially predicted HDR rescue scores for each of the 44 missense substitutions for which we had empirical HDR rescue and functional scores from models fit on the 43 remaining variants. We first compared the performance of models tested on scores from the E3 ligase and BARD1-binding assays alone or in combination. A linear model based on scores from both assays outperformed linear models based on scores from either assay alone (Figure 3, A and B). However, because we observed a nonlinear relationship between E3 ligase and BARD1-binding scores (Figure 1F), we tested whether nonlinear models would improve HDR prediction results. A support vector regression (SVR) model trained on E3 ligase scores and BARD1-binding scores yielded the best predictive power for HDR rescue (Figure 3C).
Figure 3.
Scores from massively parallel E3 ligase and BARD1-binding assays on BRCA1 RING domain variants are better predictors of the HDR activity of the full-length protein. The linear relationship of the E3 ligase scores (A), BARD1-binding scores (B), and HDR scores. (C) A support vector regression (SVR) model of HDR rescue scores from the combination of the E3 ligase and BARD1 binding functional scores. Variants are colored by database of origin. The blue line represents the least-squares fit of the displayed data. Known pathogenic splice variant R71G is marked with an asterisk. (D) Experimentally or computationally derived values for the effect of missense variants on protein function were used to predict the effect on HDR. The LOOCV R2 and RMSE for each model is indicated. The RMSE of LOOCV indicates the average distance between the HDR rescue predictions and the true HDR rescue scores, and the LOOCV R2 is the overall correlation between predicted and observed values; low RMSE and high R2 indicate better predictive power. For A-GVGD, the Grantham deviation value was used. Source of HDR predictions is indicated by color, linear model (LM), and SVR.
We next replaced our experimental data with computational predictions from several popular variant effect prediction algorithms (Grantham 1974; Ng and Henikoff 2003; Cooper et al. 2005; Tavtigian et al. 2006; Adzhubei et al. 2010; Kircher et al. 2014), which incorporate evolutionary constraints and/or chemical differences between amino acid side chains, and repeated the model training procedure. Individually, these prediction-based models performed poorly at predicting a substitution’s effect on HDR (Figure 3D, white bars, and Figure S7). Although A-GVGD was the best performing algorithm, it yielded higher error and lower correlation than all experimentally-based models (Figure 4D and Figure S7F). Furthermore, when we added the A-GVGD predictions to the experimental data and trained a hybrid model, performance was not enhanced over the experimentally based models (Figure 3D, gray bar, and Figure S7G). A plausible explanation for the comparatively poor performance of models trained on computational predictions is that they are largely based on features that are not specific to BRCA1 (e.g., Grantham chemical difference scores) or on evolutionary constraint information that captures organismal fitness over evolutionary timescales, which may poorly discriminate subtle and strong molecular effects on BRCA1 function.
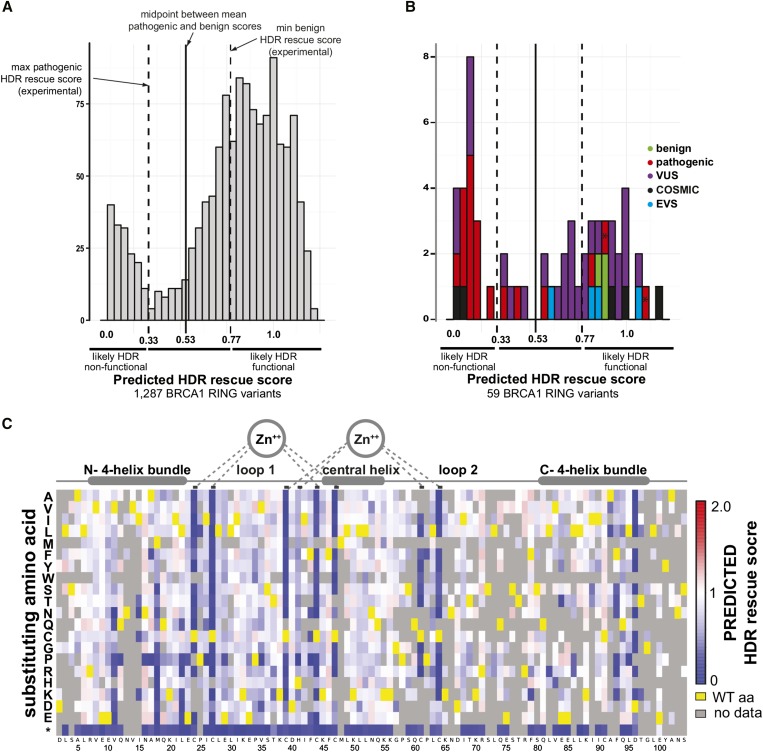
Figure 4.
Predicted HDR rescue scores for 1287 BRCA1 RING variants create a prospective map of the effect of missense substitutions. (A) A histogram of the predicted HDR scores for the 1287 BRCA1 RING variants with both high-confidence E3 ligase and BARD1-binding scores. (B) A histogram of the 59 of the 62 BRCA1 RING variants found in the human population, clinical classification, or database of origin is indicated by color. Known splice variants R71G and R71K are marked with an asterisk. (C) Sequence-function map of the predicted HDR rescue scores for BRCA1 RING variants. Colors are centered on 1.0 as wild type (white). Structural features of the BRCA1 RING domain are diagrammed above.
Because the SVR model based on combined functional data sets from the two assays was the most accurate, we used it to predict HDR scores for the 1287 BRCA1 RING domain missense variants with both high-confidence E3 ligase and BARD1-binding scores (Figure 4, A–C and Table S2), 1225 of which have not yet been reported in clinical sequencing. The distribution of predicted HDR scores is bimodal; 785 missense substitutions are predicted to have little effect on HDR, with predicted rescue scores >0.77 (Figure 4A). Conversely, 160 substitutions are predicted to be damaging to HDR, with scores <0.33; these variants would potentially increase the risk of hereditary breast and ovarian cancer. Based on this SVR modeling, only 342 variants have predicted scores in the indeterminate region between functional and nonfunctional.
As expected, predicted HDR scores for most of the 19 known pathogenic mutants in the BRCA1 RING domain are low (Figure 4B). Excluding pathogenic mutants known to affect splicing or used to train the model leaves 10 pathogenic mutants. All 10 have predicted HDR scores <0.53, the threshold for classifying a variant as functional. Nine have predicted HDR scores <0.33, the maximum empirical HDR rescue score for a known pathogenic mutant. Thus, our model demonstrates strong performance in predicting HDR activity of known variants (Figure 4B and Table S2). For 31 VUS identified in patients, predicted HDR scores range from near zero to wild-type-like, with 8 of 31 <0.53 and 5 <0.33, suggesting that a substantial fraction of individuals with VUS diagnoses may carry pathogenic BRCA1 alleles.
The data in Figure 4C represent a prospective map or look-up table for the effects of missense substitutions in the RING domain of BRCA1 on HDR function. This experimentally-derived map is more accurate than any map that could be generated using current computational tools. In terms of defining BRCA1 activity, these systematic mutational analyses uncovered positions in the four-helix bundle that show extreme sensitivity to substitution. For example, V11 does not tolerate substitutions with charged or amine-containing polar amino acids; M18 does not tolerate charged, polar, or aromatic substitutions; and F93 and D96 do not tolerate any substitutions. Our data support the idea some variants with defects in the E3 ligase activity are not compromised for HDR and tumor suppression (Reid et al. 2008; Shakya et al. 2011). The benign variants R7C and D67Y showed no binding defect with BARD1 and were able to rescue HDR but they performed poorly in the E3 ligase selections. However, they may retain enough E3 ligase activity to satisfy a possibly low threshold of requisite activity.
Our results demonstrate the power of empirical measurements to assess the impact of missense variants on complex protein functions. Thus, we envision that massively parallel experiments to measure the effects of large numbers of substitutions will meet an urgent need in the clinical translation of genetic information.
Materials and Methods
BRCA1(2-304) single codon substitution library construction by the Programmed Allelic Series method
Oligonucleotides, 90-mers, to direct the single codon mutagenesis of BRCA1 were synthesized on and released from a 12,000-feature array by Custom Array (Bothell, WA) (example in Table S3, BRCA1_00284.0). The BARD1(26–126)–GS–BRCA1(2–304) fusion open reading frame (ORF) (Christensen et al. 2007) was moved to the pGEM vector and the EcoRI site in BRCA1 was destroyed. This fragment of BRCA1 was used as a template for PALS mutagenesis (Kitzman et al. 2015). Sixteen base random barcodes (16N) were added 3′ of the stop codon in the final PCR step. The ligation of the final mutagenized and barcoded amplicon was transformed into DH10B electromax cells (NEB) and yielded 250,000 unique transformants of the pGEM_BARD1_GS_BRCA1-var_barcode library.
Subassembly to match 16N barcodes to BRCA1 variants
Since BRCA1(2–304) is too long to be sequenced in one pass by current Illumina technology, we developed a method to create randomly shortened contigs that could be grouped by barcode to use in an assembly method call tag-directed read grouping or subassembly (Hiatt et al. 2010). A total of 5 μg of the plasmid pGEM_BARD1_GS_BRCA1-var was cut at the 5′ end of the BRCA1 ORF with BamHI and purified. The purified DNA was digested using the double-strand exonuclease Bal-31 (NEB), 1 unit Bal31 per 1.6 pmol DNA ends at 30°. Aliquots were taken at 0, 3, 7, 11, 13, and 15 min and placed in the DNA-binding buffer from the Zymo clean and concentrate kit to stop the reactions. One-fifth of the digested and cleaned DNA was cut with HindIII and examined by PAGE to determine the degree of digestion. DNA from all time points was pooled and treated with End-it (Epicentre) to blunt and phosphorylate the ends. Blunt-ended, cleaned DNA was A-tailed using goTaq (NEB) and cleaned again. A double-stranded linker containing the Tru-seq Illumina Read 2 primer was ligated onto the A-tailed DNA (W-E4B-subassembly-linker and phosphorylated C-E4B-subassembly linker). The linkered and cleaned DNA was cut with SacI (NEB) to separate the ORF and barcode from the rest of the plasmid. Primers that annealed to the linker and plasmid DNA directly 3′ of the barcode that contain the p5 and p7 Illumina cluster generating sequences (newBRCA1-side_R_CG1 and BRCA1-side_R_CG2) were used to amplify the fragments and barcode for Illumina sequencing in reactions containing the high-fidelity polymerase KAPA HFHSRM and SYBR green [conditions: 95° 3:00 (98° 0:20, 63° 0:15, 72° 1:50) × 12–15]. The amplicons were sequenced on an Illumina HiSeq2000 in paired-end, 2 × 101 run mode and with an Illumina MiSeq paired-end, 2 × 250 kit.
Reads were filtered for quality and grouped by the sequence of the 16-base barcode. A Smith–Waterman algorithm was used to align the grouped reads to the wild-type BRCA1(2–304) sequence and a consensus sequence was determined for each barcode group as in Hiatt et al. (2010) and Patwardhan et aI. (2012). A minimum quality score of 20 was required for a base to contribute to an assembly. Full-length BRCA1(2–304) sequences were filtered for quality by requiring that a given base in the assembly was observed at least twice and was present at an intra-tag group allele frequency of one for positions covered by two to four reads or a frequency of at least 0.8 for positions covered by five or more reads. If these conditions were not met the assembly was discarded. We assembled 128,237 barcoded variants, of which 60,256 corresponded to 5156 single-amino-acid changes out of the possible 5757 (89% of the 19 substitutions × 303 codons) in BRCA1(2–304) (see Figure S1). A database of barcodes and their associated full-length BRCA1(2–304) assembly was created to facilitate linking barcodes sequenced from the experimental samples to the full-length subassemblies.
Phage-based E3 ligase assays
The BARD1(26–126)_glycine–serine linker_BRCA1(2–304) library was subcloned from pGEM_BARD1_GS_BRCA1-var_barcode by cutting and gel purifying the EcoRI and HindIII fragment and ligating into the genome of T7–Select 10-3b bacteriophage. Phage genomic DNA was packaged into phage particles, the number of ligation/packaging events was estimated by titer as 2.56 × 107 plaque-forming units (PFU), and the phages were amplified in E. coli strain BLT5403 according to the T7Select Cloning Kit instructions (EMD Millipore). The selections for functional BRCA1(2–304) phages were performed as in Starita et al. (2013) with these differences: amplified phage were never stored more than 24 hr before a sequential round of selection and the 50-μl ubiquitination reactions contained 20 μl (∼1 × 107 PFU) of amplified phage, 2 mM ATP, 5 mM MgCl2, 1 μM wheat E1 ubiquitin activating enzyme, 4 μM UBE2D3 (UbcH5c), and 8 μM Flag-tagged ubiquitin.
DNA from the initial amplified phage population and amplified phage from each replicate from each of five rounds of selection was purified from 200 μl of lysate by phenol chloroform extraction. Barcodes were PCR amplified in two sequential reactions. The first reaction contained primers jkA0390_BBcplxcheckF and E4B-index01-8_CG-R or T7_barcodes_common primer_R and 200 ng of phage DNA in reactions containing the high-fidelity polymerase Phusion (NEB), 2 mM added MgCl2, and SYBR green [conditions: 95° 3:00 (98° 0:20, 63° 0:15, 72° 1:50) × 10–13]. Reaction products were monitored by qPCR and removed during exponential amplification. The first reactions were purified using the Zymo clean and concentrate kit. One-tenth of that product was amplified with JK19 and one of the index containing primers E4B-index01-8_CG-R or common_index_primers such as NexV2ad2_A1 [conditions: 95° 3:00 (98° 0:20, 63° 0:15, 72° 1:50) × 4–6]. Again, reaction products were monitored by qPCR and removed during exponential amplification. Reaction products were treated with exonuclease I (Affymetrix) for 15 min at 37° then purified using the Zymo clean and concentrate kit. Samples were multiplexed and sequenced using primer jkA0390_BBcplxcheckF on an Illumina GAIIx or HiSeq2000 in single end mode.
Yeast two-hybrid-based deep mutational scan for BRCA1-BARD1 binding
The Gal4 DNA-binding domain (Gal4DBD) was amplified from pOBD2 (Cagney et al. 2000) using primers SpeI_Gal4DBD_F and SpeI_Gal4DBD_R and cloned into p414–ADH (Mumberg et al. 1995). The BRCA1(2–304) variant library was excised from pGEM_BARD1_GS_BRCA1-var_barcode library by digestion with BamHI (NEB) and PstI (NEB) and ligated into p414_Gal4DBD to create p414_Gal4DBD_BRCA1_var_barcode, yielding ∼1.16 × 105 total transformants. BARD1(26–126) was amplified from pGEM_BARD1_GS_BRCA1 using primers EcoRI_BARD1_Ln_F and NcoI_BARD1_Stop_R and cloned into pOAD (Cagney et al. 2000) containing the Gal4 transcriptional activation domain.
The S. cerevisiae strain, PJ69a (James et al. 1996), containing pOAD_BARD1 was transformed (Melamed et al. 2013) with the p414_Gal4DBD_BRCA1_var_barcode library with a yield of ∼1.26 × 106 total transformants. Transformed yeast were transferred to 40 ml SD–Leu–Trp, cultured overnight and stored in 6.7 optical density units (ODU) aliquots at −80°.
Two independent experiments (A and B) were performed, each consisting of three independent selections: 12.5 ODU (A) or four ODU (B) of cells were collected from each culture at each time point for sequencing. Each experiment began by culturing one frozen aliquot of PJ69a transformed with pOAD_BARD1 and p414_Gal4DBD_BRCA1_var_barcode in SD–Leu–Trp to logarithmic phase (A, 1.01 OD/ml; B, 0.83 OD/ml). Cells from this input population were collected for sequencing and for back dilution into the selection medium (SD–His–Leu–Trp + 10 mM 3-amino-1,2,4 triazole (Sigma), A, 5 OD to 200 ml; B, 2 OD to 100 ml) in triplicate. Each replicate was cultured to logarithmic phase (A, 18 hr, 1.1 OD/ml; B, 16 hr, 0.7 OD/ml) after which cells were collected for sequencing and back diluted into fresh selection medium (A, 1 OD to 100 ml; B, 0.6 OD to 100 ml). Each replicate was again cultured to logarithmic phase (A, 37 hr, 0.62 OD/ml; B, 40.5 hr, 0.67 OD/ml), after which cells were collected for sequencing and back diluted into fresh selection medium (A, 12.5 OD to 125 ml; B, 1.1 OD to 100 ml). Each replicate was again cultured to logarithmic phase (A, 45 hr, 0.43 OD/ml; B, 64 hr, 1.4 OD/ml) and the final time point was collected.
Plasmid DNA was isolated from the input and samples collected during the selection for growth in the -histidine media using a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research). Sequencing amplicons were prepared individually for each sample by two successive PCR reactions using Phusion high-fidelity DNA polymerase. In the first reaction, primers jkA0390_BBcplxcheckF and BRCA1-Y2H_commonLinker_R were used to amplify the barcoded region of half of the prepared p414_Gal4DBD_BRCA1_var_barcode plasmid. Of the first reaction, 4% was amplified with primers JK19 and NexV2ad2_XX to append Illumina cluster generating sequences and a unique sample index sequence. Reaction products of all PCR reactions were monitored on a mini-opticon qPCR machine (Bio-Rad) and removed during exponential amplification. Samples were purified, multiplexed, and sequenced using primer jkA0390_BBcplxcheckF on a HiSeq2000 in single-end mode.
Slope calculations and normalization
The Illumina reads that matched to barcodes associated with full-length assemblies were retained and unmatched barcodes were discarded. The matched barcodes were converted to the sequence of the full length BRCA1(2–304) assembly and the Enrich software package (Fowler et al. 2011) was used to determine locations and identity of substitutions and to tally the number of times each variant appeared in each population (Table S2).
Sequencing read counts corresponding to a given variant were equal to the sum of read counts from all barcodes matching that variant. For each time point, frequencies were calculated for all variants as the variant’s read count divided by the sum of all read counts at that time point. Variants that dropped out (cannot be found in the selected populations) had their frequencies set to the lowest possible frequency at that time point. Ratios were calculated as the variant’s frequency in the selected time point divided by its frequency in the input library. For each variant, a linear model was fit by least squares to the log ratios over time using numpy.polyfit. The inverse log of this slope corresponds to the percentage change in frequency per unit time. To obtain a normalized score, the average inverse log of the slope for all stop codons was subtracted from all inverse-log-slopes so that stop codons, on average, have a score of 0. These 0 centered values were then divided by the wild-type (WT) inverse-log-slope so that a score of 1 corresponds to WT function.
The normalized score for variant i is
where m is the number of stop codons from positions 2–103 and all slopes were fit to the log ratios at each time point. A conservative estimate of the standard deviation of the slopes was generated using a Loess curve to model the relationship between frequency in the input population and variance across all replicates (based on the assumption that the variance is related to the frequency in the input population; see Figure S3 and Figure S6). For each variant, the conservative variance was set to whichever was larger: the variance across all replicates or the value of the Loess curve evaluated at the number of input reads for that variant. This estimate was used to generate the reported confidence intervals (Table S2). Additionally, we used cutoff based on the number of input reads to determine the high-confidence data set that would be used for the final HDR predictions. The heuristic to determine the cutoff is described in Figure S3 and Figure S6. HDR predictions were made only for variants with high-confidence scores in both the E3 ligase and BARD1-binding assays. Finally, a permutation test was used to compare each variant’s slopes to the WT slopes across all replicates. The average difference between paired slopes was used as the test statistic and 10,000 permutations were performed for each variant (Table S2).
Full-length BRCA1 variant construction and HDR assays
Mutations in the BRCA1 RING domain were made by overlap-extension PCR and subcloned into the HindIII and EcoRI sites of pcDNA3–HA–BRCA1 (plasmid described in Chiba and Parvin 2001). All constructs were verified by Sanger sequencing. BRCA1 rescue of HDR assays were performed in triplicate as in Ransburgh et al. (2010). All BRCA1 HDR rescue scores are normalized to that of the wild-type protein at HDR rescue = 1. The maximum HDR score for known pathogenic variant of BRCA1 is 0.33. Seven pathogenic variants (excluding splice variant R71G) have been tested for HDR rescue with a mean score of 0.19. Of the only three known benign BRCA1 RING domain variants, all have been tested for HDR rescue and have a minimum score of 0.77 with an average score 0.88. We defined a BRCA1 HDR rescue score of 0.53—the value midway between the average HDR rescue scores for known pathogenic BRCA1 variants and the average scores for known benign variants—as the inflection point for discriminating between functional and nonfunctional variants, as was done for BRCA2 (Guidugli et al. 2014).
HDR prediction model building and testing
We obtained SIFT, Polyphen-2, GERP++, and CADD values from the CADD database (Kircher et al. 2014) and references therein (http://cadd.gs.washington.edu/download). For every possible amino acid substitution in BRCA1 (2-103), we obtained Grantham chemical difference values from Grantham(1974), and GVGD values from the A-GVGD BRCA1 web-tool (http://agvgd.iarc.fr/agvgd_input.php). Grantham deviation (GD) values were used to predict HDR rescue scores.
All models were fit and cross-validated using the R package caret (Kuhn 2008). Linear models were fit by least squares. Support vector regression models used the radial basis function kernel and were validated using a nested cross validation scheme (Cawley and Talbot 2010). Briefly, for each step of the LOOCV, an inner LOOCV loop was used to determine model performance on each C and sigma pair in the tested parameter space and the best performing model (based on root mean square error, RMSE) was used to predict the holdout in the outer loop. The range of sigma values tested in the inner loop was determined using the sigest function from the R package kernlab and the C values tested were 0.1, 1, 2, 5, 10, 100, and 1000. The final model used for HDR predictions was chosen by picking the parameter pair with the lowest average RMSE across all iterations of the outer loop (Y2H and E3 model—C = 5 and sigma = 0.1633448, Y2H, E3; GVGD model—C = 5 and sigma= 0.08220825).
Acknowledgments
We thank Deborah Nickerson, Gail Jarvik, Peter Byers, and Peter Brzovic for comments on the manuscript, Rachel Klevit and Peter Brzovic for the gift of purified E1 and E2 enzymes, Martin Kircher for providing a database of prediction algorithm scores for missense substitutions in BRCA1, Charlie Lee for assistance with Illumina sequencing, Amanda Toland for advice regarding the clinical classification of BRCA1 missense variants, and Nancy Lill for providing mutant BRCA1 expression plasmids. This work was supported by National Institutes of Health grants to S.F. (Biomedical Technology Research Resource project no. P41GM103533), J.S. (Director’s Pioneer Award no. DP1HG007811) and D.F. (no. R01GM109110). S.F. is an investigator of the Howard Hughes Medical Institute.
Author contributions: L.M.S. and J.O.K. constructed the BRCA1 allelic series. L.M.S. and J.G. performed the deep mutational scanning experiments. M.I. performed the HDR rescue experiments. L.M.S., D.L.Y., J.G., R.J.H., and D.M.F. analyzed the data. L.M.S., S.F., J.S., D.M.F., D.L.Y., J.G., and J.D.P. wrote the manuscript; all authors reviewed it. S.F., J.D.P., and J.S. supported this project.
Footnotes
Communicating editor: S. K. Sharan
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175802/-/DC1.
Literature Cited
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya C. L., Fowler D. M., Chen W., Muniez I., Kelly J. W., et al. , 2012. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc. Natl. Acad. Sci. USA 109: 16858–16863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., van der Gulden H., van der Heijden I., Drost R., Klijn C. N., et al. , 2013. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 3: 1142–1155. [DOI] [PubMed] [Google Scholar]
- Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E., 2001. Structure of a BRCA1–BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8: 833–837. [DOI] [PubMed] [Google Scholar]
- Brzovic P. S., Keeffe J. R., Nishikawa H., Miyamoto K., Fox D., et al. , 2003. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA 100: 5646–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagney G., Uetz P., Fields S., 2000. High-throughput screening for protein–protein interactions using two-hybrid assay. Methods Enzymol. 328: 3–14. [DOI] [PubMed] [Google Scholar]
- Cawley G. C., Talbot N. L. C., 2010. On over-fitting in model selection and subsequent selection bias in performance evaluation. J. Mach. Learn. Res. 11: 2079–2107. [Google Scholar]
- Chiba N., Parvin J. D., 2001. Redistribution of BRCA1 among four different protein complexes following replication blockage. J. Biol. Chem. 276: 38549–38554. [DOI] [PubMed] [Google Scholar]
- Christensen D. E., Brzovic P. S., Klevit R. E., 2007. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14: 941–948. [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Stone E. A., Asimenos G., Program N. I. S. C. C. S., Green E. D., et al. , 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15: 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost R., Bouwman P., Rottenberg S., Boon U., Schut E., et al. , 2011. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell 20: 797–809. [DOI] [PubMed] [Google Scholar]
- Fowler D. M., Fields S., 2014. Deep mutational scanning: a new style of protein science. Nat. Methods 11: 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Araya C. L., Fleishman S. J., Kellogg E. H., Stephany J. J., et al. , 2010. High-resolution mapping of protein sequence-function relationships. Nat. Methods 7: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Araya C. L., Gerard W., Fields S., 2011. Enrich: software for analysis of protein function by enrichment and depletion of variants. Bioinformatics 27: 3430–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., 1974. Amino acid difference formula to help explain protein evolution. Science 185: 862–864. [DOI] [PubMed] [Google Scholar]
- Guidugli L., Carreira A., Caputo S. M., Ehlen A., Galli A., et al. , 2014. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum. Mutat. 35: 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., et al. , 2001. The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276: 14537–14540. [DOI] [PubMed] [Google Scholar]
- Hiatt J. B., Patwardhan R. P., Turner E. H., Lee C., Shendure J., 2010. Parallel, tag-directed assembly of locally derived short sequence reads. Nat. Methods 7: 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A., 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., et al. , 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman J. O., Starita L. M., Lo R. S., Fields S., Shendure J., 2015. Massively parallel single amino acid mutagenesis. Nat. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., 2008. Building predictive models in R using the caret package. J. Stat. Softw. 28(5). [Google Scholar]
- Lindor N. M., Guidugli L., Wang X., Vallée M. P., Monteiro A. N. A., et al. , 2012. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum. Mutat. 33: 8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D., Young D. L., Gamble C. E., Miller C. R., Fields S., 2013. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA 19: 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., et al. , 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71. [DOI] [PubMed] [Google Scholar]
- Morris J. R., Pangon L., Boutell C., Katagiri T., Keep N. H., et al. , 2006. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 15: 599–606. [DOI] [PubMed] [Google Scholar]
- Moynahan M. E., Chiu J. W., Koller B. H., Jasin M., 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4: 511–518. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller R., Funk M., 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan R. P., Hiatt J. B., Witten D. M., Kim M. J., Smith R. P., et al. , 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 30: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.J., Hu P., Han M., Ellis N., Jasin M., 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15: 3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransburgh D. J. R., Chiba N., Ishioka C., Toland A. E., Parvin J. D., 2010. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 70: 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid L. J., Shakya R., Modi A. P., Lokshin M., Cheng J.-T., et al. , 2008. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. USA 105: 20876–20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R., Reid L. J., Reczek C. R., Cole F., Egli D., et al. , 2011. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334: 525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L. M., Pruneda J. N., Lo R. S., Fowler D. M., Kim H. J., et al. , 2013. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc. Natl. Acad. Sci. USA 110: E1263–E1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian S. V., Deffenbaugh A. M., Yin L., Judkins T., Scholl T., et al. , 2006. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 43: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler W. I., Zhang J., Ransburgh D. J. R., Toland A. E., Ishioka C., et al. , 2013. Analysis of BRCA1 variants in double-strand break repair by homologous recombination and single-strand annealing. Hum. Mutat. 34: 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega A., Campos B., Bressac-De-Paillerets B., Bond P. M., Janin N., et al. , 2001. The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum. Mutat. 17: 520–521. [DOI] [PubMed] [Google Scholar]
- Wu W., Sato K., Koike A., Nishikawa H., Koizumi H., et al. , 2010. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 70: 6384–6392. [DOI] [PubMed] [Google Scholar]