Abstract
Alternative splicing is prevalent in plants, but little is known about its regulation in the context of developmental and signaling pathways. We describe here a new factor that influences pre-messengerRNA (mRNA) splicing and is essential for embryonic development in Arabidopsis thaliana. This factor was retrieved in a genetic screen that identified mutants impaired in expression of an alternatively spliced GFP reporter gene. In addition to the known spliceosomal component PRP8, the screen recovered Arabidopsis RTF2 (AtRTF2), a previously uncharacterized, evolutionarily conserved protein containing a replication termination factor 2 (Rtf2) domain. A homozygous null mutation in AtRTF2 is embryo lethal, indicating that AtRTF2 is an essential protein. Quantitative RT-PCR demonstrated that impaired expression of GFP in atrtf2 and prp8 mutants is due to inefficient splicing of the GFP pre-mRNA. A genome-wide analysis using RNA sequencing indicated that 13–16% of total introns are retained to a significant degree in atrtf2 mutants. Considering these results and previous suggestions that Rtf2 represents an ubiquitin-related domain, we discuss the possible role of AtRTF2 in ubiquitin-based regulation of pre-mRNA splicing.
Keywords: alternative splicing, C2HC2 zinc finger, intron retention, Rtf2 domain, ubiquitin ligase
IT is increasingly recognized that cotranscriptional and post-transcriptional gene regulation is comparable to transcriptional regulation in intricacy and importance. Pre-mRNA splicing is a cotranscriptional process and a major determinant of transcript abundance and complexity (Reddy et al. 2013). Constitutive splicing refers to the use of only one set of splice sites to generate a single mature mRNA. By contrast, alternative splicing occurs when variable splice sites are selected, leading to the generation of more than one processed RNA product from a single pre-messenger RNA (mRNA). An individual gene can thus potentially encode multiple proteins, leading to a substantial increase in proteomic diversity (Chen and Manley 2009; Syed et al. 2012; Reddy et al. 2013).
Recent work has established that alternative splicing is common in plants, affecting ∼60% of intron-containing genes (Marquez et al. 2012). Alternative splicing has important roles in plant growth, development, abiotic stress tolerance, circadian rhythms, and pathogen defense (Staiger and Brown 2013). The most common outcome of alternative splicing in plants is intron retention (Marquez et al. 2012; Lan et al. 2013), which occurs when an intron fails to be spliced out of the pre-mRNA. Retained introns frequently contain premature termination codons (PTCs) that can channel the transcript into the nonsense-mediated decay (NMD) pathway. Intron retention provides a means for “transcriptome tuning” (Braunschweig et al. 2014) and contributes to the post-transcriptional regulation of gene expression by reducing levels of inappropriately expressed transcripts (Kalyna et al. 2012; Ge and Porse 2013).
Alternative splicing is subject to elaborate regulation that relies on general and specific trans-acting factors as well as cis-acting sequence elements, epigenetic modifications of DNA and chromatin, and post-translational modifications of splicing proteins (Chen and Manley 2009; Reddy et al. 2013). However, the mechanistic roles of diverse splicing regulators and the means by which internal and external signals are conveyed to the splicing machinery are not yet fully understood (Heyd and Lynch 2011; Reddy et al. 2013). Moreover, given the prevalence of alternative splicing, it is likely that additional proteins contributing to this process remain to be discovered (Chen and Manley 2009).
In this article, we report the finding of a new factor that influences pre-mRNA splicing and is required for embryonic development in Arabidopsis thaliana (Arabidopsis). This factor was identified during a genetic suppressor screen originally intended to detect mutations that suppress the effects of the defective in meristem silencing4-1 (dms4-1) mutation on RNA-directed DNA methylation (RdDM) and plant development (Kanno et al. 2010; Sasaki et al. 2012). During the course of this screen, however, it became apparent that the GFP reporter gene under investigation is subject to alternative splicing. Hence, in addition to authentic suppressor mutations that suppress the effects of the dms4-1 mutation on RdDM and/or development, our screen is capable of identifying mutations in genes encoding proteins required for productive splicing of the GFP pre-mRNA. We describe here the identification of two splicing factors, one known and one novel, from this genetic screen.
Materials and Methods
Plant materials and generation of transgenic plants
The sdr1-1/atrtf2-1 and sdr4-1/prp8-7 mutants were screened from an ethyl methanesulfonate (EMS) mutagenized population of the dms4-1 mutant (Sasaki et al. 2012). Mapping of the sdr1-1/atrtf2-1 and sdr4-1/prp8-7 mutations was carried out on F2 mapping populations using codominant markers as described previously (Kanno et al. 2008). Whole-genome sequencing and data analysis to locate the causal mutations in these mutants were performed according to a prior protocol (Eun et al. 2011). Two T-DNA insertion alleles, atrtf2-2 (SALK_040515) and atrtf2-3 (SALK_081659), were obtained from the Arabidopsis Biological Resource Center (Alonzo et al. 2003).
For complementation tests of the defect in GFP expression in the atrtf2-1 mutant, constructs encoding C-terminal HA-tagged AtRTF2 and HA-tagged AtRTF2ΔN—which lack amino acids 6–73 of an ∼80 amino acid, plant-specific N-terminal extension in the AtRTF2 protein—were cloned under the control of the native AtRTF2 promoter into a modified binary vector based on pCB302 (Xiang et al. 1999) and introduced into the homozygous atrtf2-1 mutant (hypomorphic allele) in a T locus background (WT DMS4, no S locus) using the floral dip method (Clough and Bent 1998). Transformed seedlings (T1) were selected on solid Murashige and Skoog (MS) medium containing 20 mg/liter phosphinothricine (PPT). Complementation of the defect in GFP expression by the HA-tagged constructs in the atrtf2-1 mutant was assessed by Western blotting using a GFP antibody in T2 or T3 progeny of selected lines.
Complementation of developmental defects conditioned by the null atrtf2-2 mutation was tested in two ways. In one approach, homozygous atrtf2-1 plants containing either the AtRTF2-HA or AtRTF2ΔN-HA construct were crossed with plants heterozygous for the atrtf2-2 null allele. The resulting F1 progeny were allowed to self-fertilize, producing F2 progeny. Normal-looking F2 seedlings were genotyped for the T-DNA insertion in the atrtf2-2 mutant. In a second approach, AtRTF2-GFP and AtRTF2ΔN-GFP constructs were introduced into the heterozygous atrtf2-2 mutant using the floral dip method. T1 plants (selected by their resistance to PPT) were allowed to self-fertilize to produce T2 progeny. Normal-looking T2 progeny were genotyped for the T-DNA insertion in the atrtf2-2 mutant. Primers used for genotyping are shown in Supporting Information, Table S1. In both cases, successful complementation by a particular transgene construct was determined by recovery of normal-looking progeny that were homozygous for the atrtf2-2 allele.
Fluorescence microscopy
Fluorescence images of GFP expression in roots of PPT-resistant seedlings expressing the AtRTF2-GFP and AtRTF2ΔN-GFP fusion genes under the control of the native AtRTF2 promoter were made using a Leica TCS LSI-III confocal microscope system.
DNA methylation analysis
DNA methylation analysis of the target enhancer by bisulfite sequencing was carried out as described previously (Kanno et al. 2008; Daxinger et al. 2009; Sasaki et al. 2012, 2014). Genomic DNA was isolated using a Plant Genomic DNA Purification kit (GeneMark, Taiwan). For bisulfite sequencing, 1 μg of genomic DNA was digested with HindIII. Purified DNA was converted using an EpiTect Bisulfite kit (Qiagen). PCR products using primers listed in Table S1 were cloned with pGEM T-Easy Vector system (Promega) and transformed into competent Escherichia coli cells. A total of 16–20 independent clones were sequenced. Exon 15 of the PHAVOLUTA gene was used as a control for complete bisulfite conversion (primers in Table S1) (Daxinger et al. 2009).
Whole-genome bisulfite sequencing
Approximately 3 μg of genomic DNA was sonicated to ∼250 bp before it was ligated to Illumina adaptors, then size selected, denatured, and treated with sodium bisulfite (BS) to reveal their methylation status. The BS-sequencing libraries were sequenced using the Illumina HiSequation 2000 platform to generate up to 100 cycles in paired ends. The reads were aligned to the reference genome (TAIR10) using BS Seeker 2 (Guo et al. 2013). To profile genome-wide DNA methylation, the methylation level for each covered cytosine in the genome is estimated as #C/(#C + #T), where #C represents the number of methylated reads and #T corresponds to the number of unmethylated reads. The methylation level per cytosine serves as an estimate of the percentage of cells containing methylation at this cytosine. The raw reads and the processed dataset for the new methylomes (dms4-1, sdr1-1, and dms4-1 sdr1-1) are publicly available from National Center for Biotechnology Information (NCBI) GEO under accession no. GSE63238. The raw reads and the processed dataset for the wild-type methylome (Sasaki et al. 2014) can be downloaded from NCBI GEO under accession no. GSE47453.
Western blotting
Protein extraction, SDS/PAGE, and Western blotting to detect GFP and tubulin proteins were carried out according to published procedures (Eun et al. 2011; Sasaki et al. 2012).
RT-PCR and Quantitative RT-PCR
Total RNAs were isolated from ∼3-week-old seedlings using a Plant Total RNA Miniprep Purification kit (GeneMark, Taiwan) and treated with RQ1 DNase (Promega) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the Transcriptor First Strand cDNA Synthesis kit (Roche) following the manufacturer’s protocol using an oligonucleotide d(T) primer and 1 μg of total RNA. One microliter of cDNA was used as a template for RT-PCR. The PCR conditions for detecting GFP transcripts were as follows: 94° for 2 min followed by 30 cycles of 94° for 10 s, 58° for 20 s, and 72° for 2 min, and finally 72° for 7 min.
Quantitative RT-PCR was performed using a 7500 Real-Time PCR system (Applied Biosystems) with the program recommended by the manufacturer using 1 μl of cDNA as a PCR template and SYBR Green PCR Master Mix (Applied Biosystems). To validate intron retention events identified from the RNA-sequencing (RNA-seq) data, several endogenous genes were selected on the basis of their P-values (Table S2) for quantitative RT-PCR analysis. Stably expressed At5G60390 was used for normalization (Wang et al. 2014). Three biological replicates were carried out for each sample. Error bars indicate standard error. Primer sets for RT-PCR and quantitative RT-PCR are shown in Table S1.
5′ RACE and cloning of GFP transcripts
To determine the transcription start site for each GFP transcript, cDNA synthesis, 5′ RACE, and cloning the RACE product into the vector were carried out using SMARTer RACE 5′/3′ kit (Clontech) according to the manufacturer’s instructions using 1 μg of total RNA as starting material. The PCR conditions for the first amplification were 35 cycles of 94° for 30 s, 68° for 30 s, and 72° for 3 min, and for the nested PCR were 20 cycles of 94° for 30 s, 68° for 30 s, and 72° for 2 min. At least three clones for each cDNA (long, middle, and short) were sequenced. Gene-specific primers are listed in Table S1.
Library preparation and RNA sequencing
Total RNA was extracted from ∼14-day-old seedlings of the T line, sdr1-1/atrtf2-1 and sdr4-1/prp-8-7 mutants (T background), and from newly germinated seedlings of the heterozygous (normal phenotype) and homozygous (arrested development) atrtf2-2 mutant using a Plant Total RNA Miniprep Purification kit (TR02; GeneMark, Taiwan). The protocol with RNA Lysis Solution B was used.
Libraries for RNA-seq were prepared following Illumina TruSeq stranded mRNA sample preparation kit (RS-122-2103) according to the manufacturer’s protocol. Briefly, 4 μg of total RNA were used for library construction. PolyA RNA was captured by oligodT beads and fragmented after elution from the beads. The first-strand cDNA was synthesized by reverse transcriptase (SuperScrip III, 18080-093, Invitrogen) using dNTP and random primers. The second-strand cDNA was generated using a dUTP mix. The double-stranded cDNA was subjected to the addition of a single “A” base to the 3′ end followed by ligation of the barcoded Truseq adapters. Finally, the products were purified and enriched with 12 cycles of PCR to create the final double-stranded cDNA library. A final size selection was performed by 2% low-range agarose (161-3107, Bio-Rad) gel electrophoresis to yield a library of inserts 250–350 bases in length. The library was extracted from the agarose gel using MinElute PCR purification kit (28004, Qiagen). Final libraries were analyzed using Agilent High Sensitivity DNA analysis chip (5067-4626, Agilent) to estimate the quantity and check the size distribution and were then quantified by qPCR using the KAPA Library Quantification kit (KK4824, KAPA). The prepared library was pooled for paired-end sequencing using IlluminaHiSequation 2500 at YourGene Bioscience Co. (New Taipei City, Taiwan) with 126-bp paired-end sequence reads. Approximately one hundred million reads per sample were sequenced. All RNA-seq FASTQ files are available from the Sequence Read Archive (SRA) at NCBI under the following accession nos.: T line (SRR1652313); atrtf2-1 (SRR1652314); prp8-7 (SRR1652315); atrtf2-2 heterozygous (SRR1652316); and atrtf2-2 homozygous (SRR1652317).
Detection of intron retention events
Reads from RNA sequencing were mapped to the TAIR10 genome using BLAT (Kent 2002) with the default setting. Under a threshold of 95% mapping identity, ∼92% of reads per sample were accepted. RackJ (http://rackj.sourceforge.net/) was then used to compute average depths of all exons and all introns.
Given a control sample and a mutant sample, the preference of an intron retention event was measured using a χ2 test for goodness of fit by comparing the depths of the intron in the two samples taking the depths of neighboring exons in the two samples as the background. Here, depth was defined as total covering read bases divided by intron (or exon) length. In so doing, an intron has been retained in a higher chance than in the other sample would be inferred, and an intron retention event was identified as significantly increased if its P-value based on the χ2 value was ≤0.05 and the ratio intron depth/exon depth in mutant was larger than that in the control. See Table S2 for the comparison tables. Selected examples of intron retention were chosen for validation by quantitative RT-PCR according to the procedure described above. Primers used are shown in Table S1.
Protein immunoprecipitation
Total proteins were extracted from 2-week-old seedlings of T, AtRTF2-GFP, and AtRTFΔN-GFP transgenic plants. The T line serves as a negative control to eliminate nonspecific interactions with GFP. Five grams of seedlings were ground in liquid nitrogen. After grinding, 10 ml of immunoprecipitation (IP) buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 2 mM DTT, 1 mM PMSF, 0.7 μg/ml pepstatin A, 10 μg/ml MG132, and protease inhibitor (Roche)] were added to extract proteins. The solution was centrifuged twice at 9000 × g for 10 min at 4° to remove insoluble cell debris. The supernatant comprised the protein extract that was used for immunoprecipitation.
One hundred microliters of GFP-trap magnetic beads (ChromoTek) were washed three times with 1 ml of IP buffer and then added to the 10 ml protein extract. The whole solution was incubated at 4° overnight with 20 rpm agitation. Following this step, the beads were collected using a magnetic stand and washed three times with 1 ml of IP buffer. The bound proteins were eluted by boiling in 30 µl of 4% SDS for 10 min.
Mass-spectrometry analysis
A linear ion trap mass spectrometer (Q Exactive MS, Dionex nanoUHPLC, Thermo Scientific) combined with an on-line nano-scale high performance liquid chromatography system (nanoACQUITY UPLC, Waters Corp.) was used for protein identification and analysis. The liquid chromatography (LC) system consists of an autosampler and a binary pump, a C18 trap (Symmetry C18, 180 µm × 2.0 cm, 5 µm, Waters Corp.) and a C18 nanoACQUITY UPLC column (BEH130 C18, 75 µm × 10 cm, 1.7 µm, Waters Corp.). The linear gradient applied to separate tryptic peptides was from 5 to 40% of acetonitril in 0.1% (v/v) formic acid. LC was run for 90 min at nanoflow (300 nl/min) rate. The C18 reverse phase column was coupled to a nano-electrospray ionization (nESI) source.
Prior to LC/MS/MS analysis, the affinity purified proteins were digested with trypsin according to the filter-aided sample preparation method (Wiśniewski et al. 2009). Four microliters of the resulting peptide solution was loaded onto the column. The data acquisition was performed with a full MS scan followed by four MS/MS scans. The MS scan range was over the mass to charge (m/z) from 400 to 1800 using the data-dependent data acquisition mode with dynamic exclusion enabled. The MS/MS data were converted to peak list files with extract_msn script (Thermo Fisher Scientific) and then analyzed by a Mascot search program (Matrix Science Inc., Boston) against the Arabidopsis database. Proteins with more than two matching peptides were retained, and the proteins with peptide score over 10 and protein score over 20 were considered as reliable hits. Only the proteins with unique peptides identified were included in further analysis. The results shown in Table S3 are derived from three independent biological replicates.
Results
DMS4 suppressor screen
We previously used a two-component transgene silencing system—target plus silencer (T+S)—to identify factors required for RdDM and transcriptional gene silencing (TGS) of a GFP reporter gene under the control of an upstream enhancer active in meristem regions (Figure 1). Forward genetic screens using this system retrieved thirteen defective in meristem silencing (dms) mutants that are deficient in Pol V-associated components of the RdDM pathway (Eun et al. 2012; Matzke and Mosher 2014). One mutant, dms4, which is disabled in a putative IWR1 (interacts with RNA polymerase II) transcription factor, is unique in displaying defects not only in RdDM/TGS but also in plant development (Kanno et al. 2010).
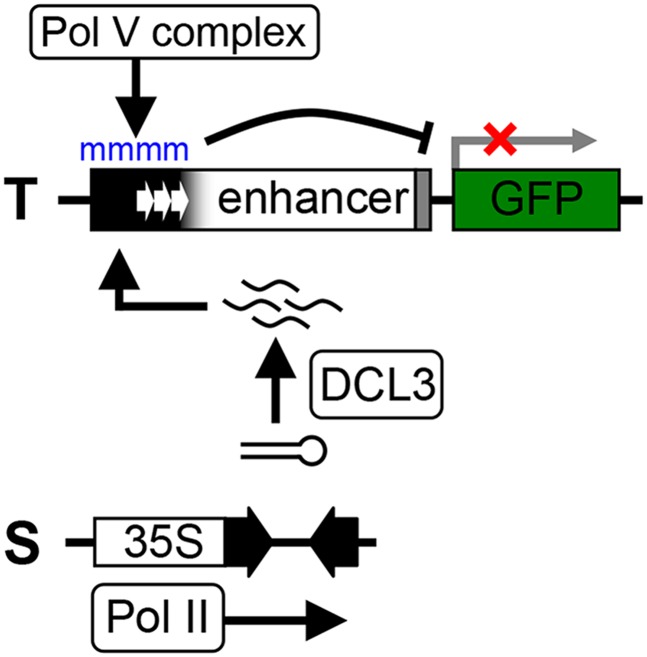
Figure 1.
T+S transgene silencing system to study RdDM. The two-component transgene silencing system consists of a target locus, T, which contains a GFP reporter gene downstream of a minimal promoter (narrow gray bar) and an upstream virus-derived enhancer (containing a short tandem repeat) that drives GFP expression in shoot and root meristem regions. The silencer locus, S, contains an inverted DNA repeat of distal enhancer sequences (black arrowheads corresponding to thick black bar in T) that is transcribed by RNA polymerase II (Pol II) from a constitutive viral promoter (35S). The resulting hairpin RNA is processed by DICER-LIKE3 (DCL3) to produce 24-nt small interfering RNAs that induce Pol V complex-mediated de novo DNA methylation (blue “m”) of the target enhancer region, leading to transcriptional silencing of GFP expression (Kanno et al. 2008, 2010; Sasaki et al. 2014). Figure is not drawn to scale.
To dissect the roles of DMS4 in RdDM and development, we initiated a genetic suppressor screen using a mutant harboring the dms4-1 allele (Sasaki et al. 2012). One type of suppressor mutation anticipated in this screen restores RdDM/TGS but not normal development (sdr, suppressor of dms4, RdDM) (Figure S1). In addition to bona fide sdr mutants displaying the expected phenotypes (restoration of GFP silencing and enhancer methylation but not normal development), we identified two unusual mutants, sdr1-1 and sdr4-1. Although these two mutants were GFP negative and resembled phenotypically the dms4-1 mutant (Figure 2, A and B), the apparent silencing of the GFP reporter gene occurred without increases in DNA methylation at the target enhancer (Figure 2C). Moreover, subsequent genetic crosses revealed that the sdr1-1 and sdr4-1 mutations were able to impair GFP expression in a DMS4 wild-type background lacking the S locus (Figure 3A) and without an increase in DNA methylation (Figure 3B, sdr1-1 shown only). Thus, the sdr1-1 and sdr4-1 mutations diminish expression of the GFP reporter gene at the T locus in a manner that is independent of DNA methylation, the dms4-1 mutation, and the S locus that triggers RdDM.
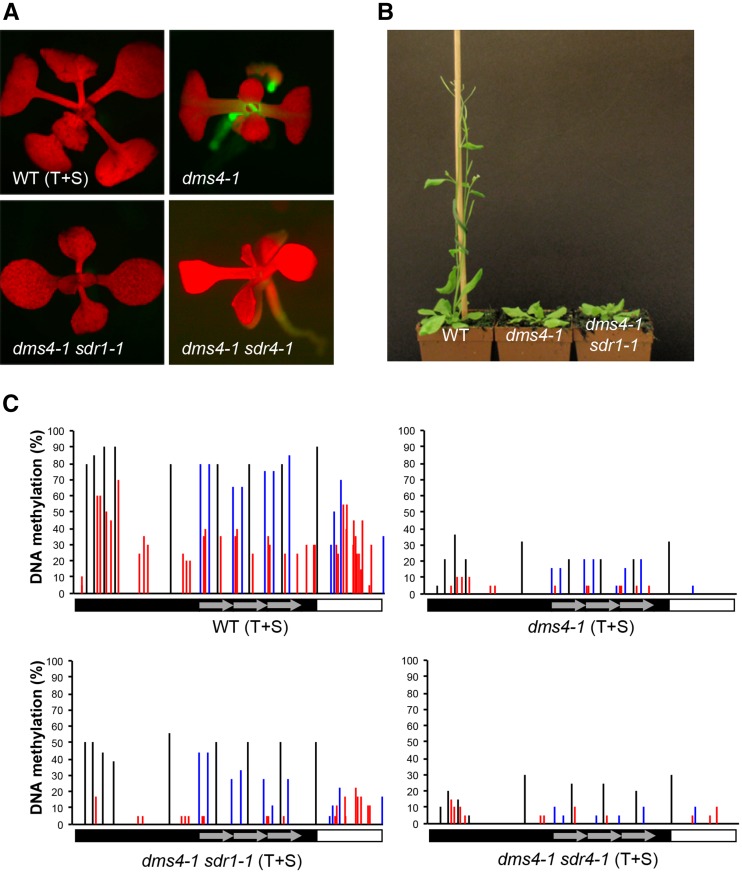
Figure 2.
Phenotypes of sdr1 and sdr4 mutants. (A) GFP expression is silenced in wild-type (WT) T+S seedlings, whereas silencing is released in the dms4-1 mutant, which is GFP positive. GFP silencing appears to be reestablished in the dms4-1 sdr1-1 and dms4-1 sdr4-1 double mutants, which are GFP negative (all mutations in T+S background). (B) The dms4-1 sdr1-1 double mutant displays delayed growth and other phenotypic features of the single dms4-1 mutant compared to the age-matched WT control. The dms4-1 sdr4-1 double mutant also appears similar to the dms4-1 single mutant (not shown). (C) Bisulfite sequencing of the target enhancer demonstrates heavy cytosine methylation in all sequence contexts (black, CG; blue CHG; red, CHH; H is A, T, or C) in WT T+S plants, which are GFP negative (A). Release of GFP silencing in the dms4-1 mutant is associated with substantial loss of methylation. The double mutants, dms4-1 sdr1-1 and dms4-1 sdr4-1, appear GFP negative (A) but this is not accompanied by restoration of the WT DNA methylation level in the target enhancer.
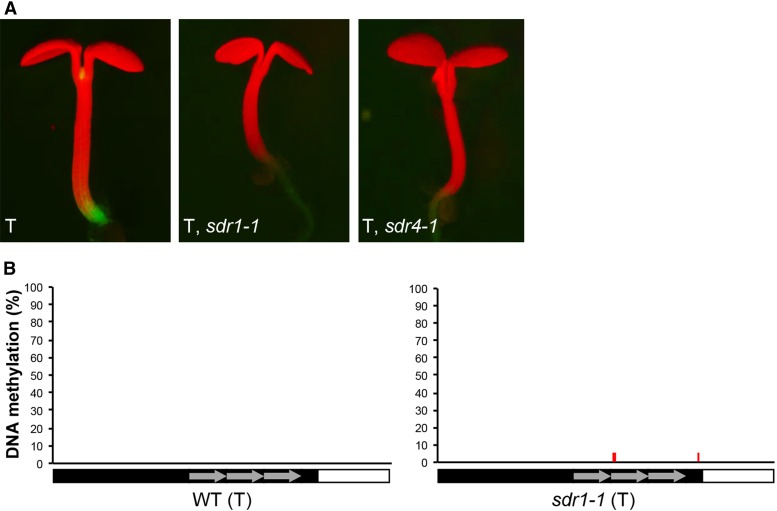
Figure 3.
Impaired GFP expression in sdr1 and sdr4 mutants in the T line. (A) The wild-type T line expresses GFP in the shoot and root meristem regions. GFP expression is impaired in sdr1-1 and sdr4-1 seedlings (T background only, WT DMS4, S locus absent). (B) Bisulfite sequencing of the target enhancer demonstrated that the impairment of GFP expression in the sdr1-1 single mutant is not accompanied by any significant DNA methylation at the target enhancer (right). The unmethylated enhancer in SDR1 wild-type plants containing the T locus is shown as a control (left). The sdr1-1 mutation also has no effect on DNA methylation genome-wide as indicated by a methylome analysis (Figure S2).
SDR1 is an Rtf2 domain-containing protein
Using classical mapping with codominant markers, we mapped the sdr1-1 mutation to the bottom arm of chromosome 5. Subsequent Illumina whole genome sequencing revealed a G-to-A nucleotide change (chr5: 23,485,838) in the gene At5g58020. This gene encodes a 354 amino acid protein that contains an Rtf2 (Replication termination factor 2) domain (Pfam PF04641), which is defined by a C2HC2 motif related to the C3HC4 RING-finger motif (Inagawa et al. 2009) (Figure 4A). Rtf2 was discovered in fission yeast, where it is needed to stabilize a paused DNA replication fork to establish imprinting at the mating type locus (Inagawa et al. 2009). Although Rtf2 proteins are found in eukaryotes ranging from fission yeast to humans (Figure 4B), the Rtf2 orthologs in plants have an N-terminal extension of ∼80 amino acids (Figure 4C). In contrast to the high amino acid sequence similarity in the Rtf2 domain among different plant species, the N-terminal extension is only weakly conserved at the amino acid sequence level (Figure 4C). The sdr1-1 mutation results in substitution of glycine for glutamic acid at position 85 (G85E), which is located at the beginning of the conserved Rtf2 domain (Figure 4, A and C). We will refer to the SDR1 protein hereafter as AtRTF2 and the sdr1-1 mutation as atrtf2-1.
Figure 4.
SDR1 is an evolutionarily conserved Rtf2 domain-containing protein. (A) SDR1 (At5g58020), which is 354 amino acids in length, contains an Rtf2 domain (amino acids 84–338) and hence renamed here AtRTF2. AtRTF2 is a single copy gene in Arabidopsis. The position of the G85E amino acid substitution resulting from the sdr1-1/atrtf2-1 point mutation identified in our screen and the sites of two T-DNA insertion alleles (atrtf2-2 and atrtf2-3) are indicated. (B) Phylogenetic tree of Rtf2 orthologs in different organisms. With the exception of budding yeast, Rtf2 orthologs are present in other eukaryotes examined. An unrooted phylogenetic tree was generated by the neighbor-joining method using ClustalW and visualized with TreeView. (C) Amino acid sequence alignments of AtRTF2 orthologs in plants show high similarity in the Rtf2 domain but only partial conservation in the plant-specific N-terminal extension, which is ∼80 amino acids in length. The red arrowhead indicates the location of the sdr1-1/atrtf2-1 G85E mutation at the beginning of the Rtf2 motif. The multiple sequence alignment by ClustalW was performed using GenomeNet (http://www.genome.jp/tools/clustalw/) and consensus amino acid residues were highlighted using BoxShade (http://www.ch.embnet.org/software/BOX_form.html).
To complement the atrtf2-1 mutation and test a functional requirement for the N-terminal extension, we generated constructs encoding HA-tagged versions of full-length AtRTF2 (AtRTF2-HA) and a truncated form lacking most of the N-terminal region (AtRTF2ΔN-HA) under the control of the endogenous AtRTF2 promoter (Figure 5A). These constructs were introduced into the atrtf2-1 mutant containing the T locus. As assessed by GFP protein accumulation using Western blotting, AtRTF2-HA but not AtRTF2ΔN-HA complemented the atrtf2-1 mutation and restored wild-type levels of GFP expression (Figure 5B). These results demonstrate that full-length AtRTF2 including the N-terminal segment is required for correct GFP expression.
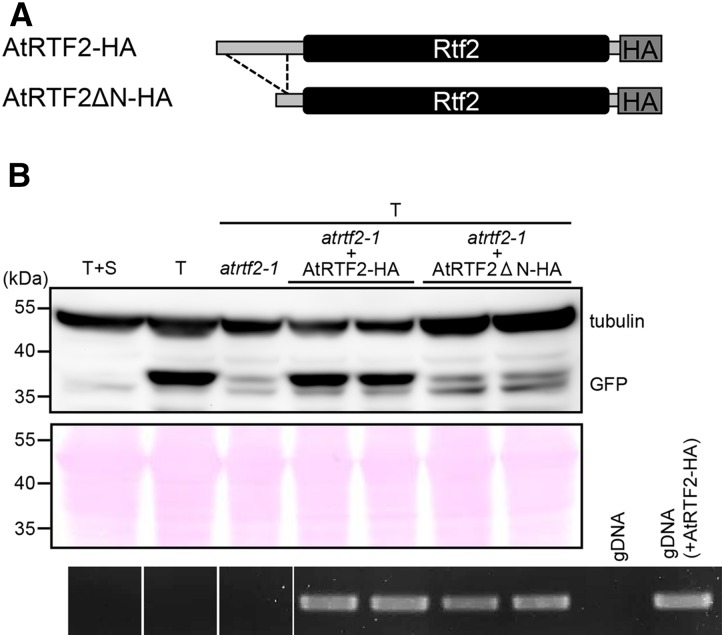
Figure 5.
Full-length AtRTF2 is required for proper GFP expression. (A) Schematic drawing of C-terminal HA-tagged constructs encoding full-length AtRTF2 (ATRTF2-HA) or a truncated version lacking amino acids 7–63 of the N-terminal extension (ATRTF2ΔN7–63-HA). These constructs were introduced into the homozygous atrtf2-1 mutant. (B) Western blots probed with a GFP antibody revealed little accumulation in the T+S line, in which GFP expression is silenced (Figure 2A) but wild-type accumulation in the T line. GFP accumulation is low in the atrtf2-1 mutant but returns to the wild-type level when the mutant is complemented with the full-length AtRTF2-HA construct (two examples shown). Levels of GFP protein remain low when using the AtRTF2ΔN-HA construct in the complementation test (two examples shown). RT-PCR (bottom) confirmed that the HA-tagged transgenes are transcribed. The blot was probed with an antibody to tubulin as a control for protein stability and loading levels. The stained membrane is also shown as a loading control. gDNA, genomic DNA (shown for plants without and with the AtRTF2-HA transgene).
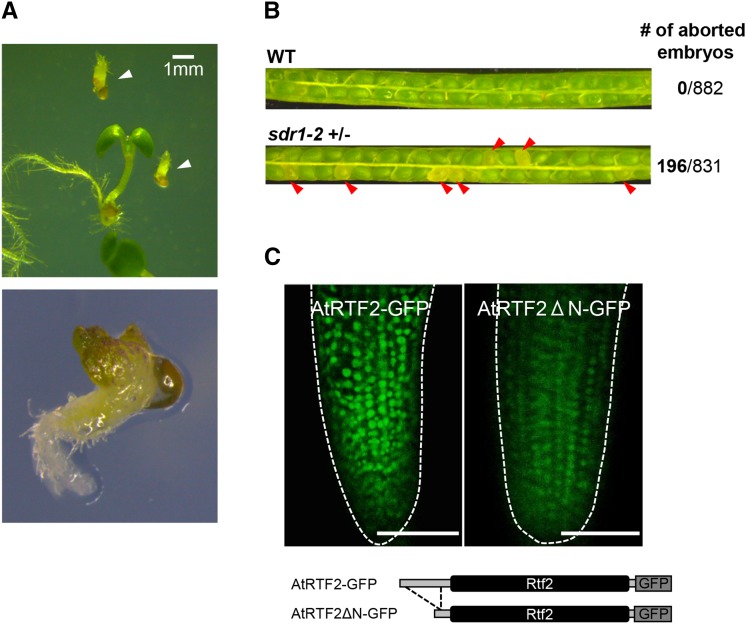
Two transfer-DNA (T-DNA) insertion alleles, atrtf2-2 and atrtf2-3, were obtained from a public stock center. Both strains harbor T-DNA insertions within the Rtf2 motif (Figure 4A). When homozygous, the putative null atrtf2-2 allele conditions defects in embryogenesis and lethality shortly after germination (Figure 6, A and B), confirming that AtRTF2 is an essential gene (Savage et al. 2013). By contrast, the other two alleles, atrtf2-1 and atrtf2-3, are hypomorphic and display only mild developmental phenotypes. The developmental defect in atrtf2-2 was complemented by transgene constructs encoding full-length AtRTF2 but not the truncated version lacking most of the N-terminal extension (Table 1). These results demonstrate that full-length AtRTF2 is essential for both normal development and for proper expression of the GFP reporter gene.
Figure 6.
AtRTF2 encodes an essential nuclear protein. (A) Self-fertilized plants heterozygous for the null atrtf2-2 mutation (Figure 4A) produce ∼25% defective seedlings (arrowheads, top and enlargement, bottom), which are homozygous for the atrtf2-2 mutation. These seedlings die shortly after germination. The developmental defect can be complemented by full-length AtRTF2 transgenes but not by truncated AtRTF2ΔN versions lacking most of the plant-specific N-terminal extension (Table 1). (B) A seed pigment defect is visible in siliques of heterozygous atrtf2-2 mutant plants (Savage et al. 2013). In our experiment, ∼10–14 days after flowering, ∼25% of the seeds (196/831 counted) appeared white (red arrowheads), whereas 100% of the seeds in an age-matched wild-type control (882/882 counted) were green. (C) AtRTF2-GFP (left) and AtRTF2ΔN-GFP (right) fusion proteins (constructs below) are located predominately in nuclei (shown here in root tip cells).
Table 1. Full-length AtRTF2 complements developmental defect in homozygous atrft2-2 mutants.
| ATRTF2/ATRTF2 | ATRTF2/atrtf2-2 | atrtf2-2/atrtf2-2 | Total | Result of χ2 test | ||
|---|---|---|---|---|---|---|
| F2 | atrtf2-2 × atrtf2-1 + AtRTF2-HA | 8 | 26 | 13 | 47 | 0.450286001 |
| F2 | atrtf2-2 × atrtf2-1 + AtRTF2ΔN-HA | 19 | 28 | 0 | 47 | 0.000194992 |
| T2 | atrtf2-2 + AtRTF2-GFP | 12 | 22 | 11 | 45 | 0.9672161 |
| T2 | atrtf2-2 + AtRTF2 ΔN-GFP | 16 | 29 | 0 | 45 | 0.000313828 |
In one complementation test, homozygous atrft2-1 plants containing either the AtRTF2-HA or AtRTF2ΔN-HA construct were crossed with plants heterozygous for the atrft2-2 null allele. The resulting F1 progeny were allowed to self-fertilize, producing F2 progeny. Normal-looking F2 seedlings were genotyped for the T-DNA insertion in atrtf2-2. Approximately 25% of the normal-looking F2 seedlings recovered from the AtRTF2-HA lines were homozygous for the atrtf2-2 mutation, indicating successful complementation of the developmental defect by the full-length AtRTF2-HA construct. By contrast, no homozygous atrtf2-2 plants were recovered from the AtRTF2ΔN-HA lines, indicating unsuccessful complementation with the truncated construct. In a second complementation test, AtRTF2-GFP and AtRTF2ΔN-GFP constructs were introduced into the heterozygous atrtf2-2 mutant using the floral dip method. T1 plants (selected by their resistance to PPT) were allowed to self-fertilize to produce T2 progeny. Normal-looking T2 progeny were genotyped for the T-DNA insertion in atrtf2-2. Consistent with the result described above, normal-looking progeny that were homozygous for the atrft2-2 mutation were only obtained with the construct encoding the full-length AtRTF2-GFP fusion protein. Results of χ2 tests carried out to determine if the segregation ratio differs from the expected 1:2:1 ratio are shown.
Although AtRTF2 has been considered a plastid-targeted protein (Savage et al. 2013) we found that an AtRTF2-GFP fusion protein under the control of the native promoter accumulates predominantly in the nucleus (Figure 6C). Expression of AtRTF2 is ubiquitous and particularly strong in developing embryos during seed maturation (Arabidopsis eFP Browser, Winter et al. 2007).
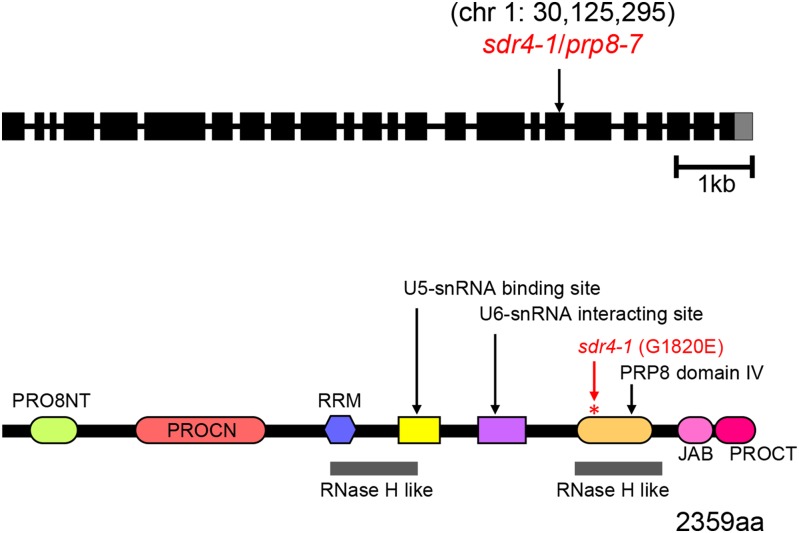
SDR4 is the core spliceosomal protein PRP8
Using classical mapping with codominant markers, we mapped the sdr4-1 mutation to a genetic interval on the bottom arm of chromosome 1. Subsequent Illumina whole genome sequencing region revealed a G-to-A mutation (chr1: 30,125,295) in the gene At1g80070. This gene encodes the core splicing factor PRP8 (pre-mRNA processing 8), which is one of the largest and most highly conserved proteins of the spliceosome (Grainger and Beggs 2005). We will refer hereafter to SDR4 as PRP8 and the sdr4-1 mutation as prp8-7 (Figure 7). The mutation we recovered results in the substitution of a highly conserved glycine residue at position 1820 to glutamic acid (G1820E), which is in the RNase H domain of the PRP8 protein (Figure 7). Similarly to AtRTF2, PRP8 is expressed ubiquitously and shows particularly strong expression during seed maturation (Arabidopsis eFP Browser, Winter et al. 2007). Homozygous null prp8 mutations result in an abnormal suspensor and embryo lethality (Schwartz et al. 1994). Plants homozygous for the prp8-7 mutation grow and reproduce normally, although they are somewhat late flowering, indicating that the prp8-7 allele is hypomorphic.
Figure 7.
SDR4 is PRP8. Intron–exon structure of the PRP8 gene (At1g80070) (top) and domain structure of PRP8 (bottom), a core spliceosomal protein of 2359 amino acids. The sdr4-1/prp8-7 mutation (G to A at position 30,125,295 on chromosome 1) creates a G1820E amino acid substitution in the RNase H-like domain. A second point mutation in this region, prp8-6 (G1891E), was reported recently (Marquardt et al. 2014). PRP8 domains were identified in Pfam (http://pfam.xfam.org/).
AtRTF2 and PRP8 are required for splicing of GFP pre-mRNA
AtRTF2 was identified in the same screen as PRP8, a known splicing factor, and GFP expression is impaired in a DNA methylation-independent manner in both atrtf2-1 and prp8-7 mutants. Moreover, both genes have similar expression patterns and null mutations are embryo lethal. These observations suggest that AtRTF2 and PRP8 act in the same pathway, prompting us to test for defects in GFP pre-mRNA splicing in the atrtf2-1 and prp8-7 mutants.
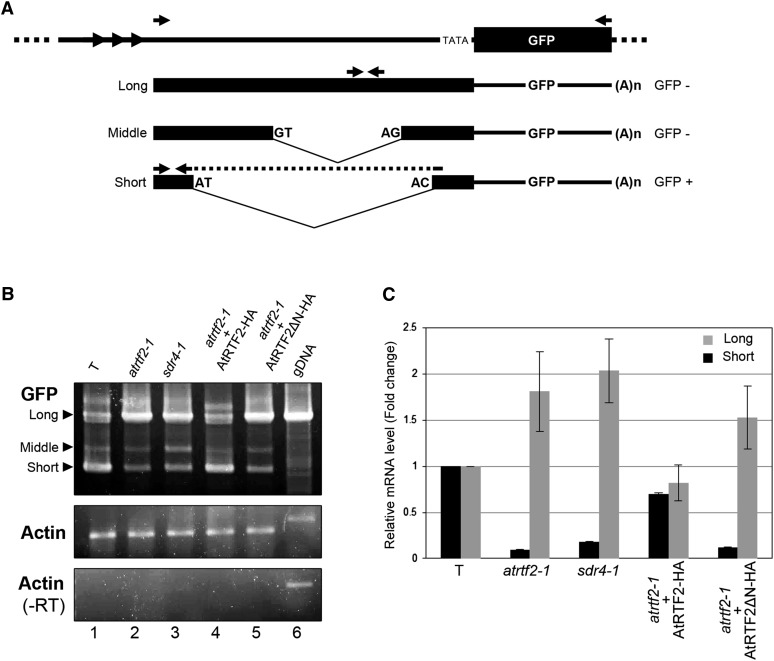
We had originally predicted from the structure of the T construct that GFP transcription would initiate in the minimal promoter upstream of the GFP coding sequence (Figure 1). In practice, however, this minimal promoter does not appear to be used and we detected instead three major GFP transcripts that are likely to result from alternative splicing of a pre-mRNA initiating upstream in the distal enhancer region (Kanno et al. 2008) (Figure 8A; Figure S3). The “short” transcript, which results from productive splicing of a cryptic intron with noncanonical donor and acceptor sites (AT-AC), can be translated into GFP protein using the first methionine codon after the transcription start site (Figure S4). Mutational analysis confirmed that the short transcript is indeed the major GFP mRNA (Figure S5). By contrast, the “long” unspliced transcript and the ‘middle” transcript, which results from unproductive splicing of a conventional GT-AG intron (Figure 8A; Figure S3), contain a number of PTCs after the initiating methionine and cannot be translated into a functional GFP protein (Figure S4).
Figure 8.
Alternative splicing of GFP pre-mRNA. (A) As shown by cDNA cloning and sequencing, the T locus encodes three polyadenylated GFP transcripts that result from alternative splicing: a long unspliced transcript; a middle transcript that results from splicing a canonical GT-AG intron; and a short transcript that results from splicing a cryptic intron with noncanonical AT-AC splice sites. Although AT-AC termini are a feature of U12 introns removed by the minor spliceosome, this intron does not contain the highly conserved U12 intron 5′ splice site or branch point sequence and therefore is not a U12 intron (Figure S3). The long and middle transcripts are not translatable into GFP protein (GFP−) owing to the presence of numerous PTCs after the initiating methionine (Figure S4). The short transcript can be translated into GFP protein (GFP+) (Figure S4) and mutational analysis indicates that it is indeed the major transcript encoding GFP protein (Figure S5). A 5′-RACE experiment demonstrated that transcription initiates in the distal enhancer region ∼45 bp downstream of the short tandem repeat in the target enhancer (arrowheads) (Figure S3). “TATA” indicates an apparently unused minimal promoter directly upstream of the GFP coding region (Figure 1). Arrows indicate the positions of the primers used for the amplification of the three GFP transcripts (B), and the individual long and short transcripts (C). We did not analyze in detail the middle transcript because it is the least abundant and accumulates inconsistently. (B) Semiquantitative RT-PCR showing accumulation of long and short GFP transcripts in the indicated genotypes. Actin is shown as a constitutively expressed control. −RT, no reverse transcriptase. (C) Quantitative RT-PCR showing accumulation of long and short GFP transcripts in the indicated genotypes. Stably expressed At5g60390 was used for normalization (Wang et al. 2014).
Consistent with a role for AtRTF2 and PRP8 in splicing the GFP pre-mRNA, the ratio of the short translatable and long untranslatable transcripts varied in wild-type and mutant plants. In GFP-positive plants (T line and atrtf2-1 mutant complemented with the wild-type AtRTF2-HA sequence; Figure 8B, lanes 1 and 4), the short translatable transcript was prominent. By contrast, in GFP-negative plants (atrtf2-1 and prp8-7 mutants, atrtf2 mutant complemented with the truncated AtRTF2ΔN-HA construct; Figure 8B, lanes 2, 3, and 5, respectively), the long untranslatable transcript was the predominant form. AtRTF2 and PRP8 are thus required to splice the AT-AC intron to generate a translatable GFP mRNA. Quantitative RT-PCR confirmed the different ratios of short and long transcripts in wild-type plants and the atrtf2-1 mutant (Figure 8C).
Genome-wide requirement for AtRTF2 in splicing
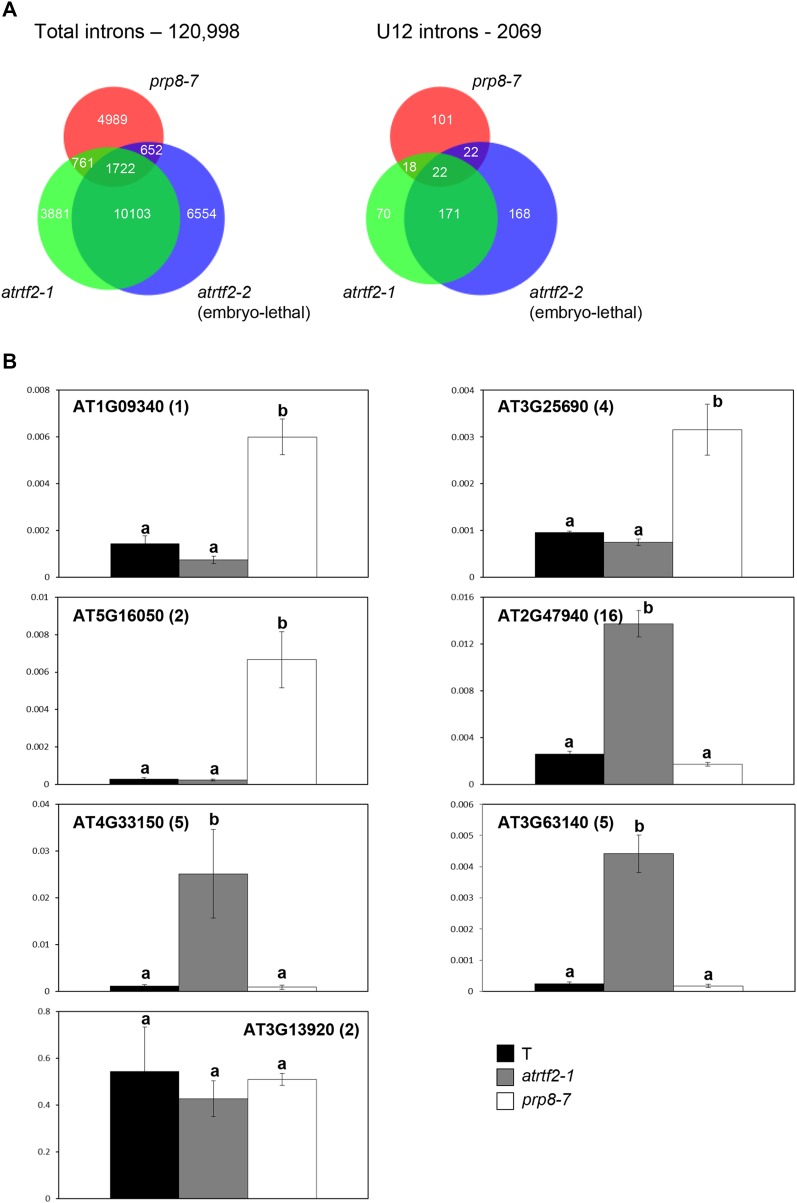
To determine whether atrtf2 mutations affect splicing of introns genome-wide, we performed RNA-seq on total RNA extracted from seedlings of the T line and homozygous atrtf2-1 and prp8-7 mutants, as well as newly germinated embryos that were either heterozygous (normal) or homozygous (arrested; Figure 6A) for the atrtf2-2 null mutation. Because the main splicing defect observed for the GFP reporter gene in the atrtf2-1 and prp8-7 mutants was intron retention (IR), which is also the most common outcome of alternative splicing in plants (Marquez et al. 2012), we focused our analysis of the RNA-seq data on IR events. In atrtf2-1, atrtf2-2, and prp8-7 mutants, 13.6, 15.7, and 6.7% of total introns, respectively, displayed a significant degree of increased intron retention (Figure 9A). Both major U2 and minor U12 introns were affected to a comparable degree. A 62.1% overlap in IR events was observed between atrtf2-1 and atrtf2-2 using the total number of IRs in atrtf2-2 as a denominator, and ∼38.6% overlap in IRs was observed between atrtf2 mutants and prp8-7, using the total number of IRs in prp8-7 as a denominator (Figure 9A; Table S2).
Figure 9.
Intron retention in atrtf2 and prp8 mutants. (A) Venn diagrams indicate significantly increased IR events in homozygous atrtf2-1 (hypomorphic allele), atrtf2-2 (null allele), and prp8-7 (hyomorphic allele) mutants. Total introns (U2 and U12) are shown at left; U12 introns only are shown at right. The full list is provided in Table S2. (B) Validation of IR events by quantitative RT-PCR. Genes were selected from Table S2 based on P-values. Types of IR events observed are exemplified by At3g63140, At2g47940, and At4g33150 (IR in atrtf2-1 only) and At1g09340, At5g16050, and At3g25690 (IR in prp8-7 only). At3g13920 is shown as a control gene that shows no statistically significant IR changes in the wild-type T line and the mutants. The y-axis indicates the relative IR level normalized to stably expressed At5g60390 (Wang et al. 2014). The primers were designed so that one was inside the target intron and the other was in an adjacent exon. The numbers in parentheses after each gene ID indicate the target intron number for validation as counted from the genomic 5′ end. The error bars indicate standard error of the mean (SEM) of three independent biological replicates. Letters above each bar indicate statistical significance tested by Tukey’s honestly significant difference test (P < 0.05). The same letter (a) indicates no statistically significant difference between the two samples. A different letter (b) indiates a statistical difference between the two samples.
Following the definition of IR fold change in a previous publication (Lan et al. 2013), the distributions of fold changes of significantly increased IR events follow the power-law distribution, which shows linear distributions in the log-log scale (Table S2, sheet 5), where the maximum fold changes were 3291.5, 10127.3, and 2718.1 for atrtf2-1, atrtf2-2, and prp8-7, respectively. The median fold changes of the three samples were all above five, which means that more than half of significantly increased IR events showed fold changes greater than five in each mutant. Additionally, >98% of significantly increased IR events represent introns fully covered by reads of the sample, suggesting that almost all increased IR events retain full introns. Our method identifies significantly increased and reduced IR events equally. However, in the mutants, increased IRs predominate among significant IR events (16,467 vs. 919 for atrtf2-1; 19,031 vs. 874 for atrtf2-2; and 8124 vs. 1875 for prp8-7), indicating that the mutations in AtRTF2 and PRP8 are associated with reduced splicing efficiency. Table S2, sheet 6 gives a summary table of significantly increased IR events. Several cases of IR events affecting endogenous genes in the atrtf2-1 and prp8-7 mutants were validated using qRT-PCR (Figure 9B).
AtRTF2-interacting proteins
To initiate studies on proteins that may associate with AtRTF2 in a complex, we carried out affinity purification of the AtRTF2-GFP fusion protein followed by mass spectrometry. This analysis revealed a number of predicted and known splicing factors, including PRP8, as well as other proteins not directly related to splicing (Table S3).
Discussion
Our study has identified a new factor, AtRTF2, which influences pre-mRNA splicing and is essential for embryo development in Arabidopsis. A splicing-related role was initially suggested by the identification of the hypomorphic atrtf2-1 mutation in the same genetic screen as the hypomorphic prp8-7 mutation, which impairs the activity of the core spliceosomal constituent PRP8. Further work showed that the atrtf2-1 and prp8-7 mutations have identical effects on the splicing pattern of GFP pre-mRNA: productive splicing of a cryptic intron with noncanonical AT-AC termini is less efficient in the two mutants, leading to lower levels of a translatable GFP mRNA and increased accumulation of an unspliced, untranslatable GFP transcript. In addition, mutations in atrtf2 disrupt splicing genome-wide, leading to a significant degree of increased retention for ∼13–16% of total introns. Mass spectrometry-based profiling suggests that AtRTF2 potentially associates with several predicted and known splicing factors, including PRP8, although these remain to be substantiated using additional approaches. Collectively, the findings implicate AtRTF2, which was functionally uncharacterized prior to our study, in pre-mRNA splicing.
In contrast to PRP8, which acts directly in the splicing reaction by providing a scaffold for spliceosome assembly as well as amino acids for catalysis (Chen and Moore 2014), AtRTF2 may have a more transient regulatory role during the spliceosome cycle. One possibility is that AtRTF2 contributes to ubiquitin-based modulation of spliceosomal proteins. The Rtf2 domain consists of a C2HC2 zinc finger that is related to the C3HC4 RING-finger motif but folds in a way to create only one functional Zn+2 ion binding site. The founding member of the Rtf2 protein family was discovered in fission yeast, where it is important for stabilizing a paused DNA replication fork during imprinting at the mating type locus, possibly by facilitating sumoylation of PCNA (Inagawa et al. 2009; Komander and Rape 2012).
With regard to pre-mRNA splicing, the Rtf2 domain has been described as an ubiquitin-related domain in a structural bioinformatics analysis of splicing factors (Korneta et al. 2012). Another example noted in the study of Korneta et al. (2012) is NOSIP (nitric oxide synthase interacting protein) (CG6179), which contains an Rtf2 domain and is a component of the Drosophila melanogaster spliceosome (Herold et al. 2009). The Rtf2 domain has been annotated in association with RING E3 ubiquitin ligases (Choy et al. 2013) and suggested to act as an ubiquitin ligase in the context of splicing (Korneta et al. 2012). A divergent cyclophilin that may be involved in splicing also contains an Rtf2 domain (Page and Winter 1998). In addition to other reversible post-translational modifications, such as acetylation, methylation, and phosphorylation, ubiquitination is increasingly recognized for its role in regulating the spliceosomal cycle (Bellare et al. 2008; Song et al. 2010; Mishra et al. 2011; Korneta et al. 2012; Ammon et al. 2014; Chen and Moore 2014; Oka et al. 2014). Notably, PRP8, which can bind ubiquitin through its conserved JAMM (JAB1/MPN/Mov34 metalloenzyme) domain, was detected as an ubiquitin conjugate in affinity-purified particles in budding yeast, suggesting a means to reversibly modulate the activity of this protein (Bellare et al. 2008). A recent advanced proteomics analysis identified PRP8 as a target of ubiquitination in Arabidopsis (Kim et al. 2013). Further work is needed to determine whether AtRTF2 has ubiquitin ligase activity, and if so, whether PRP8 and other splicing factors are among its substrates.
The embryonic lethality of null atrtf2-2 mutations is consistent with disruptions in splicing of key transcripts important for early stages of development. Further work is required to understand in more detail the nature of the splicing defects and their impact on development. Even the hypomorphic atrtf2-1 mutation decreases the efficiency of intron removal in more mature plants, demonstrating that AtRTF2 is required continuously during plant growth to maintain optimal splicing activity.
In our system, the AtRTF2-dependent, productive splicing event excises a cryptic intron with noncanonical AT-AC sites in the GFP pre-mRNA. Although AT-AC termini are a feature of U12 introns removed by the minor spliceosome, the AT-AC intron in the GFP pre-mRNA lacks additional highly conserved U12 intron sequences at the 5′ splice site and branch point (Burge et al. 1998; Lin et al. 2010; Turunen et al. 2013). Therefore, this intron is likely to be processed primarily by the major U2 spliceosome (Wu and Krainer 1997). AtRTF2 is not specialized for a particular class of intron because our genome-wide analysis of intron retention found that atrtf2 mutations affect splicing of both U2 and U12 introns to a similar extent. Although the PTC-containing long and middle GFP transcripts are potentially targets of NMD, they nevertheless accumulate to detectable levels. This observation is consistent with previous work indicating that many intron-containing transcripts are retained in the nucleus and hence not degraded by the NMD machinery, which is located in the cytoplasm (Kalyna et al. 2012).
PRP8 was identified in a previous screen for factors affecting splicing of the COOLAIR antisense transcript involved in epigenetic regulation of the FLOWERING LOCUS C (FLC) gene in Arabidopsis (Marquardt et al. 2014). Like the prp8-7 mutation we recovered (G1802E), the mutation identified by Marquardt et al. (2014), prp8-6 (G1891E), is also present in the RNaseH domain of PRP8. The physiological significance of finding two distinct hypomorphic mutations in the RNase H domain of PRP8 in independent screens is not yet clear, but these mutations should prove useful for further dissecting PRP8 function in the plant spliceosome.
Although our GFP reporter gene system illuminates a role for AtRTF2 in pre-mRNA splicing, the function of this protein may not be limited to this process. Additional roles are suggested by the identification of proteins not known to be involved in splicing in the affinity purification-mass spectrometry analysis. Moreover, the Rtf2 protein in fission yeast is involved in an activity unrelated to pre-mRNA splicing (Inagawa et al. 2009). The plant-specific N-terminal extension is essential for AtRTF2 function for reasons that are not yet known. This extension may interact with certain factors or may be modified in a way that is important for the regulation of AtRTF2 activity or stability. Given the evolutionary conservation of the RTF2 protein and the presence of the Rtf2 domain in several splicing proteins (Korneta et al. 2012), it will be interesting to determine the degree to which AtRTF2 orthologs and other Rtf2 domain-containing proteins are involved in the regulation of pre-mRNA splicing in different organisms.
The identification of AtRTF2 in our screen demonstrates the usefulness of the alternatively spliced GFP reporter gene for uncovering novel proteins involved in pre-mRNA splicing. To retrieve additional components acting in the AtRTF2 pathway, we recently initiated a new forward genetic screen based on the T line and recovered a number of putative mutants in which the wild-type GFP gene is silenced or only weakly expressed. The identification of the causal mutations conditioning weak GFP expression in these mutants has the potential to unveil more new splicing factors and provide mechanistic insights into the regulation of splicing efficiency and alternative splicing in plants.
Supplementary Material
Acknowledgments
We thank David Meinke for helpful discussions, the Proteomics Core Lab of the Institute of Plant and Microbial Biology (IPMB) at Academia Sinica (AS), for mass spectrometry analysis, the DNA Microarray Core Laboratory (IPMB, AS) for library preparation for RNA sequencing, and the sequencing services provided by the National Center for Genome Medicine of the National Core Facility Program for Biotechnology, Ministry of Science and Technology, Taiwan. We are grateful to AS and the Taiwan Ministry of Science and Technology (Project no. MOST 103-2311-B-001-004-MY3) for financial support. T.S. was supported by the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad.
Footnotes
Communicating editor: C. S. Pikaard
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.176438/-/DC1.
Literature Cited
- Alonzo J. M., Stepanova A. N., Leisse T. J., C. J. Kim, H. Chen et al, 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Ammon T., Mishra S. K., Kowalska K., Popowicz G. M., Holak T. A., et al. , 2014. The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells. J. Mol. Cell Biol. 6: 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P., Small E. C., Huang X., Wohlschlegel J. A., Staley J. P., et al. , 2008. A role for ubiquitin in the spliceosome assembly pathway. Nat. Struct. Mol. Biol. 15: 444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U., Barbosa-Morais N. L., Pan Q., Nachman E. N., Alipanahi B., et al. , 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24: 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C. B., Padgett R. A., Sharp P. A., 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2: 773–785. [DOI] [PubMed] [Google Scholar]
- Chen M., Manley J. L., 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Moore M. J., 2014. The spliceosome: disorder and dynamics defined. Curr. Opin. Struct. Biol. 24: 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy A., Severo M. S., Sun R., Girke T., Gillespie J. J., et al. , 2013. Decoding the ubiquitin-mediated pathway of arthropod disease vectors. PLoS ONE 8: e78077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Daxinger L., Kanno T., Bucher E., van der Winden J., Naumann U., et al. , 2009. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 28: 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun C., Lorkovic Z. J., Naumann U., Long Q., Havecker E. R., et al. , 2011. AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS ONE 6: e25730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun C., Lorkovic Z. J., Sasaki T., Naumann U., Matzke A. J., et al. , 2012. Use of forward genetic screens to identify genes required for RNA-directed DNA methylation in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 77: 195–204. [DOI] [PubMed] [Google Scholar]
- Ge Y., Porse B. T., 2013. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays 36: 236–243. [DOI] [PubMed] [Google Scholar]
- Grainger R. J., Beggs J. D., 2005. Prp8 protein: at the heart of the spliceosome. RNA 1: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Fiziev P., Yan W., Cokus S., Sun X., et al. , 2013. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N., Will C. L., Wolf E., Kastner B., Urlaub H., et al. , 2009. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29: 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyd F., Lynch K. W., 2011. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem. Sci. 36: 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagawa T., Yamada-Inagawa T., Eydmann T., Mian I. S., Wang T. S., et al. , 2009. Schizosaccharomyces pombe Rtf2 mediates site-specific replication termination by inhibiting replication restart. Proc. Natl. Acad. Sci. USA 106: 7927–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., Simpson C. G., Syed N. H., Lewandowska D., Marquez Y., et al. , 2012. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Bucher E., Daxinger L., Huettel B., Böhmdorfer G., et al. , 2008. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 40: 670–675. [DOI] [PubMed] [Google Scholar]
- Kanno T., Bucher E., Daxinger L., Huettel B., Kreil D. P. et al, 2010. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 11: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Scalf M., Smith L. M., Vierstra R. D., 2013. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M., 2012. The ubiquitin code. Annu. Rev. Biochem. 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Korneta I., Magnus M., Bujnicki J. M., 2012. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 40: 7046–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W., Lin W. D., Santi S., Schmidt W., 2013. Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. F., Mount S. M., Jarmołowski A., Makałowski W., 2010. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol. Biol. 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F., Sun Q., et al. , 2014. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W., Simpson C., Barta A., Kalyna M., 2012. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M., Mosher R. A., 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Mishra S. K., Ammon T., Popowicz G. M., Krajewski M., Nagel R. J., et al. , 2011. Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature 47: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Varmark H., Vitting-Seerup K., Beli P., Waage J. et al, 2014. UBL5 is essential for pre-mRNA splicing and sister chromatid cohesion in human cells. EMBO Rep. 15: 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. P., Winter A. D., 1998. A divergent multi-domain cyclophilin is highly conserved between parasitic and free-living nematode species and is important in larval muscle development. Mol. Biochem. Parasitol. 95: 215–227. [DOI] [PubMed] [Google Scholar]
- Reddy A. S., Marquez Y., Kalyna M., Barta A., 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T., U. Naumann, P. Forai, A. J. Matzke, and M. Matzke, 2012 Unusual case of apparent hypermutation in Arabidopsis thaliana. Genetics 192: 1271–1280. [DOI] [PMC free article] [PubMed]
- Sasaki T., Lee T. F., Liao W. W., Naumann U., Liao J. L., et al. , 2014. Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana. Plant J. 79: 127–138. [DOI] [PubMed] [Google Scholar]
- Savage L. J., Imre K. M., Hall D. A., Last R. L., 2013. Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS ONE 8: e73291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B. W., Leung E. C., Meinke D. W., 1994. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245. [DOI] [PubMed] [Google Scholar]
- Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., et al. , 2010. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24: 1434–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J. W., 2013. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed N. H., Kalyna M., Marquez Y., Barta A., Brown J. W., 2012. Alternative splicing in plants: coming of age. Trends Plant Sci. 17: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen J. J., Niemelä E. H., Verma B., Frilander M. J., 2013. The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 4: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang J., Jiang J., Chen S., Guan Z., et al. , 2014. Reference genes for normalizing transcription in diploid and tetraploid Arabidopsis. Sci. Rep. 4: 6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., et al. , 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J. R., Zougman A., Nagaraj N., Mann M., 2009. Universal sample preparation method for proteome analysis. Nat. Methods 6: 359–362. [DOI] [PubMed] [Google Scholar]
- Wu Q., Krainer A. R., 1997. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA 3: 586–601. [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Han P., Lutziger I., Wang K., Oliver D. J., 1999. A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.