Abstract
Obesity is an important component of the pathophysiology of chronic diseases. Identifying epigenetic modifications associated with elevated adiposity, including DNA methylation variation, may point to genomic pathways that are dysregulated in numerous conditions. The Illumina 450K Bead Chip array was used to assay DNA methylation in leukocyte DNA obtained from 2097 African American adults in the Atherosclerosis Risk in Communities (ARIC) study. Mixed-effects regression models were used to test the association of methylation beta value with concurrent body mass index (BMI) and waist circumference (WC), and BMI change, adjusting for batch effects and potential confounders. Replication using whole-blood DNA from 2377 White adults in the Framingham Heart Study and CD4+ T cell DNA from 991 Whites in the Genetics of Lipid Lowering Drugs and Diet Network Study was followed by testing using adipose tissue DNA from 648 women in the Multiple Tissue Human Expression Resource cohort. Seventy-six BMI-related probes, 164 WC-related probes and 8 BMI change-related probes passed the threshold for significance in ARIC (P < 1 × 10−7; Bonferroni), including probes in the recently reported HIF3A, CPT1A and ABCG1 regions. Replication using blood DNA was achieved for 37 BMI probes and 1 additional WC probe. Sixteen of these also replicated in adipose tissue, including 15 novel methylation findings near genes involved in lipid metabolism, immune response/cytokine signaling and other diverse pathways, including LGALS3BP, KDM2B, PBX1 and BBS2, among others. Adiposity traits are associated with DNA methylation at numerous CpG sites that replicate across studies despite variation in tissue type, ethnicity and analytic approaches.
Introduction
Epigenetics is the study of mitotically heritable modifications in chromatin structure not involving the underlying DNA sequence, and their impact on the transcriptional control of genes and cellular function. Of the different forms of epigenetic modification, DNA methylation is the most extensively studied and involves the addition and removal of methyl (-CH3) groups at CpG dinucleotides to influence regional DNA transcription (1). Although genome-wide demethylation and re-methylation occur during embryogenesis and established patterns must be set to initiate differentiation and maintain cell type-specific gene expression (2), DNA methylation and other features of the epigenome are also modifiable throughout the life course by environmental and behavioral exposures such as the nutrient content of the maternal diet (3), cigarette smoking (4) and environmental pollutants (5). Because of their role in gene expression, alterations in epigenetic patterns are a mechanism by which these and other environmental factors may increase risk of disease (6,7).
As individuals accrue excess adipose tissue, they experience chronic low-grade inflammation, associated with immunological activation and oxidative stress (8,9), as well as insulin resistance, hypertension and dyslipidemia (10). These features explain, in part, why obesity is among the strongest modifiable risk factors for diabetes, atherosclerosis and some cancers (11–19). In an epigenetic framework, obesity can be seen as an environmental factor that exposes the genome in many tissues to a suite of systemic factors [e.g. elevated circulating C-reactive protein, interleukin (IL) 6], potentially altering DNA methylation or histone protein acetylation patterns. Identifying epigenetic modifications associated with obesity may therefore point to genomic pathways that are dysregulated in numerous conditions.
As of 2015, only a small number of studies had been published showing obesity-related variation in DNA methylation, with most studies generally using either targeted repeat sequence (global methylation) or candidate gene-centric approaches. With some exceptions (20,21), such studies have not yielded results that have been replicated in independent cohorts (reviewed by Drong et al.) (7). Recent technological advances have provided platforms for systematically interrogating DNA methylation variation across the genome (22), paving the way for epigenome-wide association studies (EWASs), analogous to genome-wide association studies, to identify regions of the genome-harboring DNA methylation variation associated with disease phenotypes (6). EWASs of obesity traits have shown that methylation variants are influenced by nearby genetic variants (i.e. are haplotype specific) as in the case of the well-documented obesity gene FTO (23–26). To date, only one obesity EWAS has yielded a novel replicated locus (CpG sites in HIF3A) (27), and only one obesity EWAS has been conducted in African Americans (25), despite the fact that African-ancestry groups tend to carry higher chronic disease risk factor loads, including greater obesity, compared with European-ancestry populations (28–30).
The goal of the present investigation is to advance our understanding of the methylation signatures associated with obesity traits using leukocyte DNA samples from over 2000 African American adults, many of whom were overweight or obese at the time of DNA collection, with replication in three independent cohorts. Our findings replicate those from recent EWAS of body mass index (BMI), lipid and diabetes-related traits and identify a number of novel associations with BMI, waist circumference (WC) and BMI change. The results are a step toward understanding the pathophysiology of obesity and identifying new molecular targets to avert its negative health consequences.
Results
Description of discovery sample
The ARIC Study is a prospective cohort study of cardiovascular disease risk in White and Black adults from four US communities (31). Subjects were seen at baseline (Visit 1) in 1987–1989, with four follow-up visits (Visits 2–5) thereafter. The study sample for the present investigation (N = 2097; 64% female) includes only those with methylation data (all of whom are African American). Average age was 56 years at the time of Visit 2 when DNA methylation data and adiposity measures were both available. Subjects had mean BMI, WC and BMI change of 30.1 kg/m2, 101.3 cm and 7.0 kg/m2 (6.0), respectively. Most of the subjects were overweight (37%) or obese (44%) and 67% exceeded National Heart, Lung, and Blood Institute (NHLBI)-recommended WC limits (>88 cm for women and >102 cm for men). Prevalent diabetes was present in 26% of the participants. Imputed white blood cell (WBC) count differentials were obtained for all subjects, and the mean proportions of each cell type as well as other study covariates are provided in Table 1. A flowchart (Fig. 1) outlines the results of the subsequent analyses, detailed later.
Table 1.
Characteristics of the study sample: N = 2097 African American adults in the ARIC study
| Mean (SD) | Range | N | |
|---|---|---|---|
| Age (years) | 56.2 (5.7) | 47–70 | 2097 |
| BMI (kg/m2) | 30.1 (6.1) | 14.7–62.4 | 2096 |
| WC (cm) | 101.3 (15.1) | 61–163 | 2097 |
| BMI change (kg/m2) | 7.0 (6.0) | −25.9–36.4 | 2039 |
| Physical activity (scale of 1–5)a | 2.1 (0.6) | 1–4.25 | 2097 |
| WBC count (1000/mm3) | 5.6 (1.9) | 2.0–28.8 | 2097 |
| N (proportion) | |||
| Sex | |||
| Male | 763 (0.36) | 2097 | |
| Female | 1334 (0.64) | ||
| Field center | |||
| Forsyth County, NC | 157 (0.07) | 2097 | |
| Jackson, MS | 1940 (0.93) | ||
| BMI statusb | |||
| Underweight (BMI < 18.5 kg/m2) | 19 (0.01) | 2095 | |
| Normal weight (BMI 18.5–24.99 kg/m2) | 369 (0.18) | ||
| Overweight (BMI 25.0–29.99 kg/m2) | 789 (0.37) | ||
| Obese (≥30.0 kg/m2) | 918 (0.44) | ||
| WC statusb | |||
| Normal | 698 (0.33) | 2097 | |
| Elevated | 1399 (0.67) | ||
| Cigarette smoking | |||
| Current smoker | 512 (0.24) | 2097 | |
| Current nonsmoker | 1585 (0.76) | ||
| Alcohol use | |||
| Current drinker | 717 (0.34) | 2097 | |
| Current nondrinker | 1380 (0.66) | ||
| Education | |||
| <High school | 843 (0.40) | 2097 | |
| High school graduate | 581 (0.28) | ||
| >High school | 673 (0.32) | ||
| Household income | |||
| <$16 000 | 1087 (0.52) | 2097 | |
| $16 000–$24 999 | 379 (0.18) | ||
| $25 000–$34 999 | 278 (0.13) | ||
| $35 000–$49 999 | 202 (0.10) | ||
| >$50 000 | 151 (0.07) | ||
| Differential WBC proportionsc | |||
| Neutrophils | 0.56 | 2097 | |
| Lymphocytes | 0.36 | ||
| Monocytes | 0.05 | ||
| Eosinophils | 0.03 | ||
| Diabetes statusd | |||
| No | 1539 (0.74) | 2087 | |
| Yes | 548 (0.26) | ||
aSelf-reported leisure time physical activity using the Baecke Questionnaire at Visit 1.
bBMI status: underweight (BMI <18.5 kg/m2), normal weight (BMI 18.5–24.99 kg/m2), overweight (BMI 25.0–29.99 kg/m2) and obese (≥30.0 kg/m2). WC status: low risk (WC ≤88 cm for women and ≤102 cm for men) and high risk (WC >88 cm for women and >102 cm for men).
cDifferential WBC proportions were imputed using the method of Houseman et al. (32) using measured differentials from a subset of 179 ARIC subjects as the reference.
dPrevalent diabetes at the time of the DNA collection was defined as a self-reported physician diagnosis of or treatment for diabetes.
Figure 1.
Flowchart of experiments and summary of results.
Gene pathway analysis
To test the global hypothesis that there are significant associations of adiposity traits with methylation variation, we conducted a genome-wide gene ontology (GO) pathway analysis for BMI and WC including all CpGs, using the same statistical models as in the subsequent EWAS. The approach accounted for the degree of clustering of probes included in the HM450 in particular genes and gene regions. Results yielded significant [false discovery rate (FDR)-adjusted q-value < 0.05] enrichment of over 100 biological process pathways for BMI and 90 biological process pathways for WC (Supplementary Material, Tables S1 and S2), involving 11–204 genes per pathway. These pathways represent a diverse range of processes, particularly related to neuronal function (e.g. neuron migration and neuroblast proliferation), immune response/cytokine signaling (e.g. humoral immune response and positive regulation of IL-6 production) and energy and fatty acid metabolism [e.g. tricarboxylic acid (Krebs’) cycle and arachidonic acid secretion].
Association study
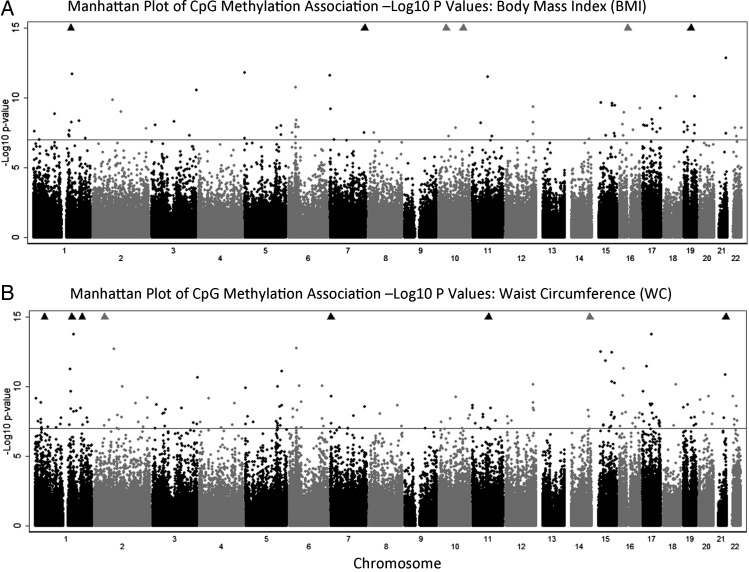
Manhattan plots showing the −log10 P-values for individual autosomal CpG probe associations for BMI and WC are provided in Figure 2. A total of 76 probes passed the threshold for genome-wide significance for BMI, and 164 probes passed this threshold for WC. For BMI change, eight probes passed this threshold, all of which were also among the statistically significant BMI and/or WC probes (P < 1 × 10−7) (Supplementary Material, Table S3), including cg15871086 (Chr. 18 intergenic, P = 8.77E − 10), cg09554443 (near CD247, P = 2.68E − -09), cg00574958 [near carnitine palmitoyltransferase-1A (CPT1A), P = 4.30E − 08] and cg16672562 [near hypoxia-inducible factor 3 (HIF3A), P = 8.60E − 08]. CPT1A methylation probe associations have been recently reported to be associated with atherogenic lipoprotein subfractions in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) cohort (33). Methylation at cg16672562 near HIF3A was recently reported to be associated with BMI in ∼2500 European adults in the Cardiogenetics Consortium, MARseille THrombosis Association and Cooperative Health Research in the Region of Augsburg cohorts (27). BMI change associations are not discussed in further detail due to their complete overlap with BMI and WC results.
Figure 2.
Manhattan plot of CpG methylation association −log10 P-values in 2107 African American adults in the ARIC study. (A) BMI and (B) WC.
Regression coefficients, standard errors, P-values and CpG marker information for the significant associations for BMI and WC in the ARIC discovery sample are provided in Supplementary Material, Tables S4 and S5, respectively. For BMI, the top CpG (cg06500161, P = 1.52E − 13) explained ∼2.6% of variation in BMI and is located in a CpG island shore in the gene body of ABCG1 [ATP-binding cassette, subfamily G (WHITE), member 1]. This gene is expressed in blood plasma and platelets and is involved in macrophage cholesterol and phospholipids transport, and cellular regulation of lipid homeostasis (34). ABCG1 promoter hypermethylation is strongly associated with coronary heart disease (CHD) (35), and this particular CpG was also associated with insulin-related traits in the GOLDN cohort (33). A second CpG site near ABCG1 also was among the top results (cg27243685, P = 3.61E − 08). The two CpGs are ∼14 kb distant from one another; their mean methylation beta values were significantly different from one another (mean beta value for cg006500161 = 0.62; mean beta value for cg27243685 = 0.85; unpaired t-test P-value < 0.0001) and were moderately inter-correlated (r = 0.32; P < 0.0001). Other loci exhibiting two or more significant CpG associations included the smoking-related methylation locus AHRR (cg23576855 and cg05575921), HIF3A (cg27146050, cg22891070 and cg16672562) with the same three CpG sites previously reported by Dick et al. (27) to be associated with BMI, the innate immune response regulator NFKBIL1 (cg21053741 and cg21587837), the histone H3 demethylase KDM2B (cg15695155, cg26995224 and cg13708645), the natural killer immune response gene LGALS3BP (cg04927537 and cg25178683) and NWD1 of unknown function (cg15845821 and cg19344626). Conditional regression models (which required switching BMI to the dependent variable) were run for loci with >1 CpG association and showed no evidence for multiple independent signals within these loci. After ABCG1 and AHRR, the third-ranked CpG association was near the CD247 gene, involved in T-cell receptor signaling, immune response and IL-12-induced interferon gamma (IFNγ) production. As a note, it is likely that the observed CpG associations near AHRR were the result of residual confounding; our EWAS was conducted using smoking coded as current/noncurrent, and when we further adjusted the results for former smoking and total pack years of smoking, the associations with BMI were greatly attenuated and no longer statistically significant (data not shown).
There was considerable overlap between results for BMI and WC; 56 of the BMI associations (74%) were also significant for WC and the top CpG was the same for both traits (cg06500161 near ABCG1, P for WC = 4.41E − 19). This CHD-associated marker explained ∼3.6% of variation in WC. The second-ranked probe in the WC analysis was cg00574958 (P = 5.79E − 17) near CPT1A, also mentioned earlier. Additional high-ranking probes in our WC EWAS included a site near LY6G6E (cg13123009, P = 1.8 × 10–13), which belongs to a cluster of leukocyte antigen-6 (LY6) genes located in the major histocompatibility complex (MHC) class III region on chromosome 6, and C7orf50, a longevity locus (36). It is generally noted that probes passing the threshold for significance for BMI and WC in ARIC explained 0.2–3.6% trait variance, included CpG sites across the spectrum of average methylation values (minimum 0.04 to maximum 0.95 mean methylation) and had intra-class correlation (ICC) coefficient values for replicates of >0.35 (Supplementary Material, Tables S4 and S5). Most CpG markers had ICC > 0.60, suggesting higher probe reliability/quality as measured by ICC increases the probability of detecting association.
Replication of association results
We took forward the 76 BMI-associated probes for replication in the Framingham Heart Study (FHS) (methylation data for which subjects were generated in one of two laboratories and considered separately here) and GOLDN. Details on the replication cohorts are found in the Supplementary Text. Direction and P-value for each BMI probe are provided by cohort, and meta-analysis P-values for the replication (FHS I + FHS II + GOLDN) are provided in Table 2. The direction of association was consistent across both replication cohorts for 47/76 BMI probes [62%, z statistic for equal proportions for null hypothesis that each of two cohorts was concordant with ARIC (null = 12.5%, or 0.5 × 0.5 × 0.5); z = 5.66, P = 0.0001]. Of these 47, meta-analysis P-value was <6.6 × 10−4 for 37 CpG probes. Top probes were those mentioned earlier near CPT1A (global meta-analysis P-value = 3.4 × 10–47), near ABCG1 (global P-value = 1.0 × 10–46), in an intergenic region on Chr 17 (cg03078551, P = 6 × 10–28) and near SREBF1 (cg11024682, P = 2.8 × 10–24) (sterol regulatory element-binding transcription factor 1), which is intimately involved in cholesterol biosynthesis, producing a product that binds the sterol regulatory element-1 (SRE-1), flanking the low density lipoprotein (LDL) receptor gene. Interestingly, the HIF3A probes initially reported by Dick et al. (27) to be associated with BMI, and that we confirmed in African Americans earlier, did not replicate in leukocyte DNA from Whites in FHS and GOLDN.
Table 2.
Replicationa of BMI–DNA methylation associations in the FHS (Cohorts I and II) and GOLDN cohorts, in order of meta-analysis P-value
| CpG marker | CHR | Closest gene | ARIC (discovery cohort) analysis |
Replication meta-analysis (FHS I + FHS II + GOLDN) |
Discovery + replication cohort meta-analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ARIC regression coefficient | ARIC P-value | Replication z statistic | Replication P-value | Direction | Summary P-value | HetISq | HetChiSq | HetPVal | |||
| cg00574958 | 11 | CPT1A | −0.0029 | 3.23E − 12 | −12.881 | 5.77E − 38 | −−−− | 3.44E − 47 | 55.9 | 6.801 | 0.0785 |
| cg06500161 | 21 | ABCG1 | 0.0081 | 1.52E − 13 | 12.456 | 1.29E − 35 | ++++ | 1.01E − 46 | 77.8 | 13.543 | 0.0036 |
| cg03078551 | 17 | NA | −0.0026 | 7.49E − 09 | −9.399 | 5.52E − 21 | −−−− | 6.03E − 28 | 1.4 | 3.042 | 0.3851 |
| cg11024682 | 17 | SREBF1 | 0.0068 | 9.58E − 09 | 8.424 | 3.63E − 17 | ++++ | 2.76E − 24 | 55.6 | 6.755 | 0.0801 |
| cg13123009 | 6 | LY6G6E;LY6G6D | 0.0069 | 1.80E − 11 | 6.894 | 5.41E − 12 | ++++ | 1.01E − 21 | 0 | 1.657 | 0.6466 |
| cg04927537 | 17 | LGALS3BP | 0.0108 | 5.91E − 10 | 7.17 | 7.48E − 13 | ++++ | 2.93E − 21 | 0 | 2.45 | 0.4845 |
| cg27243685 | 21 | ABCG1 | 0.0057 | 3.61E − 08 | 7.661 | 1.85E − 14 | ++++ | 4.23E − 21 | 61.3 | 7.747 | 0.0515 |
| cg26403843 | 5 | RNF145 | 0.0099 | 1.06E − 08 | 6.934 | 4.09E − 12 | ++++ | 2.51E − 19 | 0 | 0.567 | 0.9040 |
| cg09349128 | 22 | NA | −0.0059 | 1.44E − 08 | −6.92 | 4.51E − 12 | −−−− | 3.76E − 19 | 0 | 0.507 | 0.9173 |
| cg07814318 | 15 | KLF13 | 0.0081 | 2.33E − 10 | 5.896 | 3.73E − 09 | ++++ | 1.19E − 17 | 73.8 | 11.445 | 0.0095 |
| cg07573872 | 19 | SBNO2 | −0.0069 | 2.80E − 08 | −6.377 | 1.80E − 10 | −−−− | 3.00E − 17 | 0 | 0.683 | 0.8771 |
| cg13708645 | 12 | KDM2B | 0.0096 | 4.68E − 10 | 5.74 | 9.48E − 09 | ++++ | 6.03E − 17 | 60.7 | 7.631 | 0.0543 |
| cg25178683 | 17 | LGALS3BP | 0.0087 | 1.67E − 08 | 6.138 | 8.36E − 10 | ++++ | 9.25E − 17 | 0 | 0.869 | 0.8328 |
| cg12992827 | 3 | NA | −0.0075 | 5.31E − 09 | −5.939 | 2.87E − 09 | −−−− | 1.25E − 16 | 47.3 | 5.693 | 0.1275 |
| cg09664445 | 17 | KIAA0664 | 0.0055 | 8.89E − 09 | 5.93 | 3.03E − 09 | ++++ | 2.08E − 16 | 65.4 | 8.672 | 0.0340 |
| cg06876354 | 2 | RALB | 0.0057 | 1.02E − 09 | 5.62 | 1.91E − 08 | ++++ | 2.53E − 16 | 39 | 4.916 | 0.1780 |
| cg23998749 | 1 | NA | 0.0052 | 5.66E − 08 | 5.92 | 3.22E − 09 | ++++ | 1.14E − 15 | 40.4 | 5.035 | 0.1692 |
| cg06192883 | 15 | MYO5C | 0.0065 | 5.29E − 08 | 5.791 | 7.01E − 09 | ++++ | 2.45E − 15 | 0 | 0.979 | 0.8062 |
| cg26033520 | 10 | NA | 0.0070 | 1.44E − 08 | 5.578 | 2.43E − 08 | ++++ | 3.01E − 15 | 0 | 1.065 | 0.7856 |
| cg07136133 | 11 | PRR5L | −0.0048 | 6.70E − 09 | −5.36 | 8.34E − 08 | −−−− | 6.24E − 15 | 0 | 1.871 | 0.5996 |
| cg08972190 | 7 | MAD1L1 | 0.0054 | 6.43E − 10 | 5.054 | 4.33E − 07 | ++++ | 6.42E − 15 | 27.7 | 4.149 | 0.2458 |
| cg06946797 | 16 | NA | −0.0070 | 8.35E − 09 | −5.131 | 2.88E − 07 | −−−− | 3.04E − 14 | 17.2 | 3.622 | 0.3052 |
| cg18568872 | 15 | ZNF710 | 0.0062 | 3.74E − 10 | 4.709 | 2.49E − 06 | ++++ | 3.54E − 14 | 66.8 | 9.04 | 0.0288 |
| cg15871086 | 18 | NA | 0.0062 | 8.31E − 11 | 4.354 | 1.34E − 05 | ++++ | 1.00E − 13 | 67.5 | 9.226 | 0.0264 |
| cg08857797 | 17 | VPS25 | 0.0063 | 3.40E − 09 | 4.74 | 2.14E − 06 | ++++ | 1.55E − 13 | 39.9 | 4.99 | 0.1725 |
| cg20954977 | 2 | B3GNT7 | 0.0101 | 1.68E − 08 | 4.898 | 9.66E − 07 | ++++ | 2.13E − 13 | 21.3 | 3.811 | 0.2826 |
| cg04816311 | 7 | C7orf50 | 0.0090 | 2.56E − 12 | 3.823 | 1.32E − 04 | ++++ | 2.18E − 13 | 77.1 | 13.094 | 0.0044 |
| cg14017402 | 2 | NA | 0.0080 | 1.47E − 10 | 4.226 | 2.38E − 05 | ++++ | 3.17E − 13 | 75.9 | 12.462 | 0.0060 |
| cg17560136 | 8 | EPB49 | 0.0063 | 3.03E − 08 | 4.824 | 1.41E − 06 | ++++ | 5.27E − 13 | 25.3 | 4.018 | 0.2595 |
| cg11592786 | 15 | NA | −0.0031 | 3.60E − 08 | −4.584 | 4.56E − 06 | −−−− | 2.36E − 12 | 14.8 | 3.52 | 0.3181 |
| cg01844514 | 7 | ZNF862 | 0.0046 | 3.17E − 08 | 4.27 | 1.96E − 05 | ++++ | 1.21E − 11 | 4.5 | 3.143 | 0.3701 |
| cg18307303 | 5 | IL12B | 0.0044 | 4.67E − 08 | 4.217 | 2.48E − 05 | ++++ | 2.16E − 11 | 46.4 | 5.592 | 0.1332 |
| cg04869770 | 1 | PBX1 | 0.0057 | 5.65E − 09 | 3.805 | 1.42E − 04 | ++++ | 4.23E − 11 | 57.6 | 7.074 | 0.0696 |
| cg00863378 | 16 | BBS2 | 0.0055 | 1.91E − 08 | 3.967 | 7.27E − 05 | ++++ | 4.24E − 11 | 49.5 | 5.939 | 0.1146 |
| cg26354221 | 22 | ADORA2A | 0.0032 | 1.39E − 08 | 3.725 | 1.95E − 04 | ++++ | 1.20E − 10 | 52.7 | 6.343 | 0.0961 |
| cg15695155 | 12 | KDM2B | 0.0079 | 4.03E − 08 | 3.802 | 1.43E − 04 | ++++ | 1.71E − 10 | 49 | 5.883 | 0.1175 |
| cg27614723 | 15 | SLCO3A1 | 0.0071 | 5.62E − 08 | 3.62 | 2.94E − 04 | ++++ | 5.44E − 10 | 39.4 | 4.947 | 0.1757 |
aSuccessful replication was defined as consistent direction of association across all four studies (ARIC, FHS I, FHS II and GOLDN) and P < 0.05/76 (<6.6 × 10−4) in the replication meta-analysis. Meta-analysis used Stouffer's Z for trend test, which is based on meta-analysis of P-values with adjustment for cohort sample size and direction.
Of the 164 WC-associated probes taken forward for replication in GOLDN, 140 (85%) had consistent direction of association [z-statistic for equal proportions for null hypothesis of concordance with ARIC (null = 50%); z = 3.79, P = 0.0001]. Of these 140, 8 also passed the 3 × 10−4 significance threshold for replication, 1 of which was unique to WC: DHCR24 (Table 3). DHCR24 (3-beta-hydroxysterol delta-24-reductase) catalyzes reduction of sterol intermediates during the final step of cholesterol biosynthesis and is of current interest as a biomarker of nonalcoholic hepatic steatosis. DHCR24 gene expression changes track strongly with weight loss after bariatric surgery (37).
Table 3.
Replicationa of WC–DNA methylation associations in the GOLDN cohort (N = 911), in order of replication P-value
| CpG marker | Chr | Nearest gene | ARIC regression coefficient (per 1 SD WC) | ARIC (P-value) | GOLDN regression coefficient (per 1 SD WC) | GOLDN (P-value) | Direction | Combined meta-analysis (P-value) |
|---|---|---|---|---|---|---|---|---|
| cg13708645 | 12 | KDM2B | 0.0098 | 7.22E − 11 | 0.0074 | 1.28E − 05 | ++ | 4.52E − 15 |
| cg11024682 | 17 | SREBF1 | 0.0080 | 3.52E − 12 | 0.0039 | 1.33E − 05 | ++ | 2.37E − 16 |
| cg03078551 | 17 | intergenic | −0.0025 | 8.16E − 09 | −0.0021 | 1.74E − 05 | −− | 6.61E − 13 |
| cg06192883 | 15 | MYO5C | 0.0083 | 1.50E − 12 | 0.0031 | 2.56E − 05 | ++ | 2.15E − 16 |
| cg26403843 | 5 | RNF145 | 0.0101 | 2.38E − 09 | 0.0044 | 7.53E − 05 | ++ | 7.92E − 13 |
| cg13123009 | 6 | LY6G6E | 0.0074 | 1.81E − 13 | 0.0032 | 8.25E − 05 | ++ | 1.07E − 16 |
| cg06500161 | 21 | ABCG1 | 0.0096 | 4.41E − 19 | 0.0043 | 9.39E − 05 | ++ | 1.09E − 21 |
| cg17901584 | 1 | DHCR24 | −0.0080 | 8.34E − 08 | −0.0049 | 2.39E − 04 | −− | 8.15E − 11 |
aSuccessful replication was defined as consistent direction of association in GOLDN and ARIC and P-value of <0.05/164 (<3 × 10−4) in the replication cohort. Meta-analysis used Stouffer's Z for trend test, which is based on meta-analysis of P-values with adjustment for cohort sample size and direction.
Influence of cis-acting SNPs
As a global test of whether these associations are potentially confounded by any cis-acting single nucleotide polymorphisms (SNPs), we identified SNPs in a 500-kb window (250 kb up and down stream) of each of the 37 replicated BMI probes (see below) and determined their associations with BMI in an existing large GWAS meta-analysis conducted in ∼40 000 African American adults (38). Supplementary Material, Table S6 lists the SNP name having the lowest P-value for association with BMI in each 500-kb region (referred to as the ‘reference SNP’). All such SNPs were associated with BMI at P = 1 × 10−5 or higher. This analysis shows that replicated methylation associations reported here are unlikely to be confounded by the effect of common cis-acting SNPs on BMI in African Americans.
Secondary Analysis 1: impact of prevalent diabetes on replicated associations
Because increased adiposity is a risk factor for type 2 diabetes, and because diabetes and its treatment may impact methylation, we tested the degree to which association results were altered by covariate adjustment for concurrent diabetes status, a possible mediator of the adiposity–DNA methylation relationship. Supplementary Material, Table S7 presents a side-by-side comparison of beta values and P-values when diabetes is, and is not, included as a covariate for the 37 replicated BMI probes and the 1 additional replicated WC probe (above). Generally, regression coefficients were reduced by 5–25% with adjustment, but 19/37 of BMI-associated probes and 0/1 of the WC probes remained significant at P < 1 × 10−7. These results show that about half of the BMI–DNA methylation associations we report are independent of concurrent diabetes, and the others remain strongly associated even with adjustment for this condition.
Secondary Analysis 2: replication in adipose tissue samples
The 37 BMI probes and 1 WC methylation probe that independently replicated in FHS and GOLDN were subjected to cross-tissue association study using adipose tissue DNA (Table 4). Replication was based on P-value only, because it was assumed that the direction of effect of the same exposure on DNA methylation may differ in different tissue types. Twenty-eight of the 38 probes passed quality control (QC) in Multiple Tissue Human Expression Resource (MuTHER), and of these, 18/28 (64%) were associated with BMI in adipose tissue at P < 1.9 × 10−3, including markers near the previously identified CPT1A and ABCG1 loci, but also a large number of novel adiposity-related loci, including LYS6GE, KDM2B, RALB, PRRL5, LGALS3BP, C7orf50, PBX1, EPB49 and BBS2. BBS2 [Bardet–Biedl syndrome (BBS) 2] is one of the BBS genes involved in cilia formation, cell movement and cell signaling. Autosomal recessive variants in BBS2 are among those responsible for BBS, which is characterized by severe obesity and numerous developmental aberrations. Overall, the results show wide cross-tissue agreement in BMI– and WC–methylation associations.
Table 4.
Replicated BMI CpG sites significantly associated in both leukocyte and adipose tissue DNA from 648 females in the MuTHER, in order of MuTHER P-valuea
| CpG marker | Nearest gene | MuTHER regression coefficientb | Same direction as in blood? | MuTHER P-valuec | Function |
|---|---|---|---|---|---|
| BMI | |||||
| cg17560136 | EPB49 | 0.00233 | Yes | 4.64E − 33 | Erythrocyte membrane protein band 4.9: gene product essential for the maintenance of erythrocyte shape and membrane stability |
| cg04869770 | PBX1 | 0.00222 | Yes | 8.53E − 25 | Pre-B-cell leukemia transcription factor involved in pancreatic development and function, candidate gene SNPs associated with obesity, variants may influence porcine adipose tissue fatty acid composition |
| cg25178683 | LGALS3BP | 0.00305 | Yes | 1.68E − 18 | Lectin, galactoside-binding, soluble, 3 binding protein: a macrophage inflammatory marker, arterial expression upregulated in response to obesity in animal models |
| cg01844514 | ZNF862 | 0.00111 | Yes | 4.72E − 16 | Zinc finger protein 862: protein coding gene of unknown function, may be involved in transcriptional regulation |
| cg07136133 | PRR5L | −0.00392 | Yes | 7.85E − 15 | Proline rich 5-like: regulates cytoskeleton organization and interacts with mTOR, a central controller of cell growth, to increase apoptosis |
| cg06876354 | RALB | −0.00129 | No | 9.40E − 14 | RAS-related small GTP-ase B: involved in a variety of cellular processes including gene expression and has role in colorectal cancer oncogenesis |
| cg26033520 | Intergenic Chr. 10 | 0.00244 | Yes | 5.29E − 12 | Unknown |
| cg08857797 | VPS25 | −0.00172 | No | 5.85E − 12 | Vacuolar protein-sorting-associated protein 25: part of endosomal sorting complexes required for transport protein, II (ESCRT-II) complex involved in endosomal transport and possibly gene transcription |
| cg06946797 | Intergenic Chr. 16 | −0.00145 | Yes | 2.09E − 09 | Unknown |
| cg04816311 | C7orf50 | −0.00152 | No | 2.27E − 09 | Chromosome 7 open reading frame 50; unknown function, GWAS variants in this locus are associated with lipid levels and longevity |
| cg00574958 | CPT1A | −0.00032 | Yes | 3.40E − 08 | Carnitine palmitoyltransferase-1A: involved in mitochondrial uptake of long-chain fatty acids and triglyceride metabolism, previously associated with insulin-related DNA methylation |
| cg13123009 | LY6G6E | 0.00072 | Yes | 1.18E − 07 | Lymphocyte antigen-6 complex, locus G6E (pseudogene): one of LY6 genes located in the MHC class III region, immune function, acute lymphoblastic leukemia, hematopoiesis, cell adhesion |
| cg13708645 | KDM2B | 0.00184 | Yes | 1.74E − 07 | Lysine K-specific demethylase 2B: a H3K36 histone demethylase, involved in cellular senescence, tumor cell differentiation, part of a complex that represses preadipocyte differentiation |
| cg26403843 | RNF145 | −0.00203 | Yes | 5.59E − 07 | Ring finger protein 145: unknown function |
| cg00863378 | BBS2 | 0.00113 | Yes | 7.21E − 07 | BBS-2: member of the BBS gene family. BBS is an autosomal recessive disorder characterized by severe obesity, developmental abnormalities and mental retardation |
| cg09664445 | KIAA0664 | 0.00077 | Yes | 9.84E − 06 | Aka CLUH, clustered mitochondria homolog: regulates mitochondrial biogenesis |
| cg18568872 | ZNF710 | 0.00094 | Yes | 1.85E − 05 | Zinc finger 710: protein-coding gene, function unknown, may be involved in transcriptional regulation |
| cg15871086 | Intergenic Chr. 10 | 0.00094 | Yes | 1.46E − 03 | Unknown |
| WC | |||||
| cg17901584 | DHCR24 | 0.00162 | No | 9.07E − 13 | 3-Beta-hydroxysterol delta-24-reductase: catalyzes reduction of sterol intermediates during final step of cholesterol biosynthesis, biomarker of nonalcoholic hepatic steatosis, expression changes associated with weight loss after bariatric surgery |
aThe MuTHER includes genome-wide DNA methylation data (Illumina HumanMethylation450 array) from abdominal subcutaneous adipose tissue collected from 648 female twins (39,40).
bFixed-effects regression coefficient from linear mixed models (LMM), with probe DNA methylation beta value as the dependent variable and BMI as the primary independent variable, covariate adjusted for age, bisulfite conversion concentration, bisulfite conversion efficiency and experimental batch (Beadchip) (fixed effects), and family relationship (twin-pairing) and zygosity (random effects).
cReplication was based on P-value only because increased or decreased methylation may differ by tissue type. Threshold for replication was set at P < 1.8 × 10−3 (0.05/28 tests). There were 37 BMI-associated CpGs and 1 WC-associated CpG carried forward for replication in adipose tissue, and 10 were removed in QC procedures in MuTHER cohort and not tested.
Discussion
Our study presents the first EWAS of adiposity traits in African American adults, demonstrating numerous methylation variants associated with BMI and WC, including 37 CpG site associations for BMI and 1 additional association for WC that replicated in two European-ancestry cohorts. The ARIC results included sites in three loci (near HIF3A, CPT1A and ABCG1) that have been previously reported to be associated with BMI, insulin-related traits and lipid subfractions (27,33,41,42), as well as a number of novel loci. This demonstrates to our knowledge the first cross-ethnic replication of methylation signals for BMI from the HM450K array. Further, the results show that despite differences in blood cell type (GOLDN used CD4+ T cells, ARIC and FHS used whole blood), normalization approach [GOLDN used COMBAT followed by polynomial regression normalization (43), FHS used wateRmelon and Beta MIxture Quantile dilation (BMIQ) (44) and ARIC used BMIQ] and some variation in the average age of the cohorts and specific covariate adjustments, there was general consistency of results across these large, independent EWAS studies of adiposity traits. This is important because while epigenetic modification in cancer etiology has been established for more than a decade (45,46), identification of epigenetic patterns involved in cardiometabolic disease and its precursors (e.g. obesity) has yielded relatively few replicated loci (reviewed by Drong et al.) (7). There has been concern that the large potential for confounding in epigenetic studies would make successful replication difficult. What is likely more important to successful replication of adiposity associations is sample size; effect sizes observed in this study were relatively small, with the marginal variance in methylation beta value at individual CpG sites explained by a 1 SD difference in BMI or WC ranging from 0.25 to 2.4%.
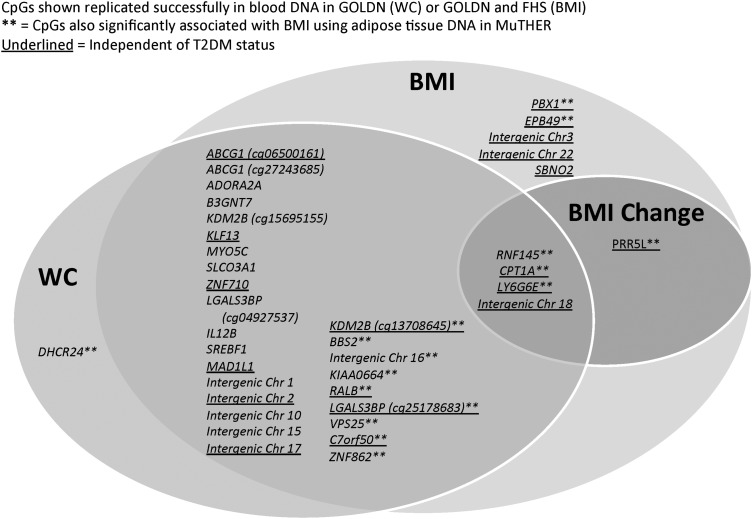
ARIC participants gained an average of 7 kg/m2 over the 30-year period from age 25 to Visit 2. BMI change was associated with eight different CpG sites, including the highly cited methylation variant near CPT1A, showing that numerous novel methylation loci identified in our study do not only index current weight status or its correlates but may also be involved in changes in body weight over time. Generally, we observed extensive overlap in the methylation associations for the three adiposity traits investigated; of the 38 replicated CpG associations for BMI/WC, 4 were shared by all 3 traits, and 27 were shared by BMI and WC (see Fig. 3 for a diagram of overlap across the 3 traits).
Figure 3.
Overlap in EWAS results for BMI, WC and BMI change in ARIC African American adults.
Significant obesity or weight gain/weight loss associations have been reported for CpG methylation sites near SLC6A4 (47), MEST (48), NPY (49), POMC (49), PGC-1α and PDK4 (50). Existing studies tend to be small (generally <200 subjects per group), which likely explains the lack of replication. Exceptions include replicated methylation associations near POMC (20) and RXRA (21). None of these CpG sites from previously reported regional or candidate gene association studies was significantly associated with BMI, WC or BMI change in the present analysis. We also did not replicate the methylation variants recently found to change in response to weight loss (51) or increased physical activity (52). A recent genome-wide BMI methylation study conducted in 48 obese and 48 lean African American children did not report associations at the CpG site level but indicated that differentially methylated regions with greater inter-individual variance in methylation are enriched in obesity, as in cancer (25).
To date, the only prior EWAS to identify and robustly replicate methylation variants associated with BMI is that of Dick et al. (27). That study reported methylation at three CpG sites in intron 1 of HIF3A to be positively associated with BMI in both blood and adipose tissue DNA in European adults (27). Here, we replicate the positive association for three of these probes in blood DNA from African Americans (cg22891070, cg16672562 and cg27146050) with BMI and between cg16672562 methylation and BMI change. As described previously by Dick et al. (27), these probes are within likely regulatory elements (open chromatin regions) and are potentially functional; moreover, methylation level was inversely associated with HIF3A gene expression at one of the five expression probes examined. HIF3A is a component of the hypoxia-inducible transcription factor (HIF) involved in the physiological response to hypoxia but is also implicated in adipocyte differentiation (53) and is expressed in response to glucose and insulin changes (54). However, we found that these HIF3A methylation probes were not associated with BMI in either the FHS or the GOLDN study. The reason for this inconsistent replication is unclear but may relate to the greater obesity comorbidities (perhaps including inflammatory status) in ARIC (e.g. diabetes) and the original study population (over 50% of whom had a history of myocardial infarction), when compared with the replication cohorts in this analysis.
Our unbiased pathway analysis of the EWAS data shows that over 100 biological pathways are significantly enriched for methylation association with BMI and WC, including those involved in lipid and energy metabolism, immune function, adipocyte, neuronal and chondrocyte differentiation and development, and many others. These results suggest that further work in larger cohort studies may identify many additional methylation variants in association with adiposity and its related traits. Many of the specific loci identified in this study are known to be involved in lipid and lipoprotein metabolism, including the known differentially methylated locus CPT1A (involved in mitochondrial uptake of long-chain fatty acids and triglyceride metabolism) and ABCG1 (involved in macrophage cholesterol and phospholipids transport, and lipid homeostasis), as well as the novel BMI-related methylation site near SREBF1, which gene encodes a transcription factor that binds to the LDL receptor and other genes involved in sterol biosynthesis. Promoter variants in SREBF1 influence hepatic cholesterol and steatosis in rats (55) and with diabetes traits in humans (56,57). Immune function and inflammatory pathways are also strongly represented in our novel loci, including IL-12B, NFKBIL1, LY6G6E, SBNO2 (part of the IL-10 anti-inflammatory signaling pathway) and LGALS3BP (lectin galactoside-binding soluble 3 binding protein). LGALS3BP expression was recently found to be one of a small group of genes whose expression is strongly modulated throughout the arterial network in response to obesity (58). This study together with our findings suggests how obesity-related methylation variants could be useful as targets for cardiovascular disease treatment and prevention. To move this work forward, the functional significance of the methylation variants discovered here must be established and a determination made that they mediate the relationships of obesity to diabetes and CHD, rather than that they are downstream effects of the obesity-related disease.
In that regard, we were interested in whether the associations identified with adiposity traits were likely mediated by or otherwise dependent on the presence of diabetes, which condition was common in our study sample (26%), is strongly related to obesity and is associated with methylation variation (e.g. 26,59). Although in half of the sites, P-values were no longer genome-wide significant after covariate adjustment for prevalent diabetes, all of the associations remained strong (P < 1 × 10−5), with an average change in regression coefficient of only 11.2%. Thus, while some adiposity-related methylation signals may be due to underlying insulin resistance or diabetes, these results suggest that many may be properly characterized as fundamentally obesity related. Longitudinal analysis of weight gain in individuals without diabetes will confirm these findings.
Blood is an accessible and plentiful tissue for genomic analysis in large studies, but there are two major concerns regarding its use in epigenetic studies of obesity and cardiometabolic conditions: biological relevance and confounding by cell type differential. In terms of relevance, obesity may be characterized as a defect in appetite/satiety regulation resulting in elevated circulating free fatty acids, leading to adipocyte differentiation to sequester the excess lipids. The most biologically relevant tissue types for the analysis of obesity-related gene expression (and, ergo, methylation) might therefore include the hypothalamus, liver and adipose tissue. It is encouraging that many of the signals we detected in WBCs also replicated in adipose tissue. Not all did so, perhaps due to gender differences: MuTHER is 100% female while the studies using blood included both males and females. It has been shown that different tissues including brain, lung, thyroid, saliva and whole blood exhibit highly concordant age associations (60) and that smoking is associated with consistent AHRR methylation profiles in both lymphoblasts and pulmonary macrophages (61). Such studies suggest that DNA methylation in blood can serve as a biomarker of methylation in other tissues, as found here.
Another concern regarding the use of blood in epigenetic epidemiologic investigations is that it contains multiple cell types, each having a characteristic methylation profile, such that DNA methylation associations with disease could be confounded by the relative proportions of different cell types in the DNA sample (62). Indeed, mean methylation percent differed significantly between blood cell types for over 70 000 of the CpG sites on the HM450 (63). This was less of a concern here in that there was no association between WBC differential and obesity at Visit 1 in ARIC (when measured differentials were available for the entire cohort). In addition, covariate adjustment for imputed WBC differentials did not substantially alter our EWAS results, and we replicated our results in different tissue/cell types, both of which procedures reduce the probability that the results are solely due to confounding by cell type distribution.
Despite its disadvantages, an advantage of leukocytes for epigenetic studies of obesity is that adiposity and immunological activation are strongly causally related. When individuals gain excessive body fat, numerous changes in the immune system occur, including increases in WBCs and alterations in the production of different leukocytes, including increases in neutrophils, mast cells, CD8+ T cells and some classes of monocytes and decreases in T regulatory cells and eosinophils. These changes in immune cell distribution reduce the production of anti-inflammatory IL-10 and increase the production of pro-inflammatory IL-6, IFNγ and tumor necrosis factor alpha (TNFα), among other cytokine changes (64). After substantial weight loss, leukocyte counts and differentials normalize, as seen following bariatric surgery (65), which suggests that weight gain and loss drive changes in low-grade inflammation and leukocyte cell types rather than the reverse. Thus, methylation signals found in blood may be potentially useful biomarkers of obesity-related inflammatory damage, as indicated by the many inflammation-related loci identified in the current analysis.
The present study has a number of limitations, the most important of which is the cross-sectional design, which does not allow for clear temporal relationships between predictor (e.g. BMI) and outcome (CpG-specific methylation) to be assessed and leaves open the potential for reverse causality. Studies including non-obese individuals with DNA methylation assessment at that time point with follow-up for incident obesity and DNA methylation changes at a later point would provide clarification on methylation variants that drive the development of obesity as opposed to those that are affected by obesity. Second, gene expression data were not available in ARIC to confirm the functional relevance of the DNA methylation variation we have identified. However, the CPT1A, HIF3A and ABCG1 probes we report have been found to influence gene expression in prior studies (27,41).
The study also had numerous strengths, including a relatively large discovery sample, which may have been responsible for the larger number of significant findings than has been reported previously for BMI, a focus on African Americans, which is important due to their higher burden of obesity and related conditions but poorer coverage by current EWAS studies, and control for numerous potential confounders and batch effects, including a test of potentially cis-acting SNPs in each of the methylation regions for association with BMI, and covariate adjustment for imputed cell type differentials. Other strengths include examination of multiple adiposity traits, a relatively large replication sample (N = 3368) with additional replication in adipose tissue as well (N = 648), quantification of probe-specific measurement error/reliability and consideration of the role of diabetes in the associations.
In conclusion, this study confirmed three previously identified methylation loci suggested to be associated with obesity and related traits (CPT1A, ABCG1 and HIF3A) and identified numerous additional novel loci harboring individual DNA methylation variation in both blood and adipose tissue that are associated with adiposity traits in African American adults. Results were successfully replicated across studies despite variation in tissue type, ethnicity and analytic approaches. Experimental and longitudinal study designs, and larger multi-cohort analyses, are needed to assess causality and to move the growing field of epigenetic epidemiology toward richer insight into the biology of obesity, as well as new therapies to reduce or reverse its downstream effects on health.
Methods and Procedures
Population
The ARIC Study is a prospective cohort study of cardiovascular disease risk in four US communities (31). Between 1987 and 1989, 7082 men and 8710 women aged 45–64 years were recruited from Forsyth County, North Carolina; Jackson, Mississippi (African Americans only); suburban Minneapolis, Minnesota; and Washington County, Maryland. The ARIC Study protocol was approved by the institutional review board of each participating university. After written informed consent was obtained, including that for genetic studies, participants underwent a baseline clinical examination (Visit 1) and four subsequent follow-up clinical exams (Visits 2–5). At this time, DNA methylation data are available for African American members of the cohort only, and the present study comprises a cross-sectional analysis of these data. Specifically, a single DNA sample was chosen for methylation analysis for each subject, and BMI, WC and covariate data detailed below were from the same study visit. For these analyses, all data come from Visit 2, except as noted below for physical activity. In addition, self-reported weight at age 25 (collected only at Visit 1) was used to calculate weight change from age 25 to Visit 2 as a measure of adulthood BMI change.
Measurements and questionnaires
Anthropometrics were taken with the subject wearing a scrub suit and no shoes. BMI was calculated from measured weight and height (weight in kilograms/height in meters squared). WC was measured at the level of the umbilicus using a flexible tape. WBC count was assessed by automated particle counters within 24 h after venipuncture in the local hospital hematology laboratory. The reliability coefficient for the WBC count measurement was >0.96 (66). Measured WBC differentials were only available for a subset at Visit 2 (N = 187), but these were used in the imputation of differential WBCs for the remaining subjects (see below for description). Questionnaires assessed education (coded as less than high school degree, high school degree or equivalent and greater than high school degree), current household income, current cigarette smoking (coded as current, former and never smoked), current alcohol consumption status (coded as current/former/never) and medical history (67). Level of leisure time physical activity was assessed at Visit 1 using the Baecke questionnaire (68). Leisure time activity scores range in whole and half increments from 1 to 5, with values <2 indicative of physical inactivity (69). Prevalent diabetes was defined as a fasting glucose level of ≥126 mg/dl (70), nonfasting glucose of ≥200 mg/dl or a self-reported physician diagnosis of or treatment for diabetes.
Bisulfite conversion of DNA
Genomic DNA was extracted from peripheral blood leukocyte samples using the Gentra Puregene Blood Kit (Qiagen; Valencia, CA, USA) according to the manufacturer's instructions (www.qiagen.com). Bisulfite conversion of 1 ug genomic DNA was performed using the EZ-96 DNA Methylation Kit (Deep Well Format) (Zymo Research; Irvine, CA, USA) according to the manufacturer's instructions (www.zymoresearch.com). Bisulfite conversion efficiency was determined by PCR amplification of the converted DNA before proceeding with methylation analyses on the Illumina platform using Zymo Research's Universal Methylated Human DNA Standard and Control Primers.
Illumina infinium methylation assay
The Illumina Infinium HumanMethylation450K Beadchip array (HM450K) (described by Sandoval et al.) (71) was used to measure DNA methylation (Illumina, Inc.; San Diego, CA, USA). The platform detects methylation status of 473 788 CpG sites by sequencing-based genotyping of bisulfite-treated DNA. Bisulfite treatment converts only unmethylated cyosines to uracils, allowing for highly multiplexed genotyping with single site resolution. The array covers 96% of CpG Islands (as well as CpG shores) and 98.9% of RefSeq genes with a global average of 17.2 probes per gene region and has been shown to have high accuracy and reliability (72,73).
Bisulfite-converted DNA was used for hybridization on the HM450K BeadChip, following the Illumina Infinium HD Methylation protocol (www.illumina.com). This consisted of a whole genome amplification step followed by enzymatic end-point fragmentation, precipitation and re-suspension. The re-suspended samples were hybridized to the complete set of bead-bound probes, followed by ligation and single-base extension during which a fluorescently labeled nucleotide is incorporated and scanned. The degree of methylation is determined for each CpG cytosine by measuring the amount of incorporated label for each probe. The intensities of the images were extracted using Illumina GenomeStudio 2011.1, Methylation module 1.9.0 software. The methylation score for each CpG was represented as a beta (β) value according to the fluorescent intensity ratio. Beta values may take any value between 0 (nonmethylated) and 1 (completely methylated). Background subtraction was conducted with the GenomeStudio software using built-in negative control bead types on the array.
Normalization
The HM450K uses two different probe types (I and II). Due to differences in design, probes using the Illumina Type II assay are less sensitive for the detection of extreme methylation values (i.e. 0 and 1) than the Type I assay and have greater average variance between technical replicates (74). BMIQ (75) was used in this analysis to adjust the beta values of type 2 design probes into a statistical distribution characteristic of type 1 probes. BMIQ has been shown to more effectively reduce probe set bias and technical error across replicates compared with some other peak-based and quantitative normalization procedures (76). An emerging conclusion is that in general, the improvements offered by different normalization approaches are modest, with very high concordance in association results across different methods (77). Further, in this study, we conducted all analyses at the single probe level, and therefore, any differences in probe type should not strongly influence the results.
Quality control
Positive and negative controls and sample replicates were included on each 96-well plate assayed. After exclusion of controls, replicates and samples with integrity issues or failed bisulfite conversion, a total of 2841 study participants had HM450K data available for further QC analyses. We removed poor-quality samples with pass rate of <99%, that is, if the sample had at least 1% of CpG sites with detection P-value > 0.01 or missing (N = 37), indicative of lower DNA quality or incomplete bisulfite conversion, and samples with a possible gender mismatch based on evaluation of selected CpG sites on the Y chromosome (N = 2), leaving a total of 2802 samples available for analysis. At the target level, we flagged poor-quality CpG sites with average detection P-value of >0.01 and calculated the percentage of samples having detection P-value of >0.01 for each autosomal and X chromosome CpG site. There were 9399 autosomal and X chromosomal markers where >1% of samples showed detection P-value of >0.01, and these sites were excluded. In addition, we filtered 370 CpG sites on the Y chromosome with average detection P-value of >0.01, leaving a total of 473 788 CpG sites for analysis.
Technical error analysis
To obtain a measure of probe-specific technical error and the reliability of the methylation measures, technical replicates were included for 130 samples (total n = 265 with 5 samples replicated 3 times), from which ICC coefficients for methylation beta values were calculated for all probes (78). Further information on the method is provided in the Supplementary Text.
Statistical analysis
Of the 2802 samples available after methylation QC, 705 did not have complete covariate data needed for confounder adjustment, leaving a final sample size of N = 2097 (2096 with BMI and BMI change data and 2097 with WC data). Mean, standard deviation and range, or frequencies, are provided to describe continuously distributed and categorical variables, respectively. Methylation data were tested and reported in terms of beta values, ranging from 0 to 100%. While beta values have non-constant variance across different CpG sites, they have the clear advantage of representing the percentage methylation for each site and are therefore more easily interpretable than M values, which are the log2 ratio of the intensities of the methylated versus unmethylated probes. Further, it has been shown that in large sample sizes as in ARIC, test statistics are similar for M and beta values (79).
Pathway analysis
Complete methylation and phenotype data were first subjected to pathway analysis for an a priori test of genomic pathway enrichment for the association of BMI or WC with DNA methylation. The Illumina 450 K probe annotation file contains (non-unique) mappings of 75% of the CpG sites to 21 160 genes, based upon the closest gene to each methylation probe. Reflecting our purpose in detecting entire genomic pathways that are enriched for individually small methylation signals, and long-standing practice in pathway analysis of GWAS SNP data, gene-level methylation signal measurements were first summarized by averaging across all probes annotated to each gene (80). Bioconductor was used to assign genes to the GO domains (molecular function, cellular component and biological process), for a total of 6700 GO pathways tested. Pathway enrichment methods that are not resampling based have been shown to be highly anti-conservative (81). We performed pathway testing while rigorously controlling false positives by using the safeExpress R package (82), controlling for the same covariates as specified below. We used the safeExpress test statistic D, which is a competitive statistic contrasting genes in each pathway versus the complement (82). For each GO domain, the output provided pathway global statistics and P-values, where correction for multiple comparisons was performed using Benjamini–Hochberg FDR q-values (83). The FDR is relatively robust to positive correlation structures (84), which are often strong in pathway analysis and thus enables effective multiple test correction in a manner that is not overly conservative. FDR q < 0.05 was considered statistically significant.
Association study
Batch effect adjustment is critical in the analysis of HM450K data, and ComBat is an Empirical Bayes method frequently used to adjust gene expression and other microarray data for potential batch effects (85). In preliminary analyses, we found very high concordance in EWAS results for BMI with and without ComBat adjustment of the beta values in our linear mixed-effect regression models (LMMs) to address batch effects, where batch effect was accounted for by adding plate number (1–34) and chip row number (1–6) as fixed effects and chip number (1–244) as a random effect. Therefore, we used the simpler LMM without ComBat adjustment for subsequent analyses. We specified the regression models with probe methylation beta value as the dependent variable, and with adiposity traits and all covariates as the independent variables. We chose this approach because the technical (e.g. batch) effects we wished to adjust for pertain to the beta values, not to BMI, and also because in our study of older African American adults, weight gain and obesity are long-standing characteristics of the participants over their lives, which we posit may have influences on DNA methylation (rather than being influenced by DNA methylation).
Cross-sectional LMM were tested using the R package lme4 with methylation beta values as the dependent variable, and with chip specified as a random effect and the following variables specified as fixed effects: standardized adiposity variable (BMI, WC and BMI change with mean = 0 and SD = 1), sex, age, study center, total WBC, education, household income, current cigarette smoking, current alcohol consumption, leisure time physical activity and 10 PCs from the Illumina Infinium HumanExome Beadchip genotype array (86), to account for potential confounding by genetic ancestry. The regression models were further adjusted for leukocyte cell type proportions (neutrophils, lymphocytes, monocytes and eosinophils) as additional fixed effects. Cell-type proportions were imputed using the algorithm developed by Houseman (40), which utilizes the known cell type specificity of methylation at selected CpG probes on the HM450 to impute cell type proportions, and based on the measured differential cell counts available for a subset of ARIC participants at Visit 2. The choice of covariates was based upon known or suspected confounding, as described in the directed acyclic graph presented in Supplementary Material, Figure 1S. The Wald test was applied to test the hypothesis that BMI, WC or BMI change (depending on the model tested) was associated with CpG methylation.
Following standard practice in association analysis (87), multiple test corrections were used to control the family-wise error at 0.05. Applying a standard Bonferroni correction for the 473 788 CpG sites gives P < 1 × 10−7 as the significance threshold. Significant EWAS results for each trait were then filtered to remove known cross-reactive probes and polymorphic CpGs (88).
A secondary analysis addressed whether the adiposity–methylation associations were independent of diabetes by including prevalent diabetes status (yes/no) in the above regression model. To account for multiple comparisons, a Bonferroni-corrected P-value of < 1 × 10−7 was used as the threshold for determining whether CpG methylation remained significantly associated with adiposity independent of diabetes status.
Replication analysis
CpG sites with association P-values of <1 × 10−7 for BMI and WC were carried forward for replication in 991 members of the GOLDN cohort (mean age = 49 years, mean BMI = 28 kg/m2, female = 52%, prevalent diabetes = 7.6%, ancestry = 100% European-American, tissue = CD4 +T cells) and 2377 members of the FHS Offspring (mean age = 67 years, mean BMI = 28 kg/m2, % female = 56%, prevalent diabetes = 13.4%, ancestry = 100% European-American, tissue = whole blood). Both studies utilized blood DNA and the same HM450 platform for methylation analysis as in ARIC. Methylation assays were performed at two different laboratories in the FHS and thus are considered as two independent cohorts for replication. Further details of the two replication cohorts are provided in the Supplementary Text. Given the different analytic strategies and blood cell types used among the discovery and replication studies (detailed in the Supplementary Text), the meta-analysis was conducted on P-values (not beta values) using a sample size-weighted method (Stouffer's Z trend) that also incorporated the direction of the beta coefficients (89) and was implemented in R. For BMI, replication in blood DNA was defined as consistent direction of the beta coefficient in all four cohorts, and a Bonferroni-corrected meta-analysis P-value for the replication cohorts (FHS I, FHS II and GOLDN) of <0.05/76 or P < 6.6 × 10−4. For WC, replication was only available in the GOLDN cohort, and probe associations were considered to have positively replicated based on consistency of the direction of effect and a Bonferroni-corrected P-value based on the number of probes tested (164 probes, 0.05/164 = P < 3 × 10−4).
Test for confounding by cis-acting genetic variants
Methylation SNPs, defined as SNPs within probe sequences, were filtered as stated earlier. As a more general means of excluding the possibility that our results are confounded by cis-acting SNPs, we examined the 500-kb regions surrounding each of our top BMI-associated methylation probes to search for SNP associations with BMI using data from a recent large meta-analysis and report the lowest P-values for each region. This meta-analysis examined the association of >3.2 million SNPs (imputed using the 1000 Genomes Project) with BMI in 39 144 men and women of African ancestry (38).
Cross-tissue replication
Concerns regarding tissue specificity of CpG site methylation and the relevance of peripheral blood leukocytes for obesity were addressed in a secondary analysis by testing the association of the replicated CpG probes identified for BMI (N = 37) and WC (N = 1) using adipose tissue DNA. The MuTHER contains genome-wide DNA methylation data using the HM450 array in subcutaneous abdominal adipose tissue collected from 648 European-ancestry female twins and singletons (97 MZ pairs, 162 DZ pairs and 130 singletons) (39,40). The subjects had a mean age of ∼60 years and a mean BMI of 26.6 kg/m2. Further details on the MuTHER cohort materials and methods are found in the Supplementary Text. The threshold for replication was set at Bonferroni-corrected P < 1.9 × 10−3 (0.05/28 tests). There were 38 BMI- or WC-associated CpGs carried forward for replication in adipose tissue, and 10 were removed in QC procedures in MuTHER cohort and not tested.
Supplementary Material
Funding
The Atherosclerosis Risk in Communities study is carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C). Funding support for ‘Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium’ was provided by the National Institutes of Health (NIH) through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). The work on the GOLDN study has been funded by the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI) (grant U01HL072524-04). The FHS is funded by National Institutes of Health contract (N01-HC-25195). The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute and the Center for Information Technology, National Institutes of Health, Bethesda, MD. The MuTHER study was funded by the Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas’ NHS Foundation Trust in partnership with King's College London.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of the ARIC study and GOLDN for their important contributions. T.S. is holder of an ERC Advanced Principal Investigator award. SNP genotyping was performed by the Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR.
Conflict of Interest statement. None declared.
References
- 1.Fan S., Zhang X. (2009) CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem. Biophys. Res. Commun., 383, 421–425. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong L. (2012) Epigenetic control of embryonic stem cell differentiation. Stem Cell Rev., 8, 67–77. [DOI] [PubMed] [Google Scholar]
- 3.Dolinoy D.C., Weidman J.R., Waterland R.A., Jirtle R.L. (2006) Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect., 114, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. (2013) Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet., 22, 843–851. [DOI] [PubMed] [Google Scholar]
- 5.Baccarelli A., Wright R.O., Bollati V., Tarantini L., Litonjua A.A., Suh H.H., Zanobetti A., Sparrow D., Vokonas P.S., Schwartz J. (2009) Rapid DNA methylation changes after exposure to traffic particles. Am. J. Respir. Crit. Care Med., 179, 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakyan V.K., Down T.A., Balding D.J., Beck S. (2011) Epigenome-wide association studies for common human diseases. Nat. Rev. Genet., 12, 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drong A.W., Lindgren C.M., McCarthy M.I. (2012) The genetic and epigenetic basis of type 2 diabetes and obesity. Clin. Pharmacol. Ther., 92, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund G., Zaina S. (2009) Atherosclerosis risk factors can impose aberrant DNA methylation patterns: a tale of traffic and homocysteine. Curr. Opin. Lipidol., 20, 448–449. [DOI] [PubMed] [Google Scholar]
- 9.Hotamisligil G.S. (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell, 140, 900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reaven G.M. (1988) Banting lecture 1988. Role of insulin resistance in human disease. Diabetes, 37, 1595–1607. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., Shen W., Withers R., Hemysfield S. (2005) In Hemysfield S., Lohman T., Wang Z. (eds), Human Body Composition. Human Kinetics, Champaign, Illinois, pp. 163–176. [Google Scholar]
- 12.Carey V.J., Walters E.E., Colditz G.A., Solomon C.G., Willett W.C., Rosner B.A., Speizer F.E., Manson J.E. (1997) Body fat distribution and risk of non-insulin-dependent diabetes mellitus in women. The Nurses’ Health Study. Am. J. Epidem., 145, 614–619. [DOI] [PubMed] [Google Scholar]
- 13.Sasai H., Sairenchi T., Iso H., Irie F., Otaka E., Tanaka K., Ota H., Muto T. (2010) Relationship between obesity and incident diabetes in middle-aged and older Japanese adults: the Ibaraki Prefectural Health Study. Mayo Clin. Proc., 85, 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubert H.B., Feinleib M., McNamara P.M., Castelli W.P. (1983) Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation, 67, 968–977. [DOI] [PubMed] [Google Scholar]
- 15.Folsom A.R., Stevens J., Schreiner P.J., McGovern P.G. (1998) Body mass index, waist/hip ratio, and coronary heart disease incidence in African Americans and whites. Atherosclerosis Risk in Communities Study Investigators. Am. J. Epidem., 148, 1187–1194. [DOI] [PubMed] [Google Scholar]
- 16.Rexrode K.M., Carey V.J., Hennekens C.H., Walters E.E., Colditz G.A., Stampfer M.J., Willett W.C., Manson J.E. (1998) Abdominal adiposity and coronary heart disease in women. JAMA, 280, 1843–1848. [DOI] [PubMed] [Google Scholar]
- 17.Rexrode K.M., Buring J.E., Manson J.E. (2001) Abdominal and total adiposity and risk of coronary heart disease in men. Int. J. Obes. Relat. Metab. Disord., 25, 1047–1056. [DOI] [PubMed] [Google Scholar]
- 18.Wilson P.W., D'Agostino R.B., Sullivan L., Parise H., Kannel W.B. (2002) Overweight and obesity as determinants of cardiovascular risk: the Framingham experience. Arch. Intern. Med., 162, 1867–1872. [DOI] [PubMed] [Google Scholar]
- 19.Calle E.E., Rodriguez C., Walker-Thurmond K., Thun M.J. (2003) Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. New Engl. J. Med., 348, 1625–1638. [DOI] [PubMed] [Google Scholar]
- 20.Kuehnen P., Mischke M., Wiegand S., Sers C., Horsthemke B., Lau S., Keil T., Lee Y.A., Grueters A., Krude H. (2012) An Alu element-associated hypermethylation variant of the POMC gene is associated with childhood obesity. PLoS Genet., 8, e1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godfrey K.M., Sheppard A., Gluckman P.D., Lillycrop K.A., Burdge G.C., McLean C., Rodford J., Slater-Jefferies J.L., Garratt E., Crozier S.R., et al. (2011) Epigenetic gene promoter methylation at birth is associated with child's later adiposity. Diabetes, 60, 1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird P.W. (2010) Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet., 11, 191–203. [DOI] [PubMed] [Google Scholar]
- 23.Almen M.S., Jacobsson J.A., Moschonis G., Benedict C., Chrousos G.P., Fredriksson R., Schioth H.B. (2012) Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics, 99, 132–137. [DOI] [PubMed] [Google Scholar]
- 24.Bell C.G., Finer S., Lindgren C.M., Wilson G.A., Rakyan V.K., Teschendorff A.E., Akan P., Stupka E., Down T.A., Prokopenko I., et al. (2010) Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PloS One, 5, e14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Su S., Barnes V.A., De Miguel C., Pollock J., Ownby D., Shi H., Zhu H., Snieder H., Wang X. (2013) A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics, 8, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toperoff G., Aran D., Kark J.D., Rosenberg M., Dubnikov T., Nissan B., Wainstein J., Friedlander Y., Levy-Lahad E., Glaser B., et al. (2012) Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum. Mol. Genet., 21, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aissi D., Wahl S., Meduri E., Morange P.E., Gagnon F., Grallert H., et al. (2014) DNA methylation and body-mass index: a genome-wide analysis. Lancet, 383, 1990–1998. [DOI] [PubMed] [Google Scholar]
- 28.Harris M.I. (1990) Noninsulin-dependent diabetes mellitus in black and white Americans. Diabetes Metab. Rev., 6, 71–90. [DOI] [PubMed] [Google Scholar]
- 29.Brancati F.L., Kao W.H., Folsom A.R., Watson R.L., Szklo M. (2000) Incident type 2 diabetes mellitus in African American and white adults: the Atherosclerosis Risk in Communities Study. JAMA, 283, 2253–2259. [DOI] [PubMed] [Google Scholar]
- 30.El-Sayed A.M., Scarborough P., Galea S. (2011) Ethnic inequalities in obesity among children and adults in the UK: a systematic review of the literature. Obes. Rev., 12 e516–e534. [DOI] [PubMed] [Google Scholar]
- 31.The ARIC Investigators. (1989) The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol., 129, 687–702. [PubMed] [Google Scholar]
- 32.Houseman E., Accomando W., Koestler D., Christensen B., Marsit C., Nelson H., Wiencke J., Kelsey K. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf., 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hidalgo B., Irvin M.R., Sha J., Zhi D., Aslibekyan S., Absher D., Tiwari H.K., Kabagambe E.K., Ordovas J.M., Arnett D.K. (2014) Epigenome-wide Association Study of Fasting Measures of Glucose, Insulin, and HOMA-IR in GOLDN. Diabetes, 63, 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yvan-Charvet L., Wang N., Tall A.R. (2010) Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol., 30, 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng P., Wang L., Yang X., Huang X., Ba Y., Chen X., Guo J., Lian J., Zhou J. (2014) A preliminary study of the relationship between promoter methylation of the ABCG1, GALNT2 and HMGCR genes and coronary heart disease. PloS one, 9, e102265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yashin A.I., Wu D., Arbeev K.G., Ukraintseva S.V. (2010) Joint influence of small-effect genetic variants on human longevity. Aging, 2, 612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berisha S.Z., Serre D., Schauer P., Kashyap S.R., Smith J.D. (2011) Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PloS one, 6, e16729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monda K.L., Chen G.K., Taylor K.C., Palmer C., Edwards T.L., Lange L.A., Ng M.C.Y., Adeyemo A.A., Allison M.A., Bielak L.F., et al. (2013) A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat. Genet., 45, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.-P., Meduri E., Barrett A., et al. (2012) Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet., 44, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundberg E., Meduri E., Sandling J.K., Hedman Å.K., Keildson S., Buil A., Busche S., Yuan W., Nisbet J., Sekowska M., et al. (2013) Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet., 93, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irvin M.R., Zhi D., Joehanes R., Mendelson M., Aslibekyan S., Claas S.A., Thibeault K.S., Patel N., Day K., Waite Jones L., et al. (2014) Epigenome-wide association study of fasting blood lipids in the genetics of lipid lowering drugs and diet network study. Circulation, 130, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gagnon F., Aissi D., Carrie A., Morange P.E., Tregouet D.A. (2014) Robust validation of methylation levels association at CPT1A locus with lipid plasma levels. J. Lipid Res., 55, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Absher D.M., Li X., Waite L.L., Gibson A., Roberts K., Edberg J., Chatham W.W., Kimberly R.P. (2013) Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet., 9, e1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pidsley R., CC Y.W., Volta M., Lunnon K., Mill J., Schalkwyk L.C. (2013) A data-driven approach to preprocessing Illumina 450 K methylation array data. BMC Genomics. 10.1186/1471–2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feinberg A.P., Tycko B. (2004) The history of cancer epigenetics. Nat. Rev. Cancer, 4, 143–153. [DOI] [PubMed] [Google Scholar]
- 46.Herman J.G., Baylin S.B. (2003) Gene silencing in cancer in association with promoter hypermethylation. New Engl. J. Med., 349, 2042–2054. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J., Goldberg J., Vaccarino V. (2013) Promoter methylation of serotonin transporter gene is associated with obesity measures: a monozygotic twin study. Int. J. Obes., 37, 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Hajj N., Pliushch G., Schneider E., Dittrich M., Muller T., Korenkov M., Aretz M., Zechner U., Lehnen H., Haaf T. (2013) Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes, 62, 1320–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crujeiras A.B., Campion J., Diaz-Lagares A., Milagro F.I., Goyenechea E., Abete I., Casanueva F.F., Martinez J.A. (2013) Association of weight regain with specific methylation levels in the NPY and POMC promoters in leukocytes of obese men: a translational study. Regul. Pept., 186, 1–6. [DOI] [PubMed] [Google Scholar]
- 50.Barres R., Kirchner H., Rasmussen M., Yan J., Kantor F.R., Krook A., Naslund E., Zierath J.R. (2013) Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep., 3, 1020–1027. [DOI] [PubMed] [Google Scholar]
- 51.Moleres A., Campion J., Milagro F.I., Marcos A., Campoy C., Garagorri J.M., Gomez-Martinez S., Martinez J.A., Azcona-Sanjulian M.C., Marti A. (2013) Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON Study. FASEB J., 27, 2504–2512. [DOI] [PubMed] [Google Scholar]
- 52.Ronn T., Volkov P., Davegardh C., Dayeh T., Hall E., Olsson A.H., Nilsson E., Tornberg A., Dekker Nitert M., Eriksson K.F., et al. (2013) A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet., 9, e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatanaka M., Shimba S., Sakaue M., Kondo Y., Kagechika H., Kokame K., Miyata T., Hara S. (2009) Hypoxia-inducible factor-3alpha functions as an accelerator of 3T3-L1 adipose differentiation. Biol. Pharm. Bull., 32, 1166–1172. [DOI] [PubMed] [Google Scholar]
- 54.Heidbreder M., Qadri F., Johren O., Dendorfer A., Depping R., Frohlich F., Wagner K.F., Dominiak P. (2007) Non-hypoxic induction of HIF-3alpha by 2-deoxy-D-glucose and insulin. Biochem. Biophy. Res. Commun., 352, 437–443. [DOI] [PubMed] [Google Scholar]
- 55.Pravenec M., Kazdova L., Landa V., Zidek V., Mlejnek P., Simakova M., Jansa P., Forejt J., Kren V., Krenova D., et al. (2008) Identification of mutated Srebf1 as a QTL influencing risk for hepatic steatosis in the spontaneously hypertensive rat. Hypertension, 51, 148–153. [DOI] [PubMed] [Google Scholar]
- 56.Grarup N., Stender-Petersen K.L., Andersson E.A., Jorgensen T., Borch-Johnsen K., Sandbaek A., Lauritzen T., Schmitz O., Hansen T., Pedersen O. (2008) Association of variants in the sterol regulatory element-binding factor 1 (SREBF1) gene with type 2 diabetes, glycemia, and insulin resistance: a study of 15,734 Danish subjects. Diabetes, 57, 1136–1142. [DOI] [PubMed] [Google Scholar]
- 57.Bouchard-Mercier A., Rudkowska I., Lemieux S., Couture P., Perusse L., Vohl M.C. (2014) SREBF1 gene variations modulate insulin sensitivity in response to a fish oil supplementation. Lipids Health and Dis., 13, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Padilla J., Jenkins N.T., Thorne P.K., Martin J.S., Rector R.S., Davis J.W., Laughlin M.H. (2014) Identification of genes whose expression is altered by obesity throughout the arterial tree. Physiol. Genomics, 46, 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ribel-Madsen R., Fraga M.F., Jacobsen S., Bork-Jensen J., Lara E., Calvanese V., Fernandez A.F., Friedrichsen M., Vind B.F., Hojlund K., et al. (2012) Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PloS One, 7, e51302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horvath S. (2013) DNA methylation age of human tissues and cell types. Genome Biol., 14, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. (2012) Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. Part B, 159B, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlen S.E., Greco D., Soderhall C., Scheynius A., Kere J. (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS One, 7, e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jaffe A.E., Irizarry R.A. (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol., 15, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carvalheira J.B., Qiu Y., Chawla A. (2013) Blood spotlight on leukocytes and obesity. Blood, 122, 3263–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dixon J.B., O'Brien P.E. (2006) Obesity and the white blood cell count: changes with sustained weight loss. Obes. Surg,, 16, 251–257. [DOI] [PubMed] [Google Scholar]
- 66.Nieto F.J., Szklo M., Folsom A.R., Rock R., Mercuri M. (1992) Leukocyte count correlates in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Am. J. Epidemiol., 136, 525–537. [DOI] [PubMed] [Google Scholar]
- 67.The ARIC Investigators. (2008) ARIC Study Website http://www2.cscc.unc.edu/aric/.
- 68.Baecke J.A., Burema J., Frijters J.E. (1982) A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am. J. Clin. Nutr., 36, 936–942. [DOI] [PubMed] [Google Scholar]
- 69.Meyer A.M., Evenson K.R., Couper D.J., Stevens J., Pereria M.A., Heiss G. (2008) Television, physical activity, diet, and body weight status: the ARIC cohort. Int. J. Behav. Nutr. Phys. Act., 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. (1997) Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care, 20, 1183–1197. [DOI] [PubMed] [Google Scholar]
- 71.Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M. (2011) Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics, 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 72.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. (2011) High density DNA methylation array with single CpG site resolution. Genomics, 98, 288–295. [DOI] [PubMed] [Google Scholar]
- 73.Dedeurwaerder S., Defrance M., Calonne E., Denis H., Sotiriou C., Fuks F. (2011) Evaluation of the Infinium Methylation 450 K technology. Epigenomics, 3, 771–784. [DOI] [PubMed] [Google Scholar]
- 74.Dedeurwaerder S., Desmedt C., Calonne E., Singhal S.K., Haibe-Kains B., Defrance M., Michiels S., Volkmar M., Deplus R., Luciani J., et al. (2011) DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med., 3, 726–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. (2013) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics, 29, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marabita F., Almgren M., Lindholm M.E., Ruhrmann S., Fagerstrom-Billai F., Jagodic M., Sundberg C.J., Ekstrom T.J., Teschendorff A.E., Tegner J., et al. (2013) An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics, 8, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu M.C., Joubert B.R., Kuan P.F., Haberg S.E., Nystad W., Peddada S.D., London S.J. (2014) A systematic assessment of normalization approaches for the Infinium 450 K methylation platform. Epigenetics, 9, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bose M., Wu C., Pankow J.S., Demerath E., Bressler J., Fornage M., Grove M.L., Mosley T., Hicks C., North K.E., et al. (2014) Evaluation of microarray-based DNA methylation measurement using technical replicates: the Atherosclerosis Risk in Communities (ARIC) Study. BMC Bioinf., 15, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhuang J., Widschwendter M., Teschendorff A.E. (2012) A comparison of feature selection and classification methods in DNA methylation studies using the Illumina Infinium platform. BMC Bioinf., 13, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders A.D., Stevenson C., Pearson J., Burt M., McGeoch G., Hudson B., Eglinton T.W. (2013) A novel pathway for investigation of colorectal symptoms with colonoscopy or computed tomography colonography. New Zealand Med. J., 126, 45–57. [PubMed] [Google Scholar]
- 81.Gatti D.M., Barry W.T., Nobel A.B., Rusyn I., Wright F.A. (2010) Heading down the wrong pathway: on the influence of correlation within gene sets. BMC Genomics. 10.1186/1471-2164-11-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Y.H., Barry W.T., Wright F.A. (2013) Empirical pathway analysis, without permutation. Biostatistics, 14, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statis. Soc. Series B, 57, 289–300. [Google Scholar]
- 84.Benjamini Y., Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann. Statis., 29, 1165–1188. [Google Scholar]
- 85.Johnson W.E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. [DOI] [PubMed] [Google Scholar]
- 86.Grove M.L., Yu B., Cochran B.J., Haritunians T., Bis J.C., Taylor K.D., Hansen M., Borecki I.B., Cupples L.A., Fornage M., et al. (2013) Best practices and joint calling of the HumanExome BeadChip: The CHARGE Consortium. PloS One, 8, e68095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duggal P., Gillanders E., Holmes T., Bailey-Wilson J. (2008) Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics, 9, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. (2013) Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics, 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whitlock M.C. (2005) Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol., 18, 1368–1373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.