Abstract
The evolutionarily conserved Dock proteins function as unconventional guanine nucleotide exchange factors (GEFs). Upon binding to engulfment and cell motility (ELMO) proteins, Dock–ELMO complexes activate the Rho family of small GTPases to mediate a diverse array of biological processes, including cell motility, apoptotic cell clearance, and axon guidance. Overlapping expression patterns and functional redundancy among the 11 vertebrate Dock family members, which are subdivided into four families (Dock A, B, C, and D), complicate genetic analysis. In both vertebrate and invertebrate systems, the actin dynamics regulator, Rac, is the target GTPase of the Dock-A subfamily. However, it remains unclear whether Rac or Rap1 are the in vivo downstream GTPases of the Dock-B subfamily. Drosophila melanogaster is an excellent genetic model organism for understanding Dock protein function as its genome encodes one ortholog per subfamily: Myoblast city (Mbc; Dock A) and Sponge (Spg; Dock B). Here we show that the roles of Spg and Mbc are not redundant in the Drosophila somatic muscle or the dorsal vessel. Moreover, we confirm the in vivo role of Mbc upstream of Rac and provide evidence that Spg functions in concert with Rap1, possibly to regulate aspects of cell adhesion. Together these data show that Mbc and Spg can have different downstream GTPase targets. Our findings predict that the ability to regulate downstream GTPases is dependent on cellular context and allows for the fine-tuning of actin cytoskeletal or cell adhesion events in biological processes that undergo cell morphogenesis.
Keywords: Drosophila, Dock proteins, GTPase, musculature, dorsal vessel
THE Rho GTPases are enzymes that bind and hydrolyze GTP, allowing for physical interactions with downstream proteins to activate pathways involved in cell morphogenesis, including cell migration, cell adhesion, and phagocytosis (Gadea and Blangy 2014; Laurin and Côté 2014). Normal development and tissue homeostasis require proper regulation of the GTP hydrolysis cycle to tightly control cytoskeletal cell shape changes and cell–cell adhesion events (Cherfils and Zeghouf 2013). Inappropriate control of cell morphogenesis can manifest in abnormal cellular behaviors. For example, cancer cells may detach from their original location, undergo cytoskeletal rearrangement, and alter membrane adhesion dynamics to migrate through complex extracellular environments in tumor metastasis. Many of the same molecules are essential for cell morphogenic events in both normal and abnormal cells (Steeg 2006; Friedl and Gilmour 2009). Thus, determining the normal function of proteins that regulate GTP activity may also reveal how abnormal misregulation of cellular events results in genetic birth defects or disease progression in different biological contexts.
The Rho GTPases are key regulators in cell morphogenesis, cycling between an “off” and an “on” state (Tetlow and Tamanoi 2013; Cook et al. 2014; Goicoechea et al. 2014). GTPase-activating proteins (GAPs) promote GTP hydrolysis and inactivate the GTPase. In contrast, guanine nucleotide exchange factors (GEFs) assist in the exchange of GDP for GTP to activate GTPases and allow for binding to effector proteins. This GDP exchange of Rho GTPases is facilitated by either the Dbl or Dock family of GEFs. Proteins of the Dbl family contain conserved tandem Dbl homology (DH) and Pleckstrin homology (PH) sequences (Gadea and Blangy 2014; Laurin and Côté 2014). Functionally, the DH domain catalyzes GEF activity, while the PH domain allows for interactions with other proteins to control subcellular localization (Rossman et al. 2005; Cook et al. 2014). The “atypical” Dock GEFs lack the canonical DH domain, but utilize an internal Dock homology region 2 (DHR2) for GTPase-binding and exchange activity. Dock GEFs also contain a separate SH3 domain that interacts with the adaptor protein engulfment and cell motility (ELMO) for the regulation of Dock protein localization and GTPase activation (Gadea and Blangy 2014; Laurin and Côté 2014). This bipartite Dock–ELMO complex is required for optimal in vivo activation of Rac (Komander et al. 2008; Laurin and Côté 2014). The current model suggests that ELMO and Dock exist in an autoinhibitory state in the cytoplasm and, upon stimulation by external cues, this inhibition is relieved for ELMO–Dock complex membrane recruitment and interaction with downstream GTPase targets (Laurin and Côté 2014).
There are 11 Dock proteins in mammals that are subdivided into four categories: Dock A–D. To date, Dock-A, -B, and -C family members can all activate the Rho GTPase Rac, while Dock-C and -D proteins also exhibit specificity for Cdc42 (Gadea and Blangy 2014; Laurin and Côté 2014). For example, the Dock-A family member Dock1/Dock180 acts through Rac to mediate vertebrate neuronal pathfinding and endothelial cell migration and functions in concert with a second Dock-A member, Dock5, to control myoblast fusion in vertebrate muscle development (Tachibana et al. 1998; Moore et al. 2007; Laurin et al. 2008; Li et al. 2008; Sanematsu et al. 2010). Expanding the repertoire of GTPase targets, the Dock-B subgroup member Dock4 has also been shown to activate the Ras-like small GTPase Rap1 (Yajnik et al. 2003; Pannekoek et al. 2009; Eguchi et al. 2013).
Less is known about the developmental roles of the mammalian Dock-B family. Two of the subfamily members, Dock3 (also called modifier of cell adhesion, or MOCA) and Dock4, are expressed in nervous system tissue, and Dock4 expression is also detected in smooth muscle cells (Biersmith et al. 2011; Kang et al. 2012; Ueda et al. 2013). Both Dock3 and Dock4 are implicated in actin reorganization through the activation of Rac in neurite outgrowth and dendritic spine morphology, respectively (Chen et al. 2005; Hiramoto et al. 2006; Ueda et al. 2013). Notably, Dock3−/− mice exhibit neuronal degeneration (Chen et al. 2009). The association of Dock3 or Dock4 in neurological disorders, including Alzheimer’s disease, schizophrenia, and autism spectrum disorders, suggests a broader role in neuroprotection (Pagnamenta et al. 2010; Shi 2013; Ueda et al. 2013; Gadea and Blangy 2014; Namekata et al. 2014). An additional role for Dock protein was demonstrated in tumorigenesis. A representational difference analysis screen using mice-derived tumors identified a single Dock4 point mutation in two different cancer cell lines (Yajnik et al. 2003). This same study showed that Dock4-mediated Rap activation was required for cells to maintain their cell–cell adhesion junctions. Clearly, these studies show the importance of Dock proteins in disease progression. Overlapping expression patterns and functional redundancy in vertebrate models complicates interpretation of the biological roles of Dock proteins. Fortunately, the less complex fly model provides an excellent system to dissect the cellular roles of this protein family.
Redundancy is simplified in flies with only one Dock homolog per subfamily. In Drosophila, the Dock-A counterpart, Myoblast city (Mbc), is required for Rac-mediated processes, such as myoblast fusion and border cell migration, which both require modulation of the actin cytoskeleton (Erickson et al. 1997; Duchek et al. 2001). Mbc also functions redundantly with the Dock-B homolog, Sponge (Spg), in border cell migration (Bianco et al. 2007). Spg has an independent role in the early blastoderm development where it is required for actin cap formation (Postner et al. 1992). However, it is unclear if these two GEFs function redundantly in other developmental processes in Drosophila where spg and mbc exhibit either overlapping or exclusive messenger RNA (mRNA) expression patterns. For example, the mbc transcript is enriched in the somatic muscle and mbc mutants show myoblast fusion defects (Erickson et al. 1997; Balagopalan et al. 2006; Geisbrecht et al. 2008). spg mRNA is not detectable in the developing musculature, and thus far no muscle phenotypes have been observed (Biersmith et al. 2011). In contrast, spg, but not mbc, transcripts are abundant in the developing CNS. However, mutations in either gene result in axon guidance or outgrowth phenotypes. While both spg and mbc are essential for CNS development, spg exhibits a genetic interaction with the cell adhesion molecule N-cadherin, while mbc does not (Biersmith et al. 2011). These data, taken together, suggest that Dock family proteins may exhibit differential roles in development. One prediction of these different roles may be activation of different downstream GTPases. This is supported by a recent report where Spg is required for Rap-mediated photoreceptor differentiation in the Drosophila eye (Eguchi et al. 2013).
Here, we use the genetically tractable model organism Drosophila melanogaster to determine if Mbc and Spg function redundantly in tissues other than border cell migration and to establish if these Dock proteins target the same or different GTPases in the dorsal vessel (dv), a tissue where both transcripts are expressed (Biersmith et al. 2011). Using genetic interaction analyses, RNA interference (RNAi) knockdown, and rescue experiments with the GAL4/UAS system, we have established that Mbc and Spg have differential functions in the development of the somatic muscle and dv. In addition, we show that the downstream GTPases of these GEFs are different in dv development. This is one of the first in vivo examples of these two related proteins having distinct targets in development.
Materials and Methods
Genetics
Fly stocks were raised on standard cornmeal medium at 25° unless otherwise indicated. Oregon R was used as the wild-type strain. The following alleles/fly stocks were used: UAS-mbc (Balagopalan et al. 2006); UAS-elmo (Geisbrecht et al. 2008); UAS-spg (Biersmith et al. 2011); UAS-spgIR13 (Eguchi et al. 2013); UAS-spgRNAi353 (Harvard TRiP Project, BL35396); spg242 (Biersmith et al. 2011); Rap1B3 (Hariharan et al. 1991); UAS-Rap1N17 (Boettner et al. 2003); UAS-Rap1V12 (Boettner et al. 2003); elmoKO (Bianco et al. 2007); and Rap1CD5 (generated by Tim Sliter and described in Asha et al. 1999). The following stocks were obtained from the Bloomington Stock Center: mbcD11.2 (BL4952); mef2-GAL4 (BL27390); UAS-trio (BL9134); C155-GAL4 (BL458); 24B-GAL4 (BL1767); twi-GAL4 (BL914); Rac1 and Rac2 (BL6677); and UAS-Rac1V12 (BL6291). The following stocks were generated by standard meiotic recombination and verified by complementation and/or PCR: UAS-spg, mbcD11.2 (for rescue); UAS-Rap1V12, UAS-spgIR13 (for rescue); 24B-GAL4, spg242; spgRNAi353; spg242, and UAS-Rap1V12; mbcD11.2 (for rescue). All rescue experiments were performed at 29° with the exception of twi-GAL4, UAS-mbcD11.2::UAS-RacV12, mbcD11.2, which was performed at 18°.
Immunostaining and statistics
Embryos were collected on agar–apple juice plates and aged at 25°. For antibody stainings, embryos were fixed and stained as described (Geisbrecht et al. 2008). Mutant embryos, including double mutants, were identified by a lack of balancer-inserted lac-Z immunostaining with anti-β−gal (1:100, Developmental Studies Hybridoma Bank; 1:10,000, Cappel). The non-lacZ embryos, or mutant embryos, were selected for further immunostaining. The musculature was visualized using anti-MHC (1:500). Secondary antibody was goat anti-mouse-HRP (1:200, Jackson). The CNS was labeled using mAb 1D4 (1:100, Developmental Studies Hybridoma Bank, University of Iowa). The dv was labeled using anti-Mef2 (kindly provided by Susan Abmayr). Fluorescent immunostaining was performed as previously described (Geisbrecht and Montell 2004) and detected using Alexa Fluor 488 or 546 at 1:400 (Molecular Probes, Carlsbad, CA). Fluorescent images were collected on Olympus Fluoview 300, Zeiss LSM 710, or Nikon Eclipse 90i, and figures were assembled using Photoshop. All raw data, phenotype quantification, and statistical analysis were performed using Graphpad Prism. P-values were determined using either Mann–Whitney or Kruskal–Wallis analyses as indicated in each figure legend.
Molecular biology
The PxxP region of Spg was determined by primary sequence alignment with Mbc, Dock180, Dock3, and Dock4 using Multalign. The following primers were designed after secondary structure prediction analysis to reduce the possibility of interfering with protein structure: forward—5′-GCCATTCCCCGGGGAGCTCCCATTC-3′ and reverse—5′-ATAGTTTAGCGGCCGCTCAGGTA-3′. The spgΔPxxP complementary DNA (cDNA) sequence was generated by amplifying the correct portion of the spg cDNA from the full-length clone (Biersmith et al. 2011) and put into the pUAST vector. Transgenic flies were produced by Genetic Services, Inc., using standard techniques.
Electron microscopy and live imaging
Embryos were prepared for electron microscopy as described (Soplop et al. 2009) and sent to the St. Louis University Microscopy Core for sectioning, low-magnification light micrographs, and high-magnification electron micrographs.
Western blotting
Fifteen embryos of the appropriate genotype were hand-selected and transferred to 6× Laemmli buffer. The protein samples were then separated by 6% SDS–PAGE, transferred to polyvinyl difluoride membranes (Pierce Biotechnology, Inc.), and probed with either guinea pig anti-Spg (1:500) (Biersmith et al. 2011) or anti-tubulin (1:100000, B-512, Sigma), followed by incubation with HRP-conjugated secondary antibodies (1:5000, GE Healthcare) and detection using the ECL Plus Western Blotting detection system (Pierce).
Results
Mbc and Spg do not function redundantly in somatic muscle development
This article addresses the in vivo roles of the Dock family members Spg and Mbc in embryogenesis. Drosophila Spg was identified as a maternal-effect mutant and is required for the formation of actin caps and metaphase furrows in the syncytial blastoderm embryos (Postner et al. 1992). We previously showed that Spg is also expressed in developing neural tissue and essential for axon outgrowth and midline crossing (Biersmith et al. 2011). In contrast, Mbc is well characterized for its role in myoblast fusion (Erickson et al. 1997; Balagopalan et al. 2006; Haralalka et al. 2011). At stage 13 in embryogenesis, specialized muscle cells termed “founder cells” are present at sites where somatic muscles will eventually form. Fusion-competent myoblasts migrate to these founder cells and undergo repeated rounds of myoblast fusion events to form multinucleated muscle fibers (Figure 1, A and B). Consistent with published literature (Erickson et al. 1997; Balagopalan et al. 2006; Haralalka et al. 2011), mutations in mbc resulted in myoblasts that were capable of migrating to the founder cells, but failed to undergo fusion (Figure 1C, arrowhead).
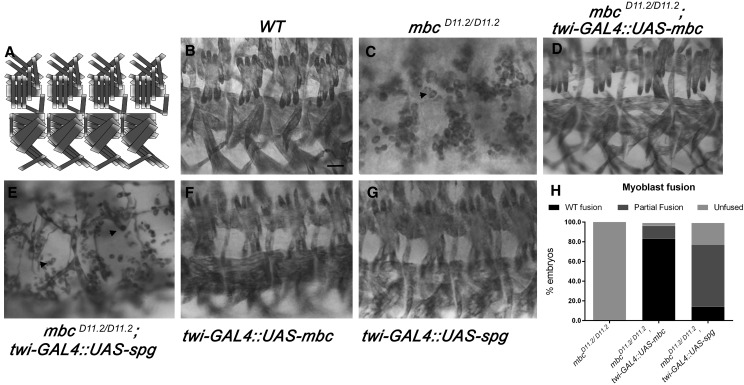
Figure 1.
Spg does not fully compensate for Mbc in myoblast fusion. (A) Schematic representation of four hemisegments of the somatic musculature in a Drosophila stage 16 embryo. (B–G) Late stage 16 embryos stained with α-MHC to visualize the final pattern of the body-wall muscles. (B) The somatic muscles in wild-type embryos are arranged in repeating, organized segments. (C) Zygotic removal of mbc results in severe myoblast fusion defects (arrowhead). (D) The reintroduction of mbc cDNA into an mbc mutant background in the mesoderm (twi-GAL4 driver) rescues the muscle fusion defects. (E) However, expression of spg by twi-GAL4 in mbc mutants does not rescue the myoblast fusion defects to the same extent as mbc. (F and G) Expression of mbc (F) or spg (G) alone does not cause myoblast fusion defects. (H) Graph showing the extent of myoblast fusion rescue upon the addition of mbc or spg. Anterior is to the left and dorsal is up for all images shown. Bar, 20 μm.
Our earlier studies showed that removal of spg in an mbc−/− mutant background does not alter the ability of myoblasts to migrate to or fuse with founder cells (Biersmith et al. 2011), despite evidence that Dock proteins can be essential in cell migration (Gadea and Blangy 2014). It is possible that the maternal contribution of spg transcript and protein (Rice and Garen 1975; Biersmith et al. 2011) masks the role of functional Spg in our double-mutant genetic analysis. We do not favor this idea; while mbc transcript is expressed in the developing somatic musculature (Erickson et al. 1997), we have never detected spg mRNA or protein in this tissue (Biersmith et al. 2011). This suggests that Spg is not present in the developing musculature to contribute to the migration or fusion of myoblasts.
To test if Dock proteins exhibit functional redundancy using an alternative strategy, we tested whether overexpression of spg could compensate for the mbc−/− myoblast fusion phenotype. In an mbc−/− mutant background, expression of UAS-mbc under control of the mesoderm-specific twist (twi)-GAL4 driver fully rescued the muscle pattern to wild type in >80% of the embryos analyzed (Figure 1, D and H). Ectopic expression of full-length spg (UAS-spg) did not rescue mbc-induced myoblast fusion defects to the same extent as expression of mbc (Figure 1, E and H). The majority of these embryos showed partial fusion, in which a small number of fully formed myotubes were present with mostly unfused myoblasts (Figure 1E). Of note, expression of UAS-mbc (Figure 2F) or UAS-spg (Figure 2G) alone did not cause myoblast fusion defects. Overall, we conclude that Mbc and Spg do not have equivalent roles in somatic muscle development.
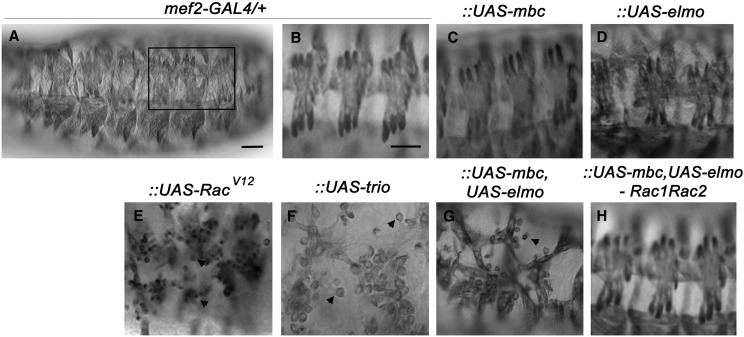
Figure 2.
The Mbc–Elmo complex functions upstream of Rac1 in myoblast fusion. (A–H) Late-stage Drosophila embryos stained with α-MHC to visualize the somatic musculature. The muscle-specific GAL4 driver, mef2, is used to drive the expression of the indicated UAS constructs. (A and B) Whole-mount view of the entire embryo (A) or a higher magnification photograph of three hemisegments (B) show the normal repeating segments of organized muscles in embryos heterozygous for the mef2-GAL4 driver. (C and D) Expression of mbc (C) or elmo (D) alone does not cause somatic muscle defects. (E and F) Expression of RacV12 (E) or ectopic induction of the Rac GEF trio (F) impairs myoblast fusion. (G and H) Co-expression of UAS-mbc and UAS-elmo (G) induces muscle fusion defects, which are suppressed upon removal of one copy of Rac1 and Rac2 (H). Anterior is to the left and dorsal is up for all embryos. Bar, 20 μm.
The Mbc–Elmo complex functions upstream of Rac1 in myoblast fusion
As mentioned in the Introduction, Mbc/Dock180 acts upstream of Rac to mediate actin cytoskeletal events. Previous experiments to rescue mbc-mediated myoblast fusion defects with the constitutively active form of Rac (Rac1V12) have been difficult to interpret due to the drastic phenotypes of activated Rac on its own (Figure 2E) and possibly additional functions of Mbc that cannot solely be rescued by a single downstream molecule (Haralalka et al. 2011). Thus, we chose a different genetic approach to examine if the Mbc–Elmo complex acts upstream of Rac in myoblast fusion, taking advantage of the GAL4/UAS system (Brand and Perrimon 1993) to drive gene expression in the muscle under control of the mef2 promoter. The mef2-GAL4 insertion alone (Figure 2, A and B), overexpression of mbc (Figure 2C), or overexpression of elmo (Figure 2D) did not induce myoblast fusion defects. However, simultaneous overexpression of mbc and elmo triggered myoblast fusion defects (Figure 2G) similar to, but less severe than, ectopic expression of the neuronal DH-containing Rac1 GEF trio (Figure 2F). The fusion defects resulting from overexpression of this Mbc–Elmo GEF complex was suppressed upon removal of a single copy of each of the downstream GTPases Rac1 and Rac2 (Figure 2H), both of which are required for myoblast fusion (Hakeda-Suzuki et al. 2002) and do not exhibit fusion defects upon loss of one copy of each gene. These data demonstrate that the Mbc–Elmo complex functions upstream of the Rac GTPase to regulate myoblast fusion in the developing Drosophila embryo. Combined with our results from Figure 1, Spg is unlikely to play a major role in muscle-specific Rac activation. Thus, we sought to examine Dock protein function in a tissue where both Mbc and Spg are expressed.
spg−/− mutants exhibit multilayered cardioblast clusters
Both mbc and spg mRNA are expressed in the developing heart tube (Biersmith et al. 2011), or dv, making this tissue a good model to further assay the cellular roles of Dock proteins. Formation of the Drosophila dv begins around stage 13 of embryonic development after heart-cell specification has delineated two rows of 52 precursor cells, or cardioblasts, on the dorsal side of the embryo (Figure 3, A and B). These rows of cardioblasts migrate toward the midline as dorsal closure proceeds. By stage 17, the cardioblast cells become paired up at the midline with a distinct posterior heart and anterior aorta region (Tao and Schulz 2007; Singh and Irvine 2012) (Figure 3, A′ and B′). This relatively simple in vivo migration and pairwise alignment of heart cells provides a simple assay for the analysis of genes important in both early (stage 13) and late (stage 17) stages of dv morphogenesis.
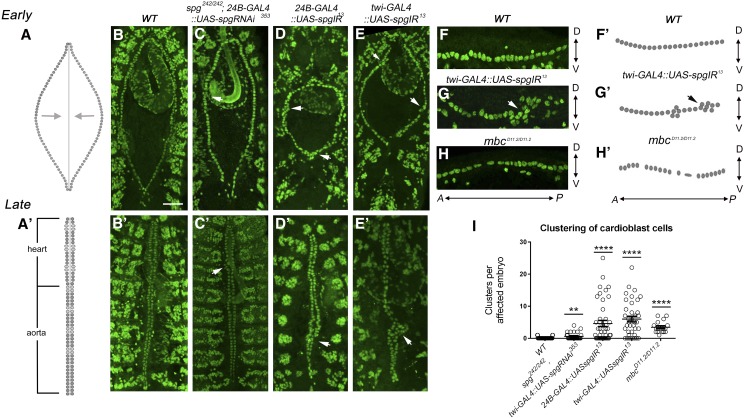
Figure 3.
Spg is required for dv patterning. (A and A′) Schematic illustration of heart tube development (dorsal view). At the beginning of dv formation (stage 13), two rows of cardioblasts begin to migrate toward the dorsal midline (A, arrows). By stage 16, the opposing cardioblast rows pair up at the dorsal midline to form a distinctive posterior heart compartment and an anterior aorta (A′). (B–E′) Dorsal views of cardioblast cells stained with the nuclear marker Mef2 in early (B–E) and late (B′–E′) dv development. (B and B′) wild-type embryos form two evenly spaced rows (B) that meet at the dorsal midline in pairs (B′). (C and C′) Expression of UAS-spgRNAi353 with 24B-GAL4 in a spg−/− mutant background occasionally shows mild clustering defects (arrowheads). (D–E′) spg RNAi using the embryonic lethal UAS-spgIR13 line. (D and D′) RNAi knockdown of spg under control of the 24B promoter results in clusters of cardioblasts that deviate from the normal cardioblast pairs observed in wild type (arrowheads). (E and E′) These large cardioblast clusters, often containing more than three nuclei, are also observed upon a decrease in Spg levels using the mesodermal twist driver (arrowheads). Bar, 50 μm. (F–H) Lateral views of approximately four hemisegments stained with α-mef2. (F′–H′) Schematics show the location of cardioblast cell nuclei. The heart cells in wild type (F and F′) and mbc−/− (H and H′) embryos form a relatively straight line compared to multilayered clustering observed in spg−/− mutants (G and G′). (I) Graph depicting the number of clusters present in each embryo of the indicated genotypes, in which spg mutants exhibit increased cardioblast-clustering defects. P-values are results of Mann–Whitney comparisons to wild type (**P < 0.005; ****P < 0.0001). Mean ± SEM. Posterior is up for B–E′. Anterior is left for F–H. Bar, 50 μm.
Previous analysis of spg−/− mutants, which contain maternally contributed spg mRNA and protein, revealed only mild defects in CNS development (Biersmith et al. 2011). Thus we sought to further knock down Spg levels using an RNAi strategy in a spg−/− mutant background. For all experiments that examine dv development, we used the nuclear Mef2 protein as a marker for cardioblast cells. Expression of UAS-spgRNAi353 in the developing musculature (24B-GAL4) of zygotic spg−/− mutants resulted in an abnormal arrangement of cardioblast cells, a phenotype that we have termed “clustering.” Rather than a single row of cells in stage 13 embryos (Figure 3A) or the paired arrangement of cardioblasts that meet up at the dorsal midline in stage 17 embryos (Figure 3B), cardioblast cells were grouped together in early stage embryos (Figure 3C, arrowheads) and groups of three or more cardioblasts were evident in late-stage embryos (Figure 3C′, arrowhead). This phenotype, while consistent (Figure 3I; Table 1A; Table 2A), was mild in spg−/− 24B-GAL4:: UAS-spgRNAi353 embryos.
Table 1. Phenotypes present in early dv development.
| Genotype | Embryos that exhibit clustering (%) | Average clusters/embryo | Cardioblast no. | n | |
|---|---|---|---|---|---|
| A | Wild type | 8.3 | 1.0 | 103.3 | 48 |
| 24B-Gal4, spg242::UAS-spgRNAi353, spg242 | 31.0 | 2.0 | 102.7 | 29 | |
| 24B-GAL4::UAS-spgIR13 | 70.6 | 7.1 | 99.8 | 34 | |
| twi-GAL4::UAS-spgIR13 | 78.9 | 7.6 | 99.8 | 38 | |
| B | mbcD11.2 | 100.0 | 3.5 | 102.4 | 17 |
| twi-GAL4, mbcD11.2::UAS-mbc, mbcD11.2 | 43.0 | 3.1 | 103.4 | 14 | |
| twi-GAL4, mbcD11.2::UAS-spg, mbcD11.2 | 88.0 | 5.6 | 104.3 | 17 | |
| twi-GAL4, mbcD11.2::UAS-spgΔPxxP-GFP, mbcD11.2 | 80.0 | 2.4 | 102.8 | 40 | |
| C | Rac1Rac2 | 43.9 | 1.4 | 102.4 | 21 |
| twi-GAL4::UAS-Rac1N17 | 59.3 | 3.3 | 96.7 | 27 | |
| elmoKO; mbcD11.2 | 75.0 | 2.8 | 99.5 | 13 | |
| Rap1B3/B3 | 62.1 | 2.7 | 101.3 | 29 | |
| twi-GAL4::UAS-Rap1N17 | 75.0 | 7.0 | 103.3 | 32 | |
| elmoKO; spg242 | 91.0 | 17.7 | 98.5 | 11 | |
| D | twi-GAL4::UAS-Rac1V12 | 100.0 | 11.5 | 101.8 | 13 |
| twi-GAL4, mbcD11.2::UAS-Rac1V12, mbcD11.2 | 53.8 | 2.8 | 102.8 | 13 | |
| twi-GAL4, mbcD11.2::UAS-Rap1V12, mbcD11.2 | 67.9 | 3.9 | 97.9 | 28 | |
| E | twi-GAL4::UAS-Rap1V12 | 40.0 | 2.3 | 101.9 | 15 |
| twi-GAL4::UAS-spgIR13, UAS-Rap1V12 | 34.5 | 2.0 | 103.4 | 29 | |
| F | spg242/242 | 41.2 | 1.6 | 104.6 | 31 |
| Rap1B3/+, spg242/242 | 66.7 | 5.3 | 100.2 | 18 | |
| Rap1CD5/B3 | 30.8 | 1.5 | 103.2 | 26 | |
| Rap1CD5/B3, spg242/+ | 52.3 | 4.7 | 102.3 | 29 |
Table 2. Phenotypes present in late dv development.
| Genotype | Single cells (%) | Embryos that exhibit clustering (%) | Average clusters/embryo | Cardioblast no. | n | |
|---|---|---|---|---|---|---|
| A | Wild type | 11.1 | 14.8 | 1.3 | 103.3 | 54 |
| 24B-GAL4, spg242::UAS-spgRNAi353, spg242 | 9.7 | 12.9 | 1.0 | 102.9 | 31 | |
| 24B-GAL4::UAS-spgIR13 | 29.4 | 35.0 | 5.5 | 102.7 | 19 | |
| twi-GAL4::UAS-spgIR13 | 21.4 | 53.6 | 5.5 | 99.6 | 28 | |
| B | mbcD11.2 | 65.4 | 86.9 | 3.3 | 94.0 | 42 |
| twi-GAL4, mbcD11.2::UAS-mbc, mbcD11.2 | 9.1 | 9.1 | 2.5 | 101.0 | 22 | |
| twi-GAL4, mbcD11.2::UAS-spg, mbcD11.2 | 40.0 | 86.7 | 4.0 | 98.7 | 13 | |
| twi-GAL4, mbcD11.2::UAS-spgΔPxxP-GFP, mbcD11.2 | 12.9 | 48.4 | 2.3 | 100.4 | 34 | |
| C | Rac1Rac2 | 0.0 | 5.3 | 3.0 | 103.6 | 19 |
| twi-GAL4::UAS-Rac1N17 | 26.5 | 41.2 | 2.6 | 98.9 | 34 | |
| elmoKO; mbcD11.2 | 50.0 | 75.0 | 2.3 | 98.8 | 17 | |
| Rap1B3/B3 | 10.7 | 17.4 | 2.9 | 100.3 | 28 | |
| twi-GAL4::UAS-Rap1N17 | 54.5 | 30.3 | 7.6 | 102.8 | 33 | |
| elmoKO; spg242 | 60.0 | 100.0 | 3.8 | 90.50 | 4a | |
| D | twi-GAL4::UAS-Rac1V12 | 100.0 | 100.0 | 7.5 | 105.5 | 2a |
| twi-GAL4, mbcD11.2::UAS-Rac1V12, mbcD11.2 | 0.0 | 62.5 | 3.2 | 103.0 | 16 | |
| twi-GAL4, mbcD11.2::UAS-Rap1V12, mbcD11.2 | 27.8 | 61.1 | 4.2 | 96.9 | 18 | |
| E | twi-GAL4::UAS-Rap1V12 | 46.2 | 100.0 | 10.2 | 106.5 | 16 |
| twi-GAL4::UAS-spgIR13, UAS-Rap1V12 | 3.2 | 25.8 | 2.5 | 102.1 | 31 | |
| F | spg242/242 | 5.0 | 20.0 | 1.5 | 103.8 | 20 |
| Rap1B3/+, spg242/242 | 1.1 | 44.4 | 5.6 | 105.1 | 9 | |
| Rap1CD5/B3 | 0.0 | 5.3 | 1.0 | 103.8 | 19 | |
| Rap1CD5/B3, spg242/+ | 8.9 | 30.0 | 6.0 | 102.3 | 17 |
These genotypes resulted in low numbers of surviving progeny after stage 13.
While our studies were being carried out, a new spg RNAi line (UAS-spgIR13) was published that resulted in embryonic lethality when expressed with a ubiquitous GAL4 driver (Eguchi et al. 2013). Before we used this line to further examine the role of spg in dv development, we first ensured that Spg protein levels were reduced in this UAS-spgIR13 line by immunostaining and Western blotting. In addition to a reduction in Spg protein levels in the dv when spg RNAi was driven by the twi promoter, we also observed a loss of Spg staining in the alary muscles, which serve to support the heart tube and facilitate hemolymph flow (Supporting Information, Figure S1). Next, we reexamined the consequences of UAS-spgIR13 expression in cardioblast clustering (arrowheads) in the developing heart tube under control of the 24B-GAL4 (Figure 3, D and D′) or the twi-GAL4 (Figure 3, E and E′) drivers. There was an increase in both the penetrance and number of cardioblasts in each cluster, where most clusters contained four or more cells (Figure 3I; Table 1A; Table 2A). While the images in Figure 3, B–E′, are maximum projections of all Z-stacks collected during data acquisition, analysis of individual sections showed that cardioblasts were stacked on top of one another. To confirm this observation, we imaged lateral views of one row of cardioblasts in stage 13 embryos. In wild-type (Figure 3, F and F′) samples, the heart cells formed a single-celled row. In contrast, the cardioblasts were multilayered in the dorsal–ventral plane in twi-GAL4::UAS-spgIR13 embryos (Figure 3, G and G′). These data suggest that Spg is essential for cardioblasts within a row of cells to maintain their contralateral alignment from the beginning of dv development. To our knowledge, this is the first report of a multilayered cell-clustering phenotype in a Drosophila dv mutant.
mbc−/− mutants affect dv morphogenesis
As mentioned above, both spg and mbc mRNA are expressed in the developing heart tube (Biersmith et al. 2011). Since our results show that loss of Spg results in defective dv patterning, namely aberrant cell clusters, we next tested if mbc−/− mutant embryos also exhibited defects in heart tube morphogenesis. Cardioblast-clustering defects were apparent in mbc−/− mutants in stage 13 (Figure 4A, arrowheads) or stage 17 (Figure 4A′, arrowhead) embryos. However, the clusters were not multilayered (compare Figure 3, G and G′ to Figure 3, H and H′), and the number of cells in each cluster was consistently less in mbc−/− mutants (average of 3.4–3.6 cells/cluster; Table 1B and Table 2B) than in spg−/− mutants (average of 5.5–7.6 cells/cluster; Table 1B and Table 2B). Furthermore, the apparent clusters in stage 13 mbc−/− mutants were often next to “gaps” between adjacent cardioblasts within each row (Figure 4A, arrows), indicating that the cluster may be a secondary effect of a loss of spacing between adjacent cardioblasts. These gaps were rarely observed in dv of spg−/− mutants. Note that mbc has been reported to affect dorsal closure (dc) and could potentially affect migration of the cardioblast rows toward the midline (Erickson et al. 1997). Indeed, we observed dc defects (Figure S2; ∼10% penetrance), but these embryos were excluded from further phenotypic or quantitative analysis.
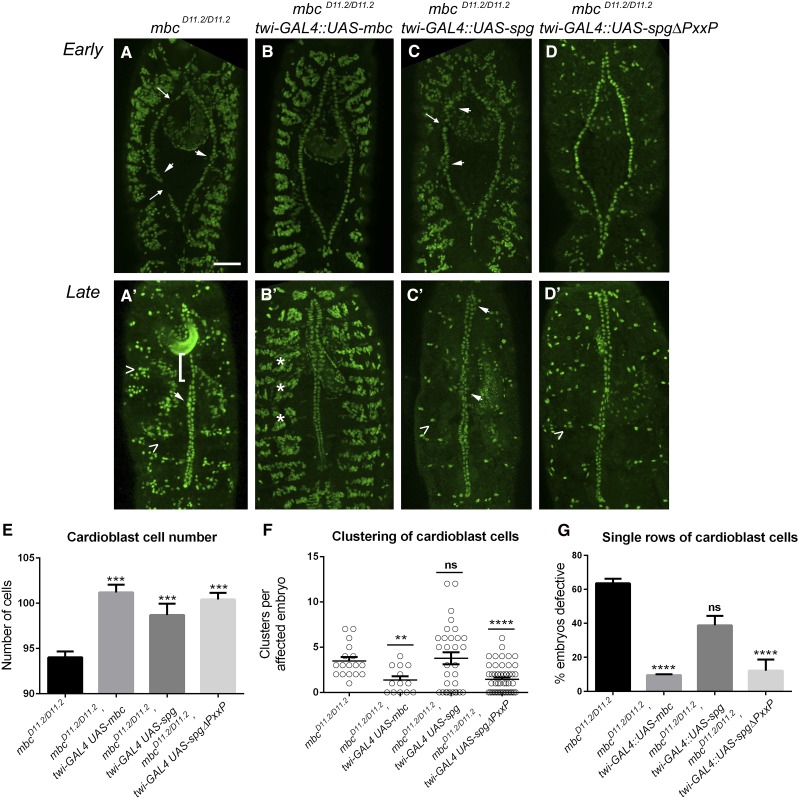
Figure 4.
spg is not capable of rescuing defects resulting from removal of mbc in the dv. (A–C′) Early and late-stage embryos fluorescently labeled with α-mef2 to visualize muscle nuclei. (A) Removal of mbc in stage 13 embryos induces clustering defects (arrowheads) that often appear next to breaks in the contralateral alignment of the cardioblasts (arrows). (A′) These clustering defects persist until later in development (arrowhead), when rows of single cells (brackets) are also observed. Note that the nuclei are clustered in the dorsal somatic muscles (carets), consistent with a failure of myoblast fusion in mbc mutant embryos. (B and B′) Expression of UAS-mbc in an mbc mutant background with twist-GAL4 ameliorates both the clustering and single-cell defects observed in early (B) and late-stage (B′) compared to mbc mutants alone (A and A′). The arrangement of nuclei in the dorsal muscles is also restored (asterisks). (C and C′) No rescue of the mbc mutant clustering or single-cell phenotypes are seen upon expression of UAS-spg. (D and D′) Expression of UAS-spgΔPxxP, which removes the C-terminal proline-rich region of Spg, alleviates the dv defects in mbc mutants, but not the myoblast fusion defects (D′, caret). (E–G) Quantitation of cardioblast cell number (E), cardioblast clustering (F), and unpaired cardioblast (G) phenotypes in mbc mutants of the indicated genotypes. Expression of UAS-spg does not rescue the clustering or single-cell phenotypes in mbc mutants, while expression of UAS-mbc or UAS-spgΔPxxP rescue the dv patterning defects to a similar extent. P-values are results of Mann–Whitney comparisons to mbc−/− alone (**P < 0.005; ***P < 0.001; ****P < 0.0001; ns, not significant). Mean ± SEM. Posterior is up for all embryos. Bar, 50 μm.
In addition to small cardioblast clusters, a second phenotype observed upon loss of mbc were regions of unpaired, single cells, within one row of cardioblasts in stage 17 embryos (Figure 4A′, bracket). This single-cell phenotype has been reported in another dv mutant, laminin-A, and is thought to result from the inability of the dv to maintain its position within the embryo, resulting in twists and breaks in the cardioblast rows (Haag et al. 1999). It is unclear if the single-cell phenotype in mbc−/− mutants is a manifestation of the gaps seen in early mbc−/− mutants or a loss of cell number during the cardioblast migration process. To test the latter hypothesis, we counted the total number of cardioblasts in early and late-stage embryos that were apparent in single sections of confocal Z-stacks. In accordance with published reports (Ponzielli et al. 2002), we observed a consistent number of ∼104 cardioblasts in wild-type embryos throughout dv development. Quantification of cardioblast number showed a loss of ∼10 heart cells in mbc−/− mutants from stage 13 to stage 17 (Figure 4E; Table 1B; Table 2B). These data, taken together, show that while loss of either spg or mbc results in cardioblast clustering, cell clusters in spg−/− mutants are more severe than in mbc−/− mutants. Moreover, mbc−/− mutants are also characterized by lateral gaps within contralateral rows of cells and a loss of cardioblast cells.
Spg is not caable of rescuing defects resulting from mbc removal in the dv
To verify and extend our observations that Mbc and Spg play independent roles in dv morphogenesis based upon their different mutant phenotypes, we utilized the GAL4/UAS system to perform rescue experiments. As expected, the reintroduction of UAS-mbc (Figure 4, B and B′) was sufficient to ameliorate the loss of cardioblast cell number (Figure 4E), cardioblast-clustering defects (Figure 4F), and the unpaired, single-cell phenotype (Figure 4G) present in mbc−/− mutants (Table 1B and Table 2B). In contrast, expression of UAS-spg (Figure 4, C and C′; arrow denotes breaks between adjacent cardioblasts and arrowheads denote clusters) did not rescue the clustering or unpaired cell phenotypes (Figure 4, E and F; Table 1B; Table 2B). Examination of myoblast fusion in mbc−/− mutants using the nuclear Mef2 marker revealed clustered nuclei in dorsal muscles that are adjacent to the dv, indicative of fusion defects (Figure 4A′, carets) (Erickson et al. 1997; Balagopalan et al. 2006). This general disorganization of the dorsal somatic nuclei was restored upon expression of UAS-mbc (Figure 4B′, asterisks), but not upon the reintroduction of UAS-spg (Figure 4C′, caret) in mbc−/− mutants. This result is consistent with data in Figure 1, E and H, showing that Spg does not rescue mbc-induced myoblast fusion defects.
The primary amino acid sequence homology between full-length Mbc and Spg proteins (33% identity/52% similarity) decreases in the C-terminal proline-rich region (16% identity/21% similarity) (Biersmith et al. 2011). This sequence divergence and alternate number of PxxP motifs (Mbc has three and Spg has five) suggests that the C-terminal portion of these Dock proteins may confer differential protein functions. To test this idea, we generated transgenic flies that contain a deleted version of Spg and lacks the entire proline-rich region (UAS-spgΔPxxP). Expression of UAS-spgΔPxxP in mbc−/− mutants rescued the cardioblast-cell number (Figure 4E), clusters of cardioblasts (Figure 4F), and single rows of cells (Figure 4G; Table 1B; Table 2B). These data suggest that removal of the proline-rich region allows this modified form of Spg to behave similarly to Mbc in dv development. Note that the mbc-mediated myoblast fusion defects (Figure 4A′, carets) are not rescued by expression of UAS-spgΔPxxP (Figure 4D′, caret), suggesting that truncated Spg can replace Mbc in heart tube formation, but not somatic muscle fusion.
GTPase activation of both Rac1 and Rap1 is required for proper dv development
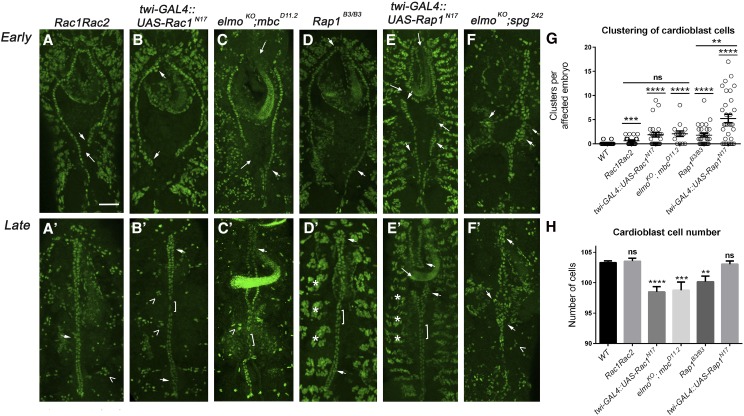
To address our central question of whether Dock family members activate different GTPase targets in vivo, we first examined if disruption of Rac or Rap1 affected dv morphogenesis. Embryos deficient in the zygotic contribution of Rac1 and Rac2 (Figure 5, A and A′) induced small cell clusters (arrowhead) and lateral gaps (arrows). Since both Rac1 and Rac2 are maternally contributed gene products (Hakeda-Suzuki et al. 2002), we expressed a dominant-negative form of Rac1 (UAS-RacN17) under control of the twi promoter to block Rac activity (Figure 5, B and B′). In addition to increased penetrance and severity of cardioblast clusters over Rac1Rac2 mutants alone (Figure 5G; Table 1C; Table 2C), we also observed regions of single cells (Figure 5B′, bracket) and a decrease in the total number of cardioblast cells (Figure 5H; Table 1C; Table 2C). Taken together, these phenotypes that are induced by expression of RacN17 mimicked the clustering, unpaired cell, and loss of cardioblast cell defects observed in mbc−/− mutants. We have shown that the Mbc–Elmo complex acts as an upstream GEF for Rac in Drosophila ommatidial development (Geisbrecht et al. 2008) and in embryonic muscle fusion (Figure 2). To further test if removal of the Mbc–Elmo complex phenocopies RacN17 mutants, we created embryos doubly mutant for both mbc and elmo. Both mutations were balanced over lacZ-containing balancer chromosomes, and mutants were selected for further analysis by a lack of lacZ staining. As expected and shown in Figure 5, cardioblast clusters (Figure 5C, arrowheads), lateral gaps (Figure 5C, arrows), regions of single cells (Figure 5C′, brackets), and a loss in cardioblast number (Figure 5H) were prevalent phenotypes, similar to a complete loss of Rac activity.
Figure 5.
GTPase activation of both Rac and Rap1 is required for correct patterning of the dv. (A–F′) Early and late-stage embryos fluorescently labeled with α-mef2 to visualize muscle nuclei. (A–C′) Inactivation of the Rac GTPase pathway using classical alleles (A and A′), dominant-negative overexpression (B and B′), or double-mutant null alleles of the upstream GEF complex (C and C′) results in gaps in the contralateral alignment of cardioblasts (arrows), clusters that deviate from the normal single rows of heart cells (arrowheads), and regions of single, unpaired cells in stage 16 embryos. Note the disorganized muscle nuclei of the dorsal muscles in late GTPase mutants (A′–C′, carets). (D–F′) More severe cardioblast-clustering defects are observed upon perturbation of the Rap1-signaling pathway. (D and D′) Removal of the zygotic contribution of Rap results in an increase in cardioblast clusters in both early (D) and late-stage (D′) embryos (arrowheads). (E and E′) Inactivation of GTPase activity by overexpression of a dominant-negative version of Rap1 also induces larger cardioblast clusters (arrowheads) and gaps (arrows) between adjacent cardioblasts in stage 13 (E) and stage 16 (E′) embryos. Note that disruption of Rap1 signaling does not impact myoblast fusion, as shown by organized muscle nuclei in the dorsal musculature (asterisks). (F and F′) Extreme examples of large cardioblast clusters (arrowheads) are found in embryos that remove only the zygotic contribution of the putative elmo; spg GEF complex. (G and H) Graphs illustrating the average number of clusters per affected embryo (G) or cardioblast number (H) in mutants that alter GTPase activity. Blocking Rap1 activity results in an increase in cardioblast clusters (G), while altering Rac function decreases the number of cardioblasts (H) normally found in wild-type embryos. P-values are results of Mann–Whitney comparisons to wild type or of the bars indicated (**P < 0.005; ***P < 0.001; ****P < 0.0001, ns, not significant). Mean ± SEM. Posterior is up for all embryos. Bar, 50 μm.
As shown in Figure 3, embryos with decreased Spg exhibit more heart cells within each cluster, suggesting a loss of adhesion between adjacent cells within a contralateral row of cardioblasts. Since Dock4 has been shown to function upstream of Rap1 in cell adhesion (Yajnik et al. 2003), we next tested if loss of Rap1 also affects putative adhesive aspects of dv morphogenesis. Overall, weak phenotypes observed in zygotic Rap1−/− mutants were increased upon expression of dominant-negative Rap1 (UAS-Rap1N17). Rap1−/− mutants showed mild clustering defects (Figure 5G; Table 1C; Table 2C) in both early (Figure 5D) and late dv development (Figure 5D′). In addition to an increase in the number of clusters per embryo (Figure 5G; Table 1C; Table 2C), gaps in the contralateral rows (Figure 5E, arrows) were also present upon expression of UAS-Rap1N17. However, loss of Rap activity did not alter cardioblast number (Figure 5H; Table 1C; Table 2C), consistent with our results for loss of spg (Table 1F and Table 2F).
Removal of the putative Spg–Elmo GEF complex was performed by examining embryos doubly mutant for both spg and elmo. This genotype also resulted in large cardioblast clusters, reminiscent of that seen in UAS-spgIR13 knockdown embryos (Figure 5, F and F′; Table 1C; Table 2C). Patterning of the dv was most affected in elmo−/−; spg−/− mutants, where cardioblasts in a portion of stage 13 embryos exhibited a “star” phenotype. The significance of this is unclear, but suggests that either the Spg–Elmo complex has addition roles in embryo development and this could be a secondary effect or that the dv is being pulled away from the central midline. Since Spg is expressed in the alary muscles (Figure S1), which are normally required for relative positioning of the dv, a third possibility is that reduction of Spg in these muscles alters the ability of the heart tube to keep its normal location. Our data clearly demonstrate a role for both Rac and Rap1 in dv morphogenesis, although the biological outputs of each GTPase differentially affect cardioblast patterning.
Mbc exhibits specificity for Rac in dv morphogenesis
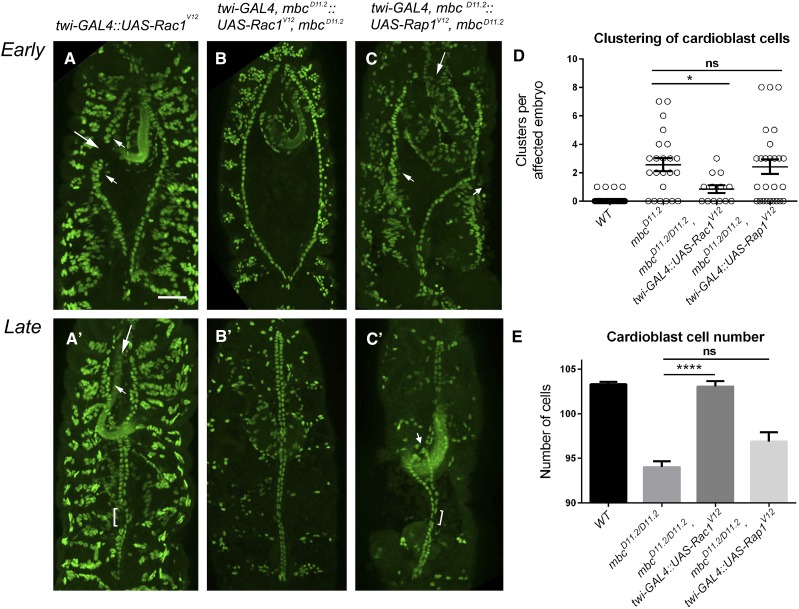
To better define the individual roles of Dock GEF function in GTPase activation, we next tested if Mbc acts upstream of Rac1 in the developing heart tube as it does in somatic muscle development (Figure 2). Expression of UAS-Rac1V12 under control of the twi promoter resulted in the same phenotypes, although with increased penetrance, as those observed in mbc−/− mutants, including cell clustering (small arrows), gaps in the cardioblast rows (long arrows), and regions of single cells (Figure 6, A and A′, bracket; Table 1D; Table 2D). Expression of UAS-Rac1V12 in an mbc−/− mutant background (Figure 6, B and B′) ameliorated these phenotypes nearly to wild type by reducing the prevalence of cardioblast clusters (Figure 6D) and rescuing the number of cardioblasts (Figure 6E). In contrast, expression of UAS-Rap1V12 in mbc mutants (Figure 6, C and C′) did not alleviate mbc-mediated cardioblast clusters (Figure 6D) and could not rescue the loss of heart cells (Figure 6E). Our data strongly suggest that Rac, but not Rap, functions downstream of Mbc in heart tissue.
Figure 6.
Mbc acts upstream of Rac in the dv. (A–C′) The nuclear marker Mef2 is used to label the cardioblast nuclei in stage 13 and stage 17 embryos upon expression of constitutively active GTPases under control of the twi promoter. (A and A′) Expression of RacV12 causes breaks in rows of adjacent cardioblasts (A, arrows), cardioblast clustering (A and A′, arrowheads), occasional posterior opening of the dv (A’, arrow) and regions of unpaired cells (A′, bracket). (B and B′) Expression of RacV12 in an mbc mutant background suppresses both early (B) and late (B′) clustering defects and eliminates the single-cell phenotype. (C and C′) Rap1V12 cannot rescue cardioblast clustering (C and C′, arrowheads), posterior gaps in the dv (C, arrow), or single cell (C′, bracket) phenotypes present in mbc mutants (compare to A and A′). (D and E) Quantitation of cardioblast clusters (D) and the total number of cardioblast cells (E) in an mbc mutant background or with the addition of activated Rac1 or Rap1. RacV12, but not RapV12, rescues mbc-mediated clustering defects and restores cardioblast cell number. Posterior is up for all embryos. P-values are results of Mann–Whitney comparisons of the bars indicated (*P < 0.05; ****P < 0.0001; ns, not significant). Mean ± SEM. Bar, 50 μm.
Spg likely acts upstream of Rap1 in dv and CNS development
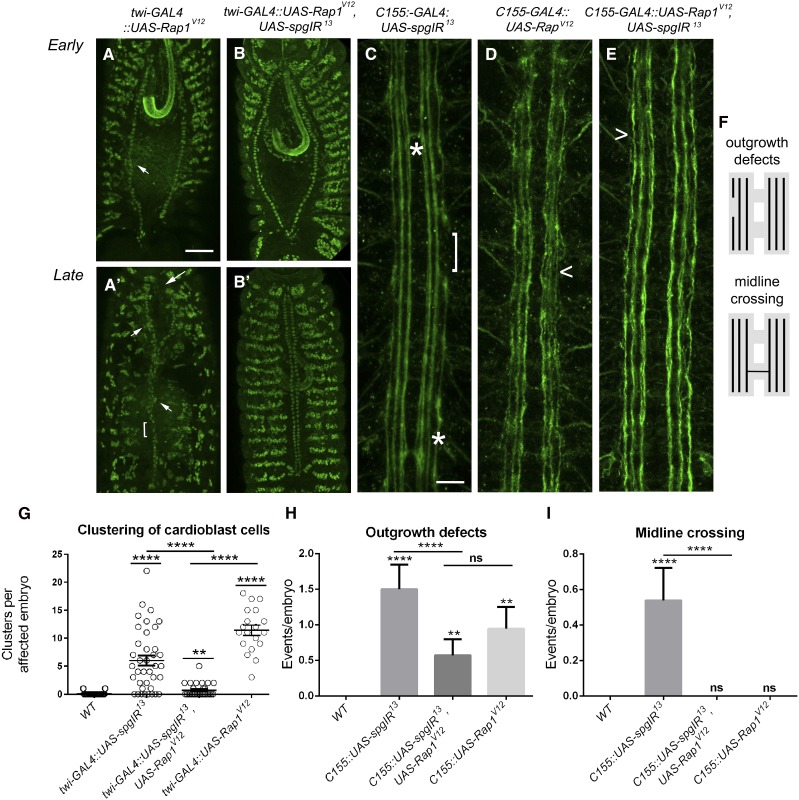
As dv defects in mbc−/− mutant embryos were rescued upon the introduction of UAS-RacV12, we wondered if we could suppress spg knockdown phenotypes upon expression of the putative downstream GEF Rap. As might be expected for either mis- or overexpression of a constitutively active form of any GTPase, patterning defects were present in the dv upon expression of UAS-Rap1V12. Specifically, we observed an increase in cardioblast clusters that persisted from early (Figure 7A, arrowheads; Table 1E) to late (Figure 7A′, arrowheads; Table 2E) stages and regions of single cells (Figure 7A′, bracket; Table 2E). Similar to the rescue of dv phenotypes in mbc mutants with the downstream GTPase Rac, expression of Rap1V12 ameliorated the cardioblast clustering phenotypes in a spg RNAi background (Figure 7, B, B′, and G; Table 1E; Table 2E; compare to spg RNAi alone in Figure 3D).
Figure 7.
Expression of RapV12 can temper spg−/− phenotypes. (A–E) Stage 13 (A and B) or stage 17 (A′, B′, C, and D) embryos labeled with α-mef2 to visualize muscle nuclei (A–B′) or α-1D4 (C–E) to label the longitudinal axons of the CNS. (A and A′) Embryos overexpressing Rap1V12 show early (A) and late (A′) clustering defects (A and A′, arrowheads), abnormal posterior openings (A, arrows), and unpaired cardioblasts (A′, bracket). (B and B′) The clustering defects observed upon loss of spg using RNAi (Figure 3, E and E′) are ameliorated with co-expression of the constitutively active form of Rap. (C) RNAi knockdown of spg (UAS-spgIR13) using the pan neuronal driver, C155-GAL4, gives rise to outgrowth defects (bracket) and midline crossovers (asterisks). (D) Overexpression of RapV12 on its own does not show guidance errors, but instead results in minor unbundling of the longitudinal axons (D, caret). (E) Outgrowth and guidance defects seen when knocking down Spg protein levels by RNAi (C, asterisks and bracket) are tempered when simultaneously expressing RapV12. (F) Schematic of outgrowth and midline crossing defects observed in spg RNAi embryos. (G–I) Graphs showing the ability of UAS-RapV12 to suppress cardioblast clustering (G), CNS outgrowth defects (H), or guidance errors due to inappropriate midline crossing (G) in a spg RNAi background. P-values are results of Mann–Whitney comparisons (G) of the bars indicated or Kruskal–Wallis tests (H and I) compared to wild type or the bars indicated (****P < 0.0001). Mean ± SEM. Posterior is up for all embryos. Bar: 50 μm (A–B′) and 10 μm (C–E).
Since previous results demonstrated a role for spg in the outgrowth of longitudinal CNS axons (Biersmith et al. 2011), we further tested if Rap1V12 could also rescue spg-induced phenotypes in this tissue. Using the neuronal C155-GAL4 driver, RNAi depletion of spg (UAS-spgIR13) resulted in increased penetrance of outgrowth defects (Figure 7C, bracket) and midline crossing errors (asterisks) compared to spg−/− mutants alone (Biersmith et al. 2011). Expression of UAS-Rap1V12 in a spg RNAi background reduced the percentage of axon outgrowth defects (Figure 7, F and H) and eliminated midline guidance defects (Figure 7, F and I). Our data in the dv and CNS support the possibility that Spg may function upstream of Rap1.
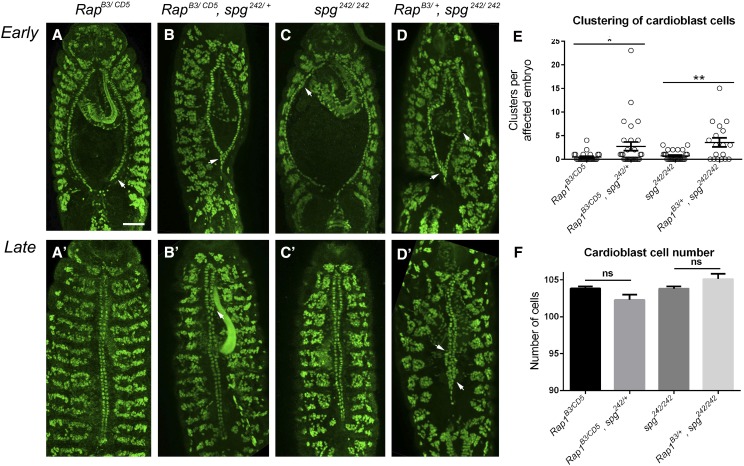
We next turned to genetic interaction analysis to further examine if spg and Rap1 may function in the same pathway. Removal of zygotic Rap−/− (Figure 8, A and A′) or spg−/− (Figure 8, C and C′) alone resulted in few cardioblast clusters (Figure 8E). This clustering phenotype was enhanced upon removal of either one copy of spg in Rap1−/− mutants (Figure 8, B, B′, and E) or a 50% reduction in Rap1 gene dosage in a spg−/− mutant background (Figure 8, D, D′, and E). Importantly, cardioblast number (Figure 8F) did not change in our genetic interaction analysis between spg and Rap1, further supporting the idea that cardioblast number is mediated by the Mbc→Rac pathway. These data provide good evidence for Spg and Rap1 acting together in dv development.
Figure 8.
Genetic interactions between spg and Rap1. (A–D′) Dorsal views of cardioblast cells stained with Mef2 in early (A–D) and late (A′–D′) dv development. The number of cell clusters (arrowheads) in Rap1−/− (A and A′) or spg−/− (C and C′) mutants is enhanced upon removal of one copy of either spg (C and C′) or Rap1 (D and D′), respectively. (E) The number of dv clusters is enhanced in Rap1B3/CD5; spg242/+ or Rap1B3/+; spg242/242 embryos compared to Rap1B3/CD5 or spg242/242 alone. (F) There is no statistically significant difference in cardioblast number upon additional removal of one copy of spg or Rap1 in Rap1 or spg mutant backgrounds, respectively. P-values in E are results of Mann–Whitney comparisons of the bars indicated (*P < 0.05; **P < 0.005; ns, not significant). Mean ± SEM. Posterior is up for all embryos. Bar, 50 μm.
mbc and spg are required for proper dv lumen formation
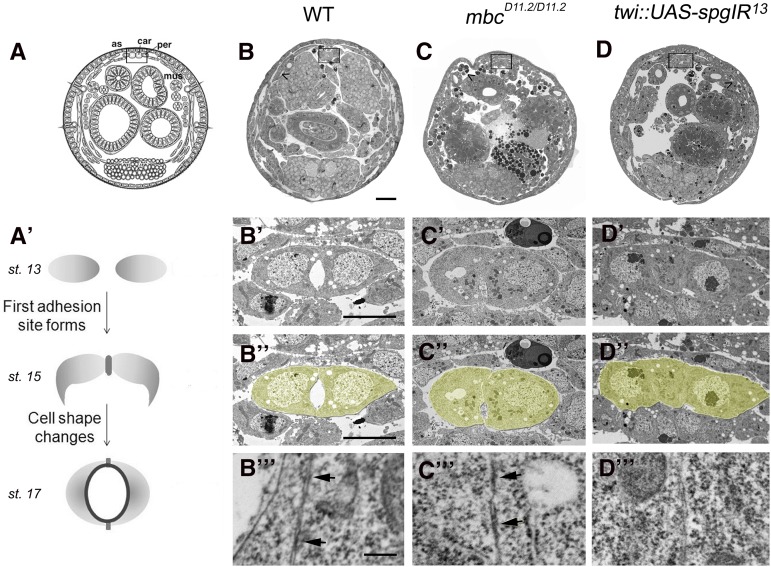
Cross sections through the Drosophila embryo allow for a detailed study of dv lumen formation, specifically to visualize cell-shape changes that occur in this in vivo two-cell system (Medioni et al. 2008; Santiago-Martínez et al. 2008). Starting in stage 13 embryos, two contralateral rows of cardioblasts migrate toward the midline to form the first junctional adhesion domain by stage 15 (Figure 9A′). The adhesion proteins β-catenin and E-cadherin accumulate at this initial adhesion site and trigger actin-mediated cell-shape changes that allow the cardioblasts to adopt a crescent-like shape to form a second, ventral adhesion site. Simultaneous with heart-cell-shape changes, cell-autonomous, repellant Slit/Robo signaling at the luminal surface of the cardioblasts ensures proper lumen formation (Helenius and Beitel 2008; Medioni et al. 2008, 2009; Santiago-Martínez et al. 2008).
Figure 9.
Mbc and Spg are required for proper dv lumen formation. (A–D′′′) Cross sections of the Drosophila dv to visualize cardioblast cell-shape changes. (A) Schematic representation of a cross section through a whole-mount embryo. The dv is located on the dorsal side just under the epidermis (boxed area). (A′) Schematic representation of dv lumen development from stage 13 to stage 17. Two cardioblasts meet at the dorsal midline to create an adhesion site, followed by cell-shape changes that allow for the formation of a ventral second adhesion site. This process is coincident with lumen formation, resulting in a linear heart tube through which hemolymph flows. (B–B′′) The dv in wild-type embryos consists of crescent-shaped cardioblasts with a large, central lumen. (C–C′′) The cardioblasts are rounded and fail to form a lumen in mbc mutants. (D–D′′) twi-GAL4::UAS-spgIR13 embryos also do not result in lumen formation, although the cardioblasts retain their crescent shape. Note the lack of myoblast fusion seen in mbc mutants (C, caret) compared to the fused muscle seen in wild type and spg RNAi (B and D, carets). (B′′′–D′′′) High-magnification electron micrographs of cardioblast junctions. (B′′′ and C′′′) Electron-dense regions (arrow) indicate the formation of adherens junctions in wild type (B′′′) and mbc mutants (C′′′). Electron-dense regions are not observed along the adjacent membranes in spg IR embryos (D′′′). Bar: (B–D) 20 μm; (B′–D′D′′) 5 μm; (B′′′–D′′′) 250 nm.
Rac activation alters actin dynamics and Rap1 activity affects adhesive properties of cells. Thus, we examined whether loss of mbc or spg resulted in aberrant cell-shape changes and/or altered the adhesion between cardioblasts. Using transmission electron microscopy (TEM), analysis of cross sections in wild-type stage 17 embryos showed two crescent-shaped cardioblasts with an internal lumen (Figure 9B′). Higher magnification revealed electron-dense regions, indicative of adhesion complexes, at the junctional domains (Figure 9B′′′, arrows). In mbc mutants, the cardioblasts were unable to change shape and remained round. In addition, the lumen was either absent or very small (Figure 9C′). Dark electron-dense regions were present in mbc mutants, suggesting that adhesion sites were present (Figure 9C′′′, arrows). In contrast, the heart cells in twiGAL4::UAS-spgIR13 mutants showed an elongated shape (Figure 9D′) that was different from the crescent-like cells observed in wild-type embryos or the round cells in mbc mutants. This genotype also lacked a lumen and the electron-dense regions seen in wild-type and mbc mutants (Figure 9D′′). These results show that both Mbc and Spg are required for lumen formation in dv morphogenesis, although Mbc and Spg differ in their ability to induce cardioblast cell-shape changes and or permit electron-dense adhesion sites.
Discussion
In the present study, we demonstrate that Mbc and Spg have independent roles in the somatic and heart muscle tissue. First, misexpression of Spg in the somatic muscle cannot compensate for loss of Mbc during myoblast fusion. However, we cannot rule out the possibility that effectiveness of rescue by Mbc and not by Spg reflects differences in protein stability rather than the effectiveness of the proteins. Second, while both Mbc and Spg are required for dv development, they affect different aspects of morphogenesis. Loss of mbc results in small clusters of cardioblast cells (3.3–3.5 cells/cluster), breaks in the contralateral rows of adjacent cells, and a total loss of ∼10 cardioblast cells per embryo. These phenotypes are distinct from knockdown of spg using RNAi, where the primary defect is larger clusters of multilayered cardioblast cells (5.5–7.6). As a better readout of cell morphogenic events, ultrastructure analysis of opposing cardioblast cells reveals that cell-shape changes do not occur in mbc−/− mutants, while a decrease in spg allows the cells to maintain an elongated shape, but lose putative electron-dense adhesion sites between cardioblasts. Taken together, these data provide strong evidence for differential roles of Mbc and Spg in vivo.
Dock protein specificity
Discrepancies concerning the downstream GTPase target(s) of the Dock family of GEFs have existed for ∼10 years (Gadea and Blangy 2014; Laurin and Côté 2014). Numerous reports demonstrate that both Dock180/Mbc and Dock3/Dock4/Spg can activate Rac1 in in vitro GTPase activation assays (Yajnik et al. 2003; Hiramoto et al. 2006; Yan et al. 2006). In vivo, it is well established that Dock180/Mbc activates Rac in all contexts examined (Erickson et al. 1997; Côté and Vuori 2002; Geisbrecht et al. 2008; Laurin et al. 2008), while the data for Dock4/Spg are less clear. A substantial body of evidence links Dock3 and Dock4 to the activation of Rac in both neuronal tissues and cancer cells (Namekata et al. 2004; Hiramoto et al. 2006; Ueda et al. 2013), primarily through Rac-dependent actin rearrangement in axon outgrowth or cellular metastasis.
Two reports suggest an alternative or additional role for Dock-B family members in the activation of Rap1. The first evidence emerged ∼10 years ago when Yajnik and colleagues showed that Dock4 is capable of activating Rap1 in GTPase activation assays (Yajnik et al. 2003). Recent studies also provide supportive evidence for Rap1 activation via Spg in the differentiation of R7 photoreceptor cells in the Drosophila eye (Eguchi et al. 2013). Duolink in situ proximity ligation assay (PLA) experiments suggest a physical interaction between Spg and Rap1 at the plasma membrane in photoreceptor cells. The authors also rule out Rac as an effector of Spg in R7 photoreceptor differentiation. A reasonable explanation for these apparently conflicting results is that Dock-B proteins may exhibit dual roles in the activation of both Rac and Rap1, depending on cellular context.
Nucleotide exchange of GDP for GTP is catalyzed by the DHR2 domain in unconventional Dock family members. A conserved valine residue within the α10 helix of DHR2 acts as a nucleotide sensor that recognizes and destabilizes bound GDP. Subsequent binding of GTP results in a conformational change and release of the activated GEF (Yang et al. 2009). However, the mechanisms that mediate GTPase specificity within each DHR2 domain is not known. There is some evidence to suggest that the C-terminal proline-rich region of Dock4 may be required for GTPase specificity other than Rap1. A homozygous mutation in Dock4 (Pro1718Leu) was identified in two independent cell lines, one derived from prostate cancer and the other from ovarian cancer. The presence of this mutation results in altered GTPase specificity for Rac and Cdc42. Furthermore, expression of this mutated version in mouse 3081 osteosarcoma cells shows a decrease in actin stress fibers and the presence of filopodia, an observation consistent with altered GTPase activation. Contact inhibition is not a normal feature of this osteosarcoma cell line, and staining of these cells with β-catenin does not show the presence of adherens junctions. While transfection with Dock4-WT results in the appearance of intercellular adherens junctions, no such effect is observed upon co-expression of Rap1N17 in this Dock4-WT background or upon independent expression of Dock4-Pro1718Leu (Yajnik et al. 2003).
Our observations are consistent with this putative role for the C-terminal proline-rich region of Spg in dv morphogenesis. Expression of Spg deleted for the entire PxxP region (UAS-spgΔPxxP) is able to suppress mbc-mediated single-cell stretching and cardioblast-clustering defects, while overexpression of full-length Spg does not. Interestingly, neither version of Spg can rescue myoblast fusion defects due to loss of mbc, suggesting that Spg exerts its effects only in tissues where both proteins are known to function. While we do not yet understand the role of the PxxP region, whether in GTPase specificity and/or binding to other SH3 domain-containing proteins, it is worth noting that Spg contains three additional putative PxxP-binding sites not present in Mbc (Biersmith et al. 2011).
Cardioblast cell-shape changes in lumen formation
We chose the dv as a two-cell system to better understand whether Mbc and Spg influence the same or independent cell morphogenic effects. Analysis of TEM cross sections through the dv highlight actin-mediated cell-shape changes and the presence of putative adherens junctions. As shown in Figure 9A, the prevailing model for cardiac lumen formation involves the coordination of both cell-shape changes and lumen formation (Santiago-Martínez et al. 2006; Medioni et al. 2008; Albrecht et al. 2011). Our current data are consistent with the canonical role of Mbc in actin cytoskeletal rearrangement through the Rac GTPase. Expression of constitutively active Rac suppressed dv patterning defects present upon loss of Mbc. Furthermore, the cardioblast cells in mbc−/− mutants properly migrate to the dorsal midline and are able to form adhesion sites, as indicated by the presence of electron-dense plaques between the cardioblast membranes. However, the cardioblast cells remain rounded, likely due to the inability of actin-mediated cytoskeletal events. Perhaps mbc mutants lack the ability to make these shape changes, thus resulting in an extended junctional domain.
We postulate that Spg is required for Rap1 activation to regulate adherens junctions formation. A genetic interaction between spg and Rap1 regulates aspects of cardioblast patterning, namely the multilayering of heart cells within a contralateral row. TEM analysis shows that spg RNAi mutants lack electron-dense accumulations along adjacent cardioblast membranes, suggesting defects in the ability to form the first junctional domain. The elongated appearance of the cardioblasts indicates that actin-mediated cell-shape changes are not affected. We cannot rule out the possibility that Spg could be affecting Slit/Robo signaling at the luminal membrane, thus resulting in an inhibition of Armadillo/DE-Cadherin accumulation at adherens junctions and an increase in the regulation of actin-mediated cytoskeletal events (Medioni et al. 2008).
Here, we have shown that the genetically tractable model organism D. melanogaster can provide an excellent in vivo system to study the cellular behavior of Dock proteins, which have already been implicated in a vast array of diseases in mammals, including developmental limb disease, congenital cognitive disorders, progressive cancers, and neurodegenerative diseases. Future experiments will be directed at identifying other proteins that regulate GEF function in tissues where both Mbc and Spg are required for cellular processes.
Supplementary Material
Acknowledgments
The authors thank Susan Abmayr and Masa Yamaguchi for providing fly stocks and reagents; Nicole Green and Jessica Kawakami for careful reading of this manuscript; the Developmental Studies Hybridoma Bank developed under the National Institute for Child Health and Human Development for antibodies; the Bloomington Stock Center for flies; Len Dobens, Richard Cripps, Susan Abmayr, Achim Pauluat, and Sunita Kramer for helpful discussion; the St. Louis University Research Core and Histology Services and members of the University of Missouri–Kansas City Histology Core, especially Leanne Szerzen and Doug Law. This work was supported by a Predoctoral Fellowship from the American Heart Association Midwest Affiliate (12PRE12050380 to B.B.) and by National Institutes of Health grant RO1AR060788 (to E.R.G.).
Footnotes
Communicating editor: I. K. Hariharan
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177063/-/DC1.
Literature Cited
- Albrecht S., Altenhein B., Paululat A., 2011. The transmembrane receptor Uncoordinated5 (Unc5) is essential for heart lumen formation in Drosophila melanogaster. Dev. Biol. 350: 89–100. [DOI] [PubMed] [Google Scholar]
- Asha H., de Ruiter N. D., Wang M., Hariharan I. K., 1999. The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J. 18: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L., Chen M. H., Geisbrecht E. R., Abmayr S. M., 2006. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol. Cell. Biol. 26: 9442–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C. M., et al. , 2007. Two distinct modes of guidance signalling during collective migration of border cells. Nature 448: 362–365. [DOI] [PubMed] [Google Scholar]
- Biersmith B., Liu Z. C., Bauman K., Geisbrecht E. R., 2011. The DOCK protein sponge binds to ELMO and functions in Drosophila embryonic CNS development. PLoS ONE 6: e16120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B., Harjes P., Ishimaru S., Heke M., Fan H. Q., et al. , 2003. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics 165: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Chen Q., Chen T. J., Letourneau P. C., Costa L. F., Schubert D., 2005. Modifier of cell adhesion regulates N-cadherin-mediated cell-cell adhesion and neurite outgrowth. J. Neurosci. 25: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Peto C. A., Shelton G. D., Mizisin A., Sawchenko P. E., et al. , 2009. Loss of modifier of cell adhesion reveals a pathway leading to axonal degeneration. J. Neurosci. 29: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., Zeghouf M., 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93: 269–309. [DOI] [PubMed] [Google Scholar]
- Cook D. R., Rossman K. L., Der C. J., 2014. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene 33: 4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J. F., Vuori K., 2002. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115: 4901–4913. [DOI] [PubMed] [Google Scholar]
- Duchek P., Somogyi K., Jékely G., Beccari S., Rørth P., 2001. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107: 17–26. [DOI] [PubMed] [Google Scholar]
- Eguchi K., Yoshioka Y., Yoshida H., Morishita K., Miyata S., et al. , 2013. The Drosophila DOCK family protein sponge is involved in differentiation of R7 photoreceptor cells. Exp. Cell Res. 319: 2179–2195. [DOI] [PubMed] [Google Scholar]
- Erickson M. R., Galletta B. J., Abmayr S. M., 1997. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J. Cell Biol. 138: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Gilmour D., 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10: 445–457. [DOI] [PubMed] [Google Scholar]
- Gadea G., Blangy A., 2014. Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 93: 466–477. [DOI] [PubMed] [Google Scholar]
- Geisbrecht E. R., Montell D. J., 2004. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 118: 111–125. [DOI] [PubMed] [Google Scholar]
- Geisbrecht E. R., Haralalka S., Swanson S. K., Florens L., Washburn M. P., et al. , 2008. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 314: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea S. M., Awadia S., Garcia-Mata R., 2014. I’m coming to GEF you: regulation of RhoGEFs during cell migration. Cell Adh. Migr. 8: 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag T. A., Haag N. P., Lekven A. C., Hartenstein V., 1999. The role of cell adhesion molecules in Drosophila heart morphogenesis: faint sausage, shotgun/DE-cadherin, and laminin A are required for discrete stages in heart development. Dev. Biol. 208: 56–69. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S., Ng J., Tzu J., Dietzl G., Sun Y., et al. , 2002. Rac function and regulation during Drosophila development. Nature 416: 438–442. [DOI] [PubMed] [Google Scholar]
- Haralalka S., Shelton C., Cartwright H. N., Katzfey E., Janzen E., et al. , 2011. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development 138: 1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan I. K., Carthew R. W., Rubin G. M., 1991. The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell 67: 717–722. [DOI] [PubMed] [Google Scholar]
- Helenius I. T., Beitel G. J., 2008. The first “Slit” is the deepest: the secret to a hollow heart. J. Cell Biol. 182: 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto K., Negishi M., Katoh H., 2006. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 312: 4205–4216. [DOI] [PubMed] [Google Scholar]
- Kang H., Davis-Dusenbery B. N., Nguyen P. H., Lal A., Lieberman J., et al. , 2012. Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J. Biol. Chem. 287: 3976–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Patel M., Laurin M., Fradet N., Pelletier A., et al. , 2008. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 19: 4837–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M., Côté J. F., 2014. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 28: 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M., Fradet N., Blangy A., Hall A., Vuori K., et al. , 2008. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc. Natl. Acad. Sci. USA 105: 15446–15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gao X., Liu G., Xiong W., Wu J., et al. , 2008. Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat. Neurosci. 11: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C., Astier M., Zmojdzian M., Jagla K., Sémériva M., 2008. Genetic control of cell morphogenesis during Drosophila melanogaster cardiac tube formation. J. Cell Biol. 182: 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C., Sénatore S., Salmand P. A., Lalevée N., Perrin L., et al. , 2009. The fabulous destiny of the Drosophila heart. Curr. Opin. Genet. Dev. 19: 518–525. [DOI] [PubMed] [Google Scholar]
- Moore C. A., Parkin C. A., Bidet Y., Ingham P. W., 2007. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 134: 3145–3153. [DOI] [PubMed] [Google Scholar]
- Namekata K., Enokido Y., Iwasawa K., Kimura H., 2004. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 279: 14331–14337. [DOI] [PubMed] [Google Scholar]
- Namekata K., Kimura A., Kawamura K., Harada C., Harada T., 2014. Dock GEFs and their therapeutic potential: neuroprotection and axon regeneration. Prog. Retin. Eye Res. 43: 1–16. [DOI] [PubMed] [Google Scholar]
- Pagnamenta A. T., Bacchelli E., de Jonge M. V., Mirza G., Scerri T. S., et al. , 2010. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol. Psychiatry 68: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekoek W. J., Kooistra M. R., Zwartkruis F. J., Bos J. L., 2009. Cell-cell junction formation: the role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim. Biophys. Acta 1788: 790–796. [DOI] [PubMed] [Google Scholar]
- Ponzielli R., Astier M., Chartier A., Gallet A., Therond P., et al. , 2002. Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development 129: 4509–4521. [DOI] [PubMed] [Google Scholar]
- Postner M. A., Miller K. G., Wieschaus E. F., 1992. Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangements in Drosophila melanogaster embryos. J. Cell Biol. 119: 1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice T. B., Garen A., 1975. Localized defects of blastoderm formation in maternal effect mutants of Drosophila. Dev. Biol. 43: 277–286. [DOI] [PubMed] [Google Scholar]
- Rossman K. L., Der C. J., Sondek J., 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6: 167–180. [DOI] [PubMed] [Google Scholar]
- Sanematsu F., Hirashima M., Laurin M., Takii R., Nishikimi A., et al. , 2010. DOCK180 is a Rac activator that regulates cardiovascular development by acting downstream of CXCR4. Circ. Res. 107: 1102–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Martínez E., Soplop N. H., Kramer S. G., 2006. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc. Natl. Acad. Sci. USA 103: 12441–12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Martínez E., Soplop N. H., Patel R., Kramer S. G., 2008. Repulsion by Slit and Roundabout prevents Shotgun/E-cadherin-mediated cell adhesion during Drosophila heart tube lumen formation. J. Cell Biol. 182: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., 2013. Dock protein family in brain development and neurological disease. Commun. Integr. Biol. 6: e26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Irvine K. D., 2012. Drosophila as a model for understanding development and disease. Dev. Dyn. 241: 1–2. [DOI] [PubMed] [Google Scholar]
- Soplop N. H., Patel R., Kramer S. G., 2009. Preparation of embryos for electron microscopy of the Drosophila embryonic heart tube. J. Vis. Exp. 34: pii: 1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg P. S., 2006. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 12: 895–904. [DOI] [PubMed] [Google Scholar]
- Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., et al. , 1998. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 393: 591–594. [DOI] [PubMed] [Google Scholar]
- Tao Y., Schulz R. A., 2007. Heart development in Drosophila. Semin. Cell Dev. Biol. 18: 3–15. [DOI] [PubMed] [Google Scholar]
- Tetlow, A. L., and F. Tamanoi, 2013 The Ras superfamily G-proteins. Enzymes 33 Pt A: 1–14. [DOI] [PubMed]
- Ueda S., Negishi M., Katoh H., 2013. Rac GEF Dock4 interacts with cortactin to regulate dendritic spine formation. Mol. Biol. Cell 24: 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajnik V., Paulding C., Sordella R., McClatchey A. I., Saito M., et al. , 2003. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell 112: 673–684. [DOI] [PubMed] [Google Scholar]
- Yan D., Li F., Hall M. L., Sage C., Hu W. H., et al. , 2006. An isoform of GTPase regulator DOCK4 localizes to the stereocilia in the inner ear and binds to harmonin (USH1C). J. Mol. Biol. 357: 755–764. [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang Z., Roe S. M., Marshall C. J., Barford D., 2009. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science 325: 1398–1402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.