Abstract
Kabuki syndrome (KS) is a rare multiple congenital anomaly syndrome characterized by distinctive facial features, global developmental delay, intellectual disability and cardiovascular and musculoskeletal abnormalities. While mutations in KMT2D have been identified in a majority of KS patients, a few patients have mutations in KDM6A. We analyzed 40 individuals clinically diagnosed with KS for mutations in KMT2D and KDM6A. Mutations were detected in KMT2D in 12 and KDM6A in 4 cases, respectively. Observed mutations included single-nucleotide variations and indels leading to frame shifts, nonsense, missense or splice-site alterations. In two cases, we discovered overlapping chromosome X microdeletions containing KDM6A. To further elucidate the functional roles of KMT2D and KDM6A, we knocked down the expression of their orthologs in zebrafish. Following knockdown of kmt2d and the two zebrafish paralogs kdm6a and kdm6al, we analyzed morphants for developmental abnormalities in tissues that are affected in individuals with KS, including craniofacial structures, heart and brain. The kmt2d morphants exhibited severe abnormalities in all tissues examined. Although the kdm6a and kdm6al morphants had similar brain abnormalities, kdm6a morphants exhibited craniofacial phenotypes, whereas kdm6al morphants had prominent defects in heart development. Our results provide further support for the similar roles of KMT2D and KDM6A in the etiology of KS by using a vertebrate model organism to provide direct evidence of their roles in the development of organs and tissues affected in KS patients.
Introduction
Kabuki syndrome (KS, OMIM # 147920) is a rare, autosomal dominant multiple congenital anomaly (MCA) syndrome. Patients with KS have a recognizable facial gestalt consisting of long palpebral fissures with eversion of the lower lateral lids, high arched eyebrows with sparse growth along the distal third, low-set and prominent ears, a shortened nasal septum and depressed nasal tip (1,2). Additional features that are frequently associated with KS include intellectual disability (ID, 92%), short stature (83%), persistent fingertip pads (93%) and skeletal abnormalities (92%) (1). The extent of ID may vary from low-normal to severe (1). Microcephaly and structural brain abnormalities including polymicrogyria and atrophy of the cerebellum and brainstem have all been described in KS (3–6). Congenital heart defects (CHDs) may have a prevalence as high as 55% and include both septal defects (atrial or ventricular) and aortic coarctation (1,7–9).
Mutations in KMT2D (previously MLL2) have been identified in 34–76% of patients with KS (10–15). The KMT2D transcript spans 19.4 Kb and codes for a 593-kDa protein product (16). KMT2D is a member of the SET-domain-containing family of histone methyltransferases, responsible for tri-methylation of histone H3 at lysine 4 (H3K4me3), an epigenetic mark associated with transcriptionally active genes (17,18). Mutations in KMT2D could not be identified in all cases of KS, suggesting that there may be other genes underlying the etiology of disease in a subset of KS patients (10). Recently, six KMT2D-mutation-negative KS patients were reported to have deletions or nonsense single-nucleotide variations (SNVs) in KDM6A (previously UTX), a histone H3 lysine 27 (H3K27)-specific demethylase (19–21). The KDM6A transcript is 5.4 Kb and is translated into a 154-kDa protein (16). KDM6A removes the H3K27me3 mark that is associated with gene silencing (22,23). KDM6A associates with KMT2D and several other proteins including RBBP5, DPY30, WDR5, MATR3, ASH2L, NSD1, PAXIP1, NCOA6 and c16orf53, in a complex which is involved in regulating the expression of a wide range of downstream genes (24).
In this study, we report the mutation analysis of KMT2D and KDM6A in 40 individuals clinically diagnosed with KS. We identified potentially pathogenic mutations in either KMT2D or KDM6A in 16 cases. Further, we have performed functional analysis of KMT2D and KDM6A by morpholino antisense oligonucleotide-based knockdown of the expression of their zebrafish orthologs. Our results demonstrate that both genes play similar and critical roles in early vertebrate development, and their reduced expression results in craniofacial, cardiac and brain abnormalities.
Results
Analysis of KMT2D and KDM6A in KS patients
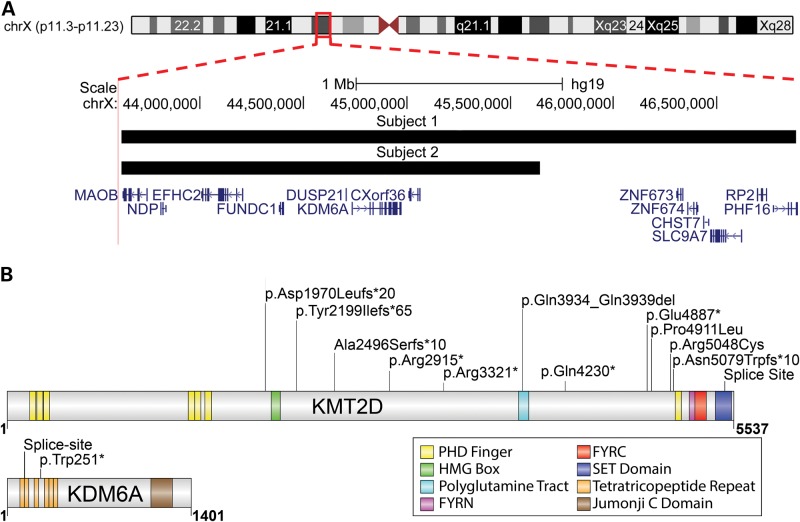
We screened 40 individuals with a clinical diagnosis of KS for mutations in KMT2D and KDM6A. Genomic DNA from these individuals was first analyzed for copy number alterations using Affymetrix 500-K SNP arrays. Two female subjects were found to have microdeletions on Chromosome X. Subject 1 had a 3.2-MB deletion at ChrXp11.3 (chrX:43620636-46881568) involving 13 genes and Subject 2 had a 2-Mb deletion (chrX:43620636-45642604) involving 7 genes nested within the larger deletion present in the other patient (Fig. 1A). The region shared between the two deletions contained KDM6A, which had recently been shown to be deleted in three KMT2D-mutation-negative KS patients (19). Furthermore, no other copy number variations (CNVs) of clinical significance were detected in any of the remaining subjects. We next performed targeted Sanger sequencing of all exons of KMT2D and KDM6A in all 40 subjects, which detected 2 additional mutations in KDM6A and 12 mutations in KMT2D (Table 1). The mutations that were detected are described further in the Supplementary Material.
Figure 1.
Spectrum of mutations in subjects diagnosed with KS. (A) An ideogram of chromosome X is shown, highlighting a region of Xp11.3–p11.23 with a red box. An expanded view of this region is shown including the genes and chromosomal coordinates. The microdeletions detected in Subjects 1 and 2 are shown as black bars. (B) The KMT2D and KDM6A proteins are shown as gray bars. The functional domains in each protein are indicated by colored bars. The location of alterations, in both proteins, detected in Subjects 3–16 are indicated by vertical black lines.
Table 1.
KMT2D and KDM6A mutations in KS subjects
| Proband | Gender | Gene affected | Chromosomal location (hg19) | Effect at cDNA level | Effect at protein level |
|---|---|---|---|---|---|
| 1 | F | KDM6A | chrX:43620636-46881568 | Microdeletion | Haploinsufficiency |
| 2 | F | KDM6A | chrX:43620636-45642604 | Microdeletion | Haploinsufficiency |
| 3 | F | KDM6A | chrX:44913077 | c.752G > A | p.Trp251*1 |
| 4 | M | KDM6A | chrX:44949177 | c.3736 + 2T > C | Splice Site |
| 5 | M | KMT2D | chr12:49425800 | c.12688C > T | p.Gln4230* |
| 6 | M | KMT2D | chr12:49431178 | c.9961C > T | p.Arg3321* |
| 7 | F | KMT2D | chr12:49432396 | c.8743C > T | p.Arg2915* |
| 8 | F | KMT2D | chr12:49421090 | c.14659G > T | p.Glu4887* |
| 9 | M | KMT2D | chr12:49436066-73 | c.5908_5915del | p.Asp1970Leufs*20 |
| 10 | M | KMT2D | chr12:49420511-14 | c.15235_15238del | p.Asn5079Trpfs*10 |
| 11 | M | KMT2D | chr12:49434958 | c.6595del | p.Tyr2199Ilefs*65 |
| 12 | F | KMT2D | chr12:49434072-73 | c.7481dup | p.Ala2496Serfs*10 |
| 13 | M | KMT2D | chr12:49415935 | c.16413-1G > C | Splice Site |
| 14 | M | KMT2D | chr12:49426675–92 | c.11796_11813del | p.Gln3934_Gln3939del |
| 15 | F | KMT2D | chr12:49420607 | c.15142C > T | p.Arg5048Cys |
| 16 | F | KMT2D | chr12:49421017 | c.14732C > T | p.Pro4911Leu |
F, female; M, male; *, premature stop codon; del, deletion; fs, frame shift; dup, duplication.
Expression patterns of zebrafish orthologs of KMT2D and KDM6A
In order to study the potential role of KMT2D and KDM6A in vertebrate development, we used the zebrafish (Danio rerio) as a model organism. There is one ortholog of KMT2D (kmt2d) in zebrafish, and two orthologs of KDM6A (kdm6a and kdm6al), which arose from an evolutionary partial genome duplication in teleosts (25). We used whole-mount in-situ hybridization to determine that kmt2d, kdm6a and kdm6al transcripts are similarly ubiquitously expressed throughout the embryo by 24-h post-fertilization (hpf), with the highest expression observed in the head (Supplementary Material, Fig. S1). By 48 hpf, transcript levels are reduced, but still present in the brain.
Knockdown of kmt2d, kdm6a and kdm6al with morpholino antisense oligonucleotides
We hypothesized that the distinctive craniofacial features observed in KS are predominantly caused by haploinsufficiency of KMT2D or KDM6A during development. To test this, we designed a combination of transcription start-site (TSS) and splice-site (SS) blocking morpholinos (MOs) to knock down the expression of kmt2d, kdm6a and kdm6al. Embryos were injected with MOs prior to the four-cell stage to knock down the expression of the target genes, after which the developmental progress of the embryos was monitored (26). Efficacy of SS blocking morpholinos was confirmed by reverse transcription PCR and sequencing of transcripts obtained from injected embryos (Supplementary Material, Fig. S2). Translation blocking and SS blocking MOs were injected independently to ensure similar phenotypes were obtained and then combined.
Analysis of craniofacial development in morphants
Alcian blue and alizarin red staining was performed on 5 day post-fertilization (dpf) morphants to visualize cartilage and bone, respectively. In kmt2d morphants (N = 41), we observed significant craniofacial defects with severe hypoplasia of the viscerocranium as compared with uninjected controls, including complete loss of branchial arches 3–7 (68%), Meckel′s cartilage (22%) and the ceratohyal (44%) from the cartilaginous structures, whereas the bony cleithrum (85%) and opercles (42%) were commonly absent as well (Fig. 2A, B and Supplementary Material, Table S1). When these structures were present, they were often incompletely formed or clefted, and in the case of the ceratohyal cartilage joint, inverted in a posterior orientation (Supplementary Material, Table S1). Development of the neurocranium appeared normal, except for a slight truncation of the ethmoid plate and trabeculae. Rescue experiments using human KMT2D transcript or the zebrafish ortholog were not possible owing to unavailability of full-length cDNA clones, likely due to the extremely large size of KMT2D transcript (>19 Kb).
Figure 2.
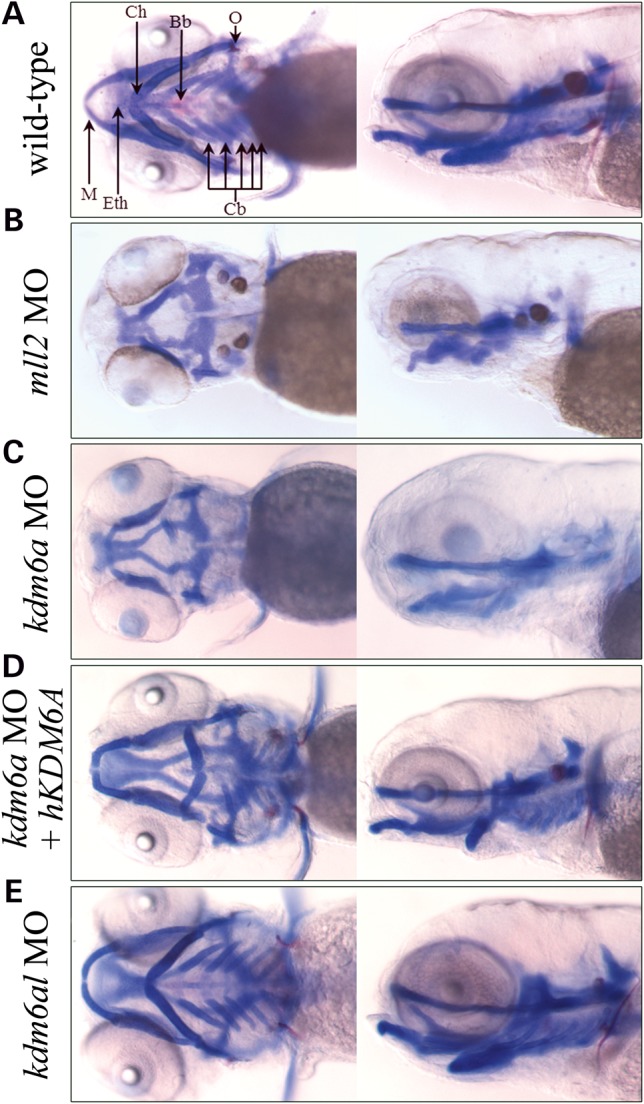
Defects in craniofacial development observed in morphant zebrafish embryos. Five day post-fertilization embryos were treated with alcian and alizarin dyes, which stain cartilage blue and bone red. Both ventral and left sagittal views focused on the visceral cranial cartilages at 10× magnification are shown. (A) Un-injected, wild-type control; (B) A typical kmt2d morphant; (C) A typical kdm6a morphant; (D) A kdm6a morphant coinjected with human KDM6A; (E) A typical kdm6al morphant. Bb, basibranchial; Cb, ceratobranchial arches 3–7; Ch, ceratohyal; O, opercle; Eth, ethmoid plate; M, Meckel's cartilage.
We observed a craniofacial phenotype comparable with kmt2d morphants in the kdm6a morphants as well (N = 131). This included hypoplastic branchial arches (92%), Meckel's cartilage (18%), ceratohyal (40%), cleithrum (72%) and opercles (87%) at similar levels to kmt2d morphants (Fig. 2C and Supplementary Material, Table S1). Coinjection of kmt2d MO with kdm6a MO was embryonic lethal prior to 4 dpf (data not shown). We also co-injected kdm6a MO with in vitro synthesized human KDM6A transcript (N = 30). This led to a partial rescue of the craniofacial phenotype, with a reduced number of embryos missing the branchial arches (13%), Meckel's cartilage (3%), ceratohyal (13%), cleithrum (7%) and opercles (30%), demonstrating that the knockdown of kdm6a is causing these specific defects and that the human homolog is sufficient to rescue the morphant phenotype (Fig. 2D and Supplementary Material, Table S1). The kdm6al morphant (N = 31) did not exhibit craniofacial defects (Fig. 2E and Supplementary Material, Table S1). Co-injection of kdm6a and kdm6al MO did not alter the severity of the phenotype compared with kdm6a morphants alone. Together, these data suggest that both kmt2d and kdm6a are necessary for patterning of the viscerocranium during embryonic development and that kdm6al does not function in this role.
Morpholino knockdown of kdm6al resulted in a posterior truncation defect (Supplementary Material, Fig. S3). However, posterior development was not affected by knockdown of kmt2d or kdm6a at concentrations that resulted in craniofacial defects (Supplementary Material, Fig. S3).
Analysis of heart development in morphants
The observation of cardiac edema in all three morphants (Supplementary Material, Fig. S3) suggested abnormalities in cardiac function. Consequently, we tested the effect of kmt2d, kdm6a and kdm6al knockdown on cardiac development in the zebrafish transgenic line Tg(szhand2:mCherry;cmlc2:gfp)co10, which expresses GFP under the cmlc2 promoter to visualize developing cardiac tissues (27). Following MO knockdown, we observed morphological defects in cardiac development at 48 hpf in kmt2d and kdm6al morphants, with a milder defect in kdm6a morphants. Morphants in all three experimental groups exhibited abnormal development of the atria and/or ventricle, as well as prominent bulging of the myocardial wall. This was most pronounced in the kmt2d morphants, whereas effects in kdm6a and kdm6al morphants were less severe.
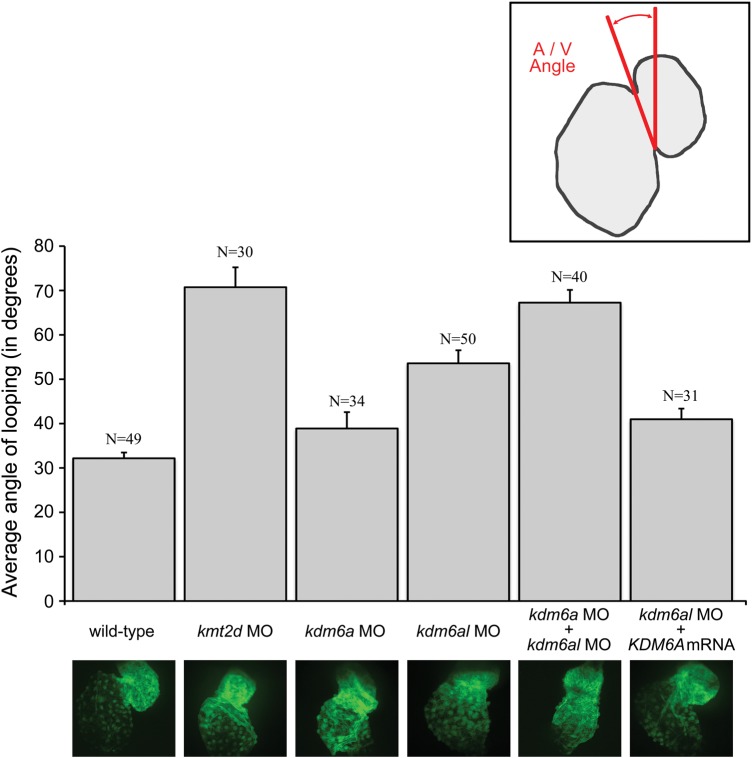
Additionally, we observed that the organization of the morphant hearts at 48 hpf was more linearly arranged along the midline when compared with wild-type embryos, whose hearts exhibited the S-shape characteristic of proper looping morphology at this time point. In order to quantify the magnitude of this abnormal cardiac morphology, we measured the extent of cardiac looping involution at 48 hpf. The ‘looping angle’ was determined by measuring the angle of the atrioventricular junction relative to the mid-sagittal plane (Fig. 3, inset). During normal heart development, the looping angle decreases over time as looping involution progresses and is used as a measure of cardiac maturation (28). At 48hpf, the wild-type embryos had an average atrioventricular angle of 32° (N = 49). We found that progression through looping morphogenesis was significantly lower in kmt2d morphants (71°, N = 30, P = 1.05 × 10−08) and kdm6al morphants (54° N = 50, P = 4.17 × 10−05) compared with wild type, but not significantly different in kdm6a morphants (39° N = 34, P = 0.096) (Fig. 3). This suggests that progression of cardiac looping involution in morphants is defective compared with wild-type embryos.
Figure 3.
Defects in heart looping observed in morphant zebrafish embryos. Images shown of hearts from 48 h post-fertilization zebrafish embryos in transgenic line Tg(szhand2:mCherry; cmlc2:gfp)co10 which expresses green fluorescent protein (GFP) under the cmlc2 promoter were analyzed. The looping angle of the heart in the embryos was measured as shown (inset). The measured looping angles are shown as a bar graph for the wild-type and morphant embryos along with a kdm6al morphant coinjected with human KDM6A mRNA. The number of embryos (n) tested in each category are shown. Error bars represent the standard error of the mean. The bottom panel shows representative GFP-expressing hearts from each of the respective categories in the graph.
To test the redundancy of kdm6a and kdm6al in cardiac development, embryos were co-injected with MOs targeting both kdm6a and kdm6al together. This resulted in further impairment of the looping progression to levels approaching kmt2d morphants (67° N = 40, P = 7.07 × 10−8) (Fig. 3). The cardiac phenotype observed in kdm6al morphants was partially rescued by coinjection with human KDM6A mRNA (41° N = 31, P = 0.001) (Fig. 3). Coinjection of human KDM6A mRNA did not significantly alter the already mild cardiac phenotype of kdm6a morphants (data not shown).These data suggest that kmt2d and kdm6al have crucial roles in cardiac development and that kdm6a and kdm6al have some functional redundancy in this role.
Analysis of brain development in morphants
In order to determine the effects of the knockdown of KS genes on brain development, we obtained transverse serial sections of the brains of wild type and morphant embryos at 48 hpf. Sections were stained with hematoxylin and eosin to observe cell morphology, and comparable sections through the hypothalamus, optic tectum and midbrain tegmentum were chosen for direct comparison (Fig. 4). When compared with wild-type embryos, the cross-sectional areas of the brains of morphants were notably reduced (Fig. 4). This phenotype was similar between kmt2d, kdm6a and kdm6al morphants and included a reduced cell layer thickness within the hypothalamus, optic tectum and midbrain tegmentum (Fig. 4B, C and E). Co-injection with hKDM6A mRNA resulted in a partially restored cell layer thickness in both kdm6a and kdm6al morphants (Fig. 4D and F). The hindbrain was not as severely affected in morphant embryos (Supplementary Material, Fig. S4). We observed a reduced cell layer thickness in sections through the hindbrain, including a decrease in the size of the medulla oblongata (Supplementary Material, Fig. S4). The decrease in hindbrain size did not affect the development of the otic vesicles in any of the morpholino injected embryos.
Figure 4.
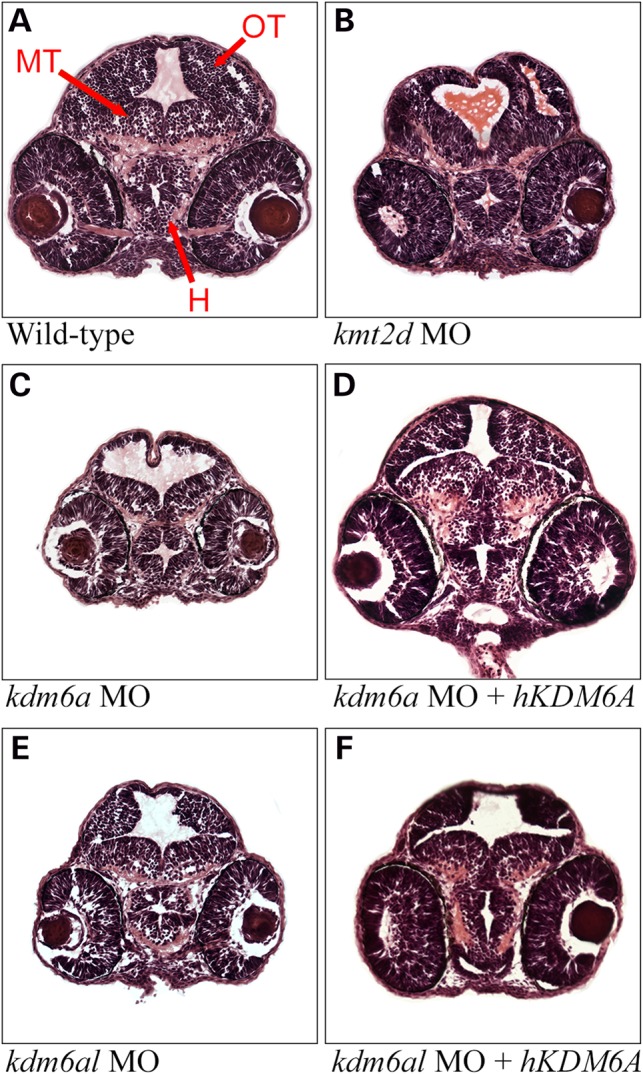
Defects in brain morphology observed in morphant zebrafish embryos. Hematoxylin and eosin (H&E) stained transverse sections of the zebrafish containing elements of both fore- and mid-brain at 48 h post-fertilization. (A) Wild-type control, (B) a typical kmt2d morphant, (C) a typical kdm6a morphant, (D) a typical kdm6a morphant coinjected with human KDM6A mRNA, (E) a typical kdm6al morphant and (F) a typical kdm6al morphant coinjected with human KDM6A mRNA. H, hypothalamus; MT, midbrain tegmentum; OT, optic tectum.
Additionally, we observed unusually elongated nuclei in cells of the midbrain along the ventricle as well as in the central region of the forebrain in all of the morphant embryos that were markedly distinct from the typically rounded nuclei present in wild-type embryos. This phenotype was not observed in the hindbrain sections (Fig. 4 and Supplementary Material, Fig. S4). Within the central nervous system, elongated nuclei are a hallmark of actively dividing neural precursor cells (NPCs) and are not observed in post-differentiated neurons (29). We postulated that a decrease in neuronal differentiation was responsible for the differences between wild-type and morphant brains. We therefore evaluated expression of the NPC marker sox2 and the post-mitotic neuronal marker huc in the brains of an additional series of 48 hpf transverse sectioned embryos (Fig. 5) (29,30). In wild-type embryos, a thin layer of sox2-positive NPCs was observed lining the ventricles of the optic tectum and midbrain tegmentum, and in the central portion of the hypothalamus. The majority of cells in these structures express huc, indicating that they have undergone differentiation into early neurons. In contrast, a much larger population of cells within the forebrain and midbrain of kmt2d, kdm6a and kdm6al morphant embryos continue to express sox2 at this time point. Only a small number of cells express huc in the morphants, indicating that NPCs are defective in their ability to differentiate. These differentiation defects were not observed in the hindbrain of morphant embryos (data not shown). These experiments highlight that kmt2d, kdm6a and kdm6al all have essential roles in neuronal differentiation during normal vertebrate brain development.
Figure 5.
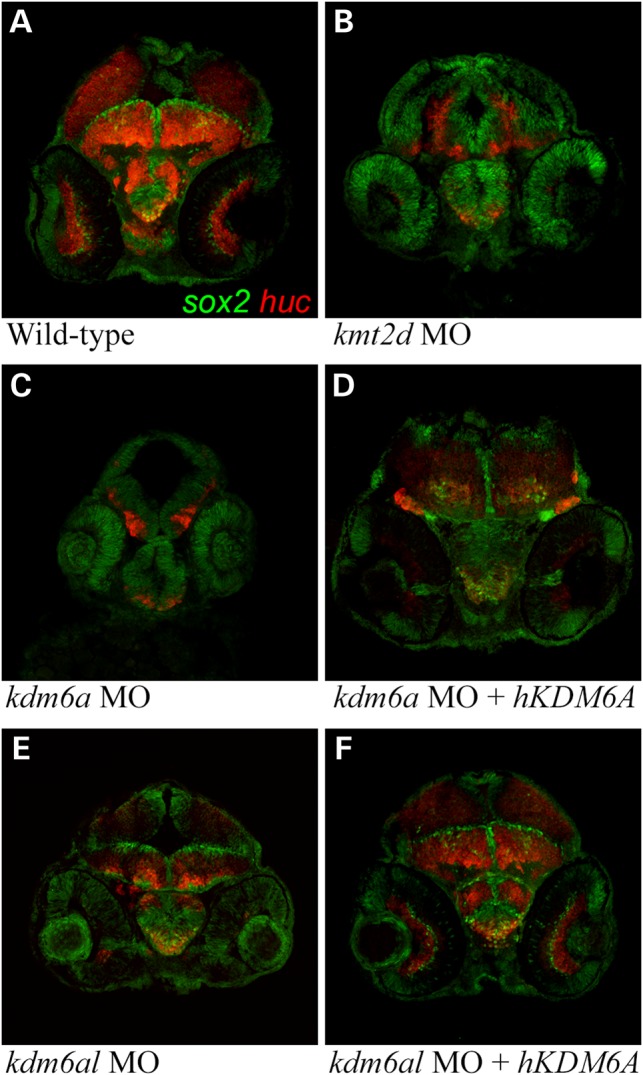
Expression of sox2 and huc in morphant zebrafish embryo brains. Transverse sections through the brain of 48 h post-fertilization zebrafish embryos immunofluorescently labeled with anti-sox2 (green) and anti-huc (red) antibodies. (A) Wild-type control, (B) a typical kmt2d morphant, (C) a typical kdm6a morphant, (D) a typical kdm6a morphant coinjected with human KDM6A mRNA, (E) a typical kdm6al morphant and (F) a typical kdm6al morphant coinjected with human KDM6A mRNA.
Discussion
We have screened individuals clinically diagnosed with KS for mutations in the candidate genes KMT2D and KDM6A. Of the 40 subjects tested, 16 had mutations in either KMT2D or KDM6A, the two genes identified thus far as causative of KS. All of the variants detected were predicted to be deleterious by computational tools that predict the effect of mutations on protein function. Previous studies have detected KMT2D mutations in 44–76% of patients, whereas KMT2D mutations were identified in 32% of subjects in this study (10–15). This difference in the detection rate of KMT2D mutations in our study is partly due to the fact that 16 of the subjects in our cohort who best fit the ‘classic’ KS presentation had been previously sequenced for KMT2D mutations and are not included in these numbers (11). The majority of these KS subjects (14/16, 88%) carried deleterious mutations in KMT2D. KS has a highly variable phenotype, and these results support the suggestion by Banka et al. (2011) that patients exhibiting a phenotype closely matching the ‘classic’ gestalt of facial and other syndromic features are more likely to have mutations in KMT2D (14).
The recent discovery of deletions encompassing KDM6A in 3/22 (14%) subjects with KS was the first evidence that mutations in a gene other than KMT2D can cause the syndrome (19). An additional screen detected point mutations in 3/32 (9%) KMT2D-mutation-negative patients, whereas another study failed to find KDM6A microdeletions/duplications in a cohort of 120 mutation-negative patients (20,31). Our detection of 4/40 subjects (10%) with variants in KDM6A is in accordance with previous findings and indicates that KDM6A mutations are not sufficient to explain all of the KMT2D-mutation-negative cases of KS. It is noteworthy that KDM6A variants were detected in both male and female KS patients in our study and by Lederer et al. (19). KDM6A has been shown to escape X inactivation independently of the pseudoautosomal regions, suggesting a dosage contribution from both alleles in females (32). Furthermore, UTY, the KDM6A paralog on the Y chromosome, compensates for the reduced KDM6A transcript abundance in males (33). This supports the view that KDM6A deletions and mutations, in both males and females, represent an X-linked dominant inheritance.
Subsequently, 24 of our suspected KS subjects did not carry detectable mutations in either KMT2D or KDM6A. There are multiple possible explanations for this result. One possibility is that some of the mutation-negative subjects have disorders other than KS, but with phenotypes similar to KS patients. Mutations in the KMT2D-related histone methyltransferase KMT2A have been identified in subjects with Wiedemann–Steiner syndrome, which can be mistaken for KS owing to some phenotypic overlap (34,35). This may explain a small number of cases but is unlikely to be the case in all of these subjects. Another possibility is that mutations in regulatory regions of KMT2D and KDM6A may account for some of the cases and were not detected by the methods used. The dominant inheritance of KS combined with the types of mutations identified to date makes this less likely.
Alternatively, there may be one or more additional KS causing genes that have not yet been discovered. This possibility is supported by the fact that in the majority of KMT2D-mutation-negative patients, a mutation in KDM6A is not identified. Many of these patients have ‘classic’ presentations and yet do not have mutations in either KMT2D or KDM6A. In addition, both KDM6A and KMT2D carry out their regulatory functions as part of a large protein complex containing several proteins, making them potential candidates for future analysis in mutation-negative cases (24). We are currently utilizing whole-exome sequencing of KMT2D and KDM6A mutation-negative patients to determine the identity of additional candidate KS genes.
The fact that modifiers of histone methylation are responsible for KS highlights an emerging trend in the genetics of MCA syndromes. There are increasing number of MCA syndromes that appear to result from aberrations in histone modifications or remodeling. These include MCA and IDD disorders associated with mutations in both histone methyltransferases [Sotos syndrome (NSD1), Wolf-Hirschhorn syndrome (NSD2), 9q sub-telomeric deletion syndrome (EHMT1)] and demethylases [X-linked intellectual disability and cleft palate (PFH8)] (36–39). Further, CHARGE syndrome (Coloboma, Heart defects, Atresia of the choanae, Retarded growth and development, Genital hypoplasia, Ear anomalies) (40–42) is primarily caused by mutations in CHD7, a DNA helicase involved in histone remodeling. CHD7 contains two chromodomains that target its binding and remodeling activity to methylated H3K4, suggesting that development may be impaired owing to disruptions in H3K4 methylation and chromatin remodeling pathways.
Although mutations in KMT2D are clearly responsible for the majority of cases of KS, its function in many developmental processes has not been characterized until now. We utilized the zebrafish as a model organism for functional studies of these candidate disease genes, which is an excellent and cost-effective alternative to other vertebrate model organisms such as the mouse. We have used morpholino-based knockdown of kmt2d, kdm6a and kdm6al in zebrafish to study their roles in the development of structures that are affected in subjects with KS. KMT2D activity is required for activation of retinoic acid responsive genes, implicating an important role in regulation of anterior-posterior patterning of the brain (43,44). Furthermore, both KMT2D and KDM6A have been shown to regulate expression of a number of Hox genes required for body patterning during development (45–47). The kdm6a and kdm6al genes have been knocked down in zebrafish previously. A reduced posterior body length and abnormal curvature of the spine has been reported in both kdm6a and kdm6al morphants, with misexpression of Hox genes and notochord degeneration (48). This may be related to spinal cord defects that are observed in some KS subjects, such as scoliosis (35%) and spina bifida occulta (19%) (49). This phenotype was observed in our kdm6al morphants as well, but was not observed in kmt2d or kdm6a morphants. A more recent study performed a detailed expression analysis of kdm6a in zebrafish embryos and demonstrated that knockdown of kdm6a led to craniofacial defects similar to those observed in our morphants (50). However, a similar role for kmt2d in craniofacial development has not been demonstrated until now.
Knockdown of kmt2d and kdm6a lead to nearly identical craniofacial defects, most notably hypoplasia of cartilaginous structures of the viscerocranium. Similarly, both kmt2d and kdm6al were required for proper cardiac looping morphogenesis, with a smaller contribution from kdm6a. The distinct roles of kdm6a in craniofacial development and kdm6al in cardiac development suggest that their functions have diverged since the ancestral gene was duplicated in the teleost lineage. Notably, both paralogs appear to be involved in neuronal development, as knockdown of either kmt2d, kdm6a or kdm6al is sufficient to induce a reduction in brain size and a reduced population of post-mitotic neurons at 48 hpf. This reduced brain volume is similar to clinical microcephaly reported in as many as 25% of KS subjects (49). Although there was an accumulation of neural precursors compared with wild-type embryos, the overall brain size of morphants was considerably reduced. The capacity of human KDM6A mRNA to partially rescue the defects caused by knocking down kdm6a or kdm6al demonstrates a high level of functional conservation between the human and zebrafish proteins. Our data provide the first direct evidence of the overlapping, functional roles of KMT2D and KDM6A in the development of tissues and organs affected in KS subjects.
Many aspects of the KS phenotype appear to be related to a global growth and developmental delay, including small stature and microcephaly. In the zebrafish, the knockdown of kmt2d, kdm6a and kdm6al lead to incomplete cardiac involution and reduced hindbrain size, both of which may result from global underdevelopment. However, the other defects we observed do not appear to be due to global underdevelopment but are likely due to perturbations in the development of specific tissues, resulting from cellular dysfunction. For example, morphants exhibit hypoplasia of structures that make up the viscerocranium, but not the neurocranium. The viscerocranium is derived from neural crest cells, whereas the neurocranium is mesodermal in origin, suggesting that neural crest derivatives may be specifically affected by haploinsufficiency of KS genes (51,52). Similarly, in the morphants, there is a defect in the differentiation NPCs only within the forebrain and midbrain, but not the hindbrain. Further, even though all morphants have markedly smaller brains, only the kdm6al morphants were reduced in overall body size (Supplementary Material, Fig. S3). Therefore, the defects in brain size in kmt2d and kdm6a morphants are likely a consequence of cellular dysfunction and not the result of underdevelopment. Furthermore, the divergence in the functional roles of kdm6a and kdm6al in the zebrafish suggests that these genes have specific regulatory roles in patterning of the craniofacial skeleton and body axis, respectively.
In summary, we have identified 16 additional mutations in the 2 known candidate genes, KMT2D and KDM6A, in patients with KS. We have further demonstrated the critical functional roles of these two candidate genes in embryonic development, using zebrafish as a model organism. We determined that knockdown of the zebrafish homologs of KMT2D and KDM6A causes defects in tissues commonly affected in patients with KS, including structures of the viscerocranial skeleton, heart and brain. We also determined that in some tissues, these genes have overlapping yet distinct functions. Thus, we provide a pathway by which candidate genes identified in subjects with genetically heterogeneous disorders such as KS can be functionally tested through biologically relevant experimentation.
Materials and Methods
Human subjects
All human subject samples used in this study were collected after obtaining informed consent. Subjects were enrolled into an IRB approved research protocol at the Children's Hospital of Philadelphia. All subjects received a clinical diagnosis of KS based on dysmorphic features characteristic of the disorder. Additional clinical features in these subjects included defects in other organ systems associated with KS, including congenital heart disease (aortic stenosis, ventricular septal defect and atrial septal defect), mild–moderate developmental delay, hypotonia, gastrointestinal abnormalities (gastroesophageal reflux), strabismus, hearing loss, genitourinary abnormalities (vesico-ureteral reflux) and microcephaly. Genomic DNA was obtained either from peripheral blood lymphocytes (PBLs) or from subject-derived cell lines.
Copy number analysis and Sanger sequencing
High resolution copy number analysis was performed using SNP 500 K arrays from Affymetrix (Santa Clara, CA, USA) as previously described (53). PCR amplification of the KMT2D and KDM6A genes was performed on an Applied Biosystems Veriti 96-well thermal cycler using previously published KMT2D primers and primers designed to tile across all exons of KDM6A (Supplementary Material, Table S2) (10). Amplicons were purified using SPRI beads and sequenced on an ABI PRISM 3730xl using BigDye Terminator v3.1 chemistry by Beckman–Coulter Genomics (Danvers, MA, USA). Alignment to the reference genes and variant calling was performed using CodonCode Aligner (CodonCode Corporation, Centerville, MA, USA), and all variants were visually inspected for quality. Additional PCR primers were designed and used to amplify regions with low-sequence, quality or low coverage calls for variant confirmation. Identified variants were submitted to ClinVar under accession numbers SCV000207387–SCV000207402.
Zebrafish transgenic strains and husbandry
Zebrafish were raised in the University of Colorado Zebrafish Core Facility as described (54). We used AB/TL wild-type zebrafish, with results reproduced in the Ekkwill line. The tp53zdf1/zdf1 transgenic line was used as a control to rule out effects of apoptosis on phenotypes (55,56). Visualization of cardiac structures was accomplished utilizing the Tg(szhand2:mCherry; cmlc2:GFP)co10 transgenic line (27). Embryos collected from pair matings were maintained in E3 medium with 0.00001% (w/v) Methylene Blue (embryo media).
Antisense morpholino and mRNA injections
All Morpholinos were acquired from Gene Tools LLC (Philomath, OR, USA). One TSS blocking antisense MO was developed per gene for kmt2d (5′ CGCAGTTTGATTTCTGCTCGTCCAT 3′), kdm6a (5′ CCGACACTCCGCACGATTTCATAGA 3′) and kdm6al (5′ CCACCGACACTCGGCACGGCTTCAT 3′). Two SS blocking MOs were developed for kmt2d (SS1: 5′ GGTATAGCAGCAATGACAAACCATT 3′, SS2: 5′ GGTCCCTAAAATGAGACAACAGCTC 3′) and one was developed for kdm6a (5′ GTGTGCTGCAAATAGAGGACAACAC 3′) and kdm6al (5′ TTTCCACAAGCATCTTTACCTTCAC 3′). Morpholinos were mixed with a 10 mg/ml Dextran dye (Invitrogen, Grand Island, NY, USA)/0.04 m KCL solution at a 1:1 ratio. 3 nL of MO mixture was injected into the yolk of embryos at the 1–4 cell stage, whereas 1-nL injections were used for mRNA rescue directly into the single-cell zygote using a PLI-100 Pico-Injector from Harvard Apparatus (Holliston, MA, USA). Poorly injected embryos were removed by sorting at 24 hpf for even distribution of fluorescent Dextran dye under a dissecting microscope. Injection of MOs into the tp53zdf1/zdf1 strain was performed to control for off-target effects (55). TSS and SS MOs were injected independently to ensure similar phenotypes were obtained before co-injection with sub-phenotypic concentrations of TSS and SS Morpholino in combination. A mixture of 3 ng of each kmt2d MO was used to knock down kmt2d. Kdm6a was knocked down with 6 ng of the TSS and 3 ng of the SS MO, whereas the kdm6al injection mixture contained 5 ng each of the TSS and SS MOs. Ten nanograms of standard control MO (5′ CCTCTTACCTCAGTTACAATTTATA 3′) were injected as a control. Co-injection with 4 ng of the Danio rerio p53 MO (5′ GCGCCATTGCTTTGCAAGAATTG 3′) was used as a control when transgenic lines were used. At no time was more than 3 nL injected into any embryo (26). Rescue experiments were performed by injecting 300 pg of human KDM6A mRNA directly into the single cell. The human KDM6A mRNA was derived from a full-length cDNA clone obtained from Open Biosystems (Lafayette, CO, USA). In vitro transcription was performed with the mMESSAGE mMACHINE SP6 Kit (Invitrogen). We verified the efficacy of SS Morpholinos by RT–PCR (Supplementary Methods and Supplementary Material, Fig. S2).
Phenotyping of morphants
Alcian blue and alizarin red staining was performed as previously described on 5 dpf wild-type and morphant embryos (57). Morphants were scored for loss of craniofacial structures or dysmorphic features, and statistical analysis of phenotypes was carried out using Welch's t-test or the Fisher's exact test in R (Supplementary Material, Table S2). Cardiac phenotypes of morphant and wild-type embryos were assessed in Tg(szhand2:mCherry;cmlc2:GFP)co10 transgenic embryos, and the looping angle was quantified as previously described (28). To determine brain phenotypes, 48 hpf embryos were embedded head-down in a heated solution consisting of 1.5% Agar and 5% Sucrose in 0.4 m phosphate buffer pH 7.4 and cut into blocks. Blocks were sunk overnight at 4°C in a 30% sucrose solution prepared with 0.1 m phosphate buffer pH 7.4. Transverse sections were obtained from frozen blocks at 7 µm of thickness for H&E staining and 15 µm of thickness for immunofluorescence on an Leica CM 1950 cryostat (Leica Biosystems, Nussloch, Germany). Hematoxylin and eosin (H&E) staining was performed as previously described. For cryosection immunofluorescence, samples were rehydrated for 30 min in PBS, blocked for 30 min in PBS supplemented with 5% BSA and 5% goat serum, incubated overnight at 4°C with primary antibodies in blocking solution, washed for 45 min with PBS, incubated for 30 min in secondary antibodies in blocking solution, washed for 45 min with PBS and coverslipped with Vectashield mounting media (Vector Laboratories). Antibodies used were rabbit anti-SOX2 (Abcam), mouse anti-HuC (Life technologies), AlexaFluor 488 goat anti-rabbit (Life technologies) and AlexaFluor 594 goat anti-mouse (Life technologies).
Imaging
Imaging experiments were performed in the University of Colorado Anschutz Medical Campus Advance Light Microscopy Core. Full-body imaging was performed on a Zeiss dissecting scope equipped with the QIClick imaging system (QImaging, Surrey, BC, USA). Imaging of the heart and brain was performed on a 3I VIVO Upright Spinning Disk confocal from Zeiss (Oberkochen, Germany). Slidebook Imaging software version 5 (Intelligent Imaging Innovations, Denver, CO, USA) was utilized to capture images and for Z-stack compression. Image J version 1.44 (NIH, Bethesda, MD, USA) and Photoshop (Adobe Systems, San Jose, CA, USA) were utilized for image manipulation including brightness, contrast and false coloring.
Web Resources
The URLs for data presented herein are as follows:
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/
Exome Variant Server: http://evs.gs.washington.edu/EVS/
GERP: http://mendel.stanford.edu/SidowLab/downloads/gerp/index.html
MaxEntScan: http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
PolyPhen-2: http://genetics.bwh.harvard.edu/pph2/
Provean: http://provean.jcvi.org/index.php
SeattleSeq: http://snp.gs.washington.edu/SeattleSeqAnnotation134/
UCSC Genome Browser: http://genome.ucsc.edu/
ClinVar: http://www.ncbi.nlm.nih.gov/clinvar/
Supplementary Material
Funding
This work was supported in part by funds from a grant to T.H.S by the National Institutes of Health (GM081519), funds and services from the Colorado Intellectual and Developmental Disabilities Research Center, and a P30 Zebrafish Core grant (NS5048154). The University of Colorado Anschutz Medical Campus Advance Light Microscopy Core is supported in part by an NIH/NCRR Colorado CTSI Grant (UL1 RR025780). J.E.M. was supported in part by a grant from the Penn Genomics Institute.
Supplementary Material
Acknowledgements
We are grateful to the patients and their families for their participation in this study. We thank Morgan Singleton and Michelle Tellez for excellent zebrafish care and for technical help. Thanks to Bruce Appel for sharing equipment and for valuable discussions, Debbie Garrity for help with cardiac phenotyping, Laura Hernandez-Lagunas and Kristi Lamonica for their technical help and for valuable discussions, and Michael Bamshad and Katy Buckingham for sharing KMT2D PCR conditions and primer sequences.
Conflict of Interest statement. None declared.
References
- 1.Niikawa N., Kuroki Y., Kajii T., Matsuura N., Ishikiriyama S., Tonoki H., Ishikawa N., Yamada Y., Fujita M., Umemoto H. (1988) Kabuki make-up (Niikawa-Kuroki) syndrome: a study of 62 patients. Am. J. Med. Genet., 31, 565–589. [DOI] [PubMed] [Google Scholar]
- 2.Adam M., Hudgins L. (2005) Kabuki syndrome: a review. Clin. Genet., 67, 209–219. [DOI] [PubMed] [Google Scholar]
- 3.Ciprero K.L., Clayton-Smith J., Donnai D., Zimmerman R.A., Zackai E.H., Ming J.E. (2005) Symptomatic Chiari I malformation in Kabuki syndrome. Am. J. Med. Genet., 132, 273–275. [DOI] [PubMed] [Google Scholar]
- 4.Yano S., Matsuishi T., Yoshino M., Kato H., Kojima K. (1997) Cerebellar and brainstem ‘atrophy’ in a patient with Kabuki make-up syndrome. Am. J. Med. Genet., 71, 486–487. [DOI] [PubMed] [Google Scholar]
- 5.Takano T., Matsuwake K., Yoshioka S., Takeuchi Y. (2010) Congenital polymicrogyria including the perisylvian region in early childhood. Congenit. Anom. (Kyoto), 50, 64–67. [DOI] [PubMed] [Google Scholar]
- 6.Di Gennaro G., Condoluci C., Casali C., Ciccarelli O., Albertini G. (1999) Epilepsy and polymicrogyria in Kabuki make-up (Niikawa-Kuroki) syndrome. Pediatr. Neurol., 21, 566–568. [DOI] [PubMed] [Google Scholar]
- 7.Kawame H., Hannibal M.C., Hudgins L., Pagon R.A. (1999) Phenotypic spectrum and management issues in Kabuki syndrome. J. Pediatr., 134, 480–485. [DOI] [PubMed] [Google Scholar]
- 8.Hughes H.E., Davies S.J. (1994) Coarctation of the aorta in Kabuki syndrome. Arch. Dis. Child., 70, 512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong L., Moneim A.A.E., Aleck K., Aughton D.J., Baumann C., Braddock S.R., Gillessen-Kaesbach G., Graham J.M., Grebe T.A., Gripp K.W., et al. (2005) Further delineation of Kabuki syndrome in 48 well-defined new individuals. Am. J. Med. Genet., 132, 265–272. [DOI] [PubMed] [Google Scholar]
- 10.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. (2010) Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet, 42, 790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannibal M.C., Buckingham K.J., Ng S.B., Ming J.E., Beck A.E., McMillin M.J., Gildersleeve H.I., Bigham A.W., Tabor H.K., Mefford H.C., et al. (2011) Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am. J. Med. Genet., 155, 1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Micale L., Augello B., Fusco C., Selicorni A., Loviglio M.N., Silengo M., Reymond A., Gumiero B., Zucchetti F., D'Addetta E.V., et al. (2011) Mutation spectrum of MLL2 in a cohort of kabuki syndrome patients. Orphanet J. Rare Dis., 6, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micale L., Augello B., Maffeo C., Selicorni A., Zucchetti F., Fusco C., De Nittis P., Pellico M.T., Mandriani B., Fischetto R., et al. (2014) Molecular analysis, pathogenic mechanisms, and read through therapy on a large cohort of kabuki syndrome patients. Hum. Mutat., 35, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banka S., Veeramachaneni R., Reardon W., Howard E., Bunstone S., Ragge N., Parker M.J., Crow Y.J., Kerr B., Kingston H., et al. (2011) How genetically heterogeneous is Kabuki syndrome?: MLL2 testing in 116 patients, review and analyses of mutation and phenotypic spectrum. Eur. J. Hum. Genet., 20, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulussen A.D.C., Stegmann A.P.A., Blok M.J., Tserpelis D., Posma-Velter C., Detisch Y., Smeets E.E.J.G.L., Wagemans A., Schrander J.J.P., van den Boogaard M.-J.H., et al. (2011) MLL2 mutation spectrum in 45 patients with Kabuki syndrome. Hum. Mutat., 32, 2018–2025. [DOI] [PubMed] [Google Scholar]
- 16.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. (2014) Ensembl 2014. Nucl. Acids Res., 42, 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith E., Lin C., Shilatifard A. (2011) The super elongation complex (SEC) and MLL in development and disease. Genes Dev., 25, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin C., Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol., 6, 838–849. [DOI] [PubMed] [Google Scholar]
- 19.Lederer D., Grisart B., Digilio M.C., Benoit V., Crespin M., Ghariani S.C., Maystadt I., Dallapiccola B., Verellen-Dumoulin C. (2012) Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with kabuki syndrome. Am. J. Hum. Genet., 90, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyake N., Mizuno S., Okamoto N., Ohashi H., Shiina M., Ogata K., Tsurusaki Y., Nakashima M., Saitsu H., Niikawa N., et al. (2013) KDM6A point mutations cause kabuki syndrome. Hum. Mutat., 34, 108–110. [DOI] [PubMed] [Google Scholar]
- 21.Hong S., Cho Y.-W., Yu L.-R., Yu H., Veenstra T.D., Ge K. (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl Acad. Sci. USA, 104, 18439–18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubner M.R., Spector D.L. (2011) Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation. Cold Spring Harb. Symp. Quant.Biol., 75, 43–49. [DOI] [PubMed] [Google Scholar]
- 23.Margueron R., Reinberg D. (2011) The polycomb complex PRC2 and its mark in life. Nature, 469, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C.M., Nakamura T., Mazo A., Eisenbach L., Canaani E. (2007) Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol., 27, 1889–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer A., Van de Peer Y. (2005) From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays, 27, 937–945. [DOI] [PubMed] [Google Scholar]
- 26.Bill B.R., Petzold A.M., Clark K.J., Schimmenti L.A., Ekker S.C. (2009) A primer for morpholino use in zebrafish. Zebrafish, 6, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iklé J.M., Artinger K.B., Clouthier D.E. (2012) Identification and characterization of the zebrafish pharyngeal arch-specific enhancer for the basic helix-loop-helix transcription factor Hand2. Dev. Biol., 368, 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chernyavskaya Y., Ebert A.M., Milligan E., Garrity D.M. (2012) Voltage-gated calcium channel CACNB2 (β2.1) protein is required in the heart for control of cell proliferation and heart tube integrity. Dev. Dyn., 241, 648–662. [DOI] [PubMed] [Google Scholar]
- 29.Gaete M., Muñoz R., Sánchez N., Tampe R., Moreno M., Contreras E.G., Lee-Liu D., Larraín J. (2012) Spinal cord regeneration in Xenopus tadpoles proceeds through activation of Sox2-positive cells. Neural Dev., 7, 13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park H.C., Kim C.H., Bae Y.K., Yeo S.Y., Kim S.H., Hong S.K., Shin J., Yoo K.W., Hibi M., Hirano T., et al. (2000) Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol., 227, 279–293. [DOI] [PubMed] [Google Scholar]
- 31.Priolo M., Micale L., Augello B., Fusco C., Zucchetti F., Prontera P., Paduano V., Biamino E., Selicorni A., Mammì C., et al. (2012) Absence of deletion and duplication of MLL2 and KDM6A genes in a large cohort of patients with Kabuki syndrome. Mol. Genet. Metab., 107, 627–629. [DOI] [PubMed] [Google Scholar]
- 32.Greenfield A., Carrel L., Pennisi D., Philippe C., Quaderi N., Siggers P., Steiner K., Tam P.P., Monaco A.P., Willard H.F., et al. (1998) The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet., 7, 737–742. [DOI] [PubMed] [Google Scholar]
- 33.Welstead G.G., Creyghton M.P., Bilodeau S., Cheng A.W., Markoulaki S., Young R.A., Jaenisch R. (2012) X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl Acad. Sci. USA, 109, 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones W.D., Dafou D., McEntagart M., Woollard W.J., Elmslie F.V., Holder-Espinasse M., Irving M., Saggar A.K., Smithson S., Trembath R.C., et al. (2012) De novo mutations in MLL cause Wiedemann-Steiner Syndrome. Am. J. Hum. Genet., 91, 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyake N., Tsurusaki Y., Koshimizu E., Okamoto N., Kosho T., Jane Brown N., Yang Tan T., Jia Jiunn Yap P., Suzumura H., Tanaka T., et al. (2015) Delineation of clinical features in Wiedemann-Steiner syndrome caused by KMT2A mutations: Clinical features in Wiedemann-Steiner syndrome. Clin. Genet. 10.1111/cge.12586. [DOI] [PubMed] [Google Scholar]
- 36.Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y., et al. (2002) Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet., 30, 365–366. [DOI] [PubMed] [Google Scholar]
- 37.Nimura K., Ura K., Shiratori H., Ikawa M., Okabe M., Schwartz R.J., Kaneda Y. (2009) A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf–Hirschhorn syndrome. Nature, 460, 287–291. [DOI] [PubMed] [Google Scholar]
- 38.Kleefstra T., Brunner H.G., Amiel J., Oudakker A.R., Nillesen W.M., Magee A., Geneviève D., Cormier-Daire V., van Esch H., Fryns J.-P., et al. (2006) Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet., 79, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laumonnier F., Holbert S., Ronce N., Faravelli F., Lenzner S., Schwartz C.E., Lespinasse J., Van Esch H., Lacombe D., Goizet C., et al. (2005) Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet., 42, 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vissers L.E.L.M., van Ravenswaaij C.M.A., Admiraal R., Hurst J.A., de Vries B.B.A., Janssen I.M., van der Vliet W.A., Huys E.H.L.P.G., de Jong P.J., Hamel B.C.J., et al. (2004) Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet., 36, 955–957. [DOI] [PubMed] [Google Scholar]
- 41.Sanlaville D., Verloes A. (2007) CHARGE syndrome: an update. Eur. J. Hum. Genet., 15, 389–399. [DOI] [PubMed] [Google Scholar]
- 42.Schnetz M.P., Bartels C.F., Shastri K., Balasubramanian D., Zentner G.E., Balaji R., Zhang X., Song L., Wang Z., LaFramboise T., et al. (2009) Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res., 19, 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo C., Chang C.C., Wortham M., Chen L.H., Kernagis D.N., Qin X., Cho Y.W., Chi J.-T., Grant G.A., McLendon R.E., et al. (2012) Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways. Proc. Natl Acad. Sci. USA, 109, 17603–17608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holder N., Hill J. (1991) Retinoic acid modifies development of the midbrain-hindbrain border and affects cranial ganglion formation in zebrafish embryos. Development, 113, 1159–1170. [DOI] [PubMed] [Google Scholar]
- 45.Agger K., Cloos P.A.C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature, 449, 731–734. [DOI] [PubMed] [Google Scholar]
- 46.Hughes C.M., Rozenblatt-Rosen O., Milne T.A., Copeland T.D., Levine S.S., Lee J.C., Hayes D.N., Shanmugam K.S., Bhattacharjee A., Biondi C.A., et al. (2004) Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell, 13, 587–597. [DOI] [PubMed] [Google Scholar]
- 47.Ansari K.I., Hussain I., Shrestha B., Kasiri S., Mandal S.S. (2011) HOXC6 is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. J. Mol. Biol., 411, 334–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature, 449, 689–694. [DOI] [PubMed] [Google Scholar]
- 49.Matsumoto N., Niikawa N. (2003) Kabuki make-up syndrome: a review. Am. J. Med. Genet., 117C, 57–65. [DOI] [PubMed] [Google Scholar]
- 50.Lindgren A.M., Hoyos T., Talkowski M.E., Hanscom C., Blumenthal I., Chiang C., Ernst C., Pereira S., Ordulu Z., Clericuzio C., et al. (2013) Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum. Genet., 132, 537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Douarin N.M., Brito J.M., Creuzet S. (2007) Role of the neural crest in face and brain development. Brain Res. Rev., 55, 237–247. [DOI] [PubMed] [Google Scholar]
- 52.Couly G.F., Coltey P.M., Le Douarin N.M. (1993) The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development, 117, 409–429. [DOI] [PubMed] [Google Scholar]
- 53.Haldeman-Englert C.R., Chapman K.A., Kruger H., Geiger E.A., McDonald-McGinn D.M., Rappaport E., Zackai E.H., Spinner B., Shaikh T.H. (2010) A de novo 8.8-Mb deletion of 21q21.1-q21.3 in an autistic male with a complex rearrangement involving chromosomes 6, 10, and 21. Am. J. Med. Genet. A, 152A, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westerfield M. (2000) The Zebrafish Book. A Guide for the Laboratory use of Zebrafish (Danio Rerio), 4th edn Univ. of Oregon Press, Eugene, Oregon. [Google Scholar]
- 55.Robu M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., Ekker S.C. (2007) p53 activation by knockdown technologies. PLoS Genet., 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berghmans S. (2005) tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl Acad. Sci. USA, 102, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker M., Kimmel C. (2007) A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem., 82, 23–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.