Abstract
Cleft lip and/or palate (CL/P) are common structural birth defects in humans. We used exome sequencing to study a patient with bilateral CL/P and identified a single nucleotide deletion in the patient and her similarly affected son—c.546_546delG, predicting p.Gln183Argfs*57 in the Distal-less 4 (DLX4) gene. The sequence variant was absent from databases, predicted to be deleterious and was verified by Sanger sequencing. In mammals, there are three Dlx homeobox clusters with closely located gene pairs (Dlx1/Dlx2, Dlx3/Dlx4, Dlx5/Dlx6). In situ hybridization showed that Dlx4 was expressed in the mesenchyme of the murine palatal shelves at E12.5, prior to palate closure. Wild-type human DLX4, but not mutant DLX4_c.546delG, could activate two murine Dlx conserved regulatory elements, implying that the mutation caused haploinsufficiency. We showed that reduced DLX4 expression after short interfering RNA treatment in a human cell line resulted in significant up-regulation of DLX3, DLX5 and DLX6, with reduced expression of DLX2 and significant up-regulation of BMP4, although the increased BMP4 expression was demonstrated only in HeLa cells. We used antisense morpholino oligonucleotides to target the orthologous Danio rerio gene, dlx4b, and found reduced cranial size and abnormal cartilaginous elements. We sequenced DLX4 in 155 patients with non-syndromic CL/P and CP, but observed no sequence variants. From the published literature, Dlx1/Dlx2 double homozygous null mice and Dlx5 homozygous null mice both have clefts of the secondary palate. This first finding of a DLX4 mutation in a family with CL/P establishes DLX4 as a potential cause of human clefts.
Introduction
Cleft lip and/or palate (CL/P) is found in an estimated 1 in 600 to 1 in 900 live births and can occur as an isolated malformation [isolated or non-syndromic (NS) CL/P; 76.8% of patients], in association with other malformations (15.9% of patients) or as part of a recognized multiple congenital anomaly syndrome (syndromic CL/P; 7.3% patients) (1,2). For the majority of CL/P patients, inheritance is multifactorial and involves the combined effects of environmental and genetic factors acting during the first weeks of pregnancy. Although several molecular pathways and genes, including interferon regulatory factor 6 (IRF6), Forkhead Box E1 (FOXE1), Muscle segment homeobox, Drosophila, homolog of, 1 (MSX1), Special AT-rich sequence-binding protein 2 (SATB2), Sonic hedgehog (SHH) and Transforming growth factor, beta-3 (TGFB3), have been strongly implicated in the pathogenesis of CL/P (3–7), the majority of cases of NS CL/P do not have identified causal mutations (7–9). The process of palatogenesis or the formation of equivalent structures has been well described in animal models. In mice, the palatal shelves emerge from the internal side of the maxillary portion of the first pharyngeal domain at embryonic day (E) 11.5, grow along the sides of the tongue at E11.5–E13.5, elevate above the tongue at E14.0 and then join from E14.5 to E16.5 (10). At closure, the medial edge epithelium fuses at the site of the approximating palatal shelves to form a midline palatal seam, termed the medial epithelial seam (MES). For successful closure, both apoptosis of the epithelial edge of the MES and epithelial to mesenchymal transition (EMT) are required (11). In Danio rerio, paired maxillary prominences extend medially and cranially to form the lateral ethmoid plate that then fuses with the median frontonasal element (12).
The Dlx genes are homeodomain-containing transcription factors that are orthologous to the Distal-less gene in Drosophila melanogaster (13,14). There are three bigene Dlx clusters with convergently transcribed, closely located gene pairs (Dlx1/Dlx2, Dlx3/Dlx4, Dlx5/Dlx6) in humans and mouse. Teleosts experienced a whole genome duplication and subsequent gene loss, resulting in eight zebrafish dlx genes: dlx1a/dlx2a, dlx2b, dlx3b/dlx4b, dlx4a, dlx5a/dlx6a (15). The dlx2, dlx3 and dlx4 proteins are structurally very similar at the homeodomain and surrounding amino acids (16). All of the murine Dlx genes are expressed differentially in the branchial region during embryogenesis (14). The murine Dlx genes are expressed in the mesenchyme derived from neural crest cells in the first pharyngeal arch or primordium of the jaw (10). Dlx1 and Dlx2 are expressed in the maxillary arch, the precursor to the upper jaw, whereas Dlx3–Dlx6 are expressed in the mandibular arch, the precursor to the lower jaw (10). In D. rerio, the dlx genes are expressed in the early development of the forebrain, migrating neural crest, branchial arches and otic and olfactory placodes (17). In humans, embryonic expression of DLX4 has not been studied and DLX4 expression is absent from most adult tissues (18). However, the long isoform of human DLX4 can be present in a variety of cancers, including acute leukemia (19,20), breast cancer (21–23), lung cancer (24), prostatic adenocarcinoma (25) and colorectal cancer (26).
Animal models of loss of Dlx gene function have established the importance of these genes in craniofacial patterning. Dlx1/Dlx2 double homozygous null mice have fully penetrant, cleft of the secondary palate (10,27) due to reduced mesenchymal cell proliferation at the initial stages of palatal shelf formation at E11.5 with severely deficient growth of the posterior palate (10). The loss of Dlx1 and Dlx2 function results in down-regulation of a signaling loop involving Shh, bone morphogenic protein 4 (Bmp4) and Fibroblast growth factor 10 (Fgf10) that is important for cell proliferation in the epithelium of the midpalate at E13.5 (10). In D. rerio, inactivation of dlx2a with dlx1a resulted in reduced cranial outgrowth and defects of pharyngeal arch derivatives, including as Meckel's cartilage and the ceratohyal elements (28). Dlx5 homozygous null mice also have a cleft of the secondary palate, with absent horizontal laminae of the palatine bones and shorter nasal and maxillary bones manifesting as a short snout (29,30). In humans, a girl with multiple anomalies including a cleft palate was reported to have a de novo chromosome deletion at 7q21.37q31.1, with a proximal breakpoint just 88 kb downstream of the DLX5 gene, leading to the hypothesis that dysregulation of this gene caused the palatal malformation (31). A missense substitution in DLX5 (c.576C > G predicting p.Ile192Met; NM_005221.5) was found in an individual with Pierre Robin sequence (CP, glossoptosis and micrognathia) and predicted to be damaging, but was inherited from the individual's unaffected mother (32) and thus of unclear significance. DLX5 mutations resulting in haploinsufficiency have also caused autosomal dominant split-hand split-foot malformation [MIM 183600] (33–36) and mutations in DLX3 cause Tricho-dento-osseous syndrome [MIM 190320] and amelogenesis imperfecta with taurodontism [MIM 104510] (37,38).
We used exome sequencing to study the mother from a parent and child pair with bilateral CL/P. Both family members had dysmorphic features comprising euryblepharon (enlargement of the palpebral fissures) and lagophthalmos (inability to close the eyelids completely) and a mild, incomplete form of blepharocheilodontic syndrome (BCDS; also known as Blepharocheilodontic dysplasia) [MIM 119580] was considered as a diagnosis (39–43), but the degree of dysmorphism was much milder than classically affected individuals. We identified a DLX4 sequence variant that was predicted to be pathogenic in the mother and that was verified in both the mother and her son by Sanger sequencing. The following report describes our studies to implicate this gene in the pathogenesis of the CL/P in this family.
Results
Exome sequencing in a mother and her son with bilateral CL/P revealed a deleterious DLX4 mutation
Exome sequencing was performed on a female patient with bilateral CL/P and a mean coverage of 51X was obtained (Supplementary Material, Table S1). In our analysis, we used wAnnovar (http://wannovar.usc.edu) and excluded sequence variants that were in the Database of Single Nucleotide Polymorphisms (http://www.ncbi.nlm.nih.gov/projects/SNP/), those that were present in segmental duplications and included only those for which minor allele frequency was <0.05 in the 1000 Genomes browser (http://www.1000genomes.org/) and the Exome Variant Server (http://evs.gs.washington.edu/EVS/). We excluded genes in which sequence variants had been identified for discordant phenotypes (for example, structural eye defects) from previous exome data and public databases (data not shown). We also examined previously reported genes in CL/P (7) for sequence variants and did not identify any pathogenic alterations (data not shown). Using this filtering strategy and examining mutations predicted to disrupt reading frame or to substitute a non-synonymous amino acid, we obtained a list of 25 genes that were predicted to disrupt reading frame (Supplementary Material, Table S2). We sought information about expression for these genes in the palate and lip from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and from NEIbank (http://neibank.nei.nih.gov/index.shtml) in view of the mild features of BCDS, although we could not find data for the majority of the genes. We also sought information regarding each gene from PubMed as a means to select candidate genes for further evaluation.
In DLX4, we noted c.546_546delG, predicting p.Gln183Argfs*57 (NM_138281.2) and resulting in a frameshift that was assessed as deleterious (Mutation Taster P = 1.0; http://mutationtaster.org/MutationTaster/). The human DLX4 gene has two isoforms—a longer isoform, henceforth termed DLX4_LI, with three exons that encodes a 240 amino acid protein (transcript NM_138281.2 and protein NP_612138.1) and a shorter isoform, henceforth termed DLX4_SI, that has two exons and encodes an 168 amino acid protein (transcript NM_001934.3 and protein NP_001925.2). Both isoforms contain a homeodomain and overlap with complete identity for the terminal 146 amino acids of the proteins. In the long isoform, the deletion of the single G nucleotide at position c.546 results in replacement of the terminal 56 amino acids of the protein with a stop codon at position 239, two codons short of the wild-type length of the protein and altering all but 7/56 (12.5%) residues of the protein after the nucleotide deletion (Supplementary Material, Table S3). The homeobox of the protein is located from amino acid 108–186 and thus the frameshift mutation affects the terminal part of this domain. The sequence variant was not present in the Exome Variant Server or 1000 Genomes browser. In the short isoform of DLX4, the sequence variant is c.330delG, predicting p.Gln111Argfs*57.
We selected DLX4 as a candidate gene because of the predicted deleteriousness of the c.546_546delG sequence alteration, its presence in both mother and child and because of the known involvement of the Dlx genes in branchial arch patterning in animal models as summarized earlier. We verified the DLX4 mutation using Sanger sequencing in the mother and found the same mutation in her son (Fig. 1). We sequenced three patients diagnosed with BCDS (42,43) for DLX4 mutations, but could not demonstrate any sequence variants (data not shown). Sequencing of 155 patients with NS CL, CL/P and CP for DLX4 mutations was also negative (data not shown).
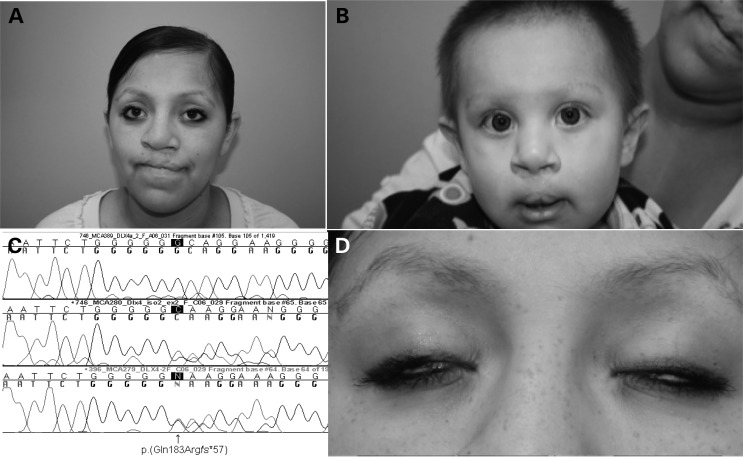
Figure 1.
Facial appearance of the patient and son with bilateral CL/P. (A) Frontal view of the patient, showing scars from bilateral cleft lip repair, hypertelorism and borderline low-set ears. (B) Frontal view of patient's son, showing scars from bilateral cleft lip repair and euryblepharon. (C) Chromatogram of the patient (third tracing) and her son (second tracing), showing c.546_546delG, predicting p.Gln183Argfs*57, in DLX4_LI, compared with wild-type sequence (top tracing). (D) Frontal view of the patient, showing lagophthalmos.
In situ hybridization showed that Dlx4 was expressed in the murine palate prior to palate closure
In situ hybridization with antisense riboprobes in frontal sections obtained from mouse upper jaw showed Dlx4 expression in the mesenchyme of the anterior murine palate at E12.5, prior to the time of palate closure (Fig. 2A and B). Whole mount staining showed a reduction in expression at later time periods after palate closure (Fig. 2C–E), consistent with a role for this gene in palatal closure.
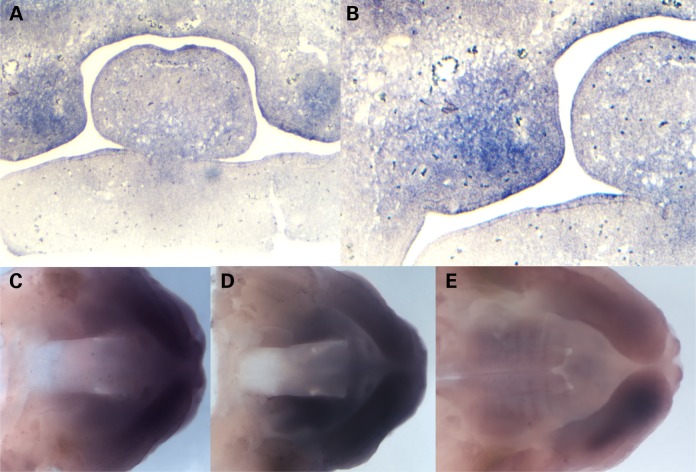
Figure 2.
In situ hybridization shows Dlx4 expression in the murine palatal shelves. (A) In situ hybridization with an antisense probe for murine Dlx4 at E12.5, showing expression in the anterior palate prior to palate closure. (B) Higher resolution view of murine Dlx4 in the palatal shelves at E12.5, showing expression in palatal mesenchyme. (C) Whole mount in situ hybridization with an antisense probe for murine Dlx4 at E12.5, showing expression in the anterior palatal shelves. (D) Whole mount in situ hybridization with an antisense probe for murine Dlx4 at E13.5, showing continued expression in the anterior palatal shelves. (E) Whole mount in situ hybridization with an antisense probe for murine Dlx4 at E14.5, showing reduced expression in the anterior palatal shelves at the time of palate closure.
Wild-type, but not mutant DLX4_c.546delG was able to activate murine conserved regulatory elements
We used a luciferase assay to demonstrate different activity between the wild-type DLX4_LI allele and the c.546delG mutant DLX4_LI allele. We showed that a positive control, murine PCAG_Dlx2_IRES_GFP, could activate the I1/2b and I5/6i murine conserved regulatory elements (CREs) (44,45) in P19 cells using a luciferase assay (Supplementary Material, Tables S4A and B). With the same assay, human wild-type DLX4_LI was able to activate both CREs, albeit less strongly. Mutant c.546delG DLX4_LI showed a 4-fold reduction in activity compared with wild-type DLX4_LI for both CREs (Supplementary Material, Table S4B; Fig. 3). These results are consistent with altered function for the mutant allele compared with wild-type DLX4_LI and suggest that DLX4 may regulate other DLX genes.
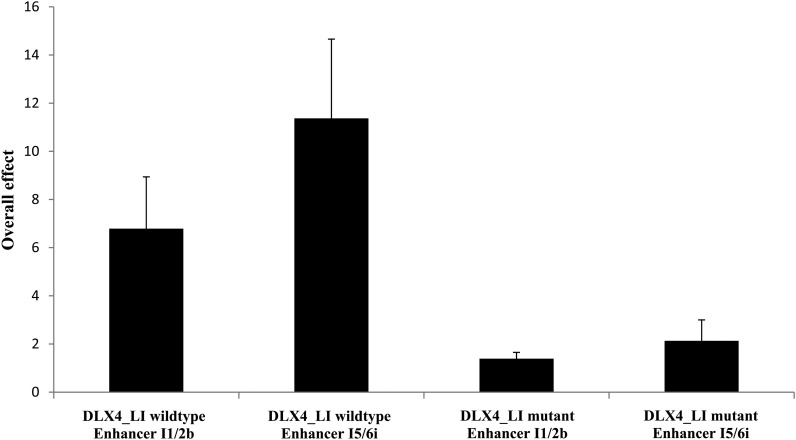
Figure 3.
Overall transcription factor and enhancer effect for activation of two murine CREs by human wild-type DLX4-long isoform and human mutant c.546delG DLX4-long isoform measured by a luciferase assay. An overall 4-fold increased activation was observed with wild-type DLX4 compared with mutant DLX4 for two CREs, I1/2b and I5/6i. All results are expressed as the mean ± standard error of the mean; sample size n = 3.
Western blotting and immunofluorescence showed no degradation of mutant DLX4 protein
We determined if the mutant DLX4 protein would be degraded or unable to localize to the nucleus similar to the wild-type protein. Immunofluorescence staining using an antibody to DLX4 in Hela cells showed that the wild-type protein was located in the cell nucleus, as expected, and that the mutant protein also localized correctly (Fig. 4). Western blotting after transient transfection of wild-type DLX4_LI and mutant DLX4_LI also showed no significant difference in wild-type and mutant protein levels (Fig. 4).
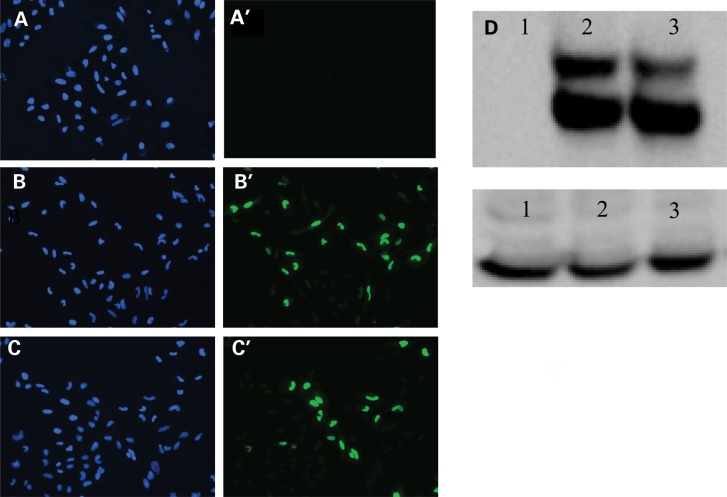
Figure 4.
Human mutant c.546delG DLX4-long isoform protein can localize to the cell nucleus and is not degraded. (A–C) Immunofluorescence study showing localization of both wild-type DLX4-long isoform (DLX4_LI) and mutant DLX4_LI proteins to the cell nucleus after transient transfection in HeLa cells. (A) Untransfected cells stained with DAPI. (A′) Untransfected cells stained with antibody to human DLX4 conjugated to AlexaFluor488. (B) Cells transfected with wild-type DLX4_LI stained with DAPI. (B′) Cells transfected with wild-type DLX4_LI stained with antibody to human DLX4 conjugated to AlexaFluor488. (C) Cells transfected with mutant DLX4_LI stained with DAPI. (C′) Cells transfected with mutant DLX4_LI stained using an antibody to human DLX4 conjugated to AlexaFluor488. (D) Western blotting with an antibody against wild-type DLX4_LI shows no significant degradation of mutant DLX4 protein compared with wild-type protein. Lane 1. HeLa cells transfected with empty vector. Lane 2. HeLa cells transfected with wild-type DLX4_LI. Lane 3. HeLa cells transfected with mutant DLX4_LI. GAPDH was used as the loading control.
Short interfering RNA and quantitative reverse-transcription-polymerase chain reaction showed that reduced DLX4 expression resulted in up-regulation of other DLX genes
We hypothesized that DLX4 was acting by altering apoptosis or dysregulating EMT required for palate closure. We overexpressed human wild-type DLX4_LI and mutant DLX4_LI by transient transfection in HeLa cells and used quantitative reverse-transcription-polymerase chain reaction (qRT-PCR) to compare expression of genes known to be involved in either apoptosis or EMT. We did not observe any significant change in the expression levels for B cell CLL/Lymphoma 2 (BCL2), BCL-XL, BCL2-Associated X protein (BAX) and Snail, Drosophila Homolog of, 1 (SNAI1) and thus could find no definite effect of DLX4 overexpression on apoptosis genes (data not shown). Similarly, transient transfection did not produce any significant changes in the genes involved in EMT, including E-CADHERIN, Matrix Metalloproteinase 2 (MMP2), Matrix Metalloproteinase 3 (MMP3), Matrix Metalloproteinase 7 (MMP7), Matrix Metalloproteinase 13 (MMP13) and Transforming Growth Factor-Beta Receptor, Type III (TGFBR3) (data not shown). Additionally, we studied Twist, Drosophila, Homolog of, 1 (TWIST1) because overexpression of DLX4 was previously shown to increase the expression of TWIST1 in cancer cell lines and to induce EMT (46). However, we found no significant difference in TWIST1 expression following DLX4 overexpression (data not shown).
We examined expression of the other DLX genes after short interfering RNA (siRNA) for DLX4 in HEK293T cells because of the results of the luciferase assay and the work of other researchers suggested that DLX4 would regulate the expression of other DLX genes. We were able to show that reduced DLX4 expression was associated with significant up-regulation of DLX3 and DLX5 and DLX6, implying possible compensatory regulation (Fig. 5). In addition, siRNA for DLX4 resulted in a small, but significant decrease in DLX2, but not DLX1 expression (Fig. 5). We also tested the SHH and BMP signaling pathways because of their known involvement in early facial morphogenesis and the implication of these genes and pathways in the clefting resulting from loss of Dlx1/Dlx2 expression. Using siRNA to reduced DLX4 expression in HeLa cells, we found a consistent and robust increase in BMP4 expression (Fig. 6), suggesting that up-regulation or altered dosage of BMP4 may be important in the action of DLX4. Transfection of wild-type and mutant DLX4_LI into HeLa cells without siRNA did not alter BMP4 expression (data not shown). However, expression differences in HeLa cells may not be relevant to embryogenesis and we could not reproduce the difference in BMP4 expression after siRNA for DLX4 in HEK293T cells (data not shown).
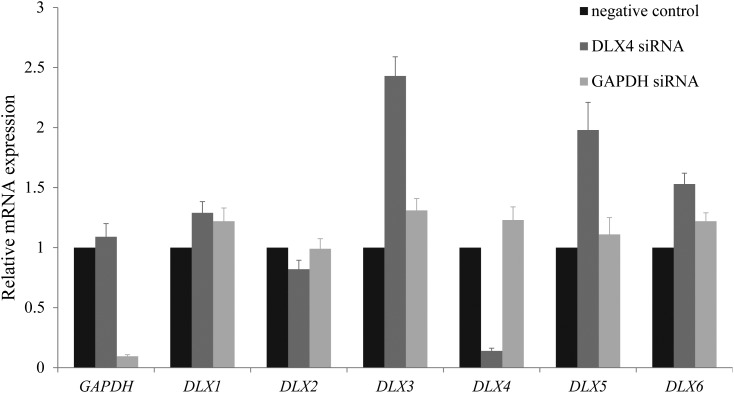
Figure 5.
siRNA and qRT-PCR in HEK293T cells show that reduced DLX4 expression results in up-regulation of DLX3, DLX5 and DLX6. Gene-specific primers are indicated on the X-axis; relative mRNA expression is indicated on the Y-axis. The first columns show data from untreated wells; expression for these wells has been normalized to 1 for comparison. The second columns show data from wells treated with 60 nm DLX4 siRNA treatment and the third columns show data from wells treated with 60 nm GAPDH siRNA. The first set of three columns confirms reduced expression of GAPDH after 60 nm GAPDH siRNA treatment (third column) using GAPDH-specific primers. Similarly, the fifth set of columns confirms reduced expression of DLX4 after 60 nm DLX4 siRNA treatment (second column) using DLX4-specific primers. The fourth, sixth and seventh sets of columns show significantly increased DLX3, DLX5 and DLX6 expression after 60 nm DLX4 siRNA treatment (second columns); all P values were <0.05. The remaining sets of columns show no significant changes in DLX1 expression and reduced expression for DLX2 (P = 0.024), after 60 nm DLX4 siRNA treatment (second columns). All results are expressed as the mean ± standard error of the mean; sample size n = 4.
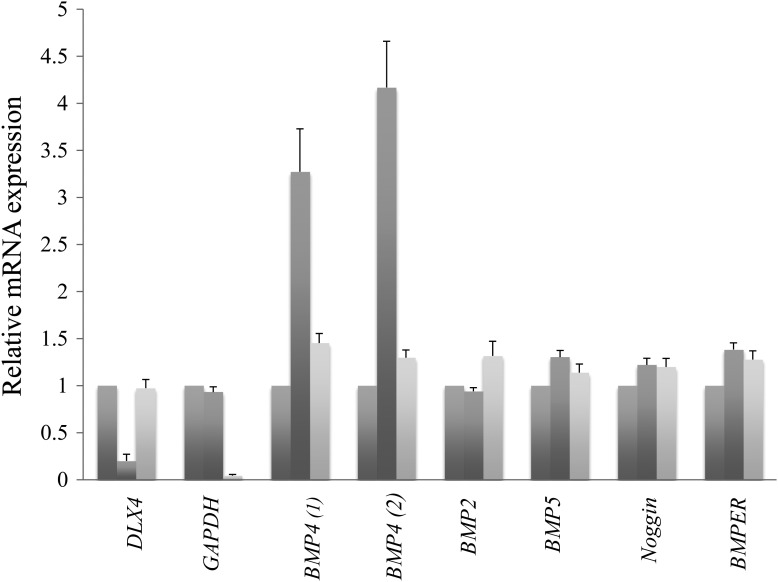
Figure 6.
siRNA and qRT-PCR in HeLa cells show that reduced DLX4 expression results in up-regulation of BMP4. Gene-specific primers are indicated on the X-axis; relative mRNA expression is indicated on the Y-axis. The first columns show data from untreated wells; expression for these wells has been normalized to 1 for comparison. The second columns show data from wells treated with 60 nm DLX4 siRNA treatment and the third columns show data from wells treated with 60 nm GAPDH siRNA. The first set of three columns confirms reduced expression of DLX4 after 60 nm DLX4 siRNA treatment (second column) using DLX4-secific primers. Similarly, the second set of columns confirms reduced expression of GAPDH after 60 nm GAPDH siRNA treatment (third column) using GAPDH-specific primers. The third and fourth sets of columns show increased BMP4 expression after 60 nm DLX4 siRNA treatment (second columns) using two primer sets for BMP4. The remaining sets of columns show no significant changes in BMP2, BMP5, Noggin and BMPER after 60 nm DLX4 siRNA treatment (second columns). All results are expressed as the mean ± standard error of the mean; sample size n = 3.
Antisense morpholino injections targeting dlx4a and dlx4b showed reduced cranial outgrowth and defects of the cartilaginous elements of the jaw
We modeled both loss and gain of function for dlx4 in zebrafish. In D. rerio, there are two orthologous dlx genes, each with three exons—dlx4a (ENSDARG00000011956) that encodes a 250 amino acid protein with 43% similarity to DLX4_LI, and dlx4b (ENSDARG00000071560) that encodes a 254 amino acid protein with 37% similarity to DLX4_LI. We first tested whether reduced expression for dlx4a and dlx4b would perturb jaw formation. We injected two dlx4a morpholino oligonucleotides (MOs), but found only minimal external differences in jaw formation and as there was increased cell death and toxicity (data not shown), these MOs were excluded from further analysis. For dlx4b, we injected both a splice site MO targeting the exon2–intron2 junction of dlx4b (dlx4b MO1) and a translational MO (dlx4b MO2) and measured cranial dimensions at 5–6 days post fertilization (dpf) after staining with Alcian blue (Supplementary Material, Fig. S1). We examined larvae for facial outgrowth as a measure of jaw development and for defects in the cartilaginous elements of the jaw (Meckel's cartilage, palatoquadrate cartilage, ceratohyal cartilage and ethmoid) as previously performed for dlx2a and dlx1a (28). For both dlx4b MO1 and dlx4b MO2 injected alone, we found minimal differences from control-injected larvae. However, dual injection of dlx4b MO1 and dlx4b MO2 resulted in defects of anterior facial protrusion such as a shortened distance from the tip of the ceratohyal cartilages to Meckel's cartilages, reduced width or deformation of Meckel's cartilages and an abnormally widened angle of the ceratohyal cartilages (Supplementary Material, Table S5; Fig. 7). We measured craniofacial distances after Alcian blue staining to calculate a length ratio (tip of Meckel's cartilages to top of ceratohyal cartilage divided by total length of cranium) and a width ratio (Supplementary Material, Fig. S1). The results showed that the length ratio of co-injected dlx4b MO1 and dlx4b MO2 larvae was increased compared with controls, with significant shortening of the posterior cranial elements compared with anterior cranial elements and that this altered ratio of lengths was rescued by co-injection of wild-type human DLX4_LI mRNA (Supplementary Material, Table S6). There were no consistent differences in cranial width ratios with dlx4b MO1 and dlx4b MO2 injections (data not shown). We verified reduced expression of dlx4b after MO1 injection using RT-PCR (Supplementary Material, Fig. S2). Injections of wild-type DLX4_LI and mutant DLX4_LI mRNA did not produce a specific external phenotype at doses up to 500 pg (Supplementary Material, Table S7), arguing against a gain of function for the mutant DLX4 allele.
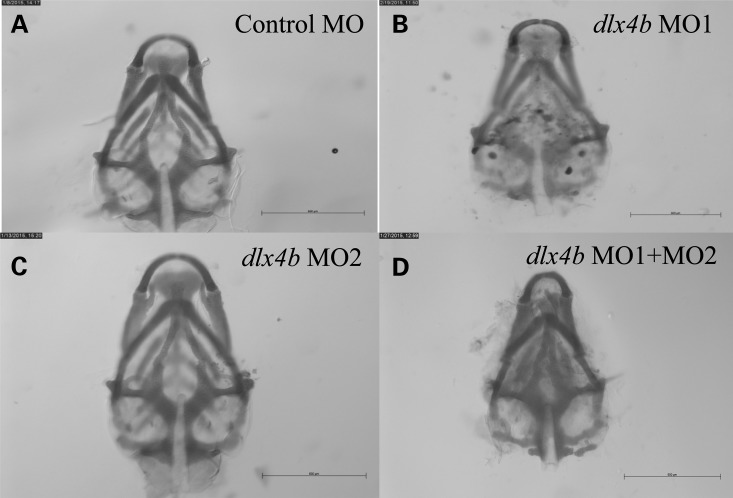
Figure 7.
Danio rerio larvae injected with two antisense morpholinos targeting dlx4b show abnormal formation of the neurocranium at 5–6 dpf. (A) Larva injected with control MO shows normal formation of the neurocranium after Alcian blue staining. (B and C) Representative larvae injected with single antisense MOs (MO1 and MO2) targeting dlx4b show minimal differences in the formation of the neurocranium after Alcian blue staining compared with larvae injected with control MO. (D) Larvae injected with two antisense MOs (MO1 and MO2) targeting dlx4b show abnormal formation of the neurocranium, in this case with narrowing of the Meckel's cartilages, after Alcian blue staining compared with larvae injected with control MO. Scale bars indicate 500 μm.
Discussion
We present a parent–child duo with bilateral CL/P who both have c.546_546delG, predicting p.Gln183Argfs*57 in DLX4, which was predicted to be pathogenic. In situ hybridization showed expression of Dlx4 in the mesenchyme of the anterior murine palate prior to fusion of the palatal shelves at E12.5. The mutation did not appear to alter DLX4 localization or stability, but mutant DLX4_LI was less able to activate two murine CREs compared with wild-type DLX4_LI, implying that the mutant allele is likely to have reduced function. Transient transfection experiments were unable to show that DLX4 altered apoptosis or EMT, but siRNA experiments reducing DLX4 expression in HEK293T cells showed up-regulation of DLX3, DLX5 and DLX6 and down-regulation of DLX2. Up-regulation of BMP4 after siRNA for DLX4 was observed in HeLa cells, implying that DLX4 may act as a regulator of this gene, but was not reproduced in HEK293T cells. Morpholino experiments targeting dlx4b in D. rerio showed abnormal cranial formation, consistent with a role for the gene in early craniofacial morphogenesis. As Dlx1/Dlx2 double homozygous null mice and Dlx5 homozygous null mice both have a cleft of the secondary palate, we consider that the clefts in this family are likely to be due to haploinsufficiency for DLX4 and that our findings have significance for craniofacial morphogenesis and for human CL/P. DLX4 is located at 17q21.33 (hg19) and genome wide association studies and linkage have previously associated with the 17q21 chromosome region with orofacial clefts in humans (47–50), although RARA at 17q21 (50) and ERBB2 at 17q21.1 (49) were suggested as the candidate genes rather than DLX4. However, the studies we have performed do not rule out a cause or contributory role for mutations in non-coding DNA or in an unidentified gene in the pathogenesis of the clefts in this family.
DLX4_LI was previously studied as Beta protein 1 (BP1) (51). Both DLX4 isoforms share the same homeodomain but have different functions, as the DLX4_LI isoform can repress the β-globin promoter by acting through the two β-globin DNA silencers in transient transfection assays, whereas DLX4_SI is unable to perform this function (51,52). In our experiments, we focused on DLX4_LI and hypothesized that mutant DLX4 would alter or up-regulate apoptosis in the developing palate, as DLX4 has previously been implicated in inhibition of apoptosis in cancer cells (53–55). However, we did not find any convincing evidence for dysregulated apoptosis in our transient transfection experiments. We also considered that mutations in DLX4 might cause orofacial clefting by a failure to induce TWIST1, the causative gene for Saethre–Chotzen syndrome [MIM101400] (56,57). An estimated 6% of patients with Saethre–Chotzen have cleft palate (57), and overexpression of DLX4 was previously shown to increase the expression of TWIST1 in cancer cell lines and to induce EMT (46). However, we could not demonstrate altered TWIST1 expression by qRT-PCR after transient transfection. Similarly, we were unable to demonstrate any differences in the genes involved in EMT using the same methods.
Using siRNA for DLX4, we were able to demonstrate dysregulation of other DLX family members, with significant up-regulation for DLX3, DLX5 and DLX6 and down-regulation of DLX2 (Fig. 6). In acute lymphoblastic leukemic cells with the t(4;11)(q21;q23) translocation, reduced expression of DLX2, DLX3 and DLX4 was associated with increased DLX5 and DLX6 (58), consistent with the results seen in these siRNA experiments. The results of our luciferase study suggest that DLX4 can regulate other Dlx genes by binding to CREs. Both Dlx1 and Dlx2 can bind to a Dlx5/Dlx6 intergenic enhancer in embryonic forebrain, with a marked reduction in Dlx5 and Dlx6 expression with Dlx1/Dlx2 knockout mice (59). As reduced DLX4 resulted in a reduction in DLX2 and Dlx1/Dlx2 homozygous null mice have had cleft palate (10,27), this may be one mechanism contributing to the observed clefts, although the magnitude of this contribution was unclear.
We also used siRNA to reduce DLX4 expression in HeLa cells and found a consistent up-regulation of BMP4. Bmp4 has previously been studied in relation to CL/P and deficiency of this gene has been shown to cause CL/P in humans and in mice (60,61). Similar to Dlx4, Bmp4 is expressed in the anterior palate of the mouse at E12.5 (62). In humans, mutations in BMP4 were identified in 1 of 30 cases of microform clefts and 5 of 968 cases of overt CL/P (60). Conditional inactivation of murine Bmp4 in the facial primordia resulted in delayed fusion of the medial nasal process and fully penetrant cleft lip (62). Mice with conditional inactivation of the Bmpr1a gene, a receptor for Bmp4, also develop completely penetrant, bilateral CL/P that is associated with diminished cell proliferation in the mesenchyme of the maxillary process (63). A link between elevated BMP4 expression and CL/P is also supported indirectly by research looking at loss of function of Bmp4 antagonists in palatogenesis. The loss of Noggin, a Bmp4 antagonist, in the palate leads to up-regulation of Bmp4, Bmp2 and Bmp7 and induces cell death in the epithelium of the palate (64). In D. rerio, treatment with dlx3b/4b MO treatment was shown to result in a transient increase in bmp4 expression and loss of cv2 expression, a bmp4 antagonist (65). However, the up-regulation of BMP4 after siRNA for DLX4 was not confirmed in HEK293T cells (data not shown) and thus the significance of this finding in HeLa cells is unclear.
Zebrafish dlx4b was previously studied as dlx7b and expression was found to be similar to its partner dlx3, with transcription in the presumptive telencephalon or developing forebrain and diencephalon at 13 h post fertilization (hpf) and in the olfactory placode at 24–48 hpf (16). In D. rerio, the pharyngeal arches can be subdivided into dorsal (forming the skeletal elements of the palatoquadrate cartilage, hyomandibular cartilage and the maxillary bone), intermediate (expressing all dlx genes except dlx2b and forming the region of the jaw joint and second arch joint, opercle and branchiostegal bones) and ventral domains (forming Meckel’'s cartilage, the ceratohyal cartilage and the dentary) (14). In the first two arches, dlx4a is the most restricted dlx gene and reveals the border of the intermediate domain that is immediately ventral to the dorsal domain (14). A zebrafish deletion mutant (b380) lacked dlx3b and dlx4b among other genes and failed to develop both otic and olfactory placodes (17). dlx3b and dlx4b were found to be responsible for the b380 placodal phenotype and co-injection of dlx4b MO and dlx3b MO had an synergistic effect, causing impaired formation of otoliths and hair cells in the majority of embryos and smaller otic vesicles at 30 hpf (17). dlx4b MO injection alone produced a severe reduction in development of the medial fin fold, but did not affect otic development or numbers of sensory hair cells and otoliths (66). In our experiments using antisense MOs, the jaw phenotype seen with loss of dlx4b function in D. rerio was similar to the defects observed in D. rerio treated with antisense MOs against dlx2a and dlx1a, in which abnormalities of anterior facial protrusion were also observed (28). Our results are in keeping with the findings observed by others who have used D. rerio to model abnormal palatal development, such as smaller pharyngeal skeletal elements, gaps in the ethmoid plate or loss of the ethmoid plate, reduced size or length of the palatoquadrate or Meckel's cartilage elements and malformations of the trabeculae, ethmoid plate and hypophyseal fenestra (28,67–69). Taken together with the reduced activation of the murine CREs, these MO findings are most consistent with likely loss of function for the human DLX4 mutation, although a gain of function effect for the mutation has not been definitively excluded.
We identified a mutation in DLX4, c.546_546delG, predicting p.Gln183Argfs*57, in a mother and child with bilateral cleft lip and palate and minor dysmorphic features. The sequence variant was absent from public databases and predicted to be deleterious and we were able to demonstrate that the mutant DLX4_c.546delG allele had reduced competence to activate two murine Dlx CREs compared with wild-type DLX4, consistent with haploinsufficiency. Co-injection of antisense MOs targeting an orthologous D. rerio gene, dlx4b, resulted in morphant larvae with altered jaw morphology. After siRNA to knockdown DLX4 in human cells, we noted a consistent increase in BMP4 expression, but this was not reproducible in another cell line; however, we also observed dysregulation of other DLX genes. Our results implicate DLX4 in jaw formation in fish and in orofacial clefting in humans for the first time and are in keeping with the known involvement of other DLX genes in murine CL/P and craniofacial morphogenesis.
Materials and Methods
Case report
The proposita (Fig. 1) was born by spontaneous vaginal delivery at 36 weeks gestation following an uncomplicated pregnancy without exposure to teratogens or maternal diabetes. Birthweight was 3544 g (97th centile) and length was 48.3 cm (25th centile). Her medical history included bilateral CL/P that was surgically repaired in early childhood. She had mild hyperopia and was reported to have hearing loss in her left ear, but no further details were available and she did not require hearing aids. She walked at 18 months, spoke her first words at 1 year of age and graduated from high school with a grade point average of 3.0–3.7. On examination at 16 years of age, her speech was hyponasal, but 90% intelligible. Her occipito-frontal-circumference (OFC) was 54.5 cm (2–50th percentile). She had a high anterior hairline with sparse eyebrows, euryblepharon and lagophthalmos (Fig. 1) and mild ectropion of the lower eyelids without distichiasis. She had hypertelorism (interpupillary distance (IPD) 6.4 cm; 97th percentile) and small epicanthic folds. There was marked midface hypoplasia with an anterior crossbite. Her ears were prominent and borderline low-set. There were scars from her CL/P repair and a small, 3–4 mm palatal fistula and she had missing lateral incisors without other missing teeth. There were bilateral single palmar creases.
In her first pregnancy at 19 years of age, a cleft lip was detected in the baby on ultrasound scan at 24 weeks of gestation. No additional anomalies were noted and a fetal echocardiogram was unremarkable; amniocentesis was declined. Her son (Fig. 1) was delivered at 40 weeks gestation with a weight of 3150 g (10–25th centile) and a length of 51.3 cm (25–50th centile). Bilateral CL/P was noted and he underwent a lip repair at 2 months of age and palate repair with release of the lingual frenulum at 13 months of age. He passed his hearing screen at birth. His developmental milestones were normal and at 15 months, he was able to walk independently and had three single words. His medical history included acute dacrocystitis of the left eye, incision and drainage of an abscess at 2 months of age and cryptorchidism and bilateral inguinal hernia repair at 15 months of age. At 7 months of age, OFC was 43 cm (3–25th centile) and IPD was 4 cm (3–25th centile). He had sparse eyebrows, upslanting palpebral fissures, euryblepharon and lagophthalmos. The lower eyelids had mild ectropion and sparse eyelashes, but there was no distichiasis. He had a bulbous nasal tip and scars from bilateral cleft lip and palate repair. The ears were slightly protruberant, and he had a right ear pit. The father of the baby was healthy and the parents had no other children. The mother had contact with two maternal half-sisters, three maternal half-brothers and her mother, but not her father. There was no further history of CL/P in either of the maternal or paternal families. Parental ethnicity was Hispanic and there was no known consanguinity.
Investigations performed on the mother revealed a normal high-resolution karyotype (46,XX) and array comparative genomic hybridization (array CGH) with an Oligo HD Scan™ (CombiMatrix, Irvine, CA) showed a normal female result. This array utilized more than 99 000 probes covering coding and non-coding genomic sequence, with an average probe resolution of 21 kb. No other clinical investigations were performed.
Exome and sanger sequencing
Ethical approval for the study was obtained from the Committee on Human Research at the University of California, San Francisco. After obtaining written, informed consent, venous blood was obtained from the mother. The mother gave permission for a buccal swab to be obtained from her son, but did not wish for venepuncture. DNA was extracted from blood and the buccal swab (Qiagen, Valencia, CA) and exome sequencing was undertaken on the mother as previously described (70,71). In brief, libraries were prepared using biotinylated DNA oligonucleotides (SeqCap EZ Human Exome Library v3.0; Roche Nimblegen, Madison, WI, USA) and sequencing was performed on a HiSeq2000 (Illumina, San Diego, CA, USA) for paired-end 100 cycles. Sequencing reads were processed with a validated and community developed variant calling pipeline (https://github.com/chapmanb/bcbio-nextgen). The sequencing reads were aligned to the hg19 reference genome using the Burrows–Wheeler Alignment tool (BWAv0.5.9) using the default parameters that allow two mismatches. Indexing, realignment and duplicate removal were performed using Picard and Samtools and variants were subsequently called using Genome Analysis Toolkit v 1.3-21-gcb284ee. We then utilized wAnnovar (http://wannovar.usc.edu/) with default parameters to analyze the sequence variants (72,73). We favored autosomal dominant inheritance because of the family pedigree with an affected mother and son, although we considered all possible modes of inheritance to analyze the data. Novel sequence variants were assessed for deleterious consequences using Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) and Mutation Taster (74). Sanger sequencing was performed as previously described to verify sequence novel variants in the mother and her son (70). Three patients with BCDS who were unrelated to this family and 155 pedigrees of Chinese ethnicity with NS CL, CL/P or CP were also sequenced for DLX4 sequence alterations. In 19 of the Asian pedigrees, two siblings were affected and in the remaining 136 pedigrees, at least two generations were affected. Primers used for DLX4 sequencing in the patients of Asian ethnicity are listed in Supplementary Material, Table S8.
Western blotting
Lysate was prepared using RIPA Lysis and Extraction buffer (Thermo Scientific, Rockford, IL) and protein concentration was determined using the Pierce™ BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). A 10–15 μg of total protein was boiled at 70°C for 10 min, run on a Novex gels (10% Bis–Tris Plus; Life Technologies, Grand Island, NY) and transferred to membrane using standard methods. DLX4 murine monoclonal antibody (M01), clone 1F11 (H00001748-M01; Abnova, Taipei City, Taiwan) was used as the primary antibody at a dilution of 1:1000 and an horseradish peroxidase-conjugated secondary antibody was used at 1:10 000 dilution.
Immunofluorescence analysis
Cells were grown on glass coverslips to 50–60% confluence and transfected the following day. At 48 h after transfection, coverslips were fixed in 4% paraformaldehyde (PFA), washed with phosphate-buffered saline (PBS) and permeabilized in 0.1%Triton X-100 at room temperature. Coverslips were incubated with DLX4 primary antibody (1:1000) overnight at 4°C, washed with PBS and incubated in Alexa-488 goat anti-mouse secondary antibody (1:500) for 1 h at room temperature prior to mounting in Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Images were collected on a CoolSNAP EZ Turbo 1394 digital camera (Photometrics, Tuscon, AZ) on a Nikon ECLIPSE 80i microscope (Nikon Instruments, Melville, NY) using a 10× objective.
qRT-PCR and siRNA
A wild-type cDNA clone for the human DLX4-long isoform (DLX4-LI_WT) was purchased (MHS6278–202756942, Accession number BC01615; Thermo Scientific, Rockford, IL). Mutagenesis was performed using the QuikChange™ Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) and the resultant mutant DLX4 with c.546delG (DLX4_LI_Mut) was verified using Sanger sequencing. Cells were transiently transfected in six well dishes with 1–2 μg DLX4 wild-type or mutant cDNA diluted in OPTI-MEM (Life Technologies, Grand Island, NY) using Lipofectamine® 3000 (Life Technologies, Grand Island, NY). RNA was obtained after 48 h using the RNeasy mini kit (Qiagen, Valencia, CA) and cDNA was prepared with SuperScript® III Reverse Transcriptase (Life Technologies, Grand Island, NY). qRT-PCR using gene-specific primers (Supplementary Material, Table S8) was run on an 7500 RT-PCR machine (Applied Biosystems, Foster City, CA) and data were analyzed according to the ΔΔCt method (75) using human TBP as the internal control. For gene silencing, we transfected siRNA (Silencer® Select s4145; Life Technologies, Grand Island, NY), targeting exon 2 of the DLX4 short isoform at basepair (bp) 1305 (NM_001934.3) and exon 3 of the DLX4-long isoform at bp 1552 (NM_138281.2) at a final concentration of 30 or 60 nm in 2 ml media in each well. Silencer® Select GAPDH Positive Control siRNA (Life Technologies, Grand Island, NY) was used as a positive control.
Luciferase assay
We obtained plasmids containing murine CREs for Dlx1/2 (I1/2b) and Dlx5/6 (I5/6i) (kindly provided by Dr John Rubenstein, UCSF) together with murine pCAG_IRES_GFP_Dlx2 as a positive control. P19 cells were cultured on 24-well plates at a density of 5 × 104 cells per well in Alpha Minimum Essential Medium (Life Technologies, Grand Island, NY). Bovine calf serum and inactivated fetal bovine serum were added to a final concentration of 7.5 and 2.5% (v:v), respectively. On the day after plating, 0.5 ng Renilla luciferase plasmid was transfected with different combinations of Dlx transcription factor, empty vector or enhancer using Lipofectamine® 3000 (Life Technologies, Grand Island, NY). We tested pCAG_Dlx2_IRES_GFP, wild-type long isoform DLX4 and mutant c.546delG long isoform DLX4 and the CREs described earlier. Two days after transfection, the cells were lysed using the Dual-Luciferase®Reporter Assay System (Promega, Madison, WI) and the luminescence of firefly luciferase activity and Renilla luciferase activity was measured. The ratio of firefly luciferase activity over Renilla luciferase activity from each well was used to calculate the enhancer and transcription factor effects as follows: Enhancer effect = (transcription factor + enhancer)/(empty transcription factor vector + enhancer); Transcription factor effect = (transcription factor + empty enhancer vector)/(empty transcription factor vector + empty enhancer vector) and Overall effect = Enhancer effect/Transcription factor effect. Each datapoint was obtained from three independent experiments of three wells each, and mean and standard error of the mean were calculated according to standard methods.
In situ hybridization
We obtained a full-length, murine cDNA clone containing Dlx4 (MMM1013-211692663, Accession number BC106967; Thermo Scientific, Rockford, IL). Database searches showed no significant cross-hybridization between the mouse clone and other murine Dlx genes, and clone orientation was verified by Sanger sequencing. Digoxygenin-labeled sense and antisense RNA probes (DIG RNA labeling kit; Roche, Indianapolis, IN) were prepared and in situ hybridization was performed according to standard protocols on jaws from E12.5 to E14.5 time periods after fixation in 4% PFA in whole mount or on 7 μm paraffin sections.
Antisense morpholino injections
We designed antisense MOs through GeneTools (http://www.gene-tools.com/) targeting the two D. rerio orthologs of DLX4–dlx4a (NM_131300.1) and dlx4b (NM_131318.1). We used dlx4a MO1, targeting the exon 1–intron 1 junction of dlx4a, 5′-TGAATGAGAGCACACTTTACCTGCA-3′ (exon underlined), dlx4a MO2, targeting the exon 2–intron 2 junction of dlx4a, 5′-CTCAAAAGCATGGCAACATACCTGT-3′ (exon underlined), dlx4b MO1, 5′-TGATGGATATTTACCTGTGTTTGCG-3′, targeting the exon 2–intron 2 boundary (exon underlined of dlx4b) and dlx4b MO2, targeting the ATG codon, 5′-ATAGACATCATTAACCGTCAAGTCC-3′ (76). BLAT searches for all MOs did not reveal any similarity to other sequences that might result in off-target effects (data not shown). We used a standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and a scrambled, 5 bp mismatch antisense MO to dlx4b E2I2 (5′-TGtTcGATAaTTACCTcTcTTTGCG-3′) as controls.
The experiments were performed under a protocol reviewed by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Francisco. Fish were maintained and bred under standard conditions at 28°C, and embryos were staged according to dpf. One to ten nanograms of antisense MO or control MO were injected into eggs at the 1–4-cell stage, and the larvae were examined daily for up to 6 dpf for external defects and for signs of MO toxicity. Photographs were taken using a light microscopy (Leica, Allendale, NJ) and SPOT imaging software (http://www.diaginc.com/software/).
RT-PCR to verify gene knockdown for D. rerio dlx4a and dlx4b was performed according to previous methods (71). RNA was obtained using Trizol and cDNA was prepared using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). We used primers dlx4aE1Fb: 5′-ACCACCATCCAGGAGCTTACCTAC-3′ with dlx4aE3Rb: 5′-CAGGTCCACTTGAGCCATGTTTC-3′ to amplify across exon 2 of D. rerio dlx4a and we used primers dlx4bE1F: 5′-ACATGCCTTACCAGCCAAC-3′and dlx4bE3R 5′-ACCTCCCAGAGCTGAGACAA-3′ to amplify across exon 2 of D. rerio dlx4b.
We performed histological staining of cartilage and bone in 6 dpf larvae according to modification of a published method (77). In brief, larvae were fixed in 4% PFA for 1 h (final concentration 2%, v:v), washed and then nutated with 0.04% Alcian Blue 8 GX (Sigma, St Louis, MO) overnight. After bleaching with 3% fresh hydrogen peroxide/0.5% potassium hydroxide, larvae were neutralized by washing in sodium borate solution for 9–12 h and then incubated with 1% trypsin (w:v) for at least 5 h. Embryos were preserved in 100% glycerol after incubation in 30% glycerol/70% of 1% potassium hydroxide with nutation for 1 day at RT and incubation in 60% glycerol/40% of 1% potassium hydroxide with nutation for 1 day at RT. Photographs were taken as described earlier.
Sense strand mRNA from both DLX4_LI_WT and DLX4_LI_Mut was transcribed using the mMessage mMachine Sp6 kit (Ambion, Austin, TX) according to the manufacturer's instructions. We injected 50–500 pg of human DLX4_LI_WT mRNA into zebrafish eggs to ensure that there was no detectable phenotype with overexpression of the gene (data not shown). A total of 50–500 pg of DLX4_LI_WT mRNA and 50–500 pg of DLX4_LI_Mut mRNA were injected together with MOs into eggs at the 1–4-cell stage, and the larvae were scored at 6 dpf. For D. rerio bmp4 mRNA injections, the full-length cDNA was obtained from RNA extracted from wild-type embryos at 2 dpf, sequence verified and then the mRNA was transcribed as described earlier.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (1R21 EY022779-01, EY019999-01 to A.S., R01-DE021420 to M.P. and O.D.K.).
Supplementary Material
Acknowledgements
The authors are very grateful to Dr John Rubenstein for providing reagents for the luciferase assay. We thank the patient and her family for participation.
Conflict of Interest statement. None declared.
References
- 1.Pavri S., Forrest C.R. (2013) Demographics of orofacial clefts in Canada from 2002 to 2008. Cleft Palate Craniofac. J., 50, 224–230. [DOI] [PubMed] [Google Scholar]
- 2.IPDTOC Working Group. (2011) Prevalence at birth of cleft lip with or without cleft palate: data from the International Perinatal Database of Typical Oral Clefts (IPDTOC). Cleft Palate Craniofac. J., 48, 66–81. [DOI] [PubMed] [Google Scholar]
- 3.Clifton-Bligh R.J., Wentworth J.M., Heinz P., Crisp M.S., John R., Lazarus J.H., Ludgate M., Chatterjee V.K. (1998) Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat. Genet., 19, 399–401. [DOI] [PubMed] [Google Scholar]
- 4.Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L., Caprau D., Christensen K., Suzuki Y., Machida J., et al. (2004) Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med., 351, 769–780. [DOI] [PubMed] [Google Scholar]
- 5.Modesto A., Moreno L.M., Krahn K., King S., Lidral A.C. (2006) MSX1 and orofacial clefting with and without tooth agenesis. J. Dent. Res., 85, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim N.Y., Kim Y.H., Park J.W., Baek S.H. (2013) Association between MSX1 SNPs and nonsyndromic cleft lip with or without cleft palate in the Korean population. J. Korean Med. Sci., 28, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Setó-Salvia N., Stanier P. (2014) Genetics of cleft lip and/or cleft palate: association with other common anomalies. Eur. J. Med. Genet., 57, 381–393. [DOI] [PubMed] [Google Scholar]
- 8.Rahimov F., Jugessur A., Murray J.C. (2012) Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac. J., 49, 73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie E.J., Marazita M.L. (2013) Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C: Semin. Med. Genet., 163C, 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong J., Cesario J., Zhao Y., Burns L., Westphal H., Rubenstein J.L. (2012) Cleft palate defect of Dlx1/2-/- mutant mice is caused by lack of vertical outgrowth in the posterior palate. Dev. Dyn., 241, 1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depew M.J., Simpson C.A., Morasso M., Rubenstein J.L. (2005) Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J. Anat., 207, 501–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougherty M., Kamel G., Grimaldi M., Gfrerer L., Shubinets V., Ethier R., Hickey G., Cornell R.A., Liao E.C. (2013) Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development, 140, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panganiban G., Rubenstein J.L. (2002) Developmental functions of the Distal-less/Dlx homeobox genes. Development, 129, 4371–4386. [DOI] [PubMed] [Google Scholar]
- 14.Talbot J.C., Johnson S.L., Kimmel C.B. (2010) hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development, 137, 2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stock D.W., Ellies D.L., Zhao Z., Ekker M., Ruddle F.H., Weiss K.M. (1996) The evolution of the vertebrate Dlx gene family. Proc. Natl Acad. Sci. U.S.A., 93, 10858–10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akimenko M.A., Ekker M., Wegner J., Lin W., Westerfield M. (1994) Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci., 14, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon K.S., Fritz A. (2002) Concerted action of two dlx paralogs in sensory placode formation. Development, 129, 3127–3136. [DOI] [PubMed] [Google Scholar]
- 18.Trinh B.Q., Barengo N., Naora H. (2011) Homeodomain protein DLX4 counteracts key transcriptional control mechanisms of the TGF-β cytostatic program and blocks the antiproliferative effect of TGF-β. Oncogene, 30, 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haga S.B., Fu S., Karp J.E., Ross D.D., Williams D.M., Hankins W.D., Behm F., Ruscetti F.W., Chang M., Smith B.D., et al. (2000) BP1, a new homeobox gene, is frequently expressed in acute leukemias. Leukemia, 14, 1867–1875. [DOI] [PubMed] [Google Scholar]
- 20.Awwad R.T., Do K., Stevenson H., Fu S.W., Lo-Coco F., Costello M., Campbell C.L., Berg P.E. (2008) Overexpression of BP1, a homeobox gene, is associated with resistance to all-trans retinoic acid in acute promyelocytic leukemia cells. Ann. Hematol., 87, 195–203. [DOI] [PubMed] [Google Scholar]
- 21.Neufing P.J., Kalionis B., Horsfall D.J., Ricciardelli C., Stahl J., Vivekanandan S., Raymond W., Tilley W.D. (2013) Expression and localization of homeodomain proteins DLX4/HB9 in normal and malignant human breast tissues. Anticancer Res., 23, 1479–1488. [PubMed] [Google Scholar]
- 22.Cavalli L.R., Man Y.G., Schwartz A.M., Rone J.D., Zhang Y., Urban C.A., Lima R.S., Haddad B.R., Berg P.E. (2008) Amplification of the BP1 homeobox gene in breast cancer. Cancer Genet. Cytogenet., 187, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kluk B.J., Fu Y., Formolo T.A., Zhang L., Hindle A.K., Man Y.G., Siegel R.S., Berg P.E., Deng C., McCaffrey T.A., et al. (2010) BP1, an isoform of DLX4 homeoprotein, negatively regulates BRCA1 in sporadic breast cancer. Int. J. Biol. Sci., 6, 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xian Y.S., Dang C.X., Yan C.X., Li H.P., Fu S.W., Wang Z.R. (2006) Clinicopathological significance of homeobox BP1 mRNA expression in lung cancer tissue. Nan Fang Yi Ke Da Xue Xue Bao, 26, 1173–1175. [PubMed] [Google Scholar]
- 25.Schwartz A.M., Man Y.G., Rezaei M.K., Simmens S.J., Berg P.E. (2009) BP1, a homeoprotein, is significantly expressed in prostate adenocarcinoma and is concordant with prostatic intraepithelial neoplasia. Mod. Pathol., 22, 1–6. [DOI] [PubMed] [Google Scholar]
- 26.Hollington P., Neufing P., Kalionis B., Waring P., Bentel J., Wattchow D., Tilley W.D. (2004) Expression and localization of homeodomain proteins DLX4, HB9 and HB24 in malignant and benign human colorectal tissues. Anticancer Res., 24, 955–962. [PubMed] [Google Scholar]
- 27.Qiu M., Bulfone A., Ghattas I., Meneses J.J., Christensen L., Sharpe P.T., Presley R., Pedersen R.A., Rubenstein J.L. (1997) Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol., 185, 165–184. [DOI] [PubMed] [Google Scholar]
- 28.Sperber S.M., Saxena V., Hatch G., Ekker M. (2008) Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev. Biol., 314, 59–70. [DOI] [PubMed] [Google Scholar]
- 29.Acampora D., Merlo G.R., Paleari L., Zerega B., Postiglione M.P., Mantero S., Bober E., Barbieri O., Simeone A., Levi G. (1999) Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development, 126, 3795–3809. [DOI] [PubMed] [Google Scholar]
- 30.Depew M.J., Liu J.K., Long J.E., Presley R., Meneses J.J., Pedersen R.A., Rubenstein J.L. (1999) Dlx5 regulates regional development of the branchial arches and sensory capsules. Development, 126, 3831–3846. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Jacobo L., Córdova-Fletes C., Ortiz-López R., Rivas F., Saucedo-Carrasco C., Rojas-Martínez A. (2013) Delineation of a de novo 7q21.3q31.1 deletion by CGH-SNP arrays in a girl with multiple congenital anomalies including severe glaucoma. Mol. Syndromol., 4, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf Z.T., Leslie E.J., Arzi B., Jayashankar K., Karmi N., Jia Z., Rowland D.J., Young A., Safra N., Sliskovic S., et al. (2014) A LINE-1 insertion in DLX6 is responsible for cleft palate and mandibular abnormalities in a canine model of Pierre Robin sequence. PLOS Genet., 10, e1004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamseldin H.E., Faden M.A., Alashram W., Alkuraya F.S. (2012) Identification of a novel DLX5 mutation in a family with autosomal recessive split hand and foot malformation. J. Med. Genet., 49, 16–20. [DOI] [PubMed] [Google Scholar]
- 34.Sowińska-Seidler A., Badura-Stronka M., Latos-Bieleńska A., Stronka M., Jamsheer A. (2014) Heterozygous DLX5 nonsense mutation associated with isolated split-hand/foot malformation with reduced penetrance and variable expressivity in two unrelated families. Birth Defects Res. A: Clin. Mol. Teratol., 100, 764–771. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Xin Q., Li L., Li J., Zhang C., Qiu R., Qian C., Zhao H., Liu Y., Shan S., et al. (2014) Exome sequencing reveals a heterozygous DLX5 mutation in a Chinese family with autosomal-dominant split-hand/foot malformation. Eur. J. Hum. Genet., 22, 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price J.A., Bowden D.W., Wright J.T., Pettenati M.J., Hart T.C. (1998) Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum. Mol. Genet., 7, 563–569. [DOI] [PubMed] [Google Scholar]
- 37.Dong J., Amor D., Aldred M.J., Gu T., Escamilla M., MacDougall M. (2005) DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. Am. J. Med. Genet., 133A, 138–141. [DOI] [PubMed] [Google Scholar]
- 38.Nieminen P., Lukinmaa P.L., Alapulli H., Methuen M., Suojärvi T. (2011) DLX3 homeodomain mutations cause tricho-dento-osseous syndrome with novel phenotypes. Cells Tissues Organs, 194, 49–59. [DOI] [PubMed] [Google Scholar]
- 39.Gorlin R.J., Zellweger H., Curtis M.W., Wiedemann H.R., Warburg M., Majewski F., Gillessen-Kaesbach G., Prahl-Andersen B., Zackai E. (1996) Blepharo-cheilo-dontic (BCD) syndrome. Am. J. Med. Genet., 65A, 109–112. [DOI] [PubMed] [Google Scholar]
- 40.Iida A., Narai S., Takagi R., Ono K., Ikeda N. (2006) Blepharo-cheilo-dontic (BCD) syndrome: case report. Cleft Palate Craniofac. J., 43, 237–243. [DOI] [PubMed] [Google Scholar]
- 41.Weaver K.N., Rutledge K.D., Grant J.H., Robin N.H. (2010) Imperforate anus is a rare associated finding in blepharocheilodontic syndrome. Am. J. Med. Genet., 152A, 438–440. [DOI] [PubMed] [Google Scholar]
- 42.Freitas E.L., Martinhago C.D., Ramos E.S., Murray J.C., Gil-da-Silva-Lopes V.L. (2007) Preliminary molecular studies on blepharocheilodontic syndrome. Am. J. Med. Genet., 143A, 2757–2759. [DOI] [PubMed] [Google Scholar]
- 43.Gil da Silva Lopes V.L., Guion-Almeida M.L., de Oliveira Rodini E.S. (2003) Blepharocheilodontic (BCD) syndrome: expanding the phenotype? Am. J. Med. Genet., 121A, 266–270. [DOI] [PubMed] [Google Scholar]
- 44.Zerucha T., Stühmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M. (2000) A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci., 20, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghanem N., Yu M., Poitras L., Rubenstein J.L., Ekker M. (2008) Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia through the activity of the I56ii enhancer. Dev. Biol., 322, 415–424. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L., Yang M., Gan L., He T., Xiao X., Stewart M.D., Liu X., Yang L., Zhang T., Zhao Y., et al. (2012) DLX4 upregulates TWIST and enhances tumor migration, invasion and metastasis. Int. J. Biol. Sci., 8, 1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juriloff D.M., Harris M.J., Dewell S.L., Brown C.J., Mager D.L., Gagnier L., Mah D.G. (2005) Investigations of the genomic region that contains the clf1 mutation, a causal gene in multifactorial cleft lip and palate in mice. Birth Defects Res. A: Clin. Mol. Teratol., 73, 103–113. [DOI] [PubMed] [Google Scholar]
- 48.Marazita M.L., Murray J.C., Lidral A.C., Arcos-Burgos M., Cooper M.E., Goldstein T., Maher B.S., Daack-Hirsch S., Schultz R., Mansilla M.A., et al. (2004) Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am. J. Hum. Genet., 75, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieira A.R., McHenry T.G., Daack-Hirsch S., Murray J.C., Marazita M.L. (2008) Candidate gene/loci studies in cleft lip/palate and dental anomalies finds novel susceptibility genes for clefts. Genet. Med., 10, 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peanchitlertkajorn S., Cooper M.E., Liu Y.E., Field L.L., Marazita M.L. (2003) Chromosome 17: gene mapping studies of cleft lip with or without cleft palate in Chinese families. Cleft Palate Craniofac. J., 40, 71–79. [DOI] [PubMed] [Google Scholar]
- 51.Chase M.B., Fu S., Haga S.B., Davenport G., Stevenson H., Do K., Morgan D., Mah A.L., Berg P.E. (2002) BP1, a homeodomain-containing Isoform of DLX4, represses the b-globin gene. Mol. Cell Biol., 22, 2505–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu S., Stevenson H., Strovel J.W., Haga S.B., Stamberg J., Do K., Berg P.E. (2001) Distinct functions of two isoforms of a homeobox gene, BP1 and DLX7, in the regulation of the beta-globin gene. Gene 278, 131–139. [DOI] [PubMed] [Google Scholar]
- 53.Stevenson H.S., Fu S.W., Pinzone J.J., Rheey J., Simmens S.J., Berg P.E. (2007) BP1 transcriptionally activates bcl-2 and inhibits TNFalpha-induced cell death in MCF7 breast cancer cells. Breast Cancer Res., 9, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y., Lu X., Yin L., Zhao F., Feng Y. (2006) Inhibition of DLX4 promotes apoptosis in choriocarcinoma cell lines. Placenta, 27, 375–383. [DOI] [PubMed] [Google Scholar]
- 55.Shimamoto T., Ohyashiki K., Takeshita K. (2000) Overexpression of the homeobox gene DLX-7 inhibits apoptosis by induced expression of intercellular adhesion molecule-1. Exp. Hematol., 28, 433–441. [DOI] [PubMed] [Google Scholar]
- 56.Gallagher E.R., Ratisoontorn C., Cunningham M.L. Saethre-Chotzen Syndrome. In Pagon R.A., Adam M.P., Bird T.D., Dolan C.R., Fong C.T., Stephens K. (eds), GeneReviews™ [Internet]. University of Washington, Seattle, Seattle: (WA: ), 1993–2013. 2003 May 16. [PubMed] [Google Scholar]
- 57.Stoler J.M., Rogers G.F., Mulliken J.B. (2009) The frequency of palatal anomalies in Saethre-Chotzen syndrome. Cleft Palate Craniofac. J., 46, 280–284. [DOI] [PubMed] [Google Scholar]
- 58.Ferrari N., Palmisano G.L., Paleari L., Basso G., Mangioni M., Fidanza V., Albini A., Croce C.M., Levi G., Brigati C. (2003) DLX genes as targets of ALL-1: DLX 2,3,4 down-regulation in t(4;11) acute lymphoblastic leukemias. J. Leukoc. Biol., 74, 302–305. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Q.P., Le T.N., Qiu X., Spencer V., de Melo J., Du G., Plews M., Fonseca M., Sun J.M., Davie J.R., et al. (2004) Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res., 32, 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki S., Marazita M.L., Cooper M.E., Miwa N., Hing A., Jugessur A., Natsume N., Shimozato K., Ohbayashi N., Suzuki Y., et al. (2009) Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am. J. Hum. Genet., 84, 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Q., Wang H., Hetmanski J.B., Zhang T., Ruczinski I., Schwender H., Liang K.Y., Fallin M.D., Redett R.J., Raymond G.V., et al. (2012) BMP4 was associated with NSCL/P in an Asian population. PLOS ONE 7, e35347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W., Sun X., Braut A., Mishina Y., Behringer R.R., Mina M., Martin J.F. (2005) Distinct functions for Bmp signaling in lip and palate fusion in mice. Development, 132, 1453–1461. [DOI] [PubMed] [Google Scholar]
- 63.Bonilla-Claudio M., Wang J., Bai Y., Klysik E., Selever J., Martin J.F. (2012) Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development, 139, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He F., Xiong W., Wang Y., Matsui M., Yu X., Chai Y., Klingensmith J., Chen Y. (2010) Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev. Biol., 347, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esterberg R., Fritz A. (2009) dlx3b/4b are required for the formation of the preplacodal region and otic placode through local modulation of BMP activity. Dev. Biol., 325, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D., Chu H., Maves L., Yan Y.L., Morcos P.A., Postlethwait J.H., Westerfield M. (2003) Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development, 130, 2213–2224. [DOI] [PubMed] [Google Scholar]
- 67.Swartz M.E., Sheehan-Rooney K., Dixon M.J., Eberhart J.K. (2011) Examination of a palatogenic gene program in zebrafish. Dev. Dyn., 240, 2204–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan Q., Chiquet B.T., Devault L., Warman M.L., Nakamura Y., Swindell E.C., Hecht J.T. (2012) Craniofacial abnormalities result from knock down of nonsyndromic clefting gene, crispld2, in zebrafish. Genesis, 50, 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCarthy N.1., Wetherill L., Lovely C.B., Swartz M.E., Foroud T.M., Eberhart J.K. (2013) Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development, 140, 3254–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slavotinek A.M., Mehrotra P., Nazarenko I., Tang P.L., Lao R., Cameron D., Li B., Chu C., Chou C., Marqueling A.L., et al. (2013) Focal facial dermal dysplasia, type IV, is caused by mutations in CYP26C1. Hum. Mol. Genet., 22, 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yahyavi M., Abouzeid H., Gawdat G., de Preux A.S., Xiao T., Bardakjian T., Schneider A., Choi A., Jorgenson E., Baier H., et al. (2013) ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum. Mol. Genet., 2, 3250–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang K., Li M., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res., 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang X., Wang K. (2012) wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet., 49, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods, 7, 575–576. [DOI] [PubMed] [Google Scholar]
- 75.Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 76.Kaji T., Artinger K.B. (2004) dlx3b and dlx4b function in the development of Rohon-Beard sensory neurons and trigeminal placode in the zebrafish neurula. Dev. Biol., 276, 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walker M.B., Kimmel C.B. (2007) A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem., 82, 23–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.