Abstract
The nuclear face of the nuclear membrane is enriched with the intermediate filament protein lamin A. Mutations in LMNA, the gene encoding lamin A, lead to a diverse set of inherited conditions including myopathies that affect both the heart and skeletal muscle. To gain insight about lamin A protein interactions, binding proteins associated with the tail of lamin A were characterized. Of 130 nuclear proteins found associated with the lamin A tail, 17 (13%) were previously described lamin A binding partners. One protein not previously linked to lamin A, matrin-3, was selected for further study, because like LMNA mutations, matrin-3 has also been implicated in inherited myopathy. Matrin-3 binds RNA and DNA and is a nucleoplasmic protein originally identified from the insoluble nuclear fraction, referred to as the nuclear matrix. Anti-matrin-3 antibodies were found to co-immunoprecipitate lamin A, and the lamin-A binding domain was mapped to the carboxy-terminal half of matrin-3. Three-dimensional mapping of the lamin A-matrin-3 interface showed that the LMNA truncating mutation Δ303, which lacks the matrin-3 binding domain, was associated with an increased distance between lamin A and matrin-3. LMNA mutant cells are known to have altered biophysical properties and the matrin-3–lamin A interface is positioned to contribute to these defects.
Introduction
The nuclear membrane is composed of an inner and outer membrane separated by the perinuclear space. The inner nuclear membrane is supported by a dense intermediate filament network known as the nuclear lamina (1). In dividing cells, the nuclear lamina contains B type lamins and to a lesser extent A type lamins. The A type lamins include lamins A and C and are encoded by the single LMNA gene. Mutations in LMNA lead to a wide array of inherited disease including skeletal and cardiac myopathies, lipodystrophy, premature aging and others (2). The mechanisms by which mutations in this single gene lead to these diverse phenotypes is likely multifold as multiple nuclear functions are affected by LMNA mutations including gene expression, nuclear shape and position, chromosomal positioning and other cellular processes (3). More than 300 LMNA mutations have been implicated in human disease (4), and autosomal dominance is the major mode of inheritance for LMNA mutations that lead to cardiac and skeletal muscle myopathy. Lamins A and C are identical over their first 556 amino acids differing only in their carboxy-terminus. Prelamin A, the precursor of mature lamin A has an additional carboxy-terminal extension, and this region contains a sequence that is farnesylated and cleaved, resulting in a 645 amino acid mature lamin A protein. These post-translational modifications allow lamin A to associate with the nuclear membrane. Lamin C is shorter and primarily adheres to the nuclear membrane through its interactions with other lamins, namely lamin A. The first 33 amino acids of Lamin A/C encode a short head like domain. The central rod domain of lamin A/C is defined by amino acids 33–383 followed by the nuclear localization signal. Residues 430–545 of lamin A/C form a globular immunoglobulin (Ig)-like fold (5).
Lamin A/C like other intermediate filament proteins, dimerizes as parallel structures mediated by the central rod domain. The antiparallel organization of the oligomerized dimers leads to the formation of intermediate filament proteins with 25 nm periodicity. Lamin A/C assembly requires the rod domain and part of the short head region (6,7). The Ig domain adopts a β sandwich configuration with nine β strands (5). The LMNA mutation R453W is associated with Emery Dreifuss Muscular Dystrophy (EDMD), a disorder with progressive skeletal muscle loss, muscle weakness and associated cardiomyopathy, and this position, was mapped to an externally facing portion of the Ig fold. In contrast, a mutation linked to Familial Dunnigan Partial Lipodystrophy was found to localize to the internal aspects of the Ig fold suggesting that greater disruption of the Ig fold may in part explain aspects of LMNA's tissue-specific effects.
In order to identify potential binding partners of lamin A, the Ig fold of lamin A was expressed and purified. To address the role of LMNA mutations in muscle-related phenotypes, potential binding proteins were identified using a nuclear protein extract from C2C12 cells, a myogenic cell line that was induced to form myotubes. One hundred and thirty proteins were found reproducibly with lamin A tails (LATs), including 17 proteins which were previously identified as known lamin A binding partners. Of these, proteins involved in nucleic acid binding were highly represented including those implicated in RNA processing and splicing. Matrin-3, a major protein component of the nucleoplasm, was identified as a potential lamin A binding partner, and the gene encoding matrin-3 was previously found to have a missense mutation in two large unrelated families with inherited myopathy (8,9). Immunoprecipitation from myogenic C2C12 cells demonstrated association between lamin A and matrin-3. The LAT bound directly to matrin-3, and one LMNA mutation, R453W, demonstrated increased binding to matrin-3. Another mutation associated with inherited myopathy, LMNAΔ303, which lacks the LAT was found to have reduced contact surfaces between matrin-3 and lamin A/C. Together these data provide a model to explain how the nuclear lamina interacts with nuclear contents.
Results
Identifying candidate lamin A binding proteins from myotubes
The LAT domain was expressed, purified and crosslinked to a sepharose column. In addition to wild-type LAT, the tail domains from two different myopathy-associated LMNA mutations, R453W and R527P, were studied (n = 4 of each). These mutations were selected because they each map to different external faces of the Ig fold (5). The nuclear lamina protein extract was isolated from differentiated C2C12 myotubes, a mouse muscle cell line, and this extract was incubated with the immobilized LAT affinity columns. We confirmed the enrichment of nuclear lamina components in the extract by immunoblotting for lamin A/C and emerin (Fig. 1). Bound proteins were eluted with high salt and subjected to LC-MS/MS analysis. Analysis of the spectra with Mascot and X! Tandem database searching and Scaffold protein identification software yielded a list of candidate LAT-interacting proteins identified by at least one high-confidence peptide in multiple experiments. This was refined by including only those proteins with a ratio ≥2 between unbound beads and LAT beads, resulting in a list of 130 nuclear protein candidates from the Mouse Genome Informatics (MGI) database (Supplementary Material, Table S1 and Fig. S1).
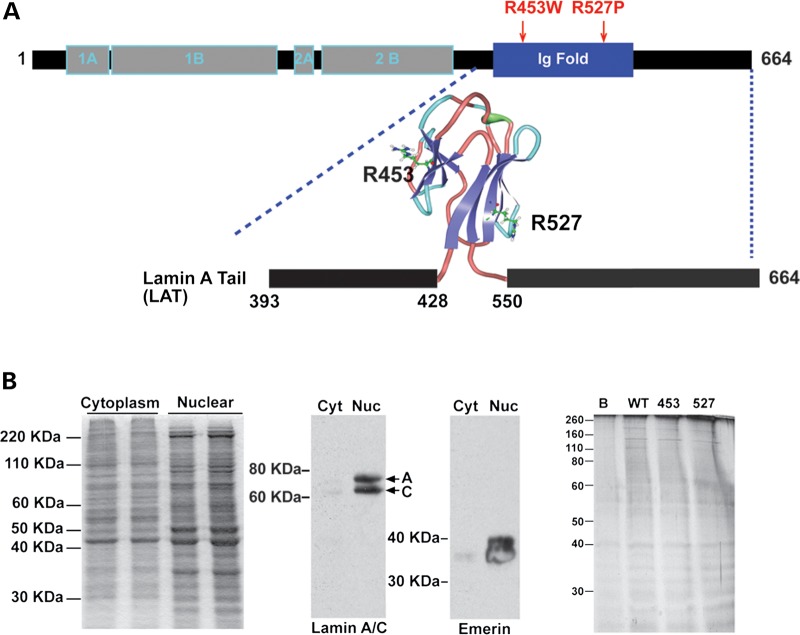
Figure 1.
Expression of LATs to identify the laminome. (A) Schematic of lamin A (664 amino acids) showing domains and the LAT domain (amino acids 393–664) including the Ig fold (amino acids 428–550). The structure of lamin A's Ig fold was previously described (5), Protein Data Base; PDB ID: 1IVT). Positions Arg453 and Arg527 map to opposite sides of the Ig fold. Wild-type LAT and two mutants associated with EDMD, R453W and R527P, were expressed and incubated with extracts from differentiated myotubes. (B) Nuclear extracts were prepared from myotubes. The insoluble non-ionic detergent and high salt resistant nuclear lamina protein extract was separated from the cytoplasmic fraction (left panel). Immunoblotting for lamin A/C and emerin, two known nuclear lamina-associated proteins, showed enrichment in the nuclear (Nuc) compared with cytoplasmic (Cyt) fractions (middle panels). Nuclear extracts were incubated with wild-type or mutant LATs, and the protein elution profile from normal (WT), R453W, R527P appeared similar in silver stained SDS gels (right panel). B refers to beads only. One hundred and thirty proteins were identified reproducibly as binding to LATs.
A summary of the proteins identified is shown in Fig. 2A. Known components of the nucleoplasm, nucleolus, Cajal Body, nuclear speckles, paraspeckles, nuclear bodies, ribonucleoprotein and spliceosomal complexes were detected using this approach. Seventeen of 130 (13%) of the identified nuclear proteins were known lamin A/C binding proteins including Barrier-to-autointegration factor (BANF1), replication-dependent and independent histones, LBR, lamin A, lamin B1, emerin, titin and tripartite motif containing 28 (Trim28) (Table 1). Twenty-six proteins were known to be associated with the nuclear lamina, nuclear pore or chromatin. Gene ontology (GO) analysis was conducted on the list of candidate lamin A binding proteins, and nucleic acid binding activity was the dominant functional classification (Supplementary Material, Fig. S2). Interestingly, proteins associated with the GO category ‘RNA processing’ (GO:0006396) were also significantly enriched, including ‘mRNA splicing process via spliceosome’ (GO:0000398). Proteins associated with ‘RNA transport’ (GO:0050658) and ‘translation’ (GO:0006412) were also found enriched in the laminome. Nearly all GO categories were shared between normal and mutant LATs. The only GO category that differed between wild-type and mutant lamin A proteins was for ‘mRNA splicing process via spliceosome’ (Fig. 2B). This difference derived from the presence of the splicing factors Transformer-2 protein homolog β (TRA2B), Serine/arginine-rich splicing factor 10 (SFRS13A) and protein Srrm1 (SRRM1) associated with LMNA R453W and R527P and not wild-type LMNA.
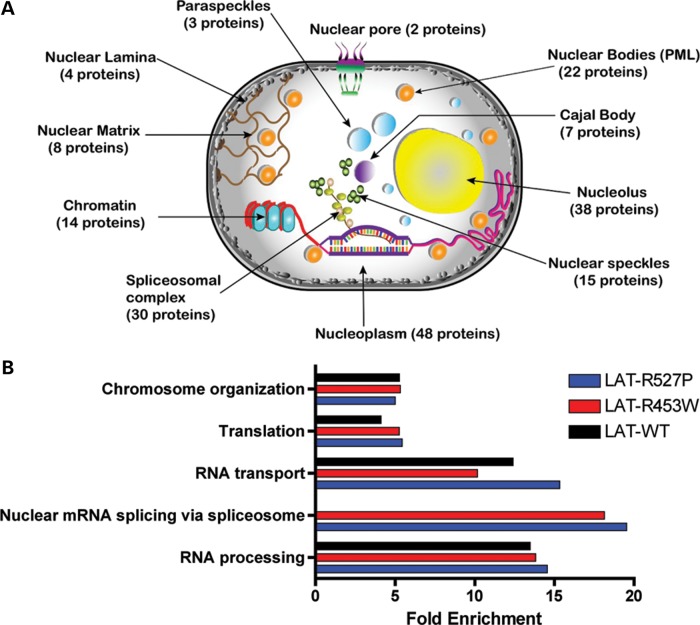
Figure 2.
Gene Ontology of 130 proteins identified as binding the LAT. (A) Cellular process annotation was performed using DAVID and MGI databases. The nuclear position of 130 proteins identified as associating with LAT is shown, and the total number of different protein is indicated in parenthesis. Several proteins have multiple nuclear locations (see also Supplementary Material, Fig. S1). (B) Biological process (BP) enrichment analysis. Using DAVID database, five processes were found significantly enriched compared with random background mouse protein list of 13588 proteins (P value < 0.001). The only biological process that differed between wild-type LAT and mutants nuclear mRNA splicing via spliceosome and this difference was due to three proteins (Transformer-2 protein homolog β (TRA2B), Serine/arginine-rich splicing factor 10 (SFRS13A) and Srrm1 (SRRM1) being associated with mutant but not wild-type LATs.
Table 1.
Known lamin-binding proteins identified in the laminome.
| Gene ID | Name | Function | Nuclear localization | References |
|---|---|---|---|---|
| BANF1 | Barrier to autointegration factor 1 | DNA binding | Chromosome | (10) |
| EMD | Emerin | Regulation of protein export from nucleus | Nuclear lamina | (11) |
| Replication-dependent histones | ||||
| HIST1H1A | Histone cluster 1, H1a (linker histone) | DNA binding | Chromosome | (12) |
| HIST1H1B | Histone cluster 1, H1b (linker histone) | DNA binding | Chromosome | (12) |
| HIST1H1C | Histone cluster 1, H1c (linker histone) | DNA binding | Chromosome | (12) |
| HIST1H2BB | Histone cluster 1, H2bb (core histone) | DNA binding | Chromosome | (13) |
| HIST1H3A | Histone cluster 1, H3a (core histone) | DNA binding | Chromosome | (13) |
| HIST2H2AB | Histone cluster 2, H2ab (core histone) | DNA binding | Chromosome | (13) |
| HIST4H4 | Histone cluster 4, H4 (core histone) | DNA binding | Chromosome | (13) |
| Replication-independent histones | ||||
| H2AFX | H2A histone family, member X | DNA binding, DNA repair | Chromosome | (14,15) |
| H2AFY | H2A histone family, member Y | DNA binding | Chromosome, Barr body | (14,15) |
| H2AFZ | H2A histone family, member Z | DNA binding | Chromosome | (14,15) |
| LBR | Lamin B receptor | DNA binding, receptor | Nuclear lamina | (16) |
| LMNA | Lamin A | Intermediate filament | Nuclear lamina, nuclear matrix | (17) |
| LMNB1 | Lamin B1 | Intermediate filament | Nuclear lamina, nuclear matrix | (17) |
| TRIM28 | Tripartite motif-containing 28 | Regulation of transcription, DNA binding | Nucleolus, nucleoplasm, chromatin | (16) |
| TTN | Titin | Protein kinase activity, actin filament binding | Nucleoplasm, nuclear pore | (18) |
Matrin-3 binds directly to lamin A
The list of potential lamin A binding candidates was surveyed for proteins implicated in inherited muscle disease and not previously considered as a lamin A binding protein. Matrin-3 was selected for further analysis because the gene encoding this protein, MATR3, was previously linked to inherited autosomal dominant distal myopathy (8,9). A single missense MATR3 mutation, S85C, has been identified in multiple large families with inherited myopathy. The Matrin-3 protein was originally identified from the insoluble nuclear fraction, and because of its nucleoplasmic distribution has been considered a nuclear matrix and scaffolding protein (19,20). Consistent with its nuclear intracellular localization, matrin-3 binds to both DNA and RNA (21,22). From the list of LAT binding proteins, 19 were previously reported as matrin-3 binding partners (Supplementary Material, Table S2).
To investigate the interaction between endogenous lamin A/C and matrin-3, immunoprecipitation with anti-matrin-3 antibodies was carried out from C2C12 cells that had been differentiated into myotubes. Two different anti-matrin-3 antibodies resulted in the co-precipitation of lamin A/C (Fig. 3A), although it appears that only a small fraction of total lamin A/C interacts with matrin-3. We also found that matrin-3 and lamin A/C co-purify. Cytoplasmic and nuclear fractions from myotubes were isolated and an amino-terminally directed matrin-3 antibody showed co-enrichment with lamin A/C in the insoluble non-ionic detergent and high salt resistant nuclear lamina protein extract (Fig. 3B). To determine if matrin-3 and lamin A interact directly, 35S-labeled matrin-3 (847 amino acids) protein was tested for its ability to bind directly to the LAT. Full-length matrin-3 specifically bound to the wild-type LAT (Fig. 3C). As a control for non-specific binding, matrin-3 did not bind to two other control proteins overexpressed at equal abundance, glutathione S-transferase (GST) and a GST fusion to the C2C domain of a membrane associated protein (24). These data demonstrate that matrin-3 binds directly to the Ig fold tail region of lamin A, amino acids 393–664.
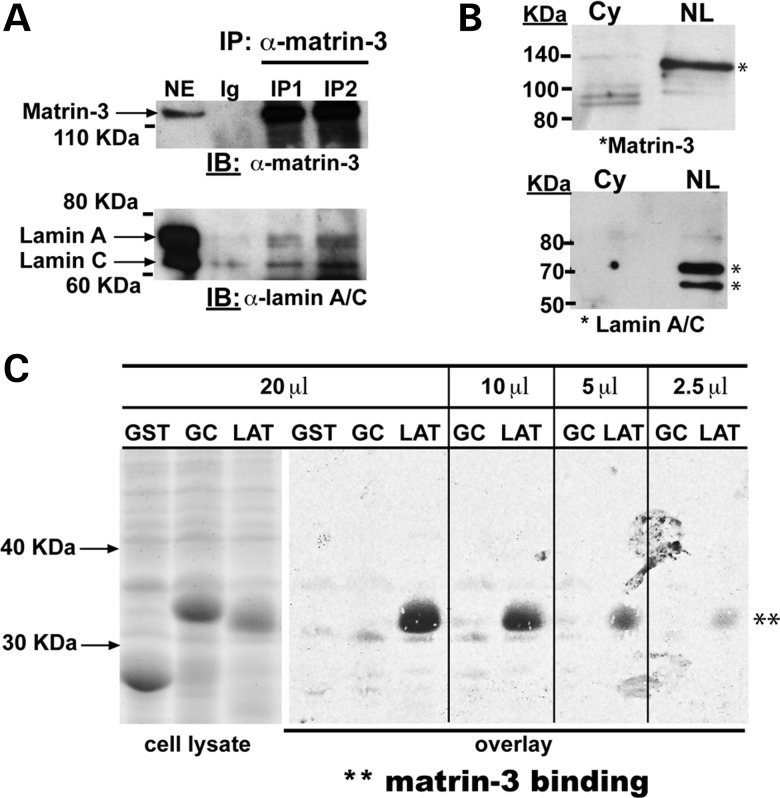
Figure 3.
The interaction of Matrin-3 and lamin A. (A) Matrin-3, a known nuclear protein, was found to associated with LATs, and MATR3 mutations associate with myopathy (8,23). To evaluate whether matrin-3 and lamin A interact, co-immunoprecipitation of endogenous proteins from differentiated myotubes was tested. Matrin-3 was immunoprecipitated (IP) using two independent anti-matrin-3 antibodies (IP1 and IP2). The immunoprecipitate was probed with anti-matrin-3 to show that matrin-3 was present. Immunoblotting with anti-lamin A/C demonstrated both isoforms of lamin A (full length and Δ10) and lamin C associate in a complex with matrin-3. Negative control ‘Ig’ lanes used normal rabbit IgG. (B) Matrin-3 and lamin A/C co-purify from myotubes. The cytoplasmic fraction (Cy) was separated from the insoluble non-ionic detergent and high salt resistant nuclear lamina protein extract (NL). Immunoblotting showed that both matrin-3 and lamins A and C were enriched in these fractions. (C) Overlay experiments to show a direct interaction between radiolabelled matrin-3 and LAT. GST, GST linked to an unrelated mammalian protein (GC, indicating the C2C domain of fer1l5 (24)) and LAT were expressed (left panels, cell lysate). Membranes containing these proteins were tested for binding to 35S-labeled matrin-3 (847 amino acids) generated using in vitro transcription/translation. Full-length matrin-3 bound to the LAT in a concentration dependent manner across all lysate volumes (20–2.5 µl), whereas no binding was detected to GST and GST-C2C (GC).
Matrin-3's carboxy-terminus binds LAT
To identify the region of matrin-3 that binds lamin A, four subdomains of matrin-3 were radiolabeled and tested for binding to LAT (Fig. 4A). Matrin-3 binding to lamin A was undetectable for matrin-3 subdomains R1 and R2 (Fig. 4B). In contrast, matrin-3 subdomains R3 and R4 both demonstrated interaction with the LAT, with potentially higher affinity binding by matrin-3 subdomain R3 than R4 (Fig. 4B), consistent with a dual binding site on matrin-3. Matrin-3 is not the only lamin A binding protein reported to carry a dual binding domain. Protein kinase PKCα and the nuclear pore protein Nup153 each harbor two distinct lamin A binding domains (25,26). The binding of full-length matrin-3 protein was tested on two different LMNA mutants R453W and R527P because these residues fall on different faces of the Ig fold (5). LMNA R453W, but not R527P, bound more matrin-3, suggesting that this face of lamin A may be more directly implicated in the matrin-3-lamin A interaction (Fig. 4C). The mean binding for LAT-R453W was significantly higher (P < 0.01) than wild-type LAT (Fig. 4D).
Figure 4.
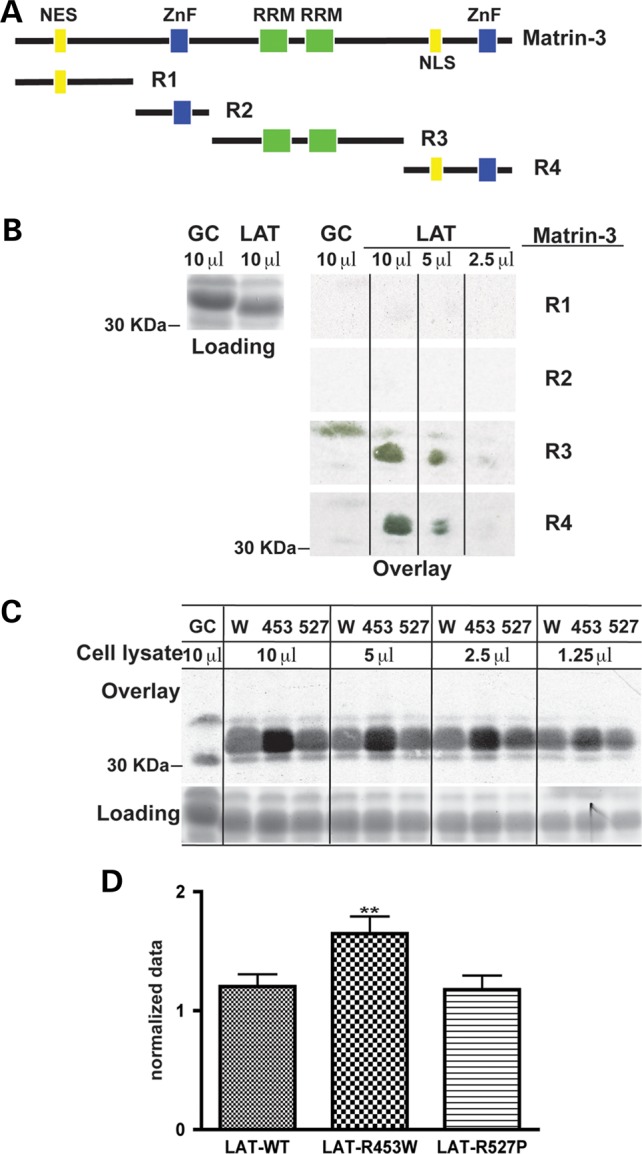
LAT binds the carboxy half of matrin-3. (A) A schematic of Matrin-3 domains is shown in the upper panel. The R1 and R2 regions include the nuclear localization signal (NES) and the first Zinc Finger (ZnF) domain, respectively. The R3 domain contains RNA binding domains (RRM), and the R4 domain includes the second ZnF domain (20). (B) 35S-Matrin-3 R1 and R2 subdomains showed no detectable binding to LAT. Matrin-3 subdomains R3 and R4 both showed binding to LAT. None of the matrin-3 subdomains showed binding to the GST-GC control fusion protein as expected. (C) LMNA mutations R453W and R527P were tested for the capacity to bind full-length 35S-matrin-3 since these two mutation are on two different faces of the lamin A Ig fold. The upper part of the panel shows that 35S- labeled matrin-3 bound LMNA R453W more than wild-type LAT (W) or LMNA R527P. The loading control is shown at the bottom. (D) The mean ratio of all combined LMNA R453W bands was significantly higher (**P < 0.01) to the mean ratios of combined LAT-control and combined LMNA 527P bands.
Matrin-3 is expressed at the nuclear membrane and in the nucleoplasm
To determine if lamin A/C and matrin-3 co-localize, we performed immunostaining on differentiated myotubes and mature skeletal muscle using antibodies to either the amino or carboxy-terminus of matrin-3. In myotubes lamin A is found as the expected ring at the nuclear membrane (27). When imaging matrin-3 in myotubes using the amino-terminally directed matrin-3 antibody, the matrin-3 signal was seen in the nuclear matrix as well as at the nuclear membrane (Fig. 5A, top). This nuclear membrane pattern overlapped with lamin A immunostaining (Fig. 5A, top). In contrast, imaging matrin-3 with a carboxy-terminal anti-matrin-3 antibody showed mainly the nucleoplasmic pattern and not the enrichment at the nuclear membrane (Fig. 5A, bottom). The binding studies between lamin A and matrin-3 localized the interaction to the carboxy-terminal half of matrin-3 (Fig. 4B), and the epitope for the carboxy-terminal antibody falls within this region (800–847 amino acids). Therefore it is likely that the epitope for matrin-3 is masked by matrin-3’s interaction with lamin A. This same pattern of matrin-3 expression, at both the nuclear membrane and in the nucleoplasm, was seen in mature skeletal muscle when using the amino-terminal anti-matrin-3 antibody (Fig. 5B). Both myotubes and mature skeletal muscle express more lamin A/C compared with undifferentiated myoblasts (28).
Figure 5.
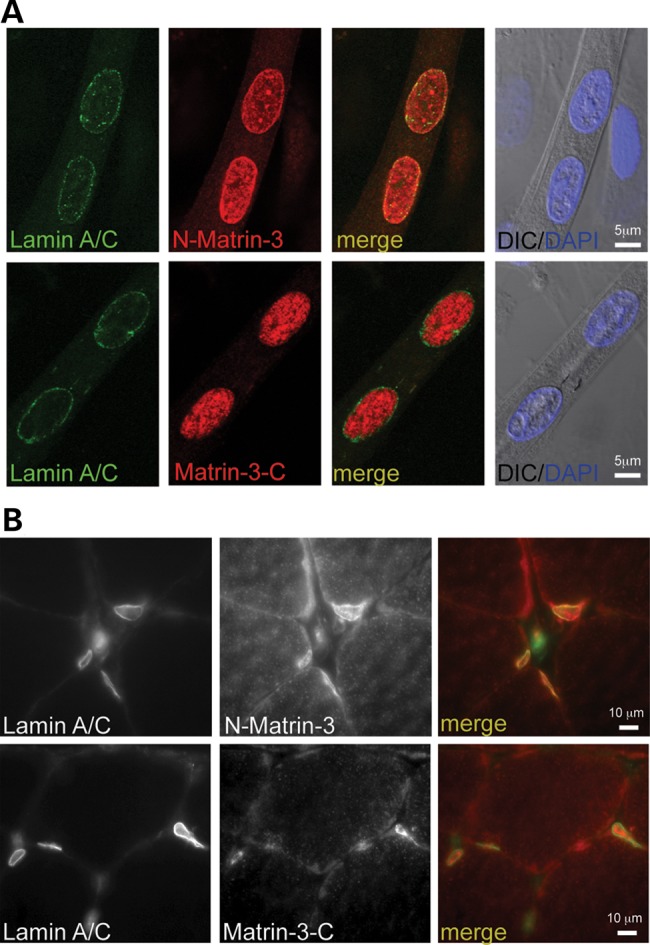
Co-localization of matrin-3 and lamin A. (A) Matrin-3 and lamin A in differentiated myotubes using two different anti-matrin-3 antibodies. The amino-terminally directed anti-matrin antibody (N-matrin) shows matrin-3 in the nucleoplasm and at the nuclear membrane. There is co-localization between matrin-3 and lamin-A at the nuclear membrane. The carboxy-terminally directed anti-matrin-3 antibody (Matrin-3-C) shows nucleoplasmic staining only. The absence of nuclear membrane staining with the carboxyl antibody may reflect altered conformational state or epitope masking by the interaction with lamin A at the nuclear membrane. (B) Similar findings in mature skeletal muscle demonstrating co-localization of matrin-3 and lamin A at the nuclear rim.
To assess the nuclear localization of matrin-3 during myogenic differentiation, the amino-terminal and carboxy-terminal anti-matrin-3 antibodies were used on undifferentiated C2C12 myoblasts. In myoblasts, which express little lamin A/C, both the amino- and carboxy-terminal directed matrin-3 antibodies showed diffuse nuclear staining (Fig. 6A). Within undifferentiated C2C12 cultures, there is a range of myogenic differentiation. The transcription factor myogenin was found to be expressed in only a subset of cells, consistent with partial entry into the myogenic lineage. In myogenin-positive myoblasts, the amino-terminal matrin-3 antibody demonstrated localization at the nuclear membrane (Fig. 6B). More than 95% of myoblasts with matrin-3 nuclear membrane rings were also myogenin-positive (Fig. 6C). The presence of myogenin indicates that these cells had entered into myogenic differentiation. However, myogenin-negative myoblasts did not show matrin-3 at the nuclear membrane. Matrin-3's presence at the nuclear rim is similar to what was previously observed for lamin A/C's expression during myogenesis in C2C12 cells (28). This colocalization suggests that matrin-3 and lamin A/C have a shared expression pattern at the nuclear rim during myogenesis.
Figure 6.
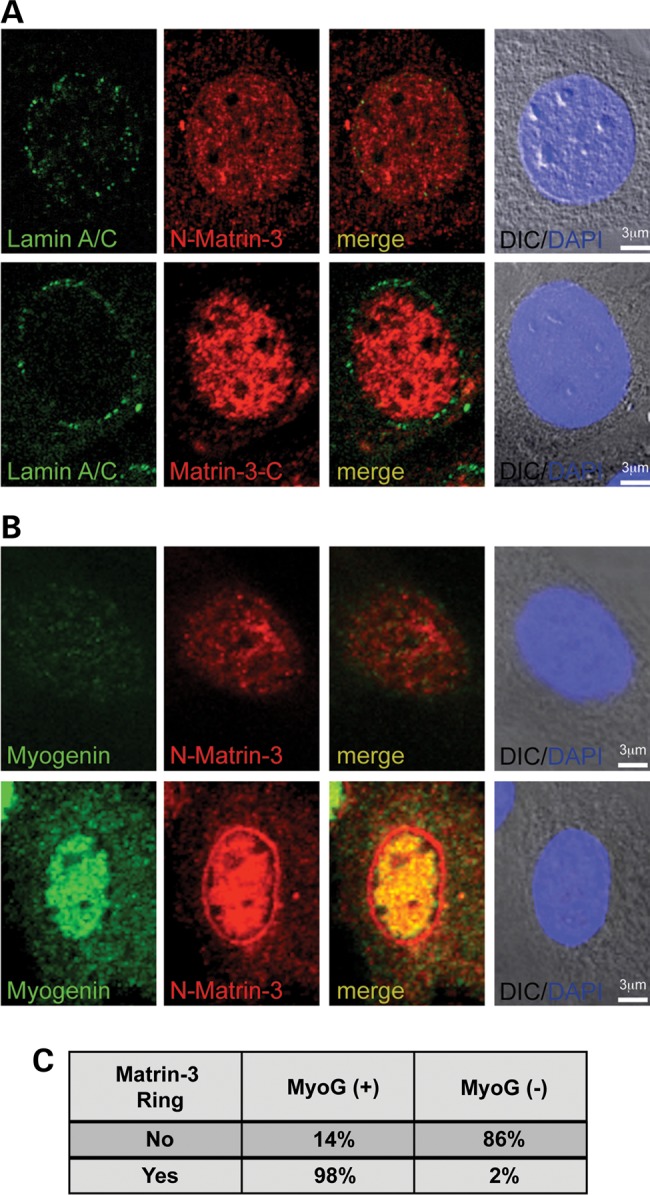
Matrin-3 localizes to the nuclear membrane upon myoblast differentiation. (A) In undifferentiated myoblasts, lamin A/C was expressed at low levels at the nuclear rim (green). Two matrin-3 antibodies, one directed at the amino-terminus and one directed at the carboxy-terminus, showed diffuse nuclear staining (red). (B) Myoblasts with low-to-no detectable myogenin staining, representing an undifferentiated state, displayed nucleoplasmic matrin-3 staining. In contrast, myoblasts that entered the myogenic lineage, marked by intense myogenin staining (green), had matrin-3 localized to both the nucleus and at the nuclear membrane, and this pattern was detected by the amino-terminal matrin-3 antibody. (C) Quantification of myoblasts revealed that the majority of nuclei positive for myogenin also have matrin-3 staining at the nuclear membrane (n = 111 nuclei).
Disruption of the normal nuclear lamina-matrix interaction in LMNAΔ303 cells
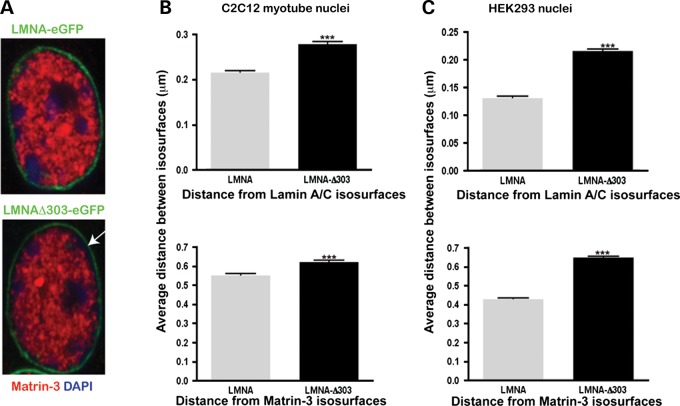
To examine whether the lamin A/matrin-3 interaction affects the structure of the nuclear lamina, fibroblasts from an individual with the LMNAΔ303 truncating mutation were studied. This mutation truncates lamin A, removing the Ig fold and matrin-3 binding domain, effectively removing the LAT domain. This LMNAΔ303 mutation causes cardiomyopathy and muscular dystrophy (29–31). For these studies, the carboxy-terminal anti-matrin-3 antibody was used to examine the distance between the nuclear membrane and nuclear matrix. This approach takes advantage of the fact that the carboxy-terminal matrin-3 antibody does not overlap with lamin A signal at the nuclear membrane. Since the amino-terminal matrin-3 antibody co-localizes with lamin A signal, it cannot be used in this analysis (Fig. 5). The LMNAΔ303 fibroblasts used for this study carry a heterozygous mutation, so the lamin A—matrin-3 interaction is not completely ablated. The three-dimensional surfaces of lamin A and matrin-3 were analyzed using IMARIS software following the methods of (32). In both control and LMNAΔ303 fibroblasts, lamin A was concentrated at the nuclear lamina, whereas a dense, interconnected network of matrin-3 signal was in the nucleoplasm (Fig. 7A). The Z-stack images showed discrete yellow punctuated signals both at the vicinity of the nuclear lamina and inside the nucleoplasm. The three-dimensional reconstruction revealed that matrin-3 was anchored to lamin A at the nuclear lamina. A few lamin A/C surfaces were detected in the nucleoplasm, and these also overlapped with matrin-3. Figure 7B shows Z-stack images of control (left panel) and Δ303 (right panel) fibroblasts. Control fibroblasts reveal overlap between lamin A and matrin-3 as visualized by discrete yellow puncta at the nuclear rim (inset). These puncta are greatly reduced in Δ303 fibroblasts, indicating an altered lamin A/matrin-3 interface. To quantify this disruption of the lamin A/matrin-3 interaction at the nuclear rim, we used MatLab to calculate the shortest distances between lamin A/C and matrin-3 isosurfaces. The distance between isosurfaces was increased in LMNAΔ303 cells compared with control cells (P < 0.0001, n = 10 cells of each genotype) (Fig. 7C). The expression level of lamin A/C and matrin-3, as marked with the carboxy-terminal anti-matrin-3 antibody, was not different between LMNAΔ303 and wild-type control cells (Fig. 7D). Therefore, the increased distance between lamin A and matrin-3 is consistent with a reduced interaction between these proteins. To further test the interaction between lamin A and matrin-3, LMNA-GFP and LMNAΔ303-GFP were introduced into C2C12 or HEK cells and the distance between lamin A and matrin-3 was examined using the same approach. In transfected cells, those expressing LMNAΔ303 also had increased distance between the nuclear membrane and nuclear matrix compared with those expressing full-length lamin A (Fig. 8).
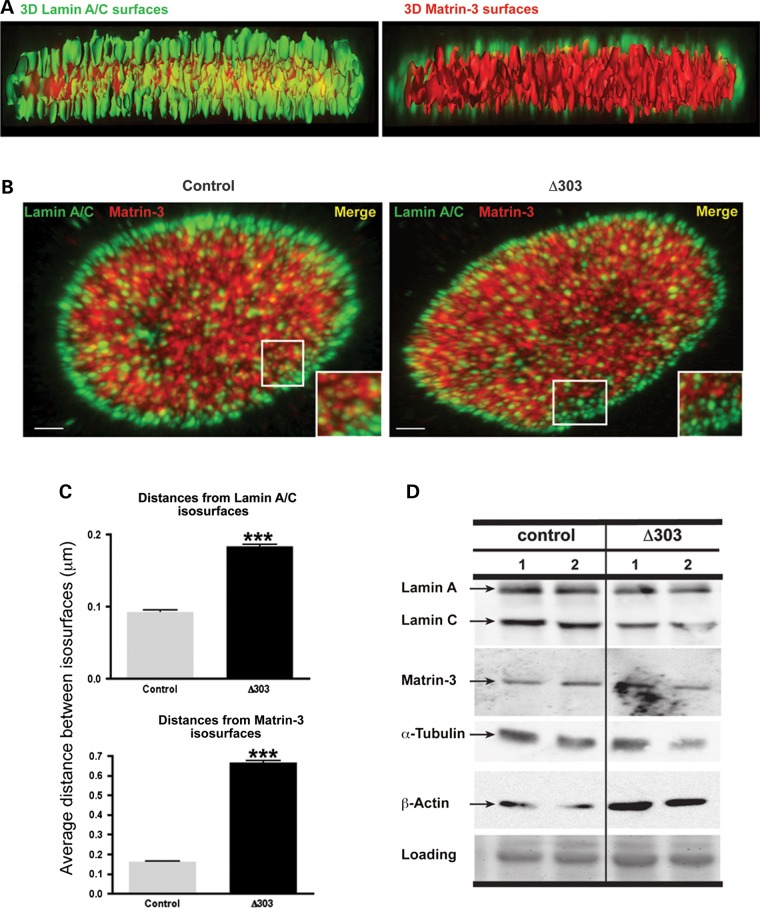
Figure 7.
The Lamin A–Matrin-3 interface disrupted in cells lacking lamin A-tail. Confocal Z-stack images, 3D surfaces of lamin A/C (green color) and matrin-3 (red color, C-term antibody) were analyzed using IMARIS software. Control and LMNA Δ303 fibroblasts were imaged. The Δ303 LMNA mutation disrupts the matrin-3 binding site; this mutation is associated with both cardiac and skeletal myopathy. (A) Lamin A/C surfaces are enriched at the nuclear lamina, whereas a dense network of interconnected matrin-3 surfaces is concentrated in the nucleoplasm. The Z-stack images show discrete yellow punctate signals both at the vicinity of the nuclear lamina and inside the nucleoplasm. Scale bar represents 1 µm. (B) Z-stack images of control and Δ303 fibroblasts immunostained with Lamin A/C (green) and Matrin-3 (red) reveal discrete yellow punta at the nuclear lamina in control fibroblasts (left, inset), which are less frequent in Δ303 fibroblasts (right, inset). Scale bar is 2 μm. (C) Matlab was used to calculate the shortest distances between lamin A/C and matrin-3 surfaces. Distances between matrin-3 and lamin A isosurfaces were calculated from lamin A to matrin-3 (upper graph) and from matrin-3 to lamin A (lower graph). The distance between lamin A and matrin-3 was larger in Δ303 cells (***P < 0.0001). (D) Immunoblot analysis of total protein extracts from control and LMNA Δ303 human fibroblasts showed that both fibroblast cell lines have similar amounts of lamins A and C (70 and 65 kDa, respectively), matrin-3 (120 kDa), α-tubulin (50 kDa) and β-actin (45 kDa) proteins.
Figure 8.
Increased distance between lamin A and matrin-3 with expression of LMNAΔ303. (A) LMNA or LMNAΔ-303 was expressed in C2C12 cells as GFP fusion proteins. Cells were imaged as described in Figure 7 and the distances between lamin A and matrin-3 were determined. LMNAΔ303 expression was associated with larger distances between the nuclear membrane and the nuclear matrix, similar to what was observed for human cells in Figure 7. (***P < 0.0001). The results were similar between expression in (B) C2C12 cells or (C) HEK cells.
Discussion
LAT-interacting proteins
A proteomics-based approach was used to identify a novel interaction between lamin A and matrin-3. Matrin-3 was first described as a residual protein component after high salt extraction of nuclei, and this same approach identified other insoluble nuclear proteins such as lamins A, B and C (19). Matrin-3 is a ∼125 kDa protein (847 amino acids) that contains two zinc finger domains and two RBM domains, domains that are implicated in binding double- and single-stranded nucleic acids, respectively. Matrin-3 has been shown to bind DNA (20,21) and also both RNA and RNA binding proteins (22). Salton et al. showed that matrin-3 bound to hnRNPK and DHX9 in an RNA-dependent manner, two proteins that were also found as part of the constellation of proteins binding LATs in this study.
The interaction between lamin A and matrin-3
There was significant overlap in the proteins bound to mutant and wild-type LATs identified by mass spectrometry. The two LMNA mutants R453W and R527P map to the Ig fold of the LAT, but are positioned on different surfaces of the folded structure (5). LMNA R453W bound matrin-3 more tightly than wild-type LAT and R527P, which suggests that one face of lamin A's Ig fold may be more critical for the interaction with matrin-3. Three arginine-serine enriched proteins, TRA2B, SFRS13A and SRRM1, linked to processing and spicing of pre-mRNAs were more enriched in the proteomic spectra associated with R527P and R453W compared with wild-type LAT. This association with splicing factors could reflect direct or indirect association between lamin A and these proteins.
A common morphological defect described in LMNA mutant cells is the presence of nuclear blebs. Nuclei from Lmna mutant mice, deleted for exons 8–11, are often grossly deformed with large evaginations seen in a significant percentage of cells (33). These same nuclear deformities are often seen in cultured cells from patients with dominant myopathy-associated LMNA mutations (34,35). It has been suggested that misshapen nuclei form more readily in Lmna mutant cells especially in those cell types that are subject to physical forces such as cardiac and muscle cells (36). The underlying cause of nuclear blebbing in LMNA mutant cells is not known, but blebbing increases with increasing cell passage (F. Depreux, M.J. Puckelwartz and E.M. McNally, unpublished data). An altered or uneven attachment between the nuclear membrane and the nuclear scaffold would predispose nuclei to form blebs. The finding that matrin-3 and lamin A are co-localized in differentiated cells may explain the susceptibility of these cells types to specific LMNA mutations.
The matrin-3 interface with lamin A in post-mitotic muscle cells
Biochemical characterization identified matrin-3 in the insoluble nuclear fraction (37), and immunolocalization and expression studies have placed matrin-3 in the nucleoplasmic space and not at the nuclear periphery (20). We now found that a fraction of matrin-3 resides at the nuclear membrane, and that this intracellular localization was detected with antibodies directed to the amino-terminus of matrin-3. The carboxy-terminus of matrin-3 binds to lamin A, and therefore may mask the nuclear rim-associated matrin-3 protein.
A series of elegant experiments described the mechanical defects attributed to lamin A. Fibroblasts from Lmna null mice subjected to biaxial strain demonstrate far greater nuclear deformation than wild-type cells with an intact and normal nuclear lamina (36). Additionally, the cytoskeletal stiffness of Lmna null fibroblasts is significantly reduced compared with what is observed in normal mouse fibroblasts (38). These mechanical defects were also shown to be associated with changes in the expression of genes known to be responsive to mechanical stimuli, iex-1 and egr-1 (36). In an alternative assay for mechanical deficits, Broers et al. (39) compressed fibroblasts from Lmna null and wild-type mice and observed greater nuclear disruption in Lmna null cells compared with control. Moreover, compression of Lmna null cells led to nuclear rupture that was further associated with leakage of nuclear contents into the cytoplasm consistent with loss of nuclear membrane integrity. Introduction of lamin A or lamin C into the Lmna cells rescued these defects (39). It is possible that mechanical weakness in Lmna mutant cells may derive from an altered attachment to the nuclear scaffold.
The nuclear lamina forms as a meshwork underlying the inner nuclear membrane (40). We hypothesize that the matrix, through matrin-3, forms a Velcro-like connection with the nuclear lamina. In this model, mutations in LMNA would alter contact points between the lamina and nucleoplasmic contents. Reduced or uneven adhesion between the lamina and the nucleoplasmic matrix could render a cell unusually susceptible to mechanical force. Most recently matrin-3 mutations were linked to amyotrophic lateral sclerosis (41). Therefore, these mechanisms may contribute to disease in other post-mitotic cells.
Materials and Methods
Constructs and mutagenesis
LAT was expressed using pET-LAT, which encodes amino acids 394–664 (18). LMNA R453W and R527P mutations were generated using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene Inc., La Jolla, CA). A human Matrin-3 cDNA (Clone 3921668; Open Biosystems Products, Huntsville, Alabama) was used as a PCR template to generate matrin fragments and for site-directed mutagenesis. All clones were confirmed by sequencing.
LAT protein expression, purification and crosslinking
Escherichia coli BL21(DE3)pLysS (Invitrogen, Carlsbad, CA) were used for expression. Expression was induced using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 30°C. Bacteria were resuspended in 50 mm NaPO4 buffer pH 8.0, 300 mm NaCl, 5 mm benzamine, 0.5 mm phenylmethanesulfonylfluoride (PMSF), 1 µg/ml pepstatin A, 42 µm leupeptin, 0.77 µm aprotinin and sonicated. Lysates were applied to an Ni-NTA Agarose bead column (Qiagen, Valencia, CA), washed with 50 mm NaPO4 buffer pH 8.0, 300 mm NaCl, 10 mm imidazole. His-tagged LAT proteins were eluted using 50 mm NaPO4 buffer pH 8.0, 300 mm NaCl, 100 mm imidazole (18). The elution buffer was exchanged with coupling citrate-carbonate buffer pH 10.0 (0.1 m Na-Citrate dehydrated, 0.05 m Na-carbonate, 0.05% Na-azide). One mg of LAT protein was coupled with 1 ml of AminoLink Plus resin (Pierce, Rockford IL) to generate LAT beads.
Nuclear protein extracts
Myogenic C2C12 cells (ATCC #CRL-1772) were differentiated to form myotubes in differentiation media (Dulbecco's modified Eagle's medium, 2% horse serum, 1% penicillin/streptomycin) for 4 days and harvested. Nuclear cell extracts were prepared as described (42,43). Cell pellets were rinsed in cold phosphate buffered saline (PBS), 2 mm DTT with proteinase inhibitors: 5 mm benzamine, 0.5 mm PMSF, 1 µg/ml pepstatin A, 42 µm leupeptin, 0.77 µm aprotinin. After centrifugation at 400g for 5 min, cells were incubated in ice-cold H20, 2 mm DTT and proteinase inhibitors for 15 min and then dounced 20 times on ice. After adjustment to 1× PBS and the addition of 2 mm MgCl2, genomic DNA was digested using Benzonase (Sigma-Aldrich, St Louis, MO; 125 U/ml) for 90 min on ice. The cells were washed three times with 1× cold PBS, 2 mm DTT with proteinase inhibitors and centrifuged at 400g for 5 min to separate into the cytoplasmic and nuclear fractions. The nuclear fraction was incubated with 1 m NaCl, 50 mm HEPES, pH 7.9, 1% Triton X-100, 2 mm DTT with proteinase inhibitors vortexing for 30 sec followed by 30 sec on ice. The solution was centrifuged at 40 000g for 30 min at 4°C. The soluble fraction was diluted to 150 mm NaCl range with 50 mm HEPES pH 7.9, 2 mm DTT with proteinase inhibitors and spun at 40 000g for 30 min at 4°C.
LAT beads were washed with 50 mm HEPES pH 7.9, 150 mm NaCl, 0.1% Triton X-100 and then incubated with 2.5 mg of C2C12 myotube nuclear extract for 4 h at 4°C. Columns were washed with 50 mm HEPES pH 7.9, 150 mm NaCl and proteins were eluted using 1 m NaCl, 50 mm HEPES pH 7.9.
Mass spectrometry for protein identification
Of note, 100 µl of eluate for each of the samples was separated by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Imperial Protein Stain (Pierce, Rockford, IL). The gels were separated into 1 mm3 pieces, washed in water and completely destained using 100 mm ammonium bicarbonate pH 7.5 in 50% acetonitrile. A reduction step was performed by addition of 100 µl 50 mm ammonium bicarbonate pH 7.5 and 10 µl of 10 mm tris(2-carboxyethyl) phosphine HCl at 37°C for 30 min. The proteins were alkylated by adding 100 µl 50 mm iodoacetamide in the dark at 20°C for 30 min. Gel sections were washed in water, dehydrated with acetonitrile and dried by SpeedVac. Trypsin digestion was carried out overnight at 37°C using a trypsin/protein ratio of 1:50 (w/w) with sequencing grade-modified trypsin (Promega, Madison WI) in 50 mm ammonium bicarbonate pH 7.5, and 20 mm CaCl2. Peptides were extracted from the gel pieces with 5% formic acid and dried. The peptide samples were loaded to a 0.25 µl OptiPak trapping cartridge (Optimize Technologies, Oregon City, OR) packed with Michrom Magic C8, 5 µm, 200 Å particles (Michrom BioResources, Inc., Auburn, CA), washed, then switched in-line with a 20 cm×75 µm C18 ‘packed spray tip’ nanocolumn packed with Michrom Magic C18AQ, 5 µm, 200 Å, for a two-step gradient.
The eluted peptides were analyzed via electrospray tandem mass spectrometry (LC-MS/MS) on a Thermo LTQ Orbitrap Hybrid FT Mass Spectrometer using a 60,000 RP Orbitrap survey scan, m/z 375-1950, with lockmasses, followed by five LTQ CAD scans on doubly and triply charged-only precursors between 375 and 1500 Da.
Analysis of LC/MS/MS spectra
All LC/MS/MS data files were analyzed on Mascot (Matrix Science Ltd, London, UK; Version 2.3.0) and X! Tandem (Version 2007; 01.01.1) and results were combined using Scaffold (Proteome Software, Portland, OR; Version 3.00.01). Mascot settings for fragment and parent ion tolerances were at 1.00 Da and 20 ppm, respectively and used to search the mouse database (IPI_mouse_20101209.fasta) assuming trypsin digestion with one missed cleavage maximum. Scaffold identification thresholds were set at 95 and 99% minimum for Peptide and Protein respectively, with a minimum of one peptide to identify a protein. The false-discovery rate for peptide and protein were set to zero.
Four independent experiments were analyzed for each LAT protein and normalized to proteins eluted from control beads prepared without LAT but similarly incubated with nuclear extracts. Proteins enriched at least 2-fold in at least one experiment were further analyzed. Gene ID and GO annotations were generated using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov) (44,45) and MGI (Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, ME, http://www.informatics.jax.org) databases. Molecular function annotation was performed using the MGI GO Slim chart. Biological Process fold was calculated using the DAVID database. Nuclear localization was generated using the Cellular Component annotation from DAVID and MGI databases. A final predicted nuclear protein list was generated based on GO annotation excluding keratin and myosin proteins.
Immunofluorescence microscopy
Myoblast and fibroblast cultures were fixed 10 min in cold methanol, and permeabilized for 20 min with 0.1% Triton X-100/PBS or with 0.5% Triton X-100/PBS for myotube cultures. Muscle was isolated from murine skeletal muscle and processed as described (46). Cells or tissue was blocked with 3% normal donkey serum in PBS (NDS/PBS) for 1 h at room temperature. Primary antibodies recognizing Matrin-3 (carboxy-terminal antibody NB100-2886, Novus Biological, Littleton CO, recognizes between 800 and 847 amino acids and the amino-terminal antibody, ab51081 from Abcam, Cambridge, MA) and Lamin A/C [XB10, Abcam, Cambridge, MA or Jol5 (Abcam)] were diluted in 3% NDS/PBS and incubated overnight at 4°C. Secondary antibodies Anti-rabbit Alexa Fluor 594 (Invitrogen) and Anti-mouse IgM Dylight 488 (Jackson ImmunoResearch, West Grove, PA) were each diluted 1/500 in 3% NDS/PBS and incubated 1 h at room temperature.
Images were captured with an Axiophot microscope and processed using Axiovision software (Carl Zeiss, Oberkochen, Germany) or a Leica SP5 II STED-CW laser scanning confocal microscope (Leica Microsystems, Buffalo Grove, IL). A Z-stack of each nucleus (10 per genotype) was generated with optical sections of 0.3 µm between images. A 3D reconstruction of each nucleus was created using IMARIS (v7.3.1, Bitplane Scientific Software, Zurich, Switzerland). From the 3D image, surface fits of both Matrin-3 (red channel) and Lamin A/C (green channel) were generated as follows: Z-stack images were cropped in 3D view and smoothed using the median filter (with a kernel of 3 × 3 × 3). For both channels, surface units were generated after background subtraction with diameter of the largest sphere of 0.3 µm. Manual threshold values were 100 and 150 gray levels for green and red channels, respectively. Distance measurements between outside unit surfaces of Lamin A/C to Matrin-3 and Matrin-3 to Lamin A/C were calculated using MATLAB extension of IMARIS software.
Immunoprecipitation
One hundred micrograms of C2C12 myotube nuclear extract protein was pre-cleared by addition of 100 µl of Protein G Plus/Protein A agarose beads (Millipore, Billerica, MA) for 20 min at 4°C. Pre-cleared nuclear extracts were incubated with 5 µg of Rabbit IgG or two different rabbit polyclonal anti-human Matrin-3 antibodies (NB100-2886 and NB100-1761, Novus Biologicals). Anti-lamin A/C (H110, Santa Cruz, Santa Cruz, CA) or Matrin-3 (NB100-1761, Novus) antibodies was used for immunoblotting. Secondary antibodies were from Jackson Immunochemicals.
Binding assays
35S-radio-labeled Matrin-3 full-length or subdomains were generated using T7 TnT coupled Reticulocyte Lysate Systems (Promega, Madison, WI). The PVDF membrane containing LATs was blocked for 2 h at 4°C in blocking buffer [0.1% gelatin, 5% bovine serum albumin (BSA), 0.1% Tween-20 in 1× PBS, pH 7.5] and then incubated overnight at 4°C in overlay buffer (150 mm NaCl, 20 mM HEPES pH 7.5, 2 mm MgCl2, 5% BSA) with 35S-radio-labeled Matrin-3 full length or subdomains (5 µl/ml of overlay buffer). The membrane was washed four times for 20 min at 4°C in overlay buffer without BSA and exposed to film overnight at −80°C (24).
Statistical tests were performed using Prism4 statistical software (GraphPad Software, La Jolla, CA).
Supplementary Material
Funding
This work was supported by a funding from NIH HL092443, HL012875, the Doris Duke Charitable Foundation, and the Chicago Biomedical Consortium.
Supplementary Material
Acknowledgements
We thank the patients for their participation.
Conflict of Interest statement. None declared.
References
- 1.Dechat T., Adam S.A., Taimen P., Shimi T., Goldman R.D. (2010) Nuclear lamins. Cold Spring Harb. Perspect. Biol., 2, a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. (2005) The nuclear lamina comes of age. Nat. Rev. Mol. Cell. Biol., 6, 21–31. [DOI] [PubMed] [Google Scholar]
- 3.Zuela N., Bar D.Z., Gruenbaum Y. (2012) Lamins in development, tissue maintenance and stress. EMBO Rep., 13, 1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand A.T., Chikhaoui K., Yaou R.B., Bonne G. (2011) Clinical and genetic heterogeneity in laminopathies. Biochem. Soc. Trans., 39, 1687–1692. [DOI] [PubMed] [Google Scholar]
- 5.Krimm I., Ostlund C., Gilquin B., Couprie J., Hossenlopp P., Mornon J.P., Bonne G., Courvalin J.C., Worman H.J., Zinn-Justin S. (2002) The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure (Camb), 10, 811–823. [DOI] [PubMed] [Google Scholar]
- 6.Heitlinger E., Peter M., Lustig A., Villiger W., Nigg E.A., Aebi U. (1992) The role of the head and tail domain in lamin structure and assembly: analysis of bacterially expressed chicken lamin A and truncated B2 lamins. J Struct Biol, 108, 74–89. [DOI] [PubMed] [Google Scholar]
- 7.Isobe K., Gohara R., Ueda T., Takasaki Y., Ando S. (2007) The last twenty residues in the head domain of mouse lamin A contain important structural elements for formation of head-to-tail polymers in vitro. Biosci. Biotechnol. Biochem., 71, 1252–1259. [DOI] [PubMed] [Google Scholar]
- 8.Senderek J., Garvey S.M., Krieger M., Guergueltcheva V., Urtizberea A., Roos A., Elbracht M., Stendel C., Tournev I., Mihailova V., et al. (2009) Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am. J. Hum. Genet., 84, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feit H., Silbergleit A., Schneider L.B., Gutierrez J.A., Fitoussi R.P., Reyes C., Rouleau G.A., Brais B., Jackson C.E., Beckmann J.S., et al. (1998) Vocal cord and pharyngeal weakness with autosomal dominant distal myopathy: clinical description and gene localization to 5q31. Am. J. Hum. Genet., 63, 1732–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montes de Oca R., Shoemaker C.J., Gucek M., Cole R.N., Wilson K.L. (2009) Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One, 4, e7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairley E.A., Kendrick-Jones J., Ellis J.A. (1999) The Emery–Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J. Cell Sci., 112 (Pt 15), 2571–2582. [DOI] [PubMed] [Google Scholar]
- 12.Yuan J., Simos G., Blobel G., Georgatos S.D. (1991) Binding of lamin A to polynucleosomes. J. Biol. Chem., 266, 9211–9215. [PubMed] [Google Scholar]
- 13.Taniura H., Glass C., Gerace L. (1995) A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol, 131, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg M., Harel A., Brandeis M., Rechsteiner T., Richmond T.J., Weiss A.M., Gruenbaum Y. (1999) The tail domain of lamin Dm0 binds histones H2A and H2B. Proc. Natl. Acad. Sci. USA, 96, 2852–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattout A., Goldberg M., Tzur Y., Margalit A., Gruenbaum Y. (2007) Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J. Cell Sci., 120, 77–85. [DOI] [PubMed] [Google Scholar]
- 16.Kubben N., Voncken J.W., Demmers J., Calis C., van Almen G., Pinto Y., Misteli T. (2010) Identification of differential protein interactors of lamin A and progerin. Nucleus, 1, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Q., Worman H.J. (1995) Protein–protein interactions between human nuclear lamins expressed in yeast. Exp. Cell. Res., 219, 292–298. [DOI] [PubMed] [Google Scholar]
- 18.Zastrow M.S., Flaherty D.B., Benian G.M., Wilson K.L. (2006) Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci., 119, 239–249. [DOI] [PubMed] [Google Scholar]
- 19.Nakayasu H., Berezney R. (1991) Nuclear matrins: identification of the major nuclear matrix proteins. Proc. Natl. Acad. Sci. USA, 88, 10312–10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibino Y., Usui T., Morita Y., Hirose N., Okazaki M., Sugano N., Hiraga K. (2006) Molecular properties and intracellular localization of rat liver nuclear scaffold protein P130. Biochim. Biophys. Acta, 1759, 195–207. [DOI] [PubMed] [Google Scholar]
- 21.Hibino Y., Ohzeki H., Sugano N., Hiraga K. (2000) Transcription modulation by a rat nuclear scaffold protein, P130, and a rat highly repetitive DNA component or various types of animal and plant matrix or scaffold attachment regions. Biochem. Biophys. Res. Commun., 279, 282–287. [DOI] [PubMed] [Google Scholar]
- 22.Salton M., Elkon R., Borodina T., Davydov A., Yaspo M.L., Halperin E., Shiloh Y. (2011) Matrin 3 binds and stabilizes mRNA. PLoS One, 6, e23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller T.J., Kraya T., Stoltenburg-Didinger G., Hanisch F., Kornhuber M., Stoevesandt D., Senderek J., Weis J., Baum P., Deschauer M., et al. (2014) Phenotype of matrin-3-related distal myopathy in 16 German patients. Ann Neurol., 76, 669–680. [DOI] [PubMed] [Google Scholar]
- 24.Posey A.D., Jr, Pytel P., Gardikiotes K., Demonbreun A.R., Rainey M., George M., Band H., McNally E.M. (2011) Endocytic recycling proteins EHD1 and EHD2 interact with fer-1-like-5 (Fer1L5) and mediate myoblast fusion. J. Biol. Chem., 286, 7379–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martelli A.M., Bortul R., Tabellini G., Faenza I., Cappellini A., Bareggi R., Manzoli L., Cocco L. (2002) Molecular characterization of protein kinase C-alpha binding to lamin A. J. Cell. Biochem., 86, 320–330. [DOI] [PubMed] [Google Scholar]
- 26.Al-Haboubi T., Shumaker D.K., Koser J., Wehnert M., Fahrenkrog B. (2011) Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus, 2, 500–509. [DOI] [PubMed] [Google Scholar]
- 27.Muralikrishna B., Dhawan J., Rangaraj N., Parnaik V.K. (2001) Distinct changes in intranuclear lamin A/C organization during myoblast differentiation. J. Cell Sci., 114, 4001–4011. [DOI] [PubMed] [Google Scholar]
- 28.Mislow J.M., Kim M.S., Davis D.B., McNally E.M. (2002) Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci., 115, 61–70. [DOI] [PubMed] [Google Scholar]
- 29.MacLeod H.M., Culley M.R., Huber J.M., McNally E.M. (2003) Lamin A/C truncation in dilated cardiomyopathy with conduction disease. BMC Med. Genet., 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Jr, Spudich S., De Girolami U., et al. (1999) Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med., 341, 1715–1724. [DOI] [PubMed] [Google Scholar]
- 31.Kass S., MacRae C., Graber H.L., Sparks E.A., McNamara D., Boudoulas H., Basson C.T., Baker P.B., 3rd, Cody R.J., Fishman M.C., et al. (1994) A gene defect that causes conduction system disease and dilated cardiomyopathy maps to chromosome 1p1–1q1. Nat. Genet., 7, 546–551. [DOI] [PubMed] [Google Scholar]
- 32.Haass-Koffler C.L., Naeemuddin M., Bartlett S.E. (2012) An analytical tool that quantifies cellular morphology changes from three-dimensional fluorescence images. J. Vis. Exp., 66, e4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell. Biol., 147, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raharjo W.H., Enarson P., Sullivan T., Stewart C.L., Burke B. (2001) Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery–Dreifuss muscular dystrophy. J. Cell. Sci., 114, 4447–4457. [DOI] [PubMed] [Google Scholar]
- 35.Muchir A., Medioni J., Laluc M., Massart C., Arimura T., van der Kooi A.J., Desguerre I., Mayer M., Ferrer X., Briault S., et al. (2004) Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve, 30, 444–450. [DOI] [PubMed] [Google Scholar]
- 36.Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., Lee R.T. (2004) Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest., 113, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belgrader P., Dey R., Berezney R. (1991) Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J. Biol. Chem., 266, 9893–9899. [PubMed] [Google Scholar]
- 38.Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., Lee R.T. (2006) Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem., 281, 25768–25780. [DOI] [PubMed] [Google Scholar]
- 39.Broers J.L., Peeters E.A., Kuijpers H.J., Endert J., Bouten C.V., Oomens C.W., Baaijens F.P., Ramaekers F.C. (2004) Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet., 13, 2567–2580. [DOI] [PubMed] [Google Scholar]
- 40.Aebi U., Cohn J., Buhle L., Gerace L. (1986) The nuclear lamina is a meshwork of intermediate-type filaments. Nature, 323, 560–564. [DOI] [PubMed] [Google Scholar]
- 41.Johnson J.O., Pioro E.P., Boehringer A., Chia R., Feit H., Renton A.E., Pliner H.A., Abramzon Y., Marangi G., Winborn B.J., et al. (2014) Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci., 17, 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schirmer E.C., Gerace L. (2005) The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci., 30, 551–558. [DOI] [PubMed] [Google Scholar]
- 43.Kubben N., Voncken J.W., Misteli T. (2010) Mapping of protein- and chromatin-interactions at the nuclear lamina. Nucleus, 1, 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang da W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 45.Huang da W., Sherman B.T., Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res., 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puckelwartz M.J., Kessler E., Zhang Y., Hodzic D., Randles K.N., Morris G., Earley J.U., Hadhazy M., Holaska J.M., Mewborn S.K., et al. (2009) Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet, 18, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.