Abstract
The p63 transcription factor, homolog to the p53 tumor suppressor gene, plays a crucial role in epidermal and limb development, as its mutations are associated to human congenital syndromes characterized by skin, craniofacial and limb defects. While limb and skin-specific p63 transcriptional targets are being discovered, little is known of the post-translation modifications controlling ΔNp63α functions. Here we show that the p300 acetyl-transferase physically interacts in vivo with ΔNp63α and catalyzes its acetylation on lysine 193 (K193) inducing ΔNp63α stabilization and activating specific transcriptional functions. Furthermore we show that Fibroblast Growth Factor-8 (FGF8), a morphogenetic signaling molecule essential for embryonic limb development, increases the binding of ΔNp63α to the tyrosine kinase c-Abl as well as the levels of ΔNp63α acetylation. Notably, the natural mutant ΔNp63α-K193E, associated to the Split-Hand/Foot Malformation-IV syndrome, cannot be acetylated by this pathway. This mutant ΔNp63α protein displays promoter-specific loss of DNA binding activity and consequent altered expression of development-associated ΔNp63α target genes. Our results link FGF8, c-Abl and p300 in a regulatory pathway that controls ΔNp63α protein stability and transcriptional activity. Hence, limb malformation-causing p63 mutations, such as the K193E mutation, are likely to result in aberrant limb development via the combined action of altered protein stability and altered promoter occupancy.
Introduction
The p63 transcription factor, highly related to the p53 and p73 transcription factors, plays a central role during development of the embryonic ectoderm and derived structures. p63 is expressed in the embryonic ectoderm and in the proliferating stem cells of the adult epidermis, breast and oral epithelium (1,2). Accordingly, p63 null mice show lack of epidermis stratification which causes death at birth, absence of nails and hairs, sweat and mammary glands and severe defects in limb and craniofacial development (3,4).
The limb defects observed in p63−/− mice are highly reminiscent of ectrodactily found in patients affected by the Ectrodactily-Ectodermal Dysplasia-Cleft palate syndrome (EEC) or in non-syndromic Ectrodactily, also known as Split-Hand Foot Malformation (SHFM) type-IV. Ectrodactily is characterized by the absence of the central rays of the limbs, resulting in a deep medial cleft, missing or hypoplastic central fingers and fusion of the remaining ones (5–8), and has been associated with developmental failure of the Apical Ectodermal Ridge (AER) a transitory pluri-stratified ectodermal region required for limb outgrowth, and for the expression of key signaling molecules (1,2,5–9).
p63 is at the center of a complex molecular network. However, its regulation and tissue distribution remain issues not fully understood. The p63 gene encodes for at least ten protein isoforms, which differ in their amino and carboxy-terminal regions as a consequence of alternative transcription start site and alternative splicing, respectively (10,11), with ΔNp63α being the most expressed isoform in the embryonic ectoderm. All p63 isoforms share with p53 and p73 homology in the DNA binding and the oligomerization domains (12–15), and indeed p53 and p63 regulate a number of common transcriptional targets, in particular those related to cell-cycle control. However, p63-specific target genes are known that justify its specific role in ectoderm development and epidermis stratification, and also explain the specific set of human diseases associated with p63 mutations (16–18).
Interestingly, while some mutations of the p63 gene occurring in the DNA Binding Domain (DBD) coding sequence (such as the R279H mutation) are causative of the EEC syndrome, which comprises ectrodactyly and several other skin and craniofacial developmental defects, others (such as the K193E mutation) result in non-syndromic ectrodactyly (or SHFM-type IV), with little or no skin/craniofacial anomalies (7,8). The logical question that arises is: why the EEC- and the SHFM-associated mutations cause limb developmental malformations, while p63 mutations found in AEC patients (i.e. the L518F mutation), localized in the SAM domain of the ΔNp63α protein, do not affect limb development? One possibility is related to the ability of the peptidyl-prolyl isomerase Pin1 to negatively regulate ΔNp63α stability, and to the activity of the key limb morphogen Fibroblast Growth Factor-8 (FGF8) (19–22) to counter-act this function (23). Mutant p63 proteins are differentially sensitive to Pin1-induced degradation (23). However, the correlation between specific p63 mutations, their stability, transcriptional activity and the onset of limb developmental anomalies remains not fully resolved.
It is becoming increasingly evident that the distinct functions of wild-type and mutant p63 protein(s) might reside not only in their specific DNA binding activity but also in their post-translational modifications such as sumoylation, phosphorylation and ubiquitylation (24–27). These modifications modulate ΔNp63α half-life, the specificity and efficiency of protein–protein interactions and overall modulate the transcriptional activity of the protein. The elucidation of these ‘upstream’ regulatory events is required for a full comprehension of the molecular network centered on p63, to explain the genotype–phenotype correlations observed in patients affected by syndromes associated to p63 mutations.
p53 and/or p73 protein activity and stability are finely regulated by several post-translational modifications (28–30), among which acetylation seems to play a pivotal role in regulating their biological functions (29,31–33). Acetylation is a reversible modification, catalyzed by histone acetyl-transferases, of lysine residues of a target protein and its function in transcriptional activation is well accepted (34). p73 is acetylated by p300 on lysines located in the DNA binding and oligomerization domains in response to DNA damage (35); acetylation enhances p73 ability to bind and activate proapototic target genes (36). Furthermore, p73–p300 interaction requires the activity of Pin1 that induces p73 conformational changes upon phosphorylation by the tyrosine kinase c-Abl (37). Acetylation of p53 is enhanced in response to DNA damage and well correlates with p53 stabilization and activation: indeed, acetylation of p53 antagonizes the MDM2 ubiquitin-ligase activity that keeps p53 protein at low levels in normal conditions. Moreover, acetylation of p53 by p300 was found to promote its sequence-specific DNA binding (31–33,38).
All considered, we set forth to examine ΔNp63α acetylation in the context of naturally occurring ΔNp63α missense mutations associated to SHFM-IV: one such mutation causes lysine 193 substitution with glutamic acid (K193E) (7,8). We noted that lysine K164 in p53, acetylated by p300 (38), correspond to K193 in ΔNp63α. Thus we raised the hypothesis that wild-type ΔNp63α could be acetylated by p300 on K193, and that mutations of this residue could prevent this post-translational modification with important developmental consequences.
Our results are consistent with this hypothesis and, for the first time, we show that FGF8 signaling participates in a regulatory pathway promoting the physical interaction of ΔNp63α with c-Abl and p300, leading to stabilization and transcriptional activation of ΔNp63α.
Results
Δnp63α is acetylated and stabilized in cultured cells
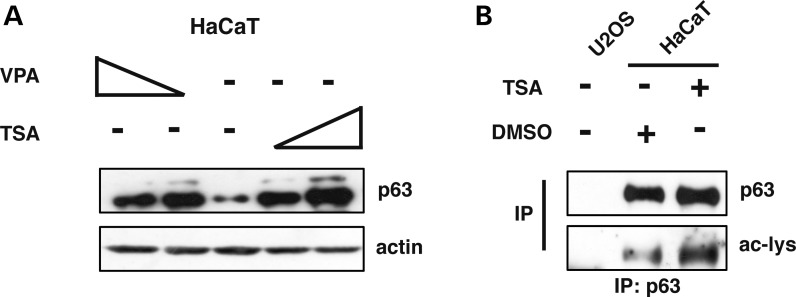
In order to assess whether p63 could be acetylated in human cells, we treated the human keratinocytes HaCaT cell line, expressing endogenous ΔNp63α, with Valproic-Acid (VPA), which selectively inhibits class I deacetylases, or with Trichostatin-A (TSA) which inhibits class I and II deacetylases. VPA and TSA treatments resulted in an increase in ΔNp63α abundance (Fig. 1A). Similar effects of ΔNp63α accumulation were also obtained when ΔNp63α was ectopically overexpressed in U2OS cells, a human osteosarcoma cell line devoid of endogenous p63 expression (Supplementary Material, Fig. S1). Then, we performed immunoprecipitation of endogenous ΔNp63α from total protein extracts of HaCaT cells treated with TSA. The level of ΔNp63α acetylation was detected using an antibody against acetylated lysines: we observed that ΔNp63α is found acetylated at a basal level, as previously reported (39), and that its acetylation increased upon TSA treatment (Fig. 1B). These results show that the ΔNp63α protein is acetylated in human cells and that the acetylation levels of ΔNp63α correlate with its accumulation in human cells following deacetyl-transferases inhibition.
Figure 1.
The ΔNp63α protein is acetylated in human keratynocytes. (A) Western Blot (WB) analysis of whole HaCaT cell extracts treated with increasing amounts of TSA (5 ng/ml and 10 ng/ml) for 5 h or VPA (0.5 mm and 1 mm) for 3 h. (B) Whole cell extracts from HaCaT cells treated with 5 ng/ml of Trichostatin (TSA) for 5 h were analyzed by immunoprecipitation of endogenous ΔNp63α with an anti-p63 antibody followed by WB analysis with an anti-acetylated lysines. U2OS cells, not expressing p63, were used as negative control.
The acetyl-transferase domain of p300 is required to induce ΔNp63α protein stabilization
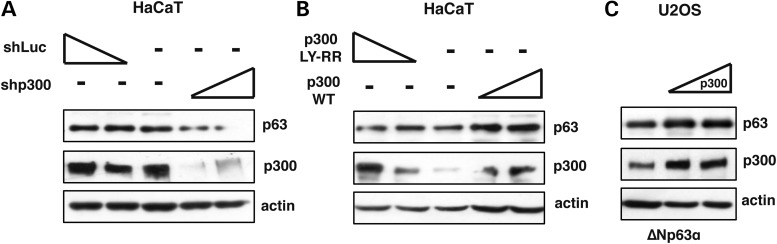
Acetylation of p53 and p73 proteins is required for their stabilization and transcriptional activation in response to DNA damage (31–33,35–38) and the p300 acetyl-transferase is known to be involved in this process (29,35,36,38). To determine whether p300 could acetylate ΔNp63α, we silenced endogenous p300 in HaCaT cells by transfecting a p300-specific shRNA plasmid. Depletion of p300 was clearly detected and, concomitant with p300 reduction, a significant decrease of ΔNp63α was also observed (Fig. 2A). Conversely, when p300 protein levels were increased by transient overexpression in HaCaT or U2OS cells, ΔNp63α protein was stabilized in a dose-dependent manner (Fig. 2B and C). In contrast, a construct expressing a mutated variant of p300, with a mutation affecting the Histone Acetyl-Transferase domain (LY-RR) (36), failed to stabilize endogenous ΔNp63α in HaCaT cells (Fig. 2B). These data clearly indicate that p300 and its acetyl-transferase activity are required for ΔNp63α protein levels regulation.
Figure 2.
The acetyl-transferase domain of p300 is required to induce ΔNp63α protein stabilization. (A) WB analysis of whole HaCaT cell extracts transiently transfected with increasing amounts (20 ng and 40 ng) of shp300 or shLuc expression vectors (B) WB analysis of HaCaT whole cell extracts transiently co-transfected with equal amount of ΔNp63α expression vectors (30 ng) and increasing amounts of p300 encoding plasmids (p300 (WT) or p300-LY-RR, mutated in the HAT domain (10 and 20 ng). (C) WB analysis of U2OS whole cell extracts transiently co-transfected with equal amount of ΔNp63α expression vectors (30 ng) and increasing amounts of p300 expression vectors (10 and 20 ng).
Accordingly, when we overexpressed ΔNp63α with p300 in U2OS cells and treated the cells with the protein synthesis inhibitor Cycloheximide (CHX), we observed an increase of ΔNp63α protein half-life (Fig. 3D). We also determined the effect of p300 silencing on ΔNp63α protein half-life in HaCaT cells, by transfecting p300-specific shRNA plasmid and treated the cells with CHX. As shown in Supplementary Material, Figure S2, the levels of ΔNp63α protein decreased upon p300 silencing with only a modest decrease of ΔNp63α half-life upon CHX addition.
Figure 3.
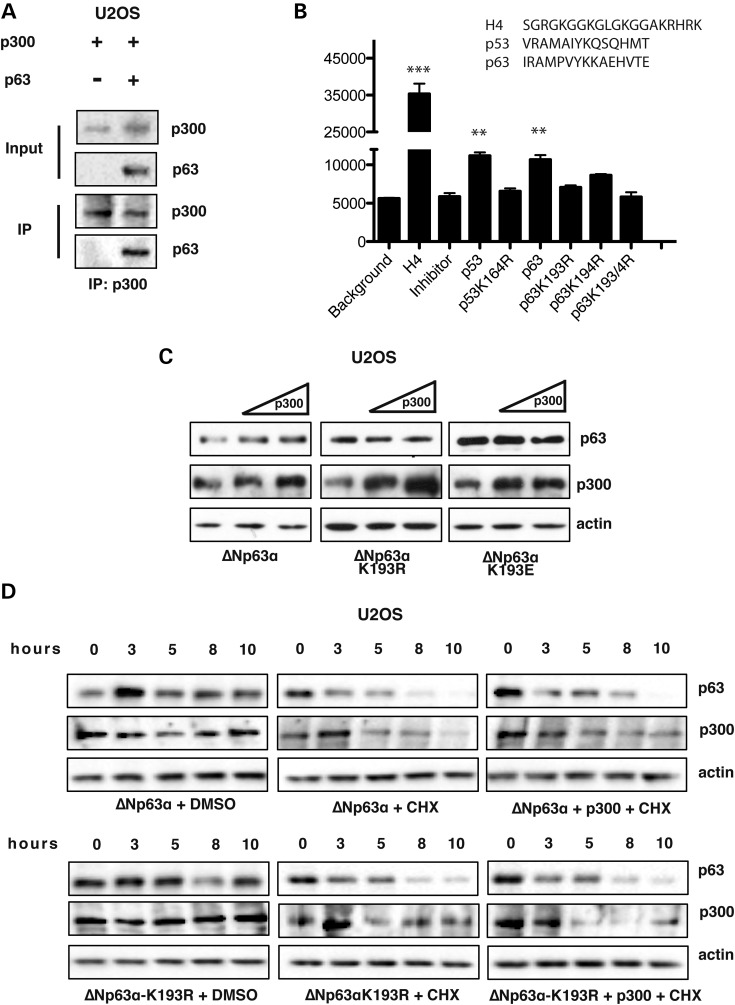
p300 interacts with ΔNp63α in human cells and catalizes in vitro acetylation of lysine K193. (A) U2OS whole cell extracts transiently co-transfected with ΔNp63α and p300 were analyzed by immunoprecipitation with an anti-p300 antibody followed by WB analysis with an anti-p63 antibody. U2OS cells, not transfected with p63 encoding plasmid were used as negative control. (B) In vitro acetylation assay performed according to the HAT assay kit protocol (Active Motif) with an H4 peptide and p53 peptides as positive controls, H4 plus anacardic acid 15 μM (an inhibitor of acetyl-transferase activity used as a negative control) and p63 peptides (peptide sequences are indicated). (C) WB analysis of U2OS whole cell extracts transiently co-transfected with ΔNp63α, ΔNp63α-K193E, ΔNp63α-K193R expression vectors (30 ng) and increasing amounts of p300 encoding plasmid (10 and 20 ng). (D) WB analysis of U2OS whole cell extracts transiently co-transfected with ΔNp63α, ΔNp63α-K193R and p300 expression vectors (30 ng and 5 ng respectively). 24 h after transfection protein half-life was measured by treating cells with 10 μg/ml of CHX.
P300 interacts with ΔNp63α in human cells and catalyzes in vitro acetylation of lysine K193
To assess whether the observed stabilization of ΔNp63α by p300 was due to a direct interaction between the two proteins, we performed co-immunoprecipitation experiment in U2OS cells. As shown in Figure 3A, ΔNp63α was found in p300 immuno-complexes, showing that the two proteins can associate in human cells.
Lysine K164 of the p53 protein, conserved in p63 and p73, is acetylated by p300 (38). In the ΔNp63α protein this residue corresponds to K193 (Supplementary Material, Fig. S3), mutated in patients affected by SHFM-IV (i.e. K193E) (7,8). We set forth to establish whether K193 was acetylated by p300, by carrying out in vitro acetylation assays using recombinant p300 protein and a set of synthetic p63 peptides centered on K193. A p53 synthetic peptide containing lysine K164 known to be acetylated by p300 (38) was used as a positive control. The results show that the p63 peptide centered on K193 was acetylated in vitro and the levels of acetylation are similar to those obtained with the p53 peptide (Fig. 3B).
In the same assay we also analyzed mutant p63 peptides carrying K193 and K194 substitutions into arginine, either one at a time or simultaneously, to determine which one (or both) could be target of the p300 acetyl-transferase activity. As shown in Figure 3B, p300 acetylates lysine K193: indeed the levels of acetylation of the p63-K193R mutant were reduced. However we cannot exclude that also K194 could be acetylated by p300 since we observed a modest decrease in the level of acetylation of the p63-K194R mutant peptide compared with the wild-type peptide. Finally, p300 overexpression in U2OS cells did not induce stabilization of the ΔNp63α-K193R and of the natural ΔNp63α-K193E mutant, while the ΔNp63α protein was stabilized (Fig. 3C), indicating that the integrity of K193 is required to induce p300-dependent stabilization of ΔNp63α. Moreover, in contrast to what observed for the wild-type ΔNp63α, the half-life of the ΔNp63α-K193R mutant was not enhanced upon p300 overexpression in U2OS cells (Fig. 3D).
FGF8 positively regulates ΔNp63α protein stability inducing its interaction with c-Abl and promoting ΔNp63α acetylation
During embryonic development, FGF8 acts as a signaling peptide essential for growth, morphogenesis and patterning of the limb buds (9,19–22). We have recently shown that FGF8 exerts a stabilizing function on the ΔNp63α protein, by preventing its interaction with Pin1 that targets ΔNp63α protein for proteasomal degradation (23). We raised the hypothesis that FGF8 may stabilize the ΔNp63α protein, via p300-mediated acetylation of ΔNp63α, and that the limb malformation-associated p63 K193E mutation may pose an obstacle to this regulation.
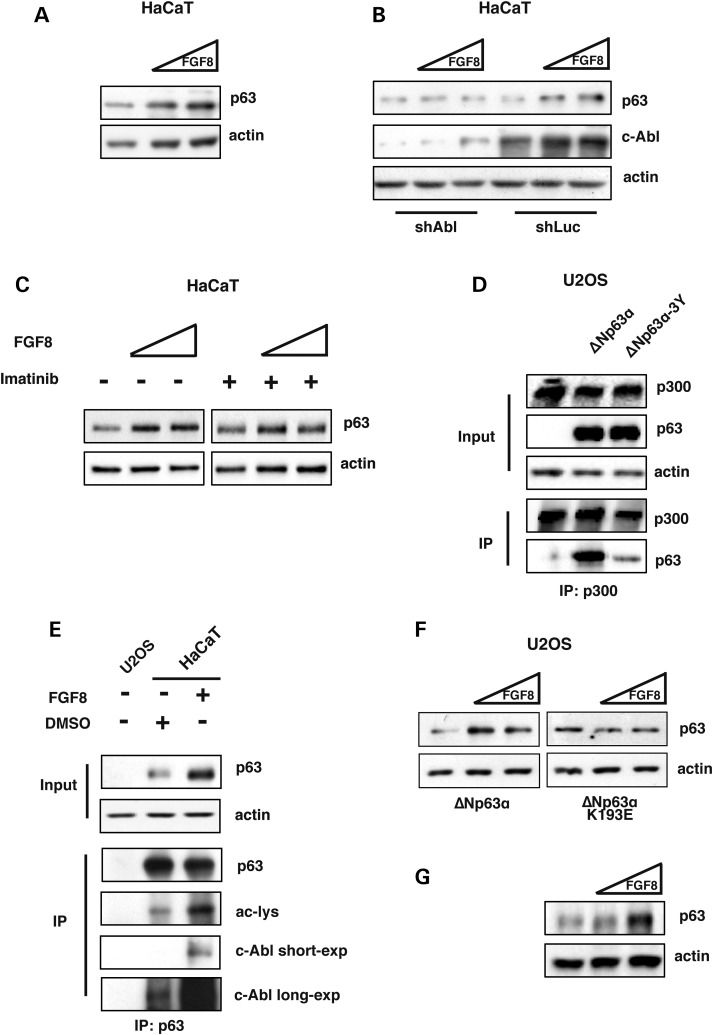
First, we treated HaCaT cells with increasing amounts of soluble FGF8 that resulted in efficient ΔNp63α protein stabilization as expected (Fig. 4A).
Figure 4.
FGF8 positively regulates ΔNp63α protein stability inducing its interaction with c-Abl and promoting ΔNp63α acetylation. (A) WB analysis of HaCaT whole cell extracts treated with increasing amounts of FGF8 (0.5 ng/ml and 1 ng/ml) for 3 h. (B) WB analysis of HaCaT whole cell extracts stably transfected with an shRNA against c-Abl or shLuc plasmids, treated with increasing amounts of FGF8 (0.5 ng/ml and 1 ng/ml) for 3 h. (C) WB analysis of HaCaT cells treated with increasing amounts of FGF8 (0.5 ng/ml and 1 ng/ml) or pre-treated for 30 min with Imatinib (10 μM) followed by FGF8 treatment for 3 h. (D) U2OS whole cell extracts transiently co-transfected with either ΔNp63α or ΔNp63α-3Y (10 μg) and p300 (5 μg), and then analyzed by immunoprecipitation with an anti-p300 antibody followed by WB analysis with an anti-p63 antibody. (E) HaCaT whole cell extracts treated with FGF8 (0.5 ng/ml) or DMSO for 3 h were analyzed by immunoprecipitation with anti-p63 antibodies followed by WB analysis with the indicated antibodies. U2OS cells, not expressing p63 were used as negative control. (F) WB analysis of U2OS whole cell extracts transiently transfected with ΔNp63α or ΔNp63α-K193E encoding plasmids (30 ng). 24 h after transfection U2OS cells were treated with increasing amounts of FGF8 for 2 h (0.5 ng/ml and 1 ng/ml). (G) WB analysis of total proteins extracts from forelimbs isolated from wild-type mouse embryos at E11.5, cultured whole-mount for 48 h in the absence or presence of recombinant FGF8 (0.5 μg/ml and 1 μg/ml).
One of the down-stream effector of FGFs is the tyrosine kinase c-Abl: indeed, c-Abl is activated by FGF2 treatment (40). c-Abl is also a key regulator of the p53 family members (29,37,41–45). To verify whether c-Abl was required to induce the observed FGF8-mediated stabilization of ΔNp63α, we stably silenced endogenous c-Abl expression in HaCaT cells and then treated these cells with either FGF8 or FGF2. c-Abl silencing abolished ΔNp63α stabilization induced by either FGF8 (Fig. 4B) or FGF2 (data not shown), suggesting that FGFs stabilization of ΔNp63α requires the presence of the c-Abl protein. In order to verify whether the tyrosine kinase activity of c-Abl was required for the FGF8-mediated stabilization of ΔNp63α, we treated HaCaT cells with Imatinib, an inhibitor of c-Abl tyrosine kinase activity. As shown in Figure 4C, FGF8-mediated stabilization of ΔNp63α was prevented by Imatinib pre-treatment. Furthermore, the ΔNp63α-3Y mutant protein, with the three tyrosines known to be phopshorylated by c-Abl mutated into phenylalanin (44,45), was not stabilized by FGF8 treatment (Supplementary Material, Fig. S4).
To verify whether c-Abl was promoting the interaction between ΔNp63α and p300, we performed co-immunoprecipitation experiments of p300 with wild-type ΔNp63α or with the ΔNp63α-3Y mutant. As shown in Figure 4D, the ΔNp63α-3Y mutant displayed a drastically reduced interaction with p300 compared with wild-type ΔNp63α. Furthermore p300 overexpression did not modulate ΔNp63α-3Y protein levels (Supplementary Material, Fig. S5).
In order to verify whether FGF8, c-Abl and p300 were linked together in the same regulatory pathway, promoting ΔNp63α stabilization, we treated HaCaT cells with FGF8 and performed co-immunoprecipitation of ΔNp63α; we observed a great increase in ΔNp63α-c-Abl interaction and in the levels of ΔNp63α acetylation upon FGF8 treatment (Fig. 4E). Interestingly, we found that the signaling cascade activated by FGF8 was not active on the SHFM-IV-causing ΔNp63α-K193E mutant protein. Indeed as shown in Figure 4F, FGF8 treatment in U2OS cells did not induce ΔNp63α-K193E stabilization, clearly resembling the results obtained by p300 overexpression on this mutant (Fig. 3C). All these results indicate that c-Abl and p300 are linked together in a cascade, activated by FGF8, regulating ΔNp63α protein stability.
To verify if FGF8 and c-Abl are required to modulate not only ΔNp63α protein stability, but also its transcriptional activity, we performed luciferase assay in U2OS cells transiently transfected with the DLX5 promoter, a known ΔNp63α transcriptional target in the AER cells of developing limbs (46). FGF8, Imatinib and Imatinib followed by FGF8 treatment were used. Interestingly, when we inhibited c-Abl kinase activity, ΔNp63α was unable to transactivate the DLX5 promoter even in the presence of FGF8 treatment (Supplementary Material, Fig. S6). These data suggest that the c-Abl kinase activity was required to transduce the signal induced by FGF8 leading to ΔNp63α stabilization and transcriptional activation. Similar results were also obtained with the EGFR promoter (18, data not shown).
Finally, in order to assess if such mechanism could be relevant in vivo, e.g. the developing limb bud, we adopted an ex vivo method by culturing the embryonic limb buds obtained from wild-type mouse embryos at the age E10.5, and maintained whole-mount for 48 h (47). During the culture time, purified recombinant FGF8 was added to the medium at physiological doses, then the tissues were collected and analyzed by Western blot analyses for the abundance of ΔNp63α protein. Compared with untreated limbs, addition of FGF8 resulted in a clear accumulation of the ΔNp63α protein (Fig. 4G), indicating that FGF8 efficiently stabilizes, and most likely activates, ΔNp63α in the context of the limb embryonic tissue.
The K193E mutation alters ΔNp63α transcriptional activity in a promoter-specific manner
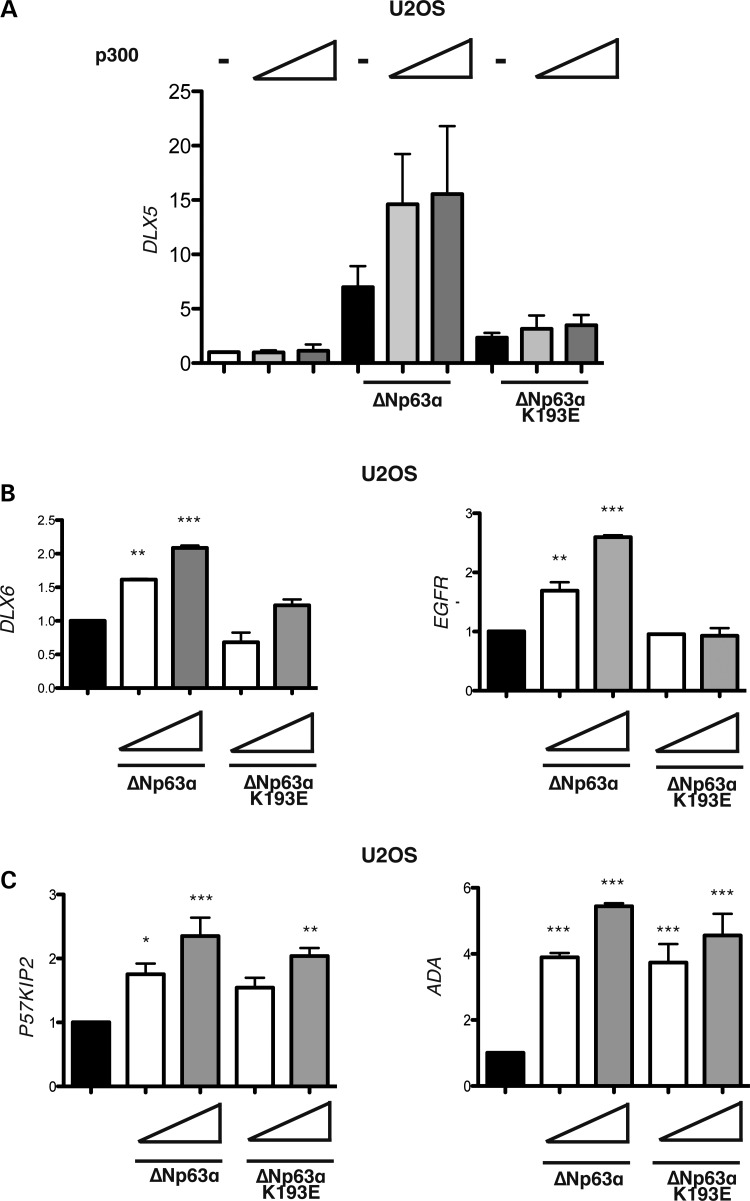
In order to verify whether p300 could act as a ΔNp63α co-activator we performed luciferase-reporter assays with the DLX5 promoter. Interestingly, we observed that p300 co-transfection greatly enhanced the transcriptional activity of ΔNp63α, while the transcriptional activity of the ΔNp63α-K193E mutant could not be enhanced by p300 overexpression (Fig. 5A).
Figure 5.
The K193E mutation alters ΔNp63α transcriptional activity in a promoter-specific manner. (A) Luciferase assay performed on U2OS cells transiently co-transfected with the −1200 bp DLX5 promoter (200 ng) in the presence of ΔNp63α or ΔNp63α-K193E (50 ng) with increasing amounts of p300 (10 and 20 ng) expression vectors. Each histogram bar represents the mean of three indipendent transfection duplicates. Standard deviation are indicated. (B) Luciferase assay performed in U2OS cells transiently co-transfected with the DLX6, and EGFR reporter promoters (200 ng) in the presence of increasing amounts of ΔNp63α or ΔNp63α-K193E (50 and 100 ng) plasmids. (C) Luciferase assay performed on U2OS cells transiently co-transfected with the p57kip2, and ADA reporter promoters (200 ng) in the presence of increasing amounts of ΔNp63α or ΔNp63α-K193E (50 and 100 ng) plasmids. For (A–C) cells were lysed 24 h after transfection and luciferase activity was determined. The basal activity of the reporter plasmid was set to 1. Data are presented as fold activation/repression relative to the sample without effector. Each histogram bar represents the mean of three independent transfection duplicates. Standard deviations are indicated.
ΔNp63α transcriptional activity was impaired by the K193E mutation also on other ΔNp63α target genes involved in development, EGFR and DLX6 (Fig. 5B) (18,46). We then examined whether the K193E mutation could alter ΔNp63α transcriptional activity on genes not directly required for limb development; for this aim we used the p57KIP2 and ADA promoters, known to be involved in p63-dependent cell-cycle regulation (48,49). Interestingly, we found that the ΔNp63α-K193E mutant behaved as the wild-type ΔNp63α protein on both promoters (Fig. 5C), suggesting that the K193E mutation selectively alters ΔNp63α transcriptional activity.
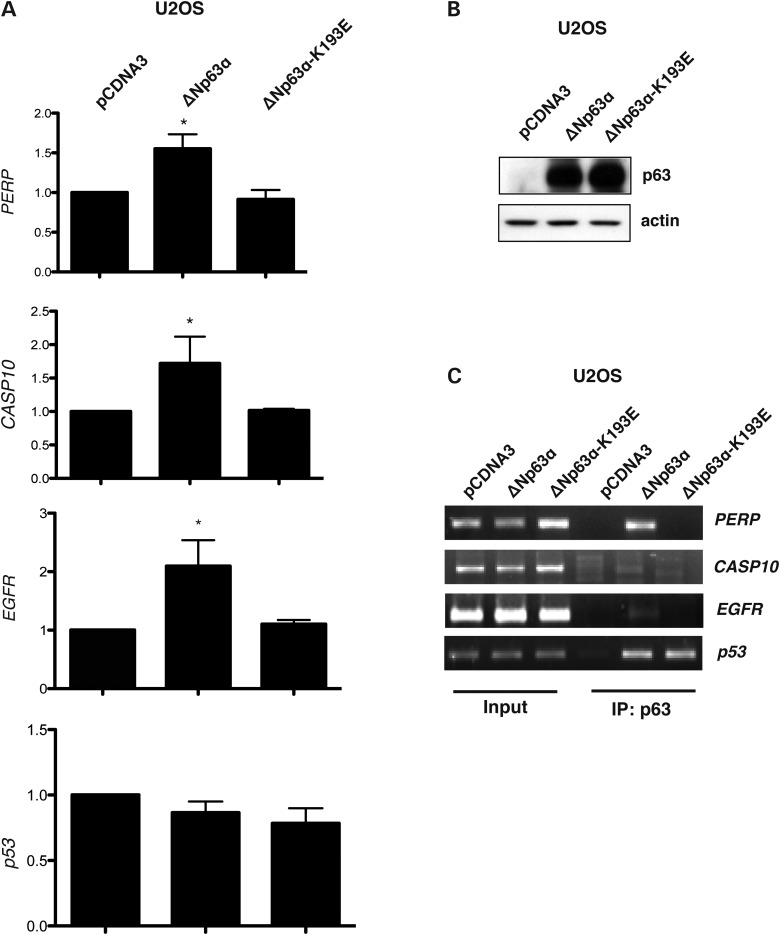
To further characterize the transcriptional activity of the ΔNp63α-K193E mutant, we performed real-time, quantitative qPCR analyses in U2OS cells stably transfected with either the wild-type ΔNp63α or the ΔNp63α-K193E expression vectors. Interestingly, we confirmed that the ΔNp63α-K193E mutant over-expression results in altered expression of ΔNp63α target genes involved in development and apoptosis such as PERP, CASP10, EGFR (18,50) while it behaves like the wild-type ΔNp63α on p53 (Fig. 6A). Taken together, these data clearly show that the K193E mutation alters the transcriptional activity of ΔNp63α in a gene-specific manner.
Figure 6.
The ΔNp63α-K193E mutant displays an altered DNA binding activity and transcriptional activity on developmental related genes. (A) Expression of CASP10, EGFR, PERP and p53 was analyzed by Real-Time qPCR in U2OS cells stably transfected with pCDNA3 (empty vector), ΔNp63α or ΔNp63α-K193E cDNAs. (B) ΔNp63α and ΔNp63α-K193E proteins expression was confirmed by WB analysis. (C) Cells used in A and B were subjected to ChIP analysis, and the recovered chromatin was amplified with PERP, EGFR, p53 and CASP10-specific primers.
Next we tested whether the ΔNp63α-K193E mutant displayed altered DNA binding ability by Chromatin Immunoprecipitation (ChIP) assay of U2OS cells stably transfected with the wild-type ΔNp63α or with the ΔNp63α-K193E expressing vectors; the proteins were correctly expressed (Fig. 6B). We observed that the ΔNp63α-K193E mutant was not efficiently recruited on the Responsive Elelments (RE) of genes relevant for developmental and apoptotic processes (PERP, CASP10 and EGFR) while it was normally recruited on RE of the p53 gene, involved in cell cycle regulation (Fig. 6C).
In conclusion, the K193E mutation alters the ability of ΔNp63α to bind specific RE sequences resulting in altered transcriptional regulation of genes involved in the regulation of development and apoptotic processes.
Discussion
The p63 transcription factor is emerging as a master regulator of development and differentiation of ectoderm derived cells and tissues. In the last few years much attention has been paid to the analysis and identification of p63 transcriptional targets, their tissue and process specificity and how mutations in p63 affect its down-stream transcriptional properties (16–18,51,52). Clearly, this is only part of the full story. Indeed, more recently several p63 post-translational modifications have been recognized, acting either during response to DNA damage, differentiation or embryonic development (24–27). The full spectrum of these modifications, likely able to regulate stability and activity of the ΔNp63α protein, are not fully understood.
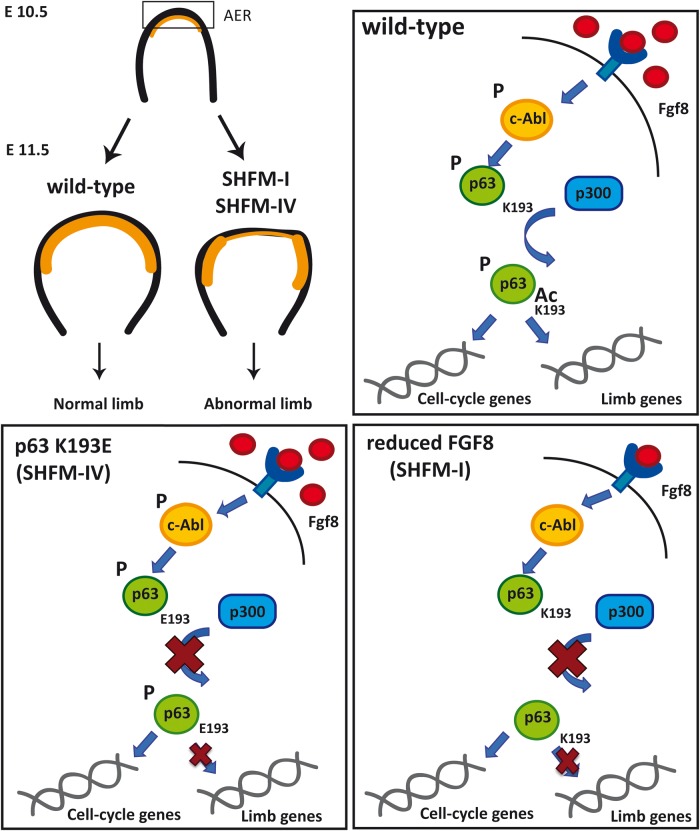
Here we report that FGF8, c-Abl, p300 and ΔNp63α are functionally linked in a molecular pathway modulating ΔNp63α activity and stability. Our data show that treatments with FGF8, a signaling molecule essential for limb outgrowth and patterning, result in increased ΔNp63α protein stability, both in cultured cells and in embryonic mouse limb buds ex vivo. Based on these data and previous findings from our team (23), we propose a model in which FGF8 promotes the interaction of c-Abl and ΔNp63α, and that this interaction is required for the consequent association of ΔNp63α with p300, leading to ΔNp63α acetylation (scheme in Fig. 7). When such acetylation is inefficient, due to reduced FGF8 expression or to mutation of the p300 target lysine K193 in ΔNp63α, limb developmental defects ensue.
Figure 7.
FGF8 positively regulates ΔNp63α protein stability in mice embryonic limb buds. FGF8, c-Abl and p300 are component of a regulatory pathway that leads to ΔNp63α stabilization and transcriptional activation in embryonic limb buds. Exposure of AER cells to FGF8 induces a signaling intracellular cascade that activates c-Abl causing ΔNp63α phosphorylation on tyrosine residues. This phosphorylation event is indispensable for the interaction of ΔNp63α with the p300 acetyl-transferases; acetylation of ΔNp63α result in its stabilization and transcriptional activation. In the absence of FGF8, or in the presence of p63 mutations, like the SHFM-associated K193E mutation, this signaling pathway is not active leading to improper expression of genes involved in limb development. This pathway could be relevant for correct AER stratification (in the scheme marked in yellow) ensuring correct limb outgrowth.
Based on our data, p300 appears to be an important regulator of ΔNp63α function during limb development, and in particular the results point to the possibility that p300 is required to selectively induce and activate with ΔNp63α a set of genes required to warrant correct limb development. No direct evidence of this is available, in fact the disruption of the p300 gene in the mouse model is embryonic lethal and p300−/− mice arrest their development prior to the limb bud stage (53). Conversely, the role of acetylation and deacetylation on ΔNp63α are better known, indeed mice double knock-out for Histone Deacetylase-1 and -2 (HDAC1/HDAC2) display developmental limb malformations similar to those observed in p63 null mice (54). HDAC1 and HDAC2 mediate the repressive function of ΔNp63α on some of its transcriptional targets (like 14-3-3σ, p16/Ink4a, p19/ARF) whose down-modulation is essential to ensure correct development (54). On the other hand, it's possible to speculate that Histone acetyl-transferases, such as p300, are needed to activate p63 target gene expression in concert with ΔNp63α. Indeed, luciferase-reporter assays indicated that p300 acetylation on K193 of ΔNp63α is required to guarantee an efficient transcription of genes involved in limb development, such as human DLX5 and DLX6 (46,55) (Fig. 5A, B).
We show here that lysine K193 of ΔNp63α is acetylated by p300, in human cells and in vitro.
This has important implications in the pathogenesis of the SHFM-IV syndrome, since this residue is found mutated into glutamic acid (K193E) in patients affected by this syndrome (7,8). Indeed, we found that the K193E mutant ΔNp63α protein is unable to activate p63 target genes required for developmental (such as PERP, EGFR) or apoptotic processes (CASP10). Indeed, programed cell death and cell differentiation are relevant to ensure correct shape of the limb (56). In particular, at early stages of limb development, the anterior and posterior necrotic zones are essential regions regulating the number of digits (57). On the other hand, the ΔNp63α-K193E mutant correctly induced the expression of genes connected to cell-cycle regulation (such as p53 and p57KIP2) efficiently as the wild-type ΔNp63α protein. We found that, the altered transactivation activity of mutant ΔNp63α-K193E on target genes involved in developmental or apoptotic processes, is possibly due to a significant decrease in the DNA binding activity on the RE of such promoters, while this natural mutant was found to bind normally to the promoter of the p53 gene. However, it is not clear how this mutation could alter the binding of ΔNp63α in a promoter-specific manner.
The role of FGF signaling in the SHFM malformation has been partly clarified. It is well established that FGF10 and FGF8 signaling are essential for AER induction and maintenance and that FGF8, expressed by the AER cells, is the key morphogen for limb bud outgrowth and patterning. Indeed FGF8 knock-out mice display severe defects in skeletal and limb development (19–22). The complete loss of p63, or the knock-in mouse model for the R279H mutation, associated to the EEC syndrome, leads to an evident downregulation of FGF8 expression in the AER cells (3,4,9,46 and data not shown). Likewise, FGF8 is downregulated in the AER of embryos carrying the combined loss of Dlx5; Dlx6; two transcription factors causally implicated in SHFM type-I (55). Dlx5 and Dlx6 proteins co-localize with ΔNp63α in the AER cells and are direct ΔNp63α targets (46). Hence, the emerging picture is that FGF8 serves a double function, (a) a morphogen driving limb growth and patterning, via its actions on mesenchymal cells (paracrine) and AER cells (autocrine), (b) as stabilizer of ΔNp63α, to ensure the transitory stratification of the AER cells and the expression of limb-related p63 target genes.
In summary, the work presented here sheds new light on an important regulatory loop activated by FGF8 essential for ΔNp63α activation and stabilization in cell cultures and in mice limb buds and on the molecular mechanism that could be at the bases of the SHFM-IV pathogenesis.
Experimental procedures
Plasmids
All expression vectors encoding ΔNp63α wild-type and mutant proteins, p300 cDNAs, c-Abl and shAbl have been previously described (36,58). The shRNA against p300 (shp300) and control shRNA (shLuc) were purchased from Origene.
Cell culture and transfection
U2OS and HaCaT cells were kept in DMEM supplemented with 10% FBS (Euroclone) at 37°C in a humified atmosphere of 5% (v/v) CO2 in air.
For transient transfection, 50 000 cells were seeded into 24-multiwell plates and on the next day transfected with Lipofectamine 2000 (Invitrogen) or Lipofectamine LTX (Invitrogen) for HaCaT cell, under the conditions suggested by the manufacturer. Transfection efficiency was checked by transfection of β-gal or GFP expression vectors. The total amount of transfected DNA (500 ng for 50 000 cells) was kept constant using empty vector as necessary.
For stable transfection 300 000 HaCaT or U2OS cells were plated in six wells and on the next day, HaCaT cells were transfected with 3 μg of shAbl or 3 μg of shLuc using Lipofectamine LTX (Invitrogen). After 24 h, cells were trypsinized and plated in a medium containing puromycin (0.8 μg/ml; Sigma). After 8 days of selection, clones were pooled and kept in puromycin. U2OS cells were transfected with 3 μg of ΔNp63α or ΔNp63α-K193E using Lipofectamine LTX (Invitrogen). After 24 h, cells were trypsinized and plated in medium containing Neomicin (G-418, 600 μg/ml). After 3 weeks of selection, clones were pooled and kept in Neomicinat 300 μg/ml.
U2OS and HaCaT cells were treated with 0.5 or 1 mm Valproic-Acid (VPA), 5 ng/ml or 10 ng/ml Trichostatin (TSA), 0.5 or 1 ng/ml FGF8 or FGF2, 10 μM CHX for the indicated times. For FGF2 or FGF8 treatments, cells were starved for 12 h before treatments using DMEM supplemented with 0.5% of FBS. HaCaT and U2OS cells were treated with 5 μM or 10 μM Imatinib, (Sigma) for the indicated times.
Western blot and antibodies
24 h after transfection, cells were lysed in 100 μl of Loading Buffer 2X (2% sodium dodecyl sulfate, 30% glycerol, 144 mm β-mercaptoethanol, 100 mm Tris–HCl pH 6.8 and 0.1% Bromo-Phenol Blue). Samples were incubated at 98°C for 10 min and resolved by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane (Protran, Millipore). The blots were incubated with the following antibodies (p63 4A4 sc-8431, Santa Cruz Biotechnology), p300 (p300 C-20 sc-585, Santa Cruz Biotechnology), c-Abl (A5844, Sigma), acetylated lysine (#9441, Cell-Signaling) and actin (A2066, Sigma). We used the following secondary antibodies: α-mouse (sc-2005, Santa Cruz Biotechnology), α-rabbit (sc-2030, Santa Cruz Biotechnology). Proteins were visualized by an enhanced chemi-luminescence method (Genespin) according to manufacturer's instructions.
Luciferase activity assay
For reporter promoter assays, cells were transiently co-transfected with the DLX5, DLX6, ADA, EGFR and p57KIP2 luciferase-reporter plasmids (23,46–48) and expression plasmids encoding for ΔNp63α, ΔNp63α-K193E and p300. Cells were seeded in 24-well plates and transfected using Lipofectamine 2000 (Invitrogen, Life Sciences). At 24 h post-transfection, cell extracts were prepared with Luciferase lysis buffer (1% Triton X-100, 25 mm Gly-Gly pH 7.8, 15 mm MgSO4, 4 mm EDTA), and the luciferase activity was measured using the Beetle Luciferin Kit (Promega Inc.) on a TD 20/20 luminometer (Turner design).
The results are expressed as relative luciferase activity after normalization with the beta-Galactosidase plasmid as internal control. Basal activity of the reporter was set to 1. Each histogram bar represents the mean of three independent transfection experiments performed in triplicate. Standard deviations are indicated.
Co-immunoprecipitation
U2OS and HaCaT cells (1.25 × 106/100 mm plate) were transfected with the indicated vectors. 24 h after transfection cells were harvested for whole-cell lysates preparation using RIPA buffer (10 mm Tris–HCl pH 8, 2 mm EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 140 mm NaCl, 1X Triton X-100, supplemented with 1 mm phenylmethylsulfonylfluoride and cocktail protease inhibitors, Sigma). Cell lysates were incubated on ice for 20 min, vortexed, then centrifuged at 6600 g for 20 min to remove cell debris. Protein concentration was determined with the Bradford Reagent (Sigma). 2 mg of cell lysates were incubated overnight at 4°C with 2 μg of anti-p63 (H-129 sc-8344, Santa Cruz Biotechnology) and anti-p300 (p300 C-20 sc-585, Santa Cruz Biotechnology). The immuno-complexes were collected by incubating with a mix of Protein A Agarose and Protein G Sepharose (Sigma) overnight at 4°C. The beads were washed three times: the first wash with RIPA buffer and the others with PBS. The beads were then re-suspended in 2X Loading buffer, heated at 98°C and loaded on a SDS polyacrylamide gel and subjected to western blotting with the indicated antibodies.
RNA extraction and real-time qPCR
For quantitative Real-Time qPCR total RNA was extracted from U2OS cells with the TRI Reagent (Sigma). 1 μg of total RNA was reverse-transcribed using SuperScriptIII cDNA Preparation Kit (Life-Technology). Real-Time quantitative PCR (qPCR) was performed with SybrGreen supermix (BIORAD). Tubulin mRNA was used for normalization. For Real-Time qPCR reaction the sequence of the primer pairs are described in Supplementary Material, Table S1.
ChIP assay
U2OS cells were used for ChIP assays performed as previously described (51). Briefly, after fixing in 1% formaldehyde, cells were lysed for 5 min in 50 mm Tris pH 8.0, 2 mm EDTA, 0.1% NP-40, 10% glycerol and supplemented with protease inhibitors (all from Sigma). Nuclei were re-suspended in 50 mm Tris pH8.0, 1% SDS and 5 mm EDTA. Chromatin was sheared by sonication, centrifuged and diluted 10-fold in 50 mm Tris, pH 8.0, 0.5% NP-40, 0.2 M NaCl and 0.5 mm EDTA. After pre-clearing with a 50% suspension of salmon sperm-saturated protein A, lysates were incubated at 4°C overnight with anti-p63 (H137 sc-8343, Santa-Cruz). Immune complexes were collected with sperm-saturated protein A, washed three times with high salt buffer (20 mm Tris pH 8.0, 0.1% SDS, 1% NP-40, 2 mm EDTA and 500 mm NaCl), and three times with Tris/EDTA (TE). Immune complexes were extracted in TE containing 1% SDS, and protein–DNA cross-links were reverted by heating at 65°C overnight. DNA was extracted by phenol–chloroform, and the immunoprecipitated DNA was used in PCR reaction. PCR reactions were performed for 25–35 cycles of denaturation at 95°C for 45 s, annealing at 55–57°C for 45 s and extension at 72°C for 45 s. Primer sequences are reported in Supplementary Material, Table S2.
In vitro acetylation assay
In vitro acetylation assay was performed following instructions provided by Fluorescent HAT Assay Kit (Active Motif, 56100). The purified recombinant p300 catalitic domain was incubated with acetyl-CoA and specific synthetic substrate peptides. All peptides were provided by GeneScript. Sequences are reported in Figure 3B. For fluorescence reading, a BF10000 Fluorocount was used.
Statistical analysis
Statistical analyses were performed with one-way ANOVA followed by Dunnett's Multiple Comparison post-test, when needed, using GraphPad PRISM version 5.0 (GraphPad, San Diego, CA, USA). In the graphs, * and ** mark statistically significant data with a P < 0.05 and <0.01, respectively. Statistically highly significant data, with a P < 0.001, are marked by ***.
Supplementary Material
Funding
This work was supported by the Telethon Foundation: grant number GGP11097 to L.G., A.C. and G.R.M. Funding to pay the Open Access publication charges for this article was provided by the Telethon Foundation.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
References
- 1.Koster M.I., Kim S., Mills A.A., DeMayo F.J., Roop D.R. (2004) p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev., 18, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dötsch V., Andrews N.C., Caput D., McKeon F. (1998) p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell, 2, 305–316. [DOI] [PubMed] [Google Scholar]
- 3.Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature, 398, 708–713. [DOI] [PubMed] [Google Scholar]
- 4.Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature, 398, 714–718. [DOI] [PubMed] [Google Scholar]
- 5.Bokhoven H.V., Brunner H.G. (2002) Splitting p63. Am. J. Hum. Genet., 71, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner H.G., Hamel B.C.J., Bokhoven H.V. (2002) p63 gene mutations and human developmental syndromes. Am. J. Med. Genet., 112, 284–290. [DOI] [PubMed] [Google Scholar]
- 7.Duijf P.H., Bokhoven H.V., Brunner H.G. (2003) Pathogenesis of split-hand/split-foot malformation. Hum. Mol. Genet., 12, 51–60. [DOI] [PubMed] [Google Scholar]
- 8.Ianakiev P., Kilpatrick M.W., Toudjarska I., Basel D., Beighton P., Tsipouras P. (2000) Split-hand/split-foot malformation is caused by mutations in the p63 Gene on 3q27. Am. J. Hum. Genet., 67, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrini L., Costanzo A., Merlo G.R. (2011) A symphony of regulations centered on p63 to control development of ectoderm-derived structures. J. Biomed. Biotechnol., 864904, doi:10.1155/2011/864904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray-Zmijewski F., Lane D.P., Bourdon J.C. (2006) p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ., 13, 962–972. [DOI] [PubMed] [Google Scholar]
- 11.Mangiulli M., Valletti A., Caratozzolo M.F., Tullo A., Sbisà E., Pesole G., D'Erchia A.M. (2009) Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res., 37, 6092–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms K.L., Chen X. (2006) The functional domains in p53 family proteins exhibit both common and distinct properties. Cell Death Differ., 13, 890–897. [DOI] [PubMed] [Google Scholar]
- 13.Yang A., McKeon F. (2000) p63 and p73: p53 mimics, menaces and more. Nat. Rev. Mol. Cell. Biol., 1, 199–207. [DOI] [PubMed] [Google Scholar]
- 14.Yang A., Kaghad M., Caput D., McKeon F. (2002) On the shoulder of giants: p63, p73 and the rise of p53. Trends Genet., 18, 90–95. [DOI] [PubMed] [Google Scholar]
- 15.Irwin M.S., Kaelin W.G. (2001) p53 family update: p73 and p63 develop their own identities. Cell Growth Differ., 1, 337–349. [PubMed] [Google Scholar]
- 16.Pozzi S., Zambelli F., Merico D., Pavesi G., Robert A., Maltère P., Gidrol X., Mantovani R., Vigano M.A. (2009) Transcriptional Network of p63 in human keratinocytes. PLoS ONE, 4, e5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viganò M.A., Mantovani R. (2007) Hitting the numbers: the emerging network of p63 targets. Cell Cycle, 3, 233–239. [DOI] [PubMed] [Google Scholar]
- 18.Testoni B., Borrelli S., Tenedini E., Alotto D., Castagnoli C., Piccolo S., Tagliafico E., Ferrari S., Viganò M.A., Mantovani R. (2006) Identification of new p63 targets in human keratinocytes. Cell Cycle, 5, 2805–2811. [DOI] [PubMed] [Google Scholar]
- 19.Yu K., Ornitz D.M. (2008) FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development, 135, 483–491. [DOI] [PubMed] [Google Scholar]
- 20.Boulet A.M., Moon A.M., Arenkiel B.R., Capecchi M.R. (2004) The roles of Fgf4 and FGF8 in limb bud initiation and outgrowth. Dev. Biol., 273, 361–372. [DOI] [PubMed] [Google Scholar]
- 21.Moon A.M., Capecchi M.R. (2000) FGF8 is required for outgrowth and patterning of the limbs. Nat. Genet., 26, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewandoski M., Sun X., Martin G.R. (2004) FGF8 signalling from the AER is essential for normal limb development. Nat. Genet., 4, 460–463. [DOI] [PubMed] [Google Scholar]
- 23.Restelli M., Lopardo T., Lo Iacono N., Garaffo G., Conte D., Rustighi A., Napoli M., Del Sal G., Perez-Morga D., Costanzo A., Merlo G.R., Guerrini L. (2014) DLX5, FGF8 and the Pin1 isomerase control ΔNp63α protein stability during limb development: a regulatory loop at the basis of the SHFM and EEC congenital malformations. Hum. Mol. Genet., 23, 3830–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghioni P., D'Alessandra Y., Mansueto G., Jaffray E., Hay R.T., La Mantia G., Guerrini L. (2005) The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle, 1, 183–190. [DOI] [PubMed] [Google Scholar]
- 25.Galli F., Rossi M., D'Alessandra Y., De Simone M., Lopardo T., Haupt Y., Alsheich-Bartok O., Anzi S., Shaulian E., Calabrò V., La Mantia G., Guerrini L. (2010) MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J. Cell Sci., 123, 2423–2433. [DOI] [PubMed] [Google Scholar]
- 26.Di Costanzo A., Festa L., Duverger O., Vivo M., Guerrini L., La Mantia G., Morasso M.I., Calabrò V. (2009) Homeodomain protein Dlx3 induces phosphorylation-dependent p63 degradation. Cell Cycle, 8, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papoutsaki M., Moretti F., Lanza M., Marinari B., Sartorelli V., Guerrini L., Chimenti S., Levrero M., Costanzo A. (2005) A p38-dependent pathway regulates ΔNp63 DNA binding to p53-dependent promoters in UV-induced apoptosis of keratinocytes. Oncogene, 24, 6970–6975. [DOI] [PubMed] [Google Scholar]
- 28.Brooks C.L., Gu W. (2003) Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol., 15, 164–171. [DOI] [PubMed] [Google Scholar]
- 29.Pietsch E.C., Sykes S.M., McMahon S.B., Murphy M.E. (2008) The p53 family and programmed cell death. Oncogene, 50, 6507–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu B., Zhu W.G. (2012) Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci., 8, 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks C.L., Gu W. (2011) The impact of acetylation and deacetylation on the p53 pathway. Protein Cell, 2, 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu W., Roeder R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 33.Luo J., Li M., Tang Y., Laszkowska M., Roeder R.G., Gu W. (2004) Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A., 101, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marmorstein R., Roth S.Y. (2001) Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev., 11, 155–161. [DOI] [PubMed] [Google Scholar]
- 35.Zeng X., Li X., Miller A., Yuan Z., Yuan W., Kwok R.P., Goodman R., Lu H. (2000) The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol. Cell Biol., 20, 1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costanzo A., Merlo P., Pediconi N., Fulco M., Sartorelli V., Cole P.A., Fontemaggi G., Fanciulli M., Schiltz L., Blandino G., Balsano C., Levrero M. (2002) DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell, 9, 175–186. [DOI] [PubMed] [Google Scholar]
- 37.Mantovani F., Piazza S., Gostissa M., Strano S., Zacchi P., Mantovani R., Blandino G., Del Sal G. (2004) Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol. Cell, 14, 625–636. [DOI] [PubMed] [Google Scholar]
- 38.Tang Y., Zhao W., Chen Y., Zhao Y., Gu W. (2008) Acetylation is indispensable for p53 activation. Cell, 133, 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chae Y.S., Kim H., Kim D., Lee H., Lee H.O. (2012) Cell density-dependent acetylation of ΔNp63α is associated with p53-dependent cell cycle arrest. FEBS Lett., 8, 1128–1134. [DOI] [PubMed] [Google Scholar]
- 40.Yan W., Bentley B., Shao R. (2008) Distinct angiogenic mediators are required for basic fibroblast growth factor- and vascular endothelial growth factor-induced angiogenesis: the role of cytoplasmic tyrosine kinase c-Abl in tumor angiogenesis. Mol. Biol. Cell, 19, 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agami R., Blandino G., Oren M., Shaul Y. (1999) Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature, 399, 809–813. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Prieto R., Sanchez-Arevalo V.J., Servitja J.M., Gutkind J.S. (2002) Regulation of p73 by c-Abl through the p38 MAP kinase pathway. Oncogene, 21, 974–979. [DOI] [PubMed] [Google Scholar]
- 43.Levav-Cohen Y., Goldberg Z., Zuckerman V., Grossman T., Haupt S., Haupt Y. (2005) C-Abl as a modulator of p53. Biochem. Biophys. Res. Commun., 331, 737–749. [DOI] [PubMed] [Google Scholar]
- 44.Gonfloni S., Di Tella L., Caldarola S., Cannata S.M., Klinger F.G., Di Bartolomeo C., Mattei M., Candi E., De Felici M., Melino G., Cesareni G. (2009) Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat. Med., 15, 1179–1185. [DOI] [PubMed] [Google Scholar]
- 45.Yuan M., Luong P., Hudson C., Gudmundsdottir K., Basu S. (2010) c-Abl phosphorylation of ΔNp63α is critical for cell viability. Cell Death Dis., doi:10.1038/cddis.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo Iacono N., Mantero S., Chiarelli A., Garcia E., Mills A.A., Morasso M.I., Costanzo A., Levi G., Guerrini L., Merlo G.R. (2008) Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development, 135, 1377–1388. [DOI] [PubMed] [Google Scholar]
- 47.Lussier M., Canoun C., Ma C., Sank A., Shuler C. (1993) Interdigital soft tissue separation induced by retinoic acid in mouse limbs cultured in vitro. Int. J. Dev. Biol., 4, 555–564. [PubMed] [Google Scholar]
- 48.Beretta C., Chiarelli A., Testoni B., Mantovani R., Guerrini L. (2005) Regulation of the cyclin-dependent kinase inhibitor p57Kip2 expression by p63. Cell Cycle, 4, 1625–1631. [DOI] [PubMed] [Google Scholar]
- 49.Sbisà E., Mastropasqua G., Lefkimmiatis K., Caratozzolo M.F., D'Erchia A.M., Tullo A. (2006) Connecting p63 to cellular proliferation: the example of the adenosine deaminase target gene. Cell Cycle, 2, 205–212. [DOI] [PubMed] [Google Scholar]
- 50.Ihrie R.A., Marques M.R., Nguyen B.T., Horner J.S., Papazoglu C., Bronson R.T., Mills A.A., Attardi L.D. (2005) Perp is a p63-regulated gene essential for epithelial integrity. Cell, 120, 843–856. [DOI] [PubMed] [Google Scholar]
- 51.Marinari B., Ballaro C., Koster M.I., Giustizieri M.L., Moretti F., Crosti F., Papoutsaki M., Karin M., Alema S., Chimenti S., Roop D.R., Costanzo A. (2009) IKKalpha is a p63 transcriptional target involved in the pathogenesis of ectodermal dysplasias. J. Invest. Dermatol., 129, 60–69. [DOI] [PubMed] [Google Scholar]
- 52.Lopardo T., Lo Iacono N., Marinari B., Giustizieri M.L., Cyr D.G., Merlo G., Crosti F., Costanzo A., Guerrini L. (2008) Claudin-1 is a p63 target gene with a crucial role in epithelial development. PLoS ONE, doi:10.1371/journal.pone.0002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao T.P., Oh S.P., Fuchs M., Zhou N.D., Chng L.E., Newsome D., Bronson R.T., Li E., Livingston D.M., Eckner R. (1999) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell, 93, 361–372. [DOI] [PubMed] [Google Scholar]
- 54.LeBoeuf M., Terrell A., Trivedi S., Sinha S., Epstein J.A., Olson E.N., Morrisey E.E., Millar S.E. (2010) Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell, 19, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merlo G.R., Paleari L., Mantero S., Genova F., Beverdam A., Palmisano G.L., Barbieri O., Levi G. (2002) Mouse model of split hand/foot malformation type I. Genesis, 33, 97–101. [DOI] [PubMed] [Google Scholar]
- 56.Chimal-Monroy J., Abarca-Buis R.F., Cuervo R., Díaz-Hernández M., Bustamante M., Rios-Flores J.A., Romero-Suárez S., Farrera-Hernández A. (2011) Molecular control of cell differentiation and programmed cell death during digit development. Iubmb Life, 10, 922–929. [DOI] [PubMed] [Google Scholar]
- 57.Nomura N., Yokoyama H., Tamura K. (2014) Altered developmental events in the anterior region of the chick forelimb give rise to avian-specific digit loss. Dev. Dyn., 6, 741–752. [DOI] [PubMed] [Google Scholar]
- 58.Ghioni P., Bolognese F., Duijf P.H., Van Bokhoven H., Mantovani R., Guerrini L. (2002) Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol. Cell. Biol., 22, 8659–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.