Abstract
Although a growing number of studies have reported the importance of SUMOylation in genome maintenance and DNA double-strand break repair (DSBR), relevant target proteins and how this modification regulates their functions are yet to be clarified. Here, we analyzed SUMOylation of ZTF-8, the homolog of mammalian RHINO, to test the functional significance of this protein modification in the DSBR and DNA damage response (DDR) pathways in the Caenorhabditis elegans germline. We found that ZTF-8 is a direct target for SUMOylation in vivo and that its modification is required for DNA damage checkpoint induced apoptosis and DSBR. Non-SUMOylatable mutants of ZTF-8 mimic the phenotypes observed in ztf-8 null mutants, including reduced fertility, impaired DNA damage repair, and defective DNA damage checkpoint activation. However, while mutants for components acting in the SUMOylation pathway fail to properly localize ZTF-8, its localization is not altered in the ZTF-8 non-SUMOylatable mutants. Taken together, these data show that direct SUMOylation of ZTF-8 is required for its function in DSBR as well as DDR but not its localization. ZTF-8’s human ortholog is enriched in the germline, but its meiotic role as well as its post-translational modification has never been explored. Therefore, our discovery may assist in understanding the regulatory mechanism of this protein in DSBR and DDR in the germline.
Keywords: SUMOylation, DNA damage response, double-strand break repair, ZTF-8, Caenorhabditis elegans, germline
MEIOSIS is a specialized cell division process by which a diploid cell produces haploid gametes that are essential for sexual reproduction. During meiosis, the homologous chromosomes contributed by each parent undergo genetic recombination that enhances genetic diversity in the progeny and generates chiasmata, which are essential to proper segregation at meiosis I. This process requires precise control of homologous chromosome pairing and genetic recombination to secure accurate chromosome segregation and transferring of genetic materials to the next generation. Therefore, defects in meiotic progression cause aneuploidy, which leads to reproductive failure and congenital birth defects as evidenced by 7–10% of chromosomally abnormal pregnancies in humans (Hunt and Hassold 2008).
Although a higher frequency of deleterious consequences arise from meiotic defects, relatively fewer studies have focused on the mechanisms underlying accurate meiotic rather than mitotic cell divisions, especially in multicellular systems. A high degree of conservation is shared between Caenorhabditis elegans and humans in proteins and pathways involved in double-strand break repair (DSBR) and DNA damage response (DDR), so we take advantage of this genetically tractable multicellular nematode to understand the fundamental mechanisms of DSBR and DDR in the germline (O’Neil and Rose 2006; Craig et al. 2012).
We previously showed that ztf-8 null mutants exhibit DNA damage sensitivity to γ-irradiation (IR) and hydroxyurea (HU) and that ZTF-8’s subcellular localization is altered in response to DNA damage as well as in mutants of C. elegans ATM and ATR homologs (Kim and Colaiacovo 2014). We found that ZTF-8 is required for DSBR as exemplified by the impaired progression of DSBR both in mitotic and meiotic germline nuclei. Moreover, due to the defective localization of the 9-1-1 DDR complex, ztf-8 mutants partially fail to trigger the p53/CEP-1-dependent DNA damage checkpoint in late pachytene, also suggesting a role for ZTF-8 in DDR.
Here, we show that SUMOylation of ZTF-8 is required for its functions in both DSBR and DNA damage checkpoint activation in the C. elegans germline. We found that ZTF-8 is a direct target for SUMOylation at its consensus ΨKXE sites in vivo. Mutations of the SUMOylation sites in ZTF-8 mimic the ztf-8 null mutant phenotypes, suggesting that SUMOylation is a critical modification for proper function of ZTF-8 and is indispensable for DSBR and DNA damage-mediated checkpoint activation in the germline.
Materials and Methods
Strains and alleles
C. elegans strains were cultured at 20° under standard conditions as described in Brenner (1974). The N2 Bristol strain was used as the wild-type background. The following mutations and chromosome rearrangements were used in this study:
LGI: hus-1(op241), opIs34[hus-1::gfp], hT2[bli-4(e937) let-?(q782) qIs48](I; III)
LGIII: ztf-8(tm2176), ztf-8(rj22[ztf-8::gfp::flag]), ztf-8(rj23[ztf-8::gfp::flag + K14R K494R K518R K527R]), ztf-8(rj24[K14R]), ztf-8(rj25[K494R K518R K527R]), ztf-8(rj26[K14R K494R K518R K527R]), sDf121 and qC1[dpy-19(e1259) glp-1(q339) qIs26].
The ztf-8(tm2176) mutant was generated by the Japanese National BioResource Project for C. elegans and carries a 524-bp out-of-frame deletion that removes most of exon 6 along with exons 7–11 (Figure 1A). This deletion results in a premature stop codon and loss of the zinc-finger motifs located in the middle of ZTF-8, the four ΨKXE consensus SUMOylation sites, and a putative phosphorylation site (http://www.phosphopep.org).
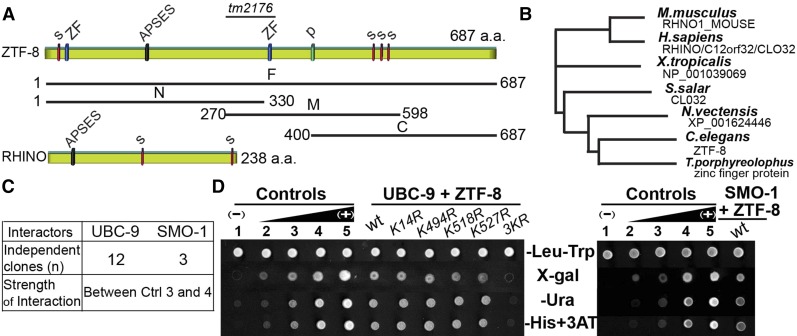
Figure 1.
ZTF-8 is a conserved protein that interacts with both UBC-9 and SMO-1. (A) Scheme of the C. elegans ZTF-8 protein and its human functional homolog, RHINO. The region deleted in the tm2176 mutant allele (codons 287–351) is indicated. Four predicted SUMOylation sites (s), two zinc-finger motifs (ZF), one APSES DNA binding site (APSES), and a phosphorylation site (p) are indicated. Full length (F) and truncations (N, M, C) of ZTF-8 used in a yeast two-hybrid screen are indicated. (B) Phylogenic tree comparing the potential orthologs of ZTF-8 with various species by ClustalW2 (EMBL-EBI). (C) Results from a yeast two-hybrid screen using the full-length and truncated versions of ZTF-8 as bait. The strength of the protein interaction is graded on the basis of comparison with one negative and four positive controls (as shown in D). (D) The yeast two-hybrid screen identified UBC-9 and SMO-1 as binding interactors for ZTF-8. Wild type as well as the indicated point mutants of ZTF-8 were tested for their interaction with UBC-9 (left) or SMO-1 (right). ZTF-8 fused to the DNA binding domain and the UBC-9/SMO-1 fused to the activation domain of GAL4 were analyzed in a yeast two-hybrid assay. One negative (no. 1) and four positive controls (nos. 2–5) were used as described in Walhout and Vidal (2001). Interactions were scored by growth on X-gal, SC–Ura, and SC–His +1 mM 3AT plates and compared to growth on the control SC–Let–Trp plates.
The following set of transgenic worms were generated with the CRISPR–Cas9 technology as described in Friedland et al. (2013) and outcrossed to wild type five times. ztf-8::gfp::flag knock-in allele is ztf-8(rj22[ztf-8::gfp::flag]). Non-SUMOylatable mutants are ztf-8(rj24[K14R]), ztf-8(rj25[K494R K518R K527R]), ztf-8(rj26[K14R K494R K518R K527R]). Non-SUMOylatable alleles with ztf-8::gfp::flag knock-in is ztf-8(rj23[ztf-8::gfp::flag + K14R K494R K518R K527R]).
Analysis of ZTF-8 protein conservation and motifs
ZTF-8 homology searches and alignments were performed using Uniprot (http://www.uniprot.org/). Pfam and Prosite (release 20.70) were used for zinc-finger motif predictions (Sonnhammer et al. 1997). SUMOylation sites and SIMs were identified by using GPS–SUMO predictor (Zhao et al. 2014).
Plasmids
The pUC57 klp-12 sgRNA and Peft-3::cas9 plasmids were described in Friedland et al. (2013). The ztf-8 sgRNA plasmids were constructed by replacing the unc-119 sgRNA sequence with a sequence corresponding to 5872–5894 bp downstream of the ztf-8 start codon genomic sequence for GFP and FLAG tagging, 1450–1472 bp downstream for 1KR and 3735–3757 bp downstream for 3KR and 4KR mutants as described in Friedland et al. (2013). To build the ztf-8::gfp::flag donor template, ztf-8 genomic DNA containing upstream and downstream 1-kb homology arms was PCR amplified and cloned into the multicloning site at pUC18 plasmid along with GFP and FLAG tag amplified from pCM1.53 and pDEST17 obtained from Addgene through Geraldine Seydoux lab. To build the ztf-8 donor vectors, ztf-8 genomic DNA containing 3943-bp homology was cloned into pUC18 vector by using the KpnI and SalI sites and mutated the SUMOylation sites by using Gibson Assembly from New England Biolabs.
DNA micro-injection
Plasmid DNA was micro-injected into the germline as described in Friedland et al. (2013) and Tzur et al. (2013). Injection solutions were prepared to contain 25 ng/μl of pCFJ90 (Pmyo-2::mCherry; Addgene), which was used as the co-injection marker, 200 ng/μl of the sgRNA vector, 175 ng/μl of the Peft-3Cas9-SV40 NLStbb-2 3′-UTR, and 200 ng/μl of the donor vector.
Quantitative analysis for RAD-51 foci
Quantitative analysis of RAD-51 foci was performed as in Colaiacovo et al. (2003). Five to nine germlines were scored for each genotype. The average number of nuclei scored per zone for a given genotype was as follows, ± SD: zone 1, n = 151.3 ± 32.3, zone 2, n = 148.0 ± 30.0, and zone 5 = 132.0 ± 37.3. Statistical comparisons between genotypes were performed using the two-tailed Mann–Whitney test, 95% confidence interval (C.I.).
In vivo SUMOylation assay
Transgenic worms harboring the GFP::FLAG fusion rj22, rj23, or control wild-type N2 were collected and immediately frozen in liquid nitrogen. N-Ethylmaleimide (Sigma) was added to inhibit de-SUMOylation of the protein and a protease inhibitor (Sigma P1860) was added (1:500 volume) to prevent degradation of the protein (Kaminsky et al. 2009). After sonication, lysates were concentrated and subjected to either Western blot analysis or immunoprecipitation by using FLAG M2 gel (Sigma) based on supplier’s protocol.
Immunofluorescence
Whole-mount preparations of dissected gonads, fixation, and immunostaining procedures were carried out as described in Colaiacovo et al. (2003). Primary antibodies were used at the following dilutions: rabbit α-ZTF-8 (1:200) for C-terminal antibody described in Kim and Colaiacovo (2014), rabbit α-RAD-51 (1:2000; SDIX), mouse α-FLAG (1:1000; Sigma), and mouse α-tubulin (1:1000; Sigma). Secondary antibodies used were: Cy3 anti-rabbit, FITC anti-rabbit, and FITC anti-mouse (Jackson Immunochemicals), each at 1:200. Immunofluorescence images were collected at 0.2-μm intervals with an IX-70 microscope (Olympus) and a CoolSNAP HQ CCD camera (Roper Scientific) controlled by the DeltaVision system (Applied Precision). Images were subjected to deconvolution by using the SoftWoRx 3.3.6 software (Applied Precision).
Quantitative analysis of germ-cell apoptosis
Germlines of age-matched (20 hr post-L4) animals were analyzed by acridine orange staining, as described in Kelly et al. (2000), utilizing a Leica DM5000B fluorescence microscope. Between 25 and 45 gonads were scored for each genotype. Statistical comparisons between genotypes were performed using the two-tailed Mann–Whitney test, 95% C.I.
Yeast two-hybrid screen
The full-length of the ztf-8 open reading frame, as well as C- (400–687), middle (270–598), and N- (1–330) terminal truncations were amplified by PCR. A cDNA library generated from mixed-stage C. elegans was used for the amplification with primers that contain Gateway compatible sequences and a gene-specific sequence as indicated in Supporting Information, Table S1. Gateway cloning, cDNA and ORFeome library screening, and X-Gal, -URA, and –HIS + 3AT assays for examining yeast two-hybrid interactions were performed as in Walhout and Vidal (2001). For supplemental experimental producures, see File S1.
Results
ZTF-8 interacts with components of the SUMOylation pathway in a manner dependent on its SUMOylation sites
ZTF-8 (open reading frame ZC395.8) encodes a 687-amino-acid protein, which contains four predicted SUMOylation sites, two C2H2-type zinc-finger-binding domains, and one APSES DNA-binding motif (Figure 1A; Kim and Colaiacovo 2014). The high degree of amino acid sequence conservation found for ZTF-8 from worms to humans, and in particular for the APSES motif required for interaction with MRT-2/Rad1, a component of the 9-1-1 complex, support conservation of the DNA damage checkpoint role throughout species (Figure 1B; Kim and Colaiacovo 2014)
To identify potential regulators of ZTF-8, we screened a C. elegans cDNA library prepared from mixed-stage worms utilizing full length and three specific regions of ZTF-8 (N1–330, M270–598, and C400–687) as baits in a yeast two-hybrid approach (Figure 1A). We identified multiple independent clones corresponding to UBC-9 (Figure 1C). UBC-9 is the only known E2 SUMO-conjugating enzyme in C. elegans (Roy Chowdhuri et al. 2006), and it interacts with ZTF-8 as evidenced by measuring three different reporter gene readouts (β-galactosidase activity, growth in medium lacking uracil, and growth in medium lacking histidine with added 3AT) (Figure 1D). In addition to UBC-9, we identified SMO-1, which encodes the C. elegans ortholog of SUMO (small ubiquitin-like moiety), as a ZTF-8 interactor (Figure 1, C and D).
Both UBC-9 and SMO-1 interact with either the full length or the C terminus of ZTF-8. These observations suggest that ZTF-8 may undergo a post-translational SUMOylation modification. SUMOylation of ZTF-8 is further supported by the presence of four consensus SUMOylation sites (ΨKXE, where Ψ is a large hydrophobic amino acid; Hay 2005) at amino acids 14, 494, 518, and 527, which in other proteins are sufficient to mediate interaction with and SUMOylation by UBC-9 (Figure 1A; Sampson et al. 2001; Bernier-Villamor et al. 2002).
To determine which of the four predicted SUMOylation sites is required for ZTF-8’s interaction with UBC-9, we generated point mutations in the four conserved SUMO conjugation sites: K14R, K494R, K518R, and K527R. Interestingly, none of the single point mutations were sufficient for disrupting ZTF-8’s interaction with the E2 SUMO ligase UBC-9 in the yeast two-hybrid assay, suggesting either the presence of multiple SUMOylation sites in ZTF-8 or SUMOylation of a nonconsensus site (Figure 1D; Zhao et al. 2014). In fact, the 3KR mutant that contains K to R replacements simultaneously at amino acids 494, 518, and 527 abrogates the interaction suggesting that more than one of the predicted sites are required for SUMOylation of the ZTF-8 protein.
The dynamic localization of ZTF-8 in germ cell nuclei requires the SUMOylation pathway
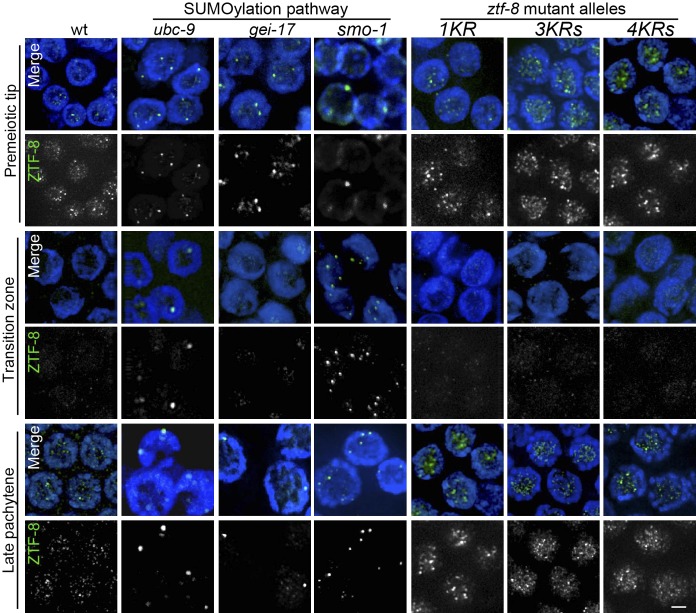
Our previous immunostaining of dissected wild-type hermaphrodite gonads with a ZTF-8-specific antibody showed that ZTF-8 exhibits a dynamic localization (Kim and Colaiacovo 2014). In brief, ZTF-8 signal is observed in mitotic nuclei at the distal tip (premeiotic tip), and this signal is then reduced upon entrance into meiosis (leptotene/zygotene stages is the transition zone) and remains weak through the midpachytene stage. The ZTF-8 signal increases once again in late pachytene nuclei and persists through late diakinesis oocytes. This dynamic pattern of localization suggests regulation of ZTF-8 during meiotic prophase.
Consistent with our yeast two-hybrid results, we found that the wild-type pattern of localization for ZTF-8 requires the E2 SUMO-conjugating enzyme UBC-9. In wild type, ZTF-8 foci are observed localizing both to the nucleolus and associating with chromatin specifically in the premeiotic tip and late pachytene nuclei. In contrast, ZTF-8 signal is sequestered in larger foci throughout all germline nuclei in ubc-9 mutants (Figure 2). To further test whether the SUMOylation pathway is required for proper localization of ZTF-8, we examined either RNAi depleted worms or mutants for other components acting in this pathway. RNAi depletion of gei-17, an E3 SUMO-ligase (Roy Chowdhuri et al. 2006), results in a similar alteration of ZTF-8 localization at the premeiotic tip and pachytene stages, but not at transition zone (Figure 2 and Figure S1). While UBC-9 is the only known E2 SUMO-conjugating enzyme in C. elegans, there are several known E3 SUMO ligases from yeast to humans, and that might also be the case in C. elegans (Johnson 2004; Hay 2005; Roy Chowdhuri et al. 2006), which could account for the more restricted windows during meiosis in which we observed an effect for the gei-17 depletion. Meanwhile, mutation of smo-1, which encodes the C. elegans ortholog of SUMO, results in an altered localization for ZTF-8 throughout all germline nuclei, similar to ubc-9 mutants (Figure 2). Taken together, these data indicate that ZTF-8 interacts with SUMOylation components and that its proper localization in the germline requires the SUMOylation pathway.
Figure 2.
The localization of ZTF-8 is altered in SUMOylation-defective mutants but not in non-SUMOylatable ZTF-8 mutants. Immunolocalization of ZTF-8 in indicated genotypes of SUMOylation pathway-deficient mutants and non-SUMOylatable ZTF-8 transgenic worms: 1KR ztf-8(rj24), 3KR ztf-8(rj25), and 4KR ztf-8(rj26). Costaining of nuclei with DAPI (blue) and an anti-ZTF-8 antibody (green). Images show premeiotic tip, transition zone (leptotene/zygotene stages), and late pachytene nuclei. Bar, 2μm.
ZTF-8 localization is not altered in the absence of its consensus SUMOylation sites
Altered localization of ZTF-8 in the SUMOylation mutants might be an indirect consequence of a defective SUMOylation pathway (i.e., lack of SUMOylation of another target, which in turn affects the localization of ZTF-8) rather than lack of SUMOylation of ZTF-8. To distinguish between these two possibilities and to examine whether ZTF-8 is a direct target for SUMOylation, we used CRISPR–CAS9 site-specific engineering to generate transgenic worms carrying mutations (K to R) in the four predicted consensus ΨKXE SUMOylation sites (amino acids 14, 494, 518, and 527) (Johnson 2004; Xue et al. 2006). Transgenic worm 1KR carries a single mutation at amino acid 14. 3KR worms contain mutations at amino acids 494, 518, and 527, whereas 4KR worms contain mutations at all four predicted SUMOylation sites, namely amino acids 14, 494, 518, and 527.
Immunolocalization with a ZTF-8-specific antibody revealed a normal localization for ZTF-8 in the ZTF-8 1KR mutants (Figure 2). Similar to wild-type and unlike SUMOylation-defective mutants, in ZTF-8 1KR mutants ZTF-8 localizes to the nucleolus and associates with chromatin in the premeiotic tip and late pachytene nuclei. Likewise, a pattern of localization for ZTF-8 indistinguishable from wild type was observed in the germlines of both 3KR and 4KR mutants. These results suggest that the absence of the consensus SUMOylation sites does not affect the localization of ZTF-8 and that therefore SUMOylation at those sites is not required for proper ZTF-8 localization.
Detection of SUMOylated ZTF-8 in vivo
Based on its interaction with components acting in the SUMOylation pathway and the presence of four consensus SUMOylation sites in ZTF-8, we hypothesize that ZTF-8 is a direct substrate for SUMOylation. To test this in vivo, we built the wild-type fusion protein ZTF-8::GFP::FLAG (rj22) and the mutant 4KR::GFP::FLAG (rj23) fusion protein in which the four predicted SUMOylation sites of ZTF-8 were mutated to arginine. Both constructs were expressed in transgenic worms under the endogenous ztf-8 promoter. The tagged wild-type version of ZTF-8 (rj22) successfully rescues the ztf-8 null mutant phenotypes and colocalizes with the anti-ZTF-8 antibody, suggesting that it is a fully functional version of the protein (Figure S2).
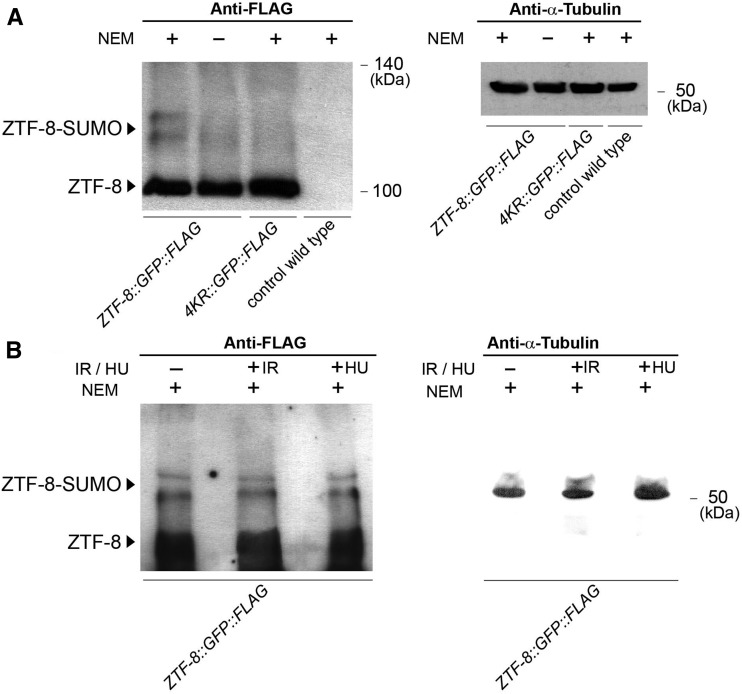
Western blot analysis revealed a band corresponding to the expected size of the tagged wild-type protein as well as two slower migrating bands with a molecular weight shift of about 10–20 kDa, consistent with SUMOylation of ZTF-8 (Figure 3A, lane 1). In contrast, 4KR mutants were missing the higher-molecular-weight bands (lane 3). These slower migrating bands were visible only in the wild-type worms and only when their lysates were supplemented with the deSUMOylation inhibitor N-ethylmaleimide (NEM; lane 1). Therefore, they were likely deSUMOylated in the absence of NEM (lane 2), which is consistent with other studies (Becker et al. 2013), suggesting that ZTF-8 is a direct substrate for SUMOylation and that the predicted ΨKXE SUMOylation sites are the primary in vivo sites of SUMOylation for ZTF-8. This is consistent with our results from liquid chromatography–mass spectrometry (LC–MS) analysis performed on pulldowns of ZTF-8 using anti-GFP agarose beads with lysates from our ZTF-8::GFP::FLAG and 4KR::GFP::FLAG transgenic lines compared to lysates from control worms expressing only GFP under the unc-17 promoter (vsIS48[Punc-17::gfp]). In agreement with the yeast two-hybrid results, we identified the E2 SUMO conjugation protein UBC-9 as a component interacting only with the wild-type ZTF-8 and not the non-SUMOylatable ZTF-8 by LC–MS analysis supporting the idea that ZTF-8 is SUMOylated (Table S1). Moreover, a separate study identified ZTF-8 as a SUMO-conjugated protein by LC–MS analysis (Kaminsky et al. 2009), thus further supporting the idea that ZTF-8 is SUMOylated in vivo.
Figure 3.
Detection of SUMOylation of ZTF-8 in vivo. (A) Lysates from transgenic worms expressing endogenous FLAG-tagged ZTF-8 (rj22), either treated or not with NEM to prevent de-SUMOylation (lanes 1 and 2, respectively), from NEM-treated K4Rs mutants (rj23, lane3), and from nontransgenic wild-type worms (lane 4, negative control) were examined on Westerns immunoblotted with an anti-FLAG antibody. An anti-α-tubulin antibody was used for a loading control (right). (B) Lysates from transgenic worms expressing FLAG-tagged ZTF-8 either untreated (−IR and −HU), after 30 min of exposure to γ-IR (50 Gy) or treated with 10 mM HU, all in the presence of NEM. The same lysates were also subjected to immunoblotting with an anti-α-tubulin antibody (right).
Finally, we also tested whether ZTF-8 is SUMOylated in response to either exogenous DSBs, by exposing worms to γ-irradiation or to replication fork arrest following hydroxyurea treatment. However, no obvious change was observed in the SUMOylated species detected between the control and exposed worms (Figure 3B), suggesting that SUMOylation of ZTF-8 may not be a direct response to either the formation of exogenous breaks or stalled replication forks.
Absence of the consensus SUMOylation sites causes reduced fertility and impairs DSBR progression
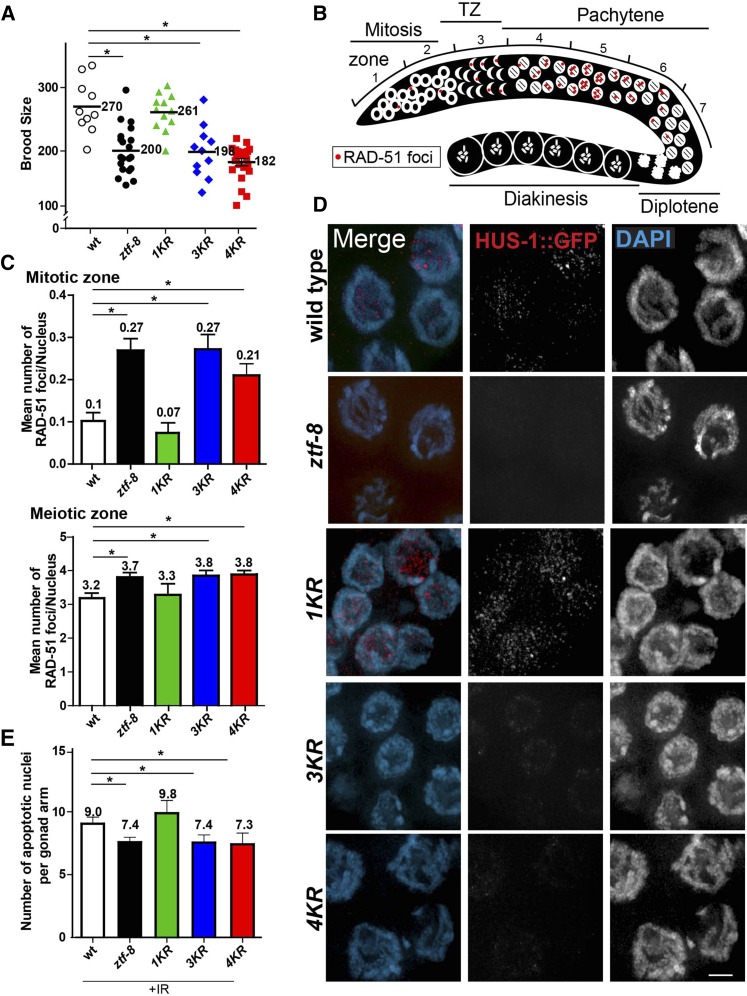
We previously showed that a ztf-8 null mutant (tm2176) exhibits a 25% reduction in brood size compared to wild type, indicative of sterility (Figure 4A and Kim and Colaiacovo 2014). We examined whether SUMOylation is important for ZTF-8’s roles in normal fertility. Interestingly, we found that both 3KR and 4KR mutants have reduced brood sizes compared to wild type (P = 0.0011 for 3KR and P < 0.0001 for 4KR, by the two-tailed Mann–Whitney test, 95% C.I.), which were similar to the level of reduction detected in ztf-8 null mutants (P = 0.8526 for 3KR and P = 0.2142 for 4KR compared to ztf-8). However, the 1KR mutant does not have a reduced brood size compared to wild type (P = 0.7713), suggesting that the predicted SUMOylation site carrying the lysine at amino acid 14 (K14) is dispensable for ZTF-8’s function, but that the other three SUMOylation consensus sites carrying lysine at amino acids 494, 518, and 527 are required for fertility in C. elegans.
Figure 4.
SUMOylation is required for proper function of ZTF-8. (A) 3KR and 4KR mutants exhibit a 25% reduction in brood size compared to wild type. (B) Diagram of a C. elegans germline indicating the position of the seven zones scored for RAD-51 foci. (C) Non-SUMOylatable ZTF-8 mutants exhibit elevated levels of RAD-51 foci similar to ztf-8 null mutants in both mitotic (zones 1 and 2) and meiotic (zone 5) nuclei. (D) Expression of a HUS-1::GFP transgene in pachytene nuclei of wild type (hus-1::gfp), ztf-8 (hus-1::gfp;ztf-8(tm2176)), 1KR (hus-1::gfp;rj24), 3KR (hus-1::gfp;rj25), and 4KR (hus-1::gfp;rj26) mutants. Bar, 2 μm. (E) Non-SUMOylatable ZTF-8 mutants exhibit impaired activation of germ cell apoptosis similar to ztf-8 null mutants.
The sterility observed in the 3KR and 4KR mutants prompted us to determine whether SUMOylation is necessary for ZTF-8’s function in DSBR. To determine if the SUMOylation of ZTF-8 is required for DSBR in both mitotic and meiotic nuclei, levels of RAD-51 foci, which mark DSBR sites (Colaiacovo et al. 2003), were quantitated in the germlines of the 1KR, 3KR, and 4KR mutants and compared to wild type and ztf-8(tm2176) mutants. Given that nuclei are positioned in a temporal-spatial manner along the germline in C. elegans (Figure 4B), proceeding from mitosis into the various stages of meiotic prophase I, levels of RAD-51 foci were assessed both in mitotic and meiotic nuclei (Figure 4C and Figure S3).
In 3KR and 4KR mutants, levels of RAD-51 foci were higher than those observed in wild type. The average number of RAD-51 foci per nucleus during mitosis is more than twofold higher in the mutants compared to wild-type germlines (Figure 4C, P = 0.0001 for 3KR and P = 0.0036 for 4KR by the two-tailed Mann–Whitney test, 95% C.I., zones 1 and 2 combined). Also, meiotic germline nuclei exhibit 20% more RAD-51 foci per nucleus than wild type (3.83 and 3.86 for 3KR and 4KR, respectively, compared to 3.18 for wild type, P = 0.0012 for 3KR and P < 0.0001 for 4KR for nuclei in zone 5). However, in 1KR mutants, levels of RAD-51 foci were indistinguishable from wild type in either mitotic or meiotic germline nuclei (P = 0.4278 for mitotic zones 1and 2 combined and P = 0.37 for meiotic zone 5), consistent with their normal level of fertility (Figure 4A). Interestingly, the levels of RAD-51 foci observed in both 3KR and 4KR mutants mimic those detected in ztf-8 mutants throughout both mitotic and meiotic nuclei, suggesting that SUMOylation is required for DSBR (Figure 4C, P = 0.1835 for mitotic zones 1 and 2 combined, and P = 0.2097 for meiotic zone 5). Taken together, these data indicate a role for SUMOylation of the three more C-terminally located SUMOylation sites on ZTF-8 for its function in DSBR.
SUMOylation of ZTF-8 is indispensable for 9-1-1 localization and normal activation of the DNA damage-mediated apoptosis pathway
It is known that accumulation of unrepaired DSBs can trigger a DNA damage checkpoint resulting in induced levels of apoptosis during late pachytene stage in the C. elegans germline (Gartner et al. 2000). We previously showed that ZTF-8 acts through the 9-1-1 complex to promote normal meiotic DNA damage checkpoint activation (Kim and Colaiacovo 2014). To determine whether SUMOylation of ZTF-8 is required for DNA damage-induced checkpoint response, we used a HUS-1::GFP transgenic line to examine the localization of this 9-1-1 complex component and assessed the level of DNA damage-mediated apoptosis during late pachytene in our non-SUMOylatable mutants compared to wild-type and ztf-8 null mutants.
We observed very weak or no HUS-1::GFP signal in the germlines of both 3KR and 4KR worms compared to wild type. This suggests that the DNA damage checkpoint operating in late pachytene is impaired in these mutants, comparable to our previous finding for ztf-8 null mutants (Figure 4D; Kim and Colaiacovo 2014). Also, consistent with preceding results, the HUS-1::GFP signal was not altered in 1KR mutants compared to wild type.
In the C. elegans germline 9-1-1 and CEP-1/p53 proteins act in the same pathway and HUS-1 is required for CEP-1/p53-dependent DNA damage-mediated apoptosis (Hofmann et al. 2002). In accordance with this, both 3KR and 4KR mutants exhibit a weak but significant reduction of apoptosis upon γ-IR exposure compared to wild type, while 1KR mutants do not (Figure 4E, P = 0.8232 for 1KR, P = 0.0058 for 3KR and P = 0.0105 for 4KR). In both the 3KR and 4KR mutants, the level of reduction is comparable to that observed for the ztf-8 null mutant (P = 0.9232 for 3KR and P = 0.7169). Taken together, all these observations support an important role for SUMOylation of ZTF-8 and suggest that SUMOylation is crucial for both the DNA damage repair and DNA damage-mediated checkpoint response functions of ZTF-8.
Discussion
SUMOylation regulates the ZTF-8 protein involved in DSBR and DDR pathways in the germline
Several types of post-translational modifications, such as phosphorylation, SUMOylation, and ubiquitylation, have been found to play a role in orchestrating protein interaction with sites of DNA damage (Morris 2010; Ulrich 2012). Since SUMO and ubiquitin can promptly and reversibly change the properties, stability, or localization of the target proteins, they are ideal controllers for fine tuning the DNA repair and DDR pathways (Ulrich 2012). SUMOylation acts in the regulation of various cellular processes such as DNA repair, nuclear transport, transcription, signal transduction, and cell-cycle progression (Johnson 2004; Cremona et al. 2012). However, many of the relevant SUMOylation target proteins along with how this modification regulates their mechanisms of function have yet to be clarified.
Mammalian RHINO was shown to interact with the Rad9–Rad1–Hus1 complex (9-1-1) and the ATR activator TopBP1 (Cotta-Ramusino et al. 2011). Our previous study showed that ZTF-8 is the functional homolog of RHINO. It interacts with the 9-1-1 complex and is required for 9-1-1 localization, both mitotic and meiotic DSBR, and the normal activation of DNA damage-checkpoint-mediated apoptosis (Kim and Colaiacovo 2014). However, the findings that direct SUMOylation of ZTF-8 is required for its functions in DSBR as well as DNA damage checkpoint activation have not been previously reported. Moreover, this study suggests a link between the lack of ZTF-8’s post-translational modification resulting in sterility to the impaired DSBR and DNA damage-mediated checkpoint activities. The absence of ZTF-8’s SUMOylation sites mimics the ztf-8 null mutant phenotypes, reiterating the significance of ZTF-8’s SUMOylation.
ZTF-8 is a direct substrate for SUMOylation at multiple predicted SUMOylation sites
ZTF-8 contains four consensus SUMOylation sites (ΨKXE) and its interaction with SUMOylation components suggests it is a potential substrate for SUMOylation. The presence of the multiple predicted sites suggests that ZTF-8 may undergo multiple SUMOylation events. No single SUMO site mutation of ZTF-8 disrupts the interaction between ZTF-8 and the E2 SUMO ligase UBC-9, whereas no interaction is found between UBC-9 and the 3KR mutant carrying all three C-terminal sites mutated, supporting the idea of multiple active SUMOylation sites (Figure 1D). Consistent with these results, mutant worms containing non-SUMOylatable mutations at either three or all four of the predicted target lysines (3KR, 4KR) mimic ztf-8 null mutant phenotypes such as sterility and defective apoptosis while the 1KR mutant, carrying a single mutated lysine (K14R) does not (Figure 4). In accordance with these observations, our LC-MS analysis discovered UBC-9 in pulldowns with wild-type ZTF-8, but not from the non-SUMOylatable 4KR transgenic line (Table S1).
Of note, although experimental data shows that 77% of known SUMOylation sites conform to the consensus motif (Xue et al. 2006), studies have shown that nonconserved sites are also infrequently sumoylated (Kamitani et al. 1998; Rodriguez et al. 2001), suggesting the possibility of additional SUMOylation sites in ZTF-8. However, this idea is not supported by the lack of remaining higher-molecular-weight SUMOylation bands in the 4KR non-SUMOylatable mutant (Figure 3).
Proper localization of ZTF-8 in the germline requires the SUMOylation pathway but not ZTF-8’s SUMOylation consensus sites
Although mutations of the components of the SUMOylation pathway alter ZTF-8’s localization, mutation of all four of ZTF-8’s predicted SUMOylation sites to non-SUMOylatable residues does not affect its localization, even though ZTF-8 is directly SUMOylated in wild-type worms (Figure 2 and Figure 3). Then, how does the SUMOylation pathway affect the proper localization of the ZTF-8 protein? Clearly, SUMOylation of ZTF-8 is not essential for its localization, but it is possible that localization of ZTF-8 relies on a SUMOylation interacting motif (SIM), which binds to other unknown SUMOylated protein(s) recruited to the sites of DNA damage (Figure 5). In fact, both SUMOylation sites and SIMs have been reported to be required for proper protein localization in other studies (Zhang et al. 2004; Lin et al. 2006; Geiss-Friedlander and Melchior 2007).
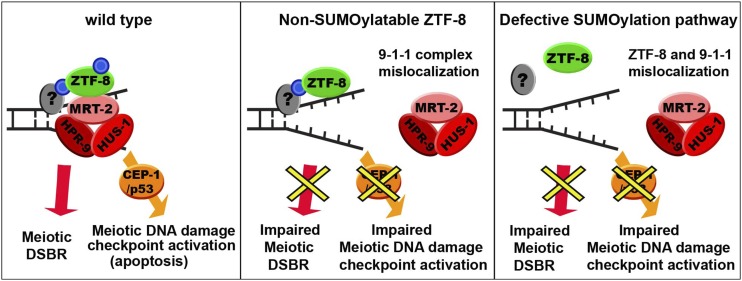
Figure 5.
Model for how SUMOylation of ZTF-8 regulates its germline functions. In wild-type animals, we propose that SUMOylation (blue circle) of ZTF-8 is required for meiotic DSBR as well as for the proper localization of the 9-1-1 complex, which results in DNA damage checkpoint activation (left). An interaction with a yet unknown SUMOylated protein(s) (gray oval with blue circle), perhaps via a SIM (SUMO interaction motif) at ZTF-8, could explain how the non-SUMOylatable ZTF-8 is still localized properly; however, it does lead to inefficient DSBR and DDR activation due to lack of interactions between ZTF-8 and the 9-1-1 complex (middle). A defective SUMOylation pathway may hinder the proper localization of ZTF-8 due to the absence of SUMOylation in another protein (gray oval), and this leads to impaired DSBR and DDR (right).
Although no predicted SIM was detected for ZTF-8 via SUMOylation prediction program analysis (http://sumosp.biocuckoo.org/), the presence of SIM sites is still plausible considering that this prediction algorithm is based only on known SIMs found in 80 proteins and needs to be further developed (Hay 2005). In addition, the presence of an interaction with SMO-1, even in the absence of SUMOylatable sites in LC–MS analysis, further supports this idea (Table S1).
SUMOylation of ZTF-8 promotes its roles in DSBR and DNA damage checkpoint activation
Roles for SUMOylation in regulating DSBR have been previously reported. For example, defects in the SUMOylation pathway cause a RAD51-dependent accumulation of recombination intermediates in response to replication stress (Branzei et al. 2006; Morris et al. 2009). How then does the SUMOylation of ZTF-8 promote its role? In our previous study, we found that ATL-1/ATR is required for the proper localization of ZTF-8, and it is plausible that SUMOylated ZTF-8 then assists in recruiting downstream repair proteins, including the 9-1-1 complex. This idea is further supported by the fact that the 9-1-1 complex is not localized properly in worms expressing non-SUMOylatable ZTF-8 (Figure 4D and Figure 5). Also the 3KR mutant ZTF-8 does not interact with MRT-2/Rad1 in a yeast two-hybrid assay while single point mutants do (Figure S4). Moreover, our pulldown combined with LC–MS analysis identified both MRT-2 and HUS-1 as interactors from worms expressing wild-type ZTF-8, although not from the non-SUMOylatable mutants suggesting that SUMOylation of ZTF-8 is required for localization of the 9-1-1 complex (Table S1). Alternatively, SUMOylated species of ZTF-8 might be recruited downstream of PCNA to bypass stalled replication forks during DNA replication. It is known from yeast studies that Rad18 is involved in the formation of X-shaped intermediates referred to as sister chromatid junctions (SCJs) during replication of damaged templates and that Ubc9 and SUMOylated PCNA are essential for bypassing SCJs (Branzei et al. 2008). The idea of ZTF-8’s role downstream of PCNA is plausible as ZTF-8 is required for processing of stalled replication forks, and it contributes to intersister repair when a homologous chromosome is not available (Kim and Colaiacovo 2014).
In summary, our study has shown that ZTF-8 is SUMOylated and that this modification is important for its functions in the germline. We analyzed SUMOylation of ZTF-8 in wild-type worms to test the functional significance of this protein modification in DSBR and DNA damage checkpoint pathway activation in the germline of C. elegans. We found that ZTF-8 is a direct target for SUMOylation in vivo and that this process is required for DNA damage-checkpoint-induced apoptosis and DSBR (Figure 5). Non-SUMOylatable mutants mimic the phenotypes of ztf-8 null mutants, such as reduced fertility suggesting that SUMOylation is crucial for its roles. Since consensus SUMOylation sites are present in RHINO, and it is expressed in both human testes and ovaries (Kim et al. 2010), this study may shed light on understanding how SUMOylation contributes to germline genomic integrity in humans as well.
Supplementary Material
Acknowledgments
We thank Sara Beese-Sims and Doris Lui for critical reading of this manuscript and members of our laboratory for discussions. This work was supported by National Institutes of Health grants R01GM072551 and R01GM105853 to M.P.C.
Footnotes
Communicating editor: S. E. Bickel
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175661/-/DC1.
Literature Cited
- Becker J., Barysch S. V., Karaca S., Dittner C., Hsiao H. H., et al. , 2013. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20: 525–531. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D., 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356. [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., et al. , 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M., 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C., McDonald E. R., 3rd, Hurov K., Sowa M. E., Harper J. W., et al. , 2011. A DNA damage response screen identifies RHINO, a 9–1-1 and TopBP1 interacting protein required for ATR signaling. Science 332: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A. L., Moser S. C., Bailly A. P., Gartner A., 2012. Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods Cell Biol. 107: 321–352. [DOI] [PubMed] [Google Scholar]
- Cremona C. A., Sarangi P., Zhao X., 2012. Sumoylation and the DNA damage response. Biomolecules 2: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F., 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956. [DOI] [PubMed] [Google Scholar]
- Hay R. T., 2005. SUMO: a history of modification. Mol. Cell 18: 1–12. [DOI] [PubMed] [Google Scholar]
- Hofmann E. R., Milstein S., Boulton S. J., Ye M., Hofmann J. J., et al. , 2002. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12: 1908–1918. [DOI] [PubMed] [Google Scholar]
- Hunt P. A., Hassold T. J., 2008. Human female meiosis: What makes a good egg go bad? Trends Genet. 24: 86–93. [DOI] [PubMed] [Google Scholar]
- Johnson E. S., 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73: 355–382. [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Denison C., Bening-Abu-Shach U., Chisholm A. D., Gygi S. P., et al. , 2009. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev. Cell 17: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T., Kito K., Nguyen H. P., Wada H., Fukuda-Kamitani T., et al. , 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273: 26675–26682. [DOI] [PubMed] [Google Scholar]
- Kelly K. O., Dernburg A. F., Stanfield G. M., Villeneuve A. M., 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiacovo M. P., 2014. ZTF-8 interacts with the 9–1-1 complex and is required for DNA damage response and double-strand break repair in the C. elegans germline. PLoS Genet. 10: e1004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. W., Fukukawa C., Ueda K., Nishidate T., Katagiri T., et al. , 2010. Involvement of C12orf32 overexpression in breast carcinogenesis. Int. J. Oncol. 37: 861–867. [DOI] [PubMed] [Google Scholar]
- Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., et al. , 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24: 341–354. [DOI] [PubMed] [Google Scholar]
- Morris J. R., 2010. More modifiers move on DNA damage. Cancer Res. 70: 3861–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., et al. , 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890. [DOI] [PubMed] [Google Scholar]
- O’Neil N. J., Rose A. M., 2006. DNA repair (January 13, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.54.1, http://www.wormbook.org. [Google Scholar]
- Rodriguez M. S., Dargemont C., Hay R. T., 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276: 12654–12659. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhuri S., Crum T., Woollard A., Aslam S., Okkema P. G., 2006. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev. Biol. 295: 664–677. [DOI] [PubMed] [Google Scholar]
- Sampson D. A., Wang M., Matunis M. J., 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276: 21664–21669. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L., Eddy S. R., Durbin R., 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420. [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H. D., 2012. Ubiquitin and SUMO in DNA repair at a glance. J. Cell Sci. 125: 249–254. [DOI] [PubMed] [Google Scholar]
- Walhout A. J., Vidal M., 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306. [DOI] [PubMed] [Google Scholar]
- Xue Y., Zhou F., Fu C., Xu Y., Yao X., 2006. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 34: W254–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Smolen G. A., Palmer R., Christoforou A., van den Heuvel S., et al. , 2004. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat. Genet. 36: 507–511. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., et al. , 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42: W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.