Abstract
Caenorhabditis elegans has emerged as a powerful model to study the genetics of feeding, food-related behaviors, and metabolism. Despite the many advantages of C. elegans as a model organism, direct measurement of its bacterial food intake remains challenging. Here, we describe two complementary methods that measure the food intake of C. elegans. The first method is a microtiter plate-based bacterial clearing assay that measures food intake by quantifying the change in the optical density of bacteria over time. The second method, termed pulse feeding, measures the absorption of food by tracking de novo protein synthesis using a novel metabolic pulse-labeling strategy. Using the bacterial clearance assay, we compare the bacterial food intake of various C. elegans strains and show that long-lived eat mutants eat substantially more than previous estimates. To demonstrate the applicability of the pulse-feeding assay, we compare the assimilation of food for two C. elegans strains in response to serotonin. We show that serotonin-increased feeding leads to increased protein synthesis in a SER-7-dependent manner, including proteins known to promote aging. Protein content in the food has recently emerged as critical factor in determining how food composition affects aging and health. The pulse-feeding assay, by measuring de novo protein synthesis, represents an ideal method to unequivocally establish how the composition of food dictates protein synthesis. In combination, these two assays provide new and powerful tools for C. elegans research to investigate feeding and how food intake affects the proteome and thus the physiology and health of an organism.
Keywords: feeding, aging, nutrition, metabolism, de novo protein synthesis
FOOD intake has proven difficult to measure in small model organisms. Yet, small model organisms such as Caenorhabditis elegans offer significant advantages for the study of nutrition, appetite, and metabolism. As a model, C. elegans is amenable to genome-wide screening, has clear human orthologs for genes involved in feeding behaviors (Luedtke et al. 2010), and has conserved mechanisms regulating feeding, fat storage, and energy expenditure (de Bono and Bargmann 1998; Sze et al. 2000; Ashrafi et al. 2003; Ashrafi 2006; Srinivasan et al. 2008).
C. elegans captures food by pumping liquids containing bacteria into its mouth. Pumping is achieved by rhythmic contractions of a neuromuscular organ called the pharynx. Measurements of pharyngeal pumping have become the standard for estimating food intake and inferring the amount of food ingested (Avery 1993; Avery and You 2012). Because C. elegans move freely, it is only possible to measure the rate of pharyngeal pumping over short intervals of time (seconds). Thus, with pharyngeal pumping, short-term measurements must be extrapolated to estimate overall food intake.
Rather than inferring the ingestion of food by measuring a behavioral change (e.g., pumping), alternative methods to estimate food intake quantify the ingestion of labeled bacteria or microbeads. These methods offer the advantage of directly measuring ingested material, but require expensive and specialized equipment and, again, can only measure the rate of feeding over short intervals of time (You et al. 2008; Avery and You 2012; Paek et al. 2012). While currently available methods allow the study of short-term changes in food intake, there is a major need for simpler methods that measure food intake over longer periods of time.
Here, we report two complementary methods that quantify C. elegans food intake over several days. The first method, termed bacterial clearance, is a 96-well microtiter plate-based assay that measures the food intake of C. elegans in liquid medium by quantifying the change in optical density of bacteria over time. Using the bacterial clearance assay, we characterized the bacterial food intake of C. elegans over the course of its development and in response to serotonin. We find that smaller animals eat less and show that the long-lived eat mutants eat substantially more than previous estimates. While other methods of bacterial clearance have been described (Voisine et al. 2007; Cabreiro et al. 2013), our assay uniquely enables the direct measurement of bacterial food intake per worm, is suitable for use over the lifespan of the animal, is not influenced or limited by the hatching of larvae, and can measure food intake for hundreds of small populations simultaneously, dramatically increasing its sensitivity. Importantly, the microtiter-based bacterial clearing assay can be expanded for medium-throughput phenotype-based chemical and genetic (RNAi, candidate mutants) screens and requires only standard laboratory equipment.
The second method, termed pulse feeding, measures the ingestion of stable isotope-labeled bacteria (15N), using mass spectrometry (LC-MS/MS) to detect newly synthesized proteins via isotope incorporation into the proteome. Simply put, it is a metabolic pulse-labeling strategy in which a 15N pulse is delivered by feeding worms 15N-labeled bacteria. Thus, any 15N in the proteome must represent food intake within the pulse period. We employ the metabolic pulse-labeling strategy to compare the assimilation of food and the subsequent de novo protein synthesis for two C. elegans strains in response to serotonin. We show that serotonin-induced feeding in C. elegans leads to changes in the de novo proteome, including increased translation and ribosome biogenesis. To our knowledge, this is the first study to measure the absorption and utilization of nutrients (i.e., amino acids) by measuring de novo protein synthesis or change the isotope enrichment of any C. elegans food source midexperiment, in a manner similar to pSILAC (Schwanhausser et al. 2009), to time stamp the incorporation of the isotope label into the worm proteome. Furthermore, as the metabolic pulse-labeling method captures de novo protein synthesis in response to any stimulus or condition, investigators can study the de novo proteome of an intact multicellular organism in both normal and diseased states.
In combination, these two methods enable a comprehensive investigation of the genes and metabolites that affect food intake and will facilitate the identification of new pharmacological interventions that affect feeding and metabolism.
Materials and Methods
Strains and genetics
All strains were maintained at 20° on NGM agar plates as previously described (Brenner 1974). C. elegans strains used were: Bristol strain (N2), DA820 eat-18(ad820sd), DA1110 eat-18(ad1110), MT15434 tph-1(mg280) II, DA1814 ser-1(ok345) X, OH313 ser-2(pk1357) X, DA1774 ser-3(ad1774) I, AQ866 ser-4(ok512) III, FX02146 ser-6(tm2146) IV, RB660 arr-1(ok401) X, AZ30 sma-1(ru18) V, CB502 sma-2(e502) III, and CB491 sma-3(e491) III. The strains DA2100 ser-7(tm1325) X, RB2277 ser-5(ok3087) I, CB205 unc-26(e205) IV, and NM204 snt-1(md290) II were outcrossed four times to Bristol N2 and renamed VV122, VV130, VV78, and VV80, respectively.
Food intake (OD600) assay
Food intake was assessed in liquid medium (Petrascheck et al. 2007; Solis and Petrascheck 2011) (S-complete medium with 50 µg ml−1 carbenicillin and 0.1 μg ml−1 fungizone) in black, flat-bottom, optically clear 96-well plates (Costar) containing 150 μl total volume per well. Plates contained 6–12 nematodes per well in 6 mg/ml Escherichia coli OP50 [1.5 × 108 colony-forming units (cfu)/ml], freshly prepared 4 days in advance unless otherwise specified. We used a carbenicillin-resistant OP50 strain to exclude growth of other bacteria. Age-synchronized nematodes were seeded as L1 larvae and grown at 20°. Plates were covered with sealers (Nunc) to prevent evaporation. To prevent self-fertilization, 5-fluoro-2′-deoxyuridine (FUDR) (0.12 mM final) (Sigma) was added 42–45 hr after seeding. Drugs were added 68 hr after seeding (day 1 of adult life) unless otherwise specified.
The absorbance at 600 nm (OD600) of each well was measured using a microplate reader (Tecan). Measurements were taken every 24 hr, beginning on day 1 of adulthood. Before measuring OD600, each plate was placed onto a plate shaker for 25 min. The fraction of animals alive per well was scored microscopically on the basis of movement on day 4 of adulthood. Each plate was placed onto a plate shaker for 1–2 min prior to counting. Strong microscope light was used to effectively stimulate movement in young and old animals, to aid counting of viable worms. Unless stated otherwise, food intake per worm was calculated as the bacterial clearance divided by the number of worms per well. For comparisons between strains, food intake was expressed as a percentage relative to control animals. A complete step-by-step protocol is available online (Nature Protocol Exchange, Petrascheck Lab).
Several technical aspects of our original bacterial clearance protocol have proven essential to the sensitivity and robustness of the assay and merit further discussion. First, in liquid medium, bacteria clump together and settle on the bottom of the assay wells. Simple shaking of the plates for 25 min on a microtiter plate shaker prior to each OD600 measurement was found to be sufficient to dissolve bacterial aggregates and resuspend the bacteria in solution. Minimizing clumps and sedimentation by shaking significantly improves the accuracy, sensitivity, and robustness of the bacterial clearance assay. Second, bacteria quickly settle to the bottom of the wells after shaking. As such, OD600 measurements become less reliable as the interval of time between the end of shaking and measurement increases. It is important to keep this interval short, preferably under 10 min, to ensure robust results. Third, after performing several control experiments, we observed that different bacterial preparations had an affect on basal food intake. Thus, it is critical to prepare the feeding bacteria in exactly the same manner for each experiment, and to measure changes in food intake relative to N2 control animals.
There are technical as well as biological explanations for the differences in basal food intake between different bacterial preparations. OD600 measures absorbance, and thus the number of particles, irrespective of their size. Bacteria harvested in the logarithmic growth phase will be much larger than bacteria in the stationary phase, because many will be in a dividing state. OD600 measurements do not distinguish size, and thus simple differences in bacterial size or state may lead to apparent basal food intake changes between bacterial preparations. Of note, we identified two major determinants of basal feeding between bacterial preparations; (i) the “freshness” of the bacteria, as inoculations from freshly streaked bacteria produced greater feeding than bacteria from prior inoculations, and (ii) the “age of the bacteria” or length of time bacteria were kept in nutrient-poor S-complete media after culture in the nutritionally rich “Terrific Broth” (TB) medium. Bacteria aged 4–6 days appeared optimal for feeding, as significantly lower basal food intake was observed when worms were fed bacteria aged for only 1 day (24 hr), independent of the C. elegans strain or genetic background.
Imaging and body length measurements
Animals were imaged in individual wells of a 96-well optically clear microtiter plate, using a Molecular Devices ImageXpress platform. WormSizer software was used to extract the body length of adult (day 4) nematodes from bright field images (Moore et al. 2013).
Statistical analysis
GraphPad Prism software was used for data analysis. Comparisons and P-value calculations were made between animals of the same or different strains, and treated and untreated animals, using Student’s t-test and ANOVA with corrections for multiple hypothesis testing. Wells containing <4 or >15 animals were excluded from statistical analyses.
Drug preparation
Serotonin hydrochloride (Alfa Aesar) was freshly prepared for each experiment and dissolved in water at ×50 final concentration before use.
Metabolic labeling of E. coli and C. elegans
Worms and bacteria were metabolically labeled with the desired nitrogen isotope (14N or 15N) as previously described (Gouw et al. 2011; Geillinger et al. 2012). Prior to each experiment, worms were transferred to nitrogen-free solid agarose plates and seeded with 14N or 15N metabolically labeled bacteria as the sole source of nitrogen. After a minimum of three generations, age-synchronized larvae were transferred to 10-cm Petri dishes and cultured in liquid medium plus metabolically labeled 14N (experimental) or 15N (external standard) bacteria (6 mg/ml). For pulse-labeling experiments, culture plates were first cleared of all 14N labeled bacteria, then reseeded with 50% 14N/50% 15N (medium, M) labeled bacteria and cultured for an additional 2 days. Unless stated otherwise, the start day of the pulse labeling was day 5 of adulthood. For tph-1 pulse-labeling experiments, ∼600 worms were harvested per strain. For serotonin pulse-labeling experiments, 600–800 worms per strain per condition were harvested for analysis by mass spectrometry. To control for differences in worm numbers between conditions within an experiment, each sample was adjusted such that an equal number of worms would be processed. All pulse-labeling experiments were performed in triplicate and independently repeated three times.
Mass spectrometry and proteomics
Lysed worms were prepared for mass spectrometry by precipitation in 13% trichloroacetic acid (TCA) overnight at 4°. The protein pellet was collected by centrifugation at maximum speed for 20 min. The pellet was washed with 10% TCA and spun again for 10 min at 4°. The pellet was then washed with ice cold acetone and spun for an additional 10 min at 4°. The protein pellet was resuspended in ∼100–200 μl of 100 mM ammonium bicarbonate with 5% (v/v) acetonitrile. Then 10% (by volume) of 50 mM DTT was added and the sample was incubated for 10 min at 65°. This incubation was followed by the addition of 10% (by volume) of 1 mM iodoacetic acid (IAA) and a 30-min incubation at 30° in the dark. A total of 5 μg of trypsin was added to each sample and incubated overnight at 37°. Trypsinized samples were then cleaned up for mass spectrometry using PepClean columns (Pierce) following the manufacturer’s directions. Clean samples were dried in a speed vacuum and then resuspended in ∼10 μl of 5% acetonitrile with 0.1% (v/v) formic acid. Samples were spun at high speed to remove particulates before placing in mass spectrometry tubes for analysis.
LC-MS
Samples were analyzed on an ABSCIEX 5600 Triple time of flight (TOF) mass spectrometer coupled to an Eksigent nano-LC Ultra equipped with a nanoflex cHiPLC system. Conditions on the ABSCIEX 5600 were as follows: The source gas conditions were optimized for each experiment and were generally set to GS1 = 8–12, GS2 = 0, and curtain gas = 25. The source temperature was set to 150°. Information dependent acquisition experiments were run with a 2-sec cycle time, with 0.5-ms accumulation time in the MS1 scan and up to 20 candidate ions per MS/MS time period. The m/z range in the MS1 scans was 400–1250 Da, and 100–1800 Da for the MS/MS scans. Target ions were excluded after two occurrences for 12 sec to increase sequencing coverage. Target ions with +2 to +5 charge were selected for sequencing once they reached a threshold of 100 counts per second.
Conditions for the Eksigent nano-HPLC were as follows: Gradients were run with a trap-and-elute setup with water plus 0.1% (v/v) formic acid as the mobile phase A and acetonitrile with 0.1% (v/v) formic acid as mobile phase B. Samples were loaded onto a 200 μm × 0.5 mm ChromXP C18-CL 3 μm 120 Å Trap column at 2 μl/min. Gradients were run from 5% mobile phase A to 40% mobile phase B over 2 hr on a 75 μm × 15 cm ChromXP C-18-CL 3 μm 120-Å analytical column. This was followed by a rapid jump to 80% B for 10 min to clean the column and a 20-min reequilibration at 95% A. Water sample blanks were run between samples to rid the column of any residual interfering peptides, including a short gradient, followed by the 80% B and 20-min reequilibration with 5% A.
LC-MS data analysis
Data were converted using the ABSCIEX conversion software to mgf format and MZML format. The peak list was generated by searching a SwissProt database using MASCOT, with the taxonomy set to C. elegans and E. coli simultaneously (Matrix Science). A MS/MS Ion search was performed using the 15N quantification, with a peptide mass tolerance of ±0.1 Da and a fragment mass tolerance of ±0.1 Da. The maximum number of missed cleavages was set to 2. MS1 scans for identified peptides were fit to three isotope distributions using ISODIST (Sperling et al. 2008; Chen et al. 2012). The percentage of labeling in the pulse was first fit using a floating variable to find the best percentage and then fit with a fixed percentage value. This value depended on the experiment and varied between 42% and 50% labeling. This value most likely varies due to the amount of 14N E. coli left in the culture after switching the food to the 50% 15N-labeled E. coli. One advantage of our fitting method is that we can easily account for these variations, as we are fitting the entire isotopic envelope. Fraction labeled was calculated as (50% 15N intensity)/(14N intensity + 50% 15N intensity).
Results
The bacterial clearing assay measures food intake of C. elegans
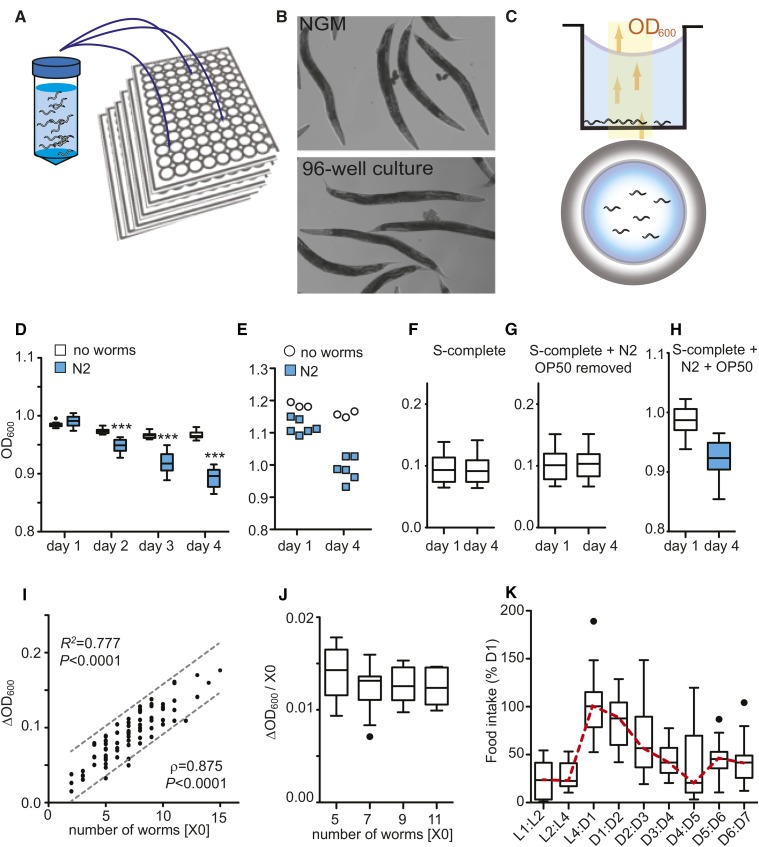
C. elegans can be grown in 96-well microtiter plates in liquid medium by seeding 4–12 developmentally synchronized L1 larvae into individual wells with E. coli as a food source. Young adults raised in this form of liquid culture are morphologically very similar to age-matched adults raised on NGM (Figure 1, A and B; Supporting Information, Figure S1). They do not show the elongated and starved appearance seen in other types of liquid culture. Incidentally, we observed that the wells of microtiter plates containing worms plus bacteria become optically clearer over the course of several days. We reasoned that, by measuring the change in bacterial concentration over time in a well, we could quantify the amount of bacteria eaten by the worms.
Figure 1.
Measurement of bacterial clearance with C. elegans. (A) A 96-well liquid culture format. (B) Morphology of day 1 (D1) adult N2 worms grown on solid NGM plates (top) or in liquid culture (bottom). (C) The bacterial clearance assay. Schematic of worms placed into wells with an optical bottom to monitor bacterial concentrations by measuring the optical density (absorbance) at 600 nm (OD600). Side and top view. (D) Bacterial clearance is only observed in the presence of worms. Tukey-style box plots. OD600 depicting four time points, comparing wild-type N2 vs. no worms, nwells = 12 biological replicates (e.g., wells). Data represent five independent experiments. ***P < 0.001, two-way ANOVA with Bonferroni post hoc test. (E) Bacterial clearance is observed for bacteria killed by irradiation. Data represent three independent experiments. Wells with N2 (nwells = 6), wells with no worms (nwells = 3). (F–H) OD600 measurements are not influenced by the presence of eggs and depend only on the presence of worms eating bacteria. OD600 on D1 and D4 for wells containing (F) S-complete only, (G) N2 and eggs in S-complete with OP50 removed, or (H) N2 plus bacteria in S-complete. Data represent three independent experiments. (I) Bacterial clearance correlates with the number of worms per well (X0). Values depict bacterial clearance over 72 hr (D1:D4). Data depict 95% confidence interval (dashed lines), goodness of fit statistic (R2), and Spearman’s correlation ρ (P < 0.0001). Data represent three independent experiments (nwells = 84). (J) Data from I normalized to worms per well. (K) Age-related changes in food intake. Food intake expressed relative to bacterial clearance within the L4:D1 interval. Data represent three independent experiments (nwells = 36).
To test this, we cultured worms in microtiter plates with an optically clear bottom to allow noninvasive monitoring of bacterial concentrations by optical density (absorbance) at 600 nm, herein referred to as OD600 (Figure 1C) (Solis and Petrascheck 2011). To maintain a constant population, viable offspring were prevented from hatching by adding FUDR at the L4 larval stage (Rangaraju et al. 2015). For our studies, L4 hermaphrodites were defined as day 0 adults.
To ensure that worms eating bacteria caused the observed changes in bacterial concentrations, we conducted OD600 measurements in the presence and absence of worms. OD600 measurements were collected by a microtiter plate reader every 24 hr over 4 days, beginning on day 1 of adulthood. In the presence of worms, bacteria cleared over time, resulting in a progressive drop in OD600 values (Figure 1D). In contrast, little bacterial clearance was observed in the absence of worms. Bacterial clearance was observed regardless of whether the worms were fed with live or dead (gamma irradiated) E. coli (Figure 1E), indicating bacterial clearance was due to the consumption of bacteria by worms rather than the death or lysis of bacteria.
After 4 days of adulthood, worms will have laid hundreds of eggs in each well. To determine the influence of laid eggs on optical density measurements, we compared OD600 values of individual wells prior to egg laying (day 1) and again after eggs had been laid (day 4). To isolate the changes in OD600 caused by the eggs from those caused by bacterial clearance, we removed all bacteria, but not worms and eggs, prior to the OD600 measurements. Despite the large numbers of eggs that were present on day 4, they did not influence OD600 measurements (Figure 1, F–H). Thus, OD600 measurements specifically detect the presence of bacteria and do not detect the presence of eggs or worms. Worms are only indirectly detected by actively eating bacteria.
If the decrease in OD600 is due to worms eating bacteria, the magnitude of bacterial clearance (ΔOD600) must depend on the number of worms per well. Operationally, we defined bacterial clearance as the difference in OD600 between two time points (ΔOD600 = OD600t1 − OD600t2). More worms should result in a larger difference than fewer worms. Indeed, we observed a strong positive correlation (R2 = 0.77) between bacterial clearance (ΔOD600) and the number of worms in each well (X0) (Figure 1I).
Next, we asked whether the food intake of an individual worm was affected by the number of worms in a given well. To accomplish this, we normalized the ΔOD600 changes to worm number per well (X0). Food intake per worm (ΔOD600/X0) can thus be defined as the bacterial clearance divided by the number of worms in the well (ΔOD600/X0 = (OD600t1 − OD600t2)/X0). As shown in Figure 1J, the number of worms present in each well did not affect the food intake per worm as long as the number of worms stayed between 3 and 14. Importantly, by measuring the food intake per worm within the range above, we can perform meaningful comparisons of food intake between wells, populations, and experimental conditions. Therefore, we used the food intake per worm as the unit of measure for food intake.
After establishing the bacterial clearance assay as a robust measure of food intake in adults, we next monitored the food intake of C. elegans throughout its development. Food intake peaked as animals transitioned from L4 larvae to young adults, coinciding with the onset of egg production. From there, food intake declined gradually with age, as expected (Huang et al. 2004) (Figure 1K). Food intake per day was greatest during the interval between day 1 and day 4 of adulthood (D1:D4), making this period ideal for detecting differences in feeding over time. In this report, we used the D1:D4 interval as the standard period to measure food intake.
Comparing the feeding rates of eat and small mutants
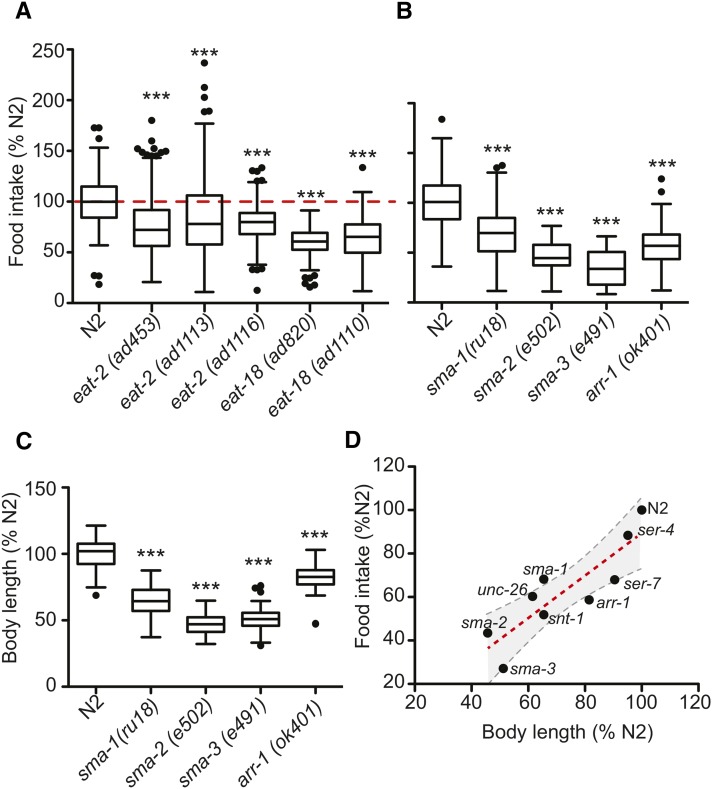
In C. elegans, mutations in the eat genes disrupt the function of the pharynx, leading to reduced pharyngeal pumping and a starved appearance (Avery 1993; Lakowski and Hekimi 1998). However, the cumulative food intake of any eat mutant has not been determined. We compared the food intake of wild-type animals with different eat-2 and eat-18 alleles. While the food intake of all eat mutants was reduced compared to wild type (Figure 2A), the magnitude of the decrease was considerably smaller than predicted, given that their pharyngeal pumping rates are ∼10% of wild type (Raizen et al. 1995; Petrascheck et al. 2007). The food intake of eat-2 and eat-18 mutants was 80% and 60% that of wild-type animals, respectively. This was much higher than we expected, but in the case of eat-2(ad1116), correlates well with its brood size (∼60% of wild type) (Hughes et al. 2007). Interestingly, the reduction in food intake we describe in long-lived eat mutants is very close to the reduction in food intake necessary to extend lifespan in mammals (Taormina and Mirisola 2014).
Figure 2.
Body size influences food intake. (A) Food intake of long-lived eat mutants. Food intake expressed relative to wild-type N2 (D1:D4). Data represent three independent experiments, nwells ≥ 88. ***P < 0.01, one-way ANOVA with Dunnett’s multiple comparison post-test. (B) Small animals eat less. Data represent three independent experiments, nwells ≥ 43. ***P < 0.01, one-way ANOVA with Dunnett’s multiple comparison post-test. (C) Body length of animals in B. Body length as measured on day 4 of adulthood. Data represent three independent experiments, nwells ≥ 28. ***P < 0.001, one-way ANOVA with Dunnett’s multiple comparison post-test. (D) Interaction of animal size and food intake. Food intake and body length measurements expressed relative to wild-type N2 on day 4 of adulthood. Data represent three independent experiments. Linear regression line (dashed red line) and the 95% confidence interval (dashed gray lines) are shown. Goodness of fit statistic, R2 = 0.753 (P < 0.002).
Feeding defects impair growth, causing the animals to be small (Morck and Pilon 2006). We asked whether the reverse is true, whether small animals eat less. Comparing the food intake of wild-type animals to several mutants with reduced body length, we observed that smaller animals do eat less. This effect was independent of the genetic pathway reducing body length (Figure 2B). Strains carrying mutations that disrupt TGFβ (sma-2 and sma-3), embryonic constriction (sma-1), global synaptic transmission (unc-26 and snt-1), or reduce pharyngeal pumping via disruption of neurotransmitter-specific synaptic transmission (eat-2, eat-18, ser-7, and arr-1) all ate less than wild-type animals (Nonet et al. 1993; McKay et al. 2004; Beaulieu et al. 2005; Steger et al. 2005; Ch’ng et al. 2008). As expected, the reduction in food intake correlated with body length (Morck and Pilon 2006) (Figure 2, C and D).
Serotonin modulates food intake via SER-7
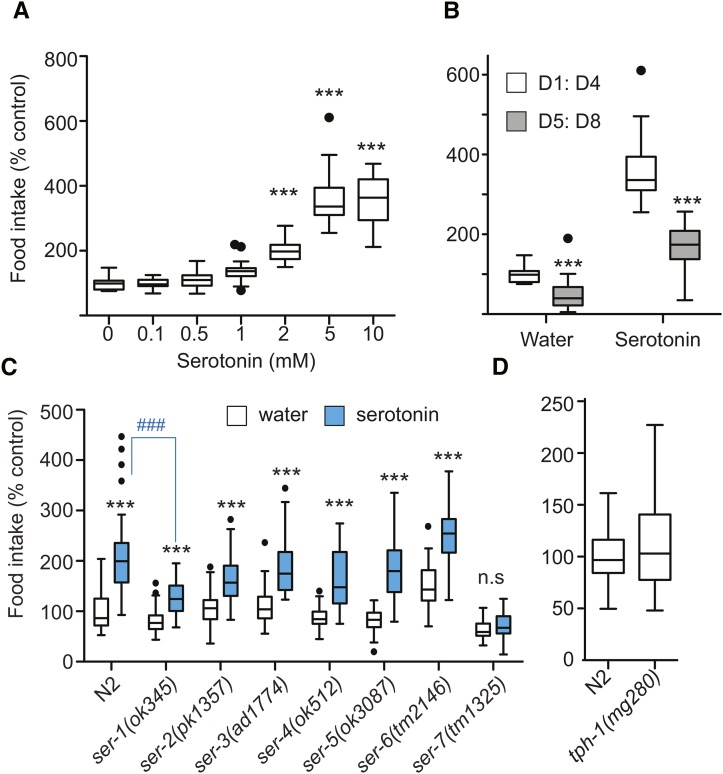
Thus far, we have established that we can determine reductions in food intake. We next set out to measure increases in food intake. The neurotransmitter serotonin (5-HT) is a conserved regulator of energy balance (Noble et al. 2013). Treatment with exogenous serotonin increases the rate of pharyngeal pumping in a manner dependent on the G-protein coupled receptors (GPCRs) ser-1, ser-5, or ser-7 (Srinivasan et al. 2008; Cunningham et al. 2012). As expected, treating C. elegans with serotonin increased bacterial clearance in a dose-dependent manner (Figure 3, A and B).
Figure 3.
Modulation of food intake by serotonergic signaling. (A) Dose–response curve for wild-type N2 animals treated with serotonin. Food intake expressed relative to control treatment (water). Tukey-style box plots, unless otherwise stated, depict food intake over the D1:D4 interval. Data are representative of three independent experiments, nwells = 20. ***P < 0.01, one-way ANOVA with Dunnett’s multiple comparison post-test. (B) Food intake of pre- and post-reproductive wild-type N2 animals treated with water or serotonin (2 mM). Food intake is expressed relative to the D1:D4 interval of control water-treated N2 animals. Data are representative of three independent experiments, n = 18. ***P < 0.0001, Student’s t-test. (C) Food intake in response to serotonin (2 mM) for wild-type N2 animals and serotonin receptor mutants. Data for each strain are representative of a minimum of three independent experiments. Data as depicted in graph represent two independent experiments, nwells ≥ 42. ***P < 0.001, two-way ANOVA with Bonferroni post-test comparing response to serotonin for each genotype. ###P < 0.001, one-way ANOVA with Dunnett’s multiple correction post-test comparing serotonin-treated animals of each genotype to wild-type serotonin-treated animals. (D) Basal food intake of serotonin-synthesis-deficient tph-1 mutants. Food intake is expressed relative to wild-type N2. Data represent three independent experiments, nwells ≥ 95. Student’s t-test was used to establish significance. Note: For a version of graphs in A and C showing S.E.M. and thus reproducibility between experiments, see Figure S2.
We next asked which of the known serotonin receptors mediates increased food intake by serotonin. Serotonin-induced feeding was completely abolished in ser-7(tm1325) mutants (Figure 3C). In contrast, ser-1(ok345) mutants displayed a partial serotonin-resistant phenotype. In ser-1(ok345) mutants, both the relative and absolute magnitude of food intake in response to serotonin was reduced, indicating a reduction in the consumption of total bacteria as compared to wild-type N2 animals. The discrepancy between pharyngeal pumping and bacterial clearance in the case of ser-5(ok3087) was unexpected and warrants further investigation (Cunningham et al. 2012).
Having confirmed that exogenous serotonin increases food intake via SER-7, we proceeded to investigate how endogenous serotonin modulates basal food intake. In the context of this article, we use the term basal food intake to describe the amount of bacteria the animals eat without stimulation by chemicals such as serotonin. The rate-limiting enzyme for serotonin synthesis is tryptophan hydroxylase (tph-1), and well-fed tph-1 mutants display multiple metabolic phenotypes (Sze et al. 2000; Cunningham et al. 2012). While mean food intake in N2 and tph-1(mg280) mutant animals was similar (Figure 3D), food intake in tph-1(mg280) mutants was more variable (Brown–Forsythe test, P = 1.5e-06), suggesting defects in the regulation of basal food intake. This is consistent with reports of increased variation in pharyngeal pumping rates of tph-1(mg280) mutants as compared to wild-type animals (Hobson et al. 2006). Thus, the metabolic phenotypes observed in tph-1(mg280) mutants are unlikely to be the result of a reduction in food intake per se, but rather of defects in food signaling.
Measuring nutrient incorporation by quantitative mass spectrometry
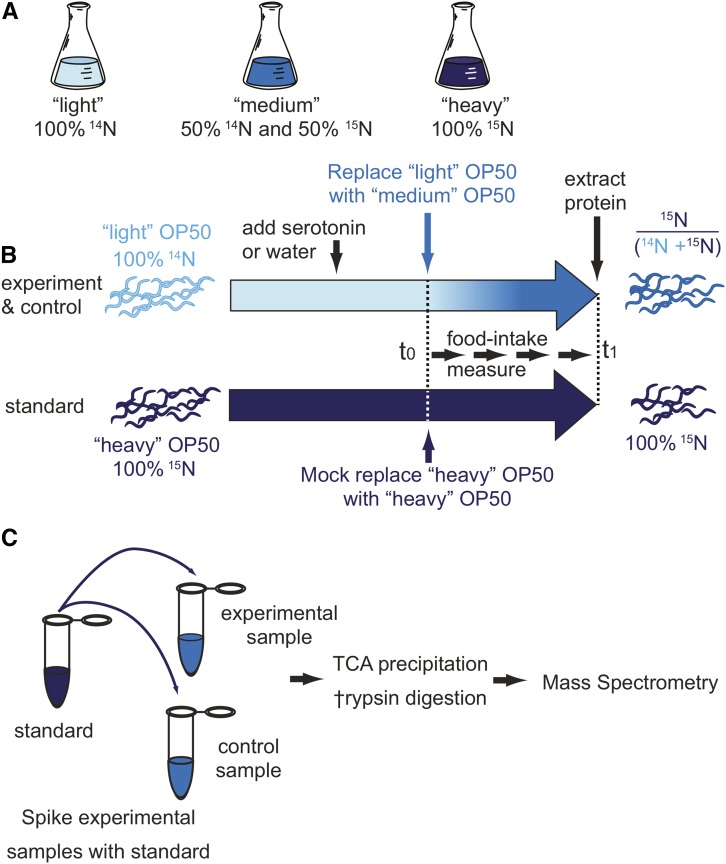
A complete understanding of feeding not only requires us to measure the amount of food eaten, but also the amount incorporated into the animal. To measure food absorption, we developed the pulse-feeding assay. In the pulse-feeding assay, a 15N pulse is delivered to C. elegans by feeding nitrogen-isotope-labeled (14N, 15N) bacteria (OP50). Nutrient (i.e., food) absorption is subsequently determined by measuring isotope incorporation into the C. elegans proteome, which is proportional to the bacteria ingested (see Supporting Information Discussion). To conduct such an experiment, C. elegans are raised on 14N-labeled bacteria. Then, 14N-labeled bacteria are exchanged with 50% 14N/50% 15N-labeled OP50 and fed to the worm throughout a period of interest (pulse interval). At the end of the pulse interval, proteins are extracted and the amount of incorporated 15N in the C. elegans proteome is determined by mass spectrometry (Figure 4, A and B).
Figure 4.
Measurement of nutrient absorption in C. elegans using metabolic labeling coupled with quantitative mass spectrometry. To metabolically label worms, (A) we first grew three different OP50 “foods” by culturing OP50 bacteria in minimal media enriched with either 100% (14NH4)2SO4, 50% (14NH4)2SO4 + 50% (15NH4)2SO4, or 100% (15NH4)2SO4. These foods were termed “light,” “medium,” and “heavy,” respectively. Second, we generated a population of heavy worms. These worms were fed heavy OP50 for three generations to ensure fully enriched 100% (15NH4)2SO4 worm proteins. (B) Third, light and heavy worms were synchronized and cultured in light and heavy food, respectively. The heavy worms were used as an external standard to facilitate the comparison of different experimental samples. 14N worms were given either the drug of choice or water at day 1. At day 5, or the start day for the pulse labeling, worms were washed and the food was switched from light to medium for drug- or water-treated worms, and from heavy to heavy for the mass-spec-standard worms. The worms were harvested on day 7 after pulse labeling and prepared for mass spectrometry analysis. (C) Preparation of samples for mass spectrometry analysis. For each experiment an external standard was generated, derived from heavy worms cultured in parallel. Each sample lysate was spiked with the external standard. Proteins were extracted by TCA precipitation, digested with trypsin, and analyzed by LC-MS/MS.
To quantitatively compare nutrient absorption between different samples, the pulse-feeding method requires three different types of labeled bacteria (Geillinger et al. 2012; Grun et al. 2014). The first type is for growing the worms, the second for pulsing them, and the third is to be added as an external standard. We used uniform 15N enrichment of ammonium ions as the source of nitrogen. To produce the three different types of feeding bacteria, OP50 were grown in M9 media containing three different concentrations of 15N-enriched ammonium sulfate (NH4SO4): only 14NH4SO4, only 15NH4SO4, or a 50:50 mixture of the two. We refer to OP50 grown on 14NH4SO4 as “light” or “L” OP50, to OP50 grown on a 50:50 mixture as “medium” or “M” OP50, and to OP50 grown on 100% 15NH4SO4 as “heavy” or “H” OP50 (Sykes et al. 2010; Gouw et al. 2011; Chen et al. 2012; Geillinger et al. 2012; Chen and Williamson 2013).
Incomplete removal of the 14N-labeled bacteria prior to the start of pulse feeding can result in the incomplete labeling of new worm proteins with 50% 15N-labeled amino acids. We were able to correct for this effect by allowing the percentage of 15N to vary as an adjustable parameter when performing the least squares fitting to the isotope distribution (Sperling et al. 2008). Typically, we found that feeding the medium OP50 (50% 15N) resulted in a relative isotope abundance of ∼45% 15N. The incomplete labeling of proteins had no effect on the feeding measurements, which only depend on the amplitude of the medium isotope component.
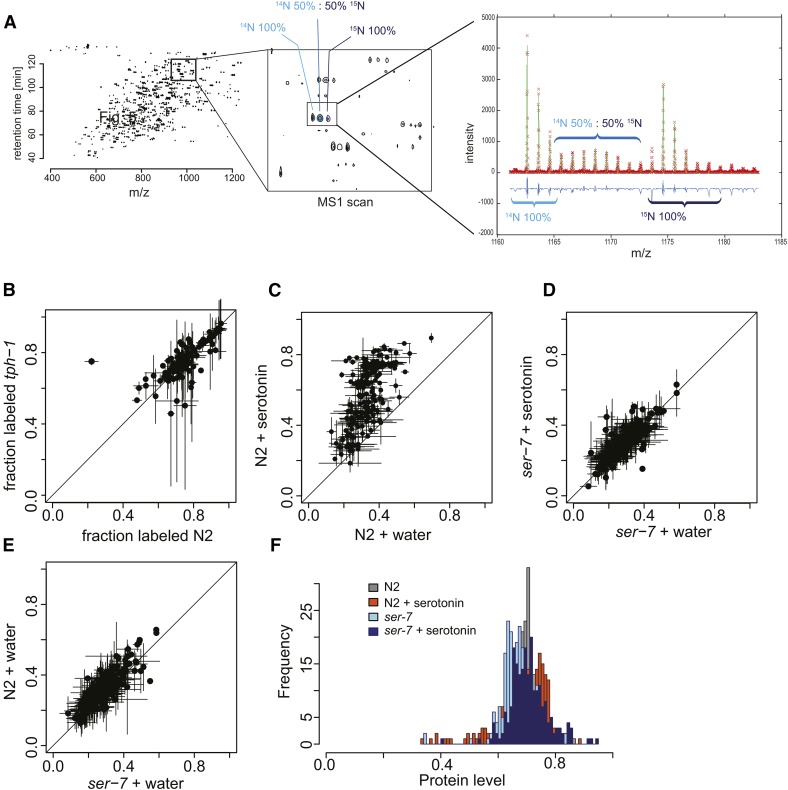
To understand how we identify individual peptides by mass spectrometry, it is necessary to address the complexity of the peptide samples. For each peptide, three isotope enrichment patterns are possible: 100% 14N-enriched (L), if it was synthesized before the pulse; 50% 15N-enriched (M), if it was synthesized during the pulse; or 100% 15N-enriched (H), if it was part of the external standard (Figure 4, A–C). Thus, each peptide in the sample can have three components in its isotope distribution in the mass spectra. Database search engines cannot identify peptides from 50% 15N-enriched parent ions and, as a consequence, only the identification of the 100% 14N- and 100% 15N-labeled peptides was carried out using MASCOT with searches for both E. coli and C. elegans (Eng et al. 1994). Because the light, medium, and heavy peptides coelute during liquid chromatography, the MS1 scan for the m/z range spanning all three labeled species can be extracted (L, M, and H) (Figure 5A). Misidentifications are readily identified because the spacing of the 14N and 15N peaks does not match the number of nitrogen atoms in the sequence. Thus, we can ensure correct identification of each peptide, despite the complexity of the isotope distribution with three components.
Figure 5.
Pulse-feeding assay. (A) Whole worm lysate analyzed on the mass spectrometer, with sample data shown in Figure 4, B and C. Each peptide in the spectra has a “light,” “medium,” and “heavy” component. The intensity of the light component depends on the amount of each peptide present before the pulse-labeling period. The intensity of the medium component depends on the amount of ingested food after the start of the pulse-labeling period. The heavy component serves as a standard sample. (B) Correlation plot of fraction-labeled values for wild-type N2 (x-axis) vs. tph-1 mutant worms (y-axis) from L4:D1. (C) Correlation plot of fraction-labeled values by protein for wild-type N2 treated with water (x-axis) or serotonin (y-axis, 5 mM) from D5:D7. (D) Correlation plot of fraction-labeled values for ser-7 mutant worms grown with water (x-axis) or treated with serotonin (y-axis, 5 mM) from D5:D7. (E) Correlation plot of fraction-labeled values for wild-type N2 (y-axis) and ser-7 mutants (x-axis) treated with water from D5:D7. (F) Histogram of protein level values for wild-type N2 and ser-7 mutants treated with water or serotonin (5 mM).
In a typical experiment, 300–400 proteins were identified based on one or more peptides. This number is less than what is achieved with deep proteomics, but is more than sufficient to determine the bulk metabolic labeling rate of the proteome. To determine isotope incorporation, and thus nutrient absorption, we calculated two values for each peptide: (i) the fraction labeled, or fraction of partial 15N labeling, (M)/(L + M) and (ii) the protein level of the sample, (L + M)/(L + M + H). The fraction labeled indicates the amount of 15N incorporated during pulse feeding for each protein and, over the entire proteome, is indicative of the amount of bacteria eaten. Importantly, 15N incorporation represents de novo protein synthesis occurring within the pulse-feeding interval and thus allows identification of both the newly synthesized and preexisting proteome. The protein level indicates the relative protein content in different experimental samples.
We first asked whether the pulse-feeding assay was able to identify a difference in nutrient absorption between N2 and tph-1(mg280) mutants. Since the bacterial clearing assay showed the greatest change in food intake to occur between L4 and D1, we chose this interval for pulse labeling. N2 or tph-1(mg280) animals were raised on light OP50, (100% 14N) until they reached the L4 stage. At L4, we began the pulse labeling by switching the feeding bacteria from light to medium OP50 (50% 14N; 50% 15N). Sixteen hours later, on day 1 of adulthood (D1), the animals were harvested to extract protein. To compare 15N incorporation between different samples, we mixed each experimental sample with an external standard. The external standard consisted of proteins extracted from worms fed 100% 15N-labeled OP50 bacteria over three generations and raised in parallel with the experimental samples. The samples were then processed for LC-MS/MS mass spectrometry.
Plotting the fraction labeled for both strains confirmed that there was no difference in feeding or nutrient absorption between N2 and tph-1(mg280), as shown by the bacterial clearing assay. The results further show that, within 16 hr of pulse labeling, ∼80% of the detected proteins become labeled (Figure 5B). We found that the ideal length of the labeling period depends on the food intake of the animals, with lower food intake requiring longer labeling periods and higher food intake requiring shorter periods. We also observed that egg laying represents a substantial “15N leak,” as eggs are almost completely de novo synthesized from 15N. As it is difficult to ensure that all eggs are harvested, we decided to start pulse feeding on day 5 in subsequent experiments, after the egg-laying period was over. This approach prevents 15N loss through egg laying and allows measurement over longer periods of time, as both food intake and protein synthesis are lower at older ages.
Serotonin increases protein synthesis
We next asked whether serotonin treatment also increased the absorption of ingested bacteria. On day 1 of adulthood, we added serotonin or water to wild-type N2 animals. On day 5, we exchanged light OP50 for medium OP50 (50% 14N; 50% 15N) to start pulse feeding. In wild-type animals treated with serotonin, the fraction labeled for each peptide increased by an average of 1.9 ± 0.5-fold as compared to water-treated control animals (Figure 5C). Hierarchical clustering of labeled peptides from wild-type N2 water-treated vs. serotonin-treated samples identified two groups: one in which the increase in fraction labeled was higher (2 ± 0.4-fold), consisting mainly of proteins involved in translation and ribosomal biogenesis (Table S1); and one in which the increase in fraction labeled was smaller (1.5 ± 0.3-fold), consisting of a variety of different proteins involved in metabolism. Of the total 153 proteins whose synthesis was increased in response to serotonin-induced food intake, 23 are likely to promote aging, as they extend lifespan when suppressed by RNAi. Thus, the increased synthesis of these proteins provides a possible link to how food intake promotes aging.
Similar to the bacterial clearing assay, the effect of added serotonin on the fraction-labeled value was abrogated in the ser-7 mutants (Figure 5, C–E). Protein levels did not change significantly between water-treated wild-type and ser-7 mutant worms, indicating that the ser-7(tm1325) allele did not have a general effect on the total amount of protein in the worm (Figure 5F). Thus, the SER-7 receptor is specifically required for the increase in food intake, nutrient absorption, and subsequent de novo protein synthesis and ribosomal biogenesis observed in serotonin-treated wild-type N2 worms.
Discussion
The present study outlines two independent methods, based on entirely different principles, which we used to measure food intake and nutrient absorption in C. elegans. These methods address two outstanding problems in the study of nutrition and metabolism in C. elegans: the absence of a direct measurement for food intake over extended periods of time and the need for information relating to how well nutrients are absorbed and assimilated upon ingestion.
The bacterial clearing assay provides a simple and direct quantitative measurement of food intake over timescales relevant to the study of organismal health and lifespan. It measures food intake by the same principles as is used in other model organisms such as flies and mice (Martin-Montalvo et al. 2013). The liquid culture format of the assay provides a scalable framework for the testing of chemical, genetic, and environmental perturbations in C. elegans. Importantly, the bacterial clearance assay is suitable for both small- and large-scale genome-wide screening and requires equipment readily available at most institutions. The assay yields robust results over a range of temperatures and bacterial concentrations and in the presence of common chemical solvents (DMSO, acetic acid, etc.) (data not shown). Under the conditions used in this study, the bacterial clearing assay consistently detected differences in food intake as low as 15%.
Studies of food intake in C. elegans have revealed deep insights into food-related behaviors, such as food choice and behavioral responses to food (Avery and You 2012). To fully understand the metabolic chain of events of feeding, we must also know the nutrient lifecycle from foraging behavior to assimilation of food into the animal. To address this, we developed the pulse-feeding assay that allows for the first time the determination of nutrient absorption into worm proteins by mass spectrometry. Our method establishes a direct link between a food intake behavior (pharyngeal pumping) and protein synthesis by measuring ingestion, absorption, and assimilation of nutrients into the proteome. Although the pulse-feeding assay is not the first to address de novo protein synthesis in C. elegans by mass spectrometry (Liang et al. 2014), our metabolic pulse-labeling approach offers significant advantages. First, metabolic labeling is nontoxic and therefore does not influence animal behavior or physiology. Second, metabolic labeling with heavy nitrogen is not subject to unwanted side reactions or chemical derivatization, nor does it require additional genetic mutations to suppress these effects (Gouw et al. 2011). Third, the pulse-feeding assay presented here allows for the quantization of de novo protein synthesis in a way that is not feasible by trace labeling with unnatural amino acids followed by mass spectrometry. With trace labeling, only labeled proteins can be compared from sample to sample. In contrast, the pulse-feeding assay uniquely labels proteins present both before and after the pulse, allowing comparison of protein levels and label incorporation of proteins prior to and after pulse labeling.
The greatest strength of the pulse-feeding method is that, while more involved than bacterial clearing, it directly establishes nutrient utilization and is independent of the culture medium. Determining nutrient utilization directly is important, especially in animals that suffer from gut defects and can provide meaningful information even when food intake appears normal. We propose that the pulse-labeling assay provides a suitable gold standard to measure the amount of food utilized by an animal. Importantly, the pulse-feeding assay can be adapted for other species, including Drosophila melanogaster, or any other organism whose food can be grown in stable-isotope-labeled media (Perez and Van Gilst 2008; Deshpande et al. 2014).
As recently shown, overabundance of protein intake is associated with poor health and accelerated aging (Solon-Biet et al. 2014). How protein intake affects subsequent protein synthesis, the proteome, and thus health, is an open question. As the pulse-feeding assay is based on measuring de novo protein synthesis, it provides an important tool to study how food composition dictates protein synthesis in the animal and thus how food composition affects the proteome, physiology, and health.
It revealed that serotonergic signals induce translation of ribosomes and other age-promoting proteins, providing a mechanistic link to how high food intake may promote aging. Thus, the two assays presented enable investigations into the complex biological interaction between food and protein synthesis and its subsequent effects on aging and health
Supplementary Material
Acknowledgments
This work was funded by a grant to M.P. from the National Institutes of Health (NIH) (DP2 OD008398), a grant from the Ellison Medical Foundation (AG-NS-0928-12), a grant to J.W. from the NIH (R37-GM-053757), a Muscular Dystrophy Association development grant to S.R., an American heart fellowship to E.V. (AHA 10POST3500084), and a National Science Foundation Graduate Research Fellowship Program fellowship to G.M.S. Strains were provided by Shigen-Japan or the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Communicating editor: O. Hobert
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175851/-/DC1.
Literature Cited
- Ashrafi K., 2006. Mapping out starvation responses. Cell Metab. 3: 235–236. [DOI] [PubMed] [Google Scholar]
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., et al. , 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268–272. [DOI] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., You Y. J., 2012. C. elegans feeding. WormBook 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J. M., Sotnikova T. D., Marion S., Lefkowitz R. J., Gainetdinov R. R., et al. , 2005. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell 122: 261–273. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974 The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F., Au C., Leung K. Y., Vergara-Irigaray N., Cocheme H. M., et al. , 2013. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng Q., Sieburth D., Kaplan J. M., 2008. Profiling synaptic proteins identifies regulators of insulin secretion and lifespan. PLoS Genet. 4: e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. S., Sperling E., Silverman J. M., Davis J. H., Williamson J. R., 2012. Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol. Biosyst. 8: 3325–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. S., Williamson J. R., 2013. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol. 425: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. A., Hua Z., Srinivasan S., Liu J., Lee B. H., et al. , 2012. AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell Metab. 16: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M., Bargmann C. I., 1998. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94: 679–689. [DOI] [PubMed] [Google Scholar]
- Deshpande S. A., Carvalho G. B., Amador A., Phillips A. M., Hoxha S., et al. , 2014. Quantifying Drosophila food intake: comparative analysis of current methodology. Nat. Methods 11: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J. K., McCormack A. L., Yates J. R., 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Geillinger K. E., Kuhlmann K., Eisenacher M., Meyer H. E., Daniel H., et al. , 2012. Dynamic changes of the Caenorhabditis elegans proteome during ontogenesis assessed by quantitative analysis with 15N metabolic labeling. J. Proteome Res. 11: 4594–4604. [DOI] [PubMed] [Google Scholar]
- Gouw J. W., Tops B. B., Krijgsveld J., 2011. Metabolic labeling of model organisms using heavy nitrogen (15N). Methods Mol. Biol. 753: 29–42. [DOI] [PubMed] [Google Scholar]
- Grun D., Kirchner M., Thierfelder N., Stoeckius M., Selbach M., et al. , 2014. Conservation of mRNA and protein expression during development of C. elegans. Cell Reports 6: 565–577. [DOI] [PubMed] [Google Scholar]
- Hobson R. J., Hapiak V. M., Xiao H., Buehrer K. L., Komuniecki P. R., et al. , 2006. SER-7, a Caenorhabditis elegans 5–HT7-like receptor, is essential for the 5-HT stimulation of pharyngeal pumping and egg laying. Genetics 172: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Xiong C., Kornfeld K., 2004. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 101: 8084–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Evason K., Xiong C., Kornfeld K., 2007. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 3: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S., 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95: 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang V., Ullrich M., Lam H., Chew Y. L., Banister S., et al. , 2014. Altered proteostasis in aging and heat shock response in C. elegans revealed by analysis of the global and de novo synthesized proteome. Cell. Mol. Life Sci. 71: 3339–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedtke S., O’Connor V., Holden-Dye L., Walker R. J., 2010. The regulation of feeding and metabolism in response to food deprivation in Caenorhabditis elegans. Invert. Neurosci. 10: 63–76. [DOI] [PubMed] [Google Scholar]
- Martin-Montalvo A., Mercken E. M., Mitchell S. J., Palacios H. H., Mote P. L., et al. , 2013. Metformin improves healthspan and lifespan in mice. Nat. Commun. 4: 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. P., Raizen D. M., Gottschalk A., Schafer W. R., Avery L., 2004. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics 166: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. T., Jordan J. M., Baugh L. R., 2013. WormSizer: high-throughput analysis of nematode size and shape. PLoS ONE 8: e57142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morck C., Pilon M., 2006. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol. 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble T., Stieglitz J., Srinivasan S., 2013. An integrated serotonin and octopamine neuronal circuit directs the release of an endocrine signal to control C. elegans body fat. Cell Metab. 18: 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Grundahl K., Meyer B. J., Rand J. B., 1993. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell 73: 1291–1305. [DOI] [PubMed] [Google Scholar]
- Paek J., Lo J. Y., Narasimhan S. D., Nguyen T. N., Glover-Cutter K., et al. , 2012. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 16: 526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C. L., Van Gilst M. R., 2008. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 8: 266–274. [DOI] [PubMed] [Google Scholar]
- Petrascheck M., Ye X., Buck L. B., 2007. An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature 450: 553–556. [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Lee R. Y., Avery L., 1995. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141: 1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S., Solis G. M., Petrascheck M., 2015. High-throughput small-molecule screening in Caenorhabditis elegans. Methods Mol. Biol. 1263: 139–155. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Gossen M., Dittmar G., Selbach M., 2009. Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9: 205–209. [DOI] [PubMed] [Google Scholar]
- Solis G. M., Petrascheck M., 2011. Measuring Caenorhabditis elegans life span in 96 well microtiter plates. J. Vis. Exp. pii 2496. 10.3791/2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet S. M., McMahon A. C., Ballard J. W., Ruohonen K., Wu L. E., et al. , 2014. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 19: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling E., Bunner A. E., Sykes M. T., Williamson J. R., 2008. Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-squares Fourier transform convolution. Anal. Chem. 80: 4906–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Sadegh L., Elle I. C., Christensen A. G., Faergeman N. J., et al. , 2008. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 7: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger K. A., Shtonda B. B., Thacker C., Snutch T. P., Avery L., 2005. The C. elegans T-type calcium channel CCA-1 boosts neuromuscular transmission. J. Exp. Biol. 208: 2191–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes M. T., Shajani Z., Sperling E., Beck A. H., Williamson J. R., 2010. Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J. Mol. Biol. 403: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Victor M., Loer C., Shi Y., Ruvkun G., 2000. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403: 560–564. [DOI] [PubMed] [Google Scholar]
- Taormina G., Mirisola M. G., 2014. Calorie restriction in mammals and simple model organisms. Biomed Res Int 2014: 308690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisine C., Varma H., Walker N., Bates E. A., Stockwell B. R., et al. , 2007. Identification of potential therapeutic drugs for huntington’s disease using Caenorhabditis elegans. PLoS ONE 2: e504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y. J., Kim J., Raizen D. M., Avery L., 2008. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 7: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.