Abstract
The Ikaros family of DNA binding proteins are critical regulators of lymphocyte differentiation. In multipotent hematopoietic progenitors, Ikaros supports transcriptional priming of genes promoting lymphocyte differentiation. Ikaros targets the Nucleosome Remodeling Deacetylase complex (NuRD) to lymphoid lineage genes, thereby increasing chromatin accessibility and transcriptional priming. After lymphoid lineage specification, Ikaros expression is raised to levels characteristic of intermediate B cell and T cell precursors, which is necessary to support maturation and prevent leukemogenesis. Loss of Ikaros in T cell precursors allows the NuRD complex to repress lymphocyte genes and extends its targeting to genes that support growth and proliferation, causing their activation and triggering a cascade of events that leads to leukemogenesis. Loss of Ikaros in B cell precursors blocks differentiation and perpetuates stromal adhesion by enhancing integrin signaling. The combination of integrin and cytokine signaling in Ikaros-deficient pre-B cells promotes their survival and self-renewal. The stages of lymphocyte differentiation that are highly dependent on Ikaros are underscored by changes in Ikaros transcription, supported by a complex network of stage-specific regulatory networks that converge upon the Ikzf1 locus. It is increasingly apparent that understanding the regulatory networks that operate upstream and downstream of Ikaros is critical not only for our understanding of normal lymphopoiesis, but also in placing the right finger on the mechanisms that support hematopoietic malignancies in mouse and human.
Keywords: Ikaros, Mi-2β, NuRD, LMPP, T-ALL, B-ALL, lymphocyte precursors, epigenetic regulation
Introduction
Ikaros is an essential regulator of lymphocyte differentiation with two major contributions in this developmental system. The first is in early hematopoietic progenitors where it provides lymphoid lineage differentiation potential. The second is at the proliferative stages of T and B cell precursor differentiation in mediating transition to a quiescent state where recombination of the second antigen receptor chain and selection of the T and B cell repertoire takes place. A stepwise increase in Ikaros expression at key developmental stages, i.e. in lymphoid lineage restricted progenitors and lymphocyte precursors, is required for Ikaros to perform its unique roles in the hemo-lymphoid pathways. Loss of Ikaros activity results in profound differentiation defects and leukemic transformation of B and T cell precursors. Studies of Ikaros function in mouse models and human GWAS studies are providing support for the involvement of Ikaros not only in normal lymphocyte differentiation but also in the development of high risk leukemias caused by IKZF1 mutations. Here we will review past and recent studies that together provide new insight into the mechanisms by which Ikaros contributes to normal lymphocyte differentiation and its aberrant manifestations.
Structure-function of Ikaros and its family members
Ikaros [1,2] and its family members are Krupple-type zinc-(Zn-) finger proteins with two highly conserved Zn-finger domains at their N- and C-terminus (Fig. 1a). The N-terminal Zn-finger domain is comprised of four highly conserved Zn-fingers encoded by exons three though five that support sequence specific DNA-binding. The second and third Zn-fingers of the N-terminal domain are required to provide sequence specific binding to a core motif A/GGGAA [3-5] and are indispensable for lymphocyte differentiation [6-8]. A recent study has revealed that the first and fourth Ikaros zinc fingers make distinct contributions to lymphoid development or leukemogenesis, suggesting that these two fingers alter the overall sequence specificity and gene targets [9].
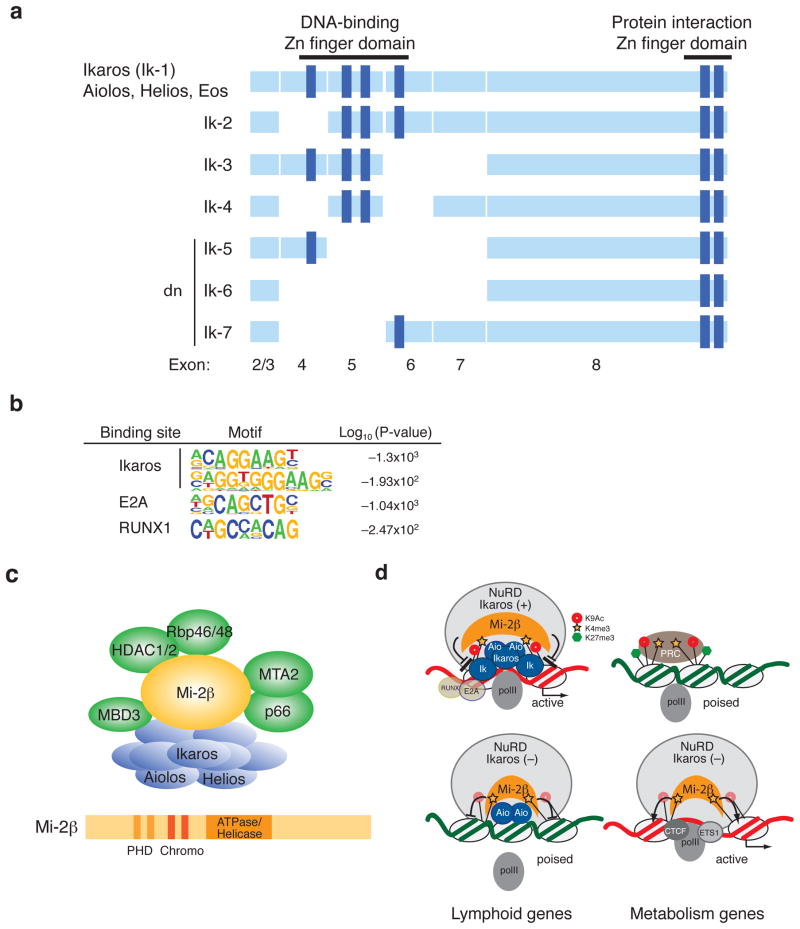
Figure 1. Schematic representation of the Ikaros isoforms, DNA binding motifs and of the Ikaros-NuRD complex and its mode of action in lymphocytes.
(a) Exon composition containing Zn finger motifs involved in DNA binding and protein dimerization is shown for Ikaros isoforms and Ikaros family members (Aiolos, Helios and Eos). Exons are shown as light blue boxes. Dark blue bars indicate zinc fingers. (b) Transcription factor binding motifs identified in the vicinity of Ikaros enrichment peaks at enhancer regions in thymocytes. Two highly enriched Ikaros binding motifs identified by de novo motif search on its chromatin binding sites. (c) Structure of the Ikaros-NuRD complex and of Mi-2β. The NuRD complex contains Class I histone deacetylases (HDAC1/2) and the ATP-dependent chromatin remodeler Mi-2β (and α). (d) A model of negative and positive regulation by the NuRD complex. Targeting of the Mi-2β–NuRD complex to permissive chromatin (H3K4me3, H3K9Ac) is restricted to lymphoid genes by the Ikaros family proteins. Our hypothesis is that NuRD’s repressive activities are poised by Ikaros extensive DNA binding at its target sites. Upon reduction in Ikaros activity either through posttranslational modification of the protein or through Ikaros inactivating mutations increases chromatin access of the Mi-2β–NuRD complex and loss of lymphoid gene expression. Upon loss of Ikaros, the NuRD complex also re-distributes to new sites associated with promoters of transcriptionally poised genes that support cell growth, proliferation and metabolism causing their activation in part by displacing the PRC2 complex. Ik, Ikaros; Aio, Aiolos; polII, RNA polymerase II; PRC, polycomb repressive complex.
Chromatin enrichment studies on the Ikaros proteins in lymphocytes provided strong selection for the core motif AGGAA that was previously predicted as an Ikaros binding site both by modeling the amino acid composition of Kruppel type Zn-fingers 2 and 3 and by in vitro DNA binding site selection studies with Ikaros proteins [3,5,10,11]. Interestingly, the Ikaros DNA binding specificity on chromatin is similar to that described for a variety of Ets factors on lymphoid specific regulatory elements [12]. Ikaros chromatin enrichment sites and motifs are found in both enhancers and promoters with relative distributions dependent on cell type. At T cell specific enhancers, the most frequent partners of Ikaros are the E-box E2A proteins and Runx1, supporting a functional interaction between these factors during T cell differentiation [10] (Fig. 1b).
The Ikaros C-terminal domain contains two Zn-fingers encoded by exon eight and is required for oligomerization with self and other family members (Aiolos, Helios and Eos) [13-16]. Oligomerization increases Ikaros DNA binding activity in vitro and is critical for Ikaros function in vivo [13,17].
A number of Ikaros isoforms can be generated by differential splicing of exons three to eight [3,4,13]. This is in part due to non-canonical splice acceptor sites at exons four, six and seven. The major Ikaros isoforms encountered from the early to the late stages of hemo-lymphopoiesis are the DNA binding isoforms Ik-1 and Ik-2 (Fig. 1a), whereas splicing variants lacking the N-terminal Zn-fingers and unable to bind DNA are normally produced at very low frequency[4,14,15]. However, genomic alterations and deletions of the Zn-finger encoding exons cause an increase in the frequency of isoforms that lack DNA binding activity (e.g. Ik-6) (reviewed by ref. [18]). These interfere with the activity of DNA-binding Ikaros isoforms and family members by competing for incorporation into the Nucleosome Remodeling Histone Deacetylase (NuRD) chromatin remodeling complex [19-24] and are frequently referred to as Ikaros dominant negative (dn) (Fig. 1a, c) [6,13].
Ikaros is an integral component of the NuRD complex in lymphocytes
Biochemical purification of Ikaros proteins has identified a stable association with the 2MDa NuRD complex in the nucleus of both lymphocytes and erythroleukemia cells [23-25] (Fig. 1c). The major components of the NuRD complex are HDAC1, HDAC2, MTA1, MTA2, MBD3, Rbp46/48, and Mi-2β. Mi-2β is a 220kDa ATP-dependent chromatin remodeler that modulates the topology of the chromatin by sliding nucleosomes relative to DNA [26,27] (Fig. 1c). Unlike Ikaros whose expression is largely limited to the hematopoietic system, components of the NuRD complex are ubiquitously expressed.
Mouse genetic studies investigating the transcriptional properties of Ikaros and Mi-2β, a unique component of the NuRD complex, have provided dual functions for the Ikaros-NuRD complex as a repressor as well as an activator of gene expression through its histone deactylase and nucleosome remodeling activities [10,25,28-31]. Although Ikaros and Mi-2β are in the same complex, antagonistic interactions have been observed such as in the regulation of Cd4 expression as well as in the cell fate decisions of the HSC [30,31]. Down-regulation of the stem cell self-renewing genes (e.g. Mpl, Tek, Ndn, Mdmdc2, Tgm2, Ebi3) and the early myeloid promoting genes (e.g. Csf1r, Egr1, Il6ra, Il6st) in Mi-2β deficient HSC correlates with the loss of self-renewal leading to transient expansion and exhaustion of the HSC as well as impaired differentiation into the myeloid lineage[31]. Given that Ikaros down-regulates the stem cell program in the lymphoid-primed multi-potent progenitor (LMPP, see the following section) and up-regulates lymphoid programs in the HSC and LMPP, Ikaros and Mi-2β may have opposing roles on the regulation of stem cell self-renewal as well as in cell fate decisions towards the lymphoid versus myeloid pathways in the earliest lympho-myeloid progenitors.
Recent studies on genome-wide chromatin mapping and gene expression profiling have provided new mechanistic insights into the function of Ikaros in the NuRD complex [10]. In wild-type DP thymocytes, Mi-2β binding sites are largely limited to where Ikaros binds to the genome. In these cells, the Ikaros-NuRD complex binds to active genes that are members of pathways that support lymphocyte differentiation. Reduction in Ikaros proteins increases chromatin access to the NuRD complex resulting in increased nucleosome remodeling and histone deacetylation with subsequent loss in lymphoid gene expression. In addition, the NuRD complex is re-distributed to transcriptionally poised genes located in permissive chromatin, causing activation of these genes, frequently by interfering with local activity of the Polycomb repressive complex. Many of these poised genes are associated with cell growth, proliferation, migration and metabolism such as those of the Notch signaling pathway [10,32](Fig. 1d). This is likely the underlying cause of the developmental block and malignant transformation of Ikaros mutant DP thymocytes and provides the mechanism by which Ikaros serves as a tumor suppressor by antagonizing NuRD complex chromatin remodeling activities both directly at lymphoid-specific genes and indirectly at growth and proliferation promoting genes.
Priming lymphoid potential in multi-potent progenitors; the first task
The first restriction that a multi-potent HSC undergoes towards becoming a lymphocyte produces an LMPP in the bone marrow (Fig. 2a). We and others have independently identified the LMPP using either the cell surface receptor Flt3 or an Ikaros-based reporter that is strongly up-regulated from the HSC to the LMPP [33-35]. The LMPP has robust lymphoid and myeloid differentiation potential and minimal potential for erythroid and megakaryocyte differentiation. Further restrictions along the lymphoid pathways give rise to the early lymphoid precursor (ELP) [36] [37], the common lymphoid progenitor (CLP) [38] and the early thymic precursor (ETP) [39]. These further differentiate into pro-B and double negative 2 (DN2) pro-T cell precursors, respectively (Fig. 2a).
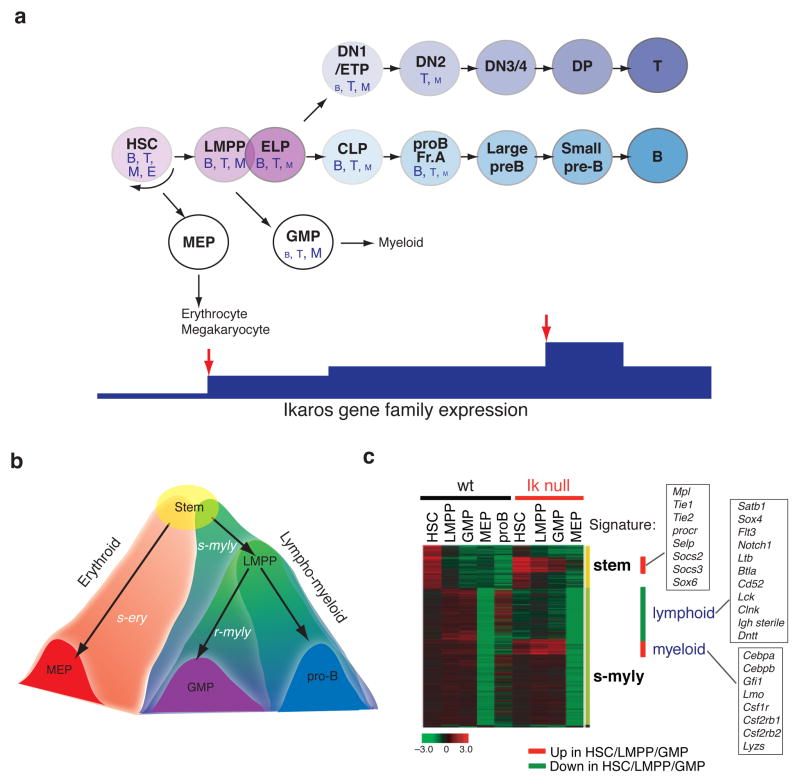
Figure 2. Cellular and transcriptional hierarchies in early hematopoiesis.
(a) The lineage potential of each progenitor in the hematopoietic hierarchy is denoted by letter size. B, B cells; T, T cells; E, erythroid; M, myeloid. Relative changes in Ikaros expression in this developmental hierarchy is shown at the bottom. Red arrows indicate critical stages where Ikaros family proteins are up-regulated. (b) The two cascades of lineage specific gene programs that originate in the multi-potent HSC and segregate within the erythroid and lympho-myeloid, pathways are shown. The early lympho-myeloid cascade is comprised of two layers of gene expression, the s-myly is primed in the HSC and further induced in the LMPP and downstream progeny whereas r-myly is primed in the LMPP. In sharp contrast, only one layer is seen in the early erythroid cascade reflecting a lack of progressive lineage restriction points. (c) Effects of Ikaros deficiency on the stem and s-myly signatures. A heatmap of gene expression of the s-myly and stem cell signatures in the HSC, LMPP, GMP, MEP and proB from WT and Ikaros null mice is shown. The signature designation is provided on the right side. The red bars indicate the up-regulated genes and the green bar indicates the down-regulated genes upon loss of Ikaros. The leg of the s-myly signature that is down-regulated is highly enriched for lymphoid promoting genes, whereas the middle section of the s-myly signature that is up-regulated is highly enriched for myeloid promoting growth factors and transcription regulators. Examples of the deregulated genes are provided in each box.
The human counterpart of the LMPP was also identified in CD34+ cord blood cells and as a source of CD34+ acute myeloid leukemia (AML), providing support for a key role in human hemo-lymphopoiesis and in leukemogenesis [40-42]. This revised road map of early hematopoiesis first described in the mouse is highly conserved in the human indicating the importance of mouse genetic models for the study of normal hematopoiesis and lymphoid disorders in the human.
Studies on mouse genetic models have revealed that while Ikaros is not required for development of the LMPP, it is critical for its ability to differentiate into lymphoid restricted progenitors such as the CLP and the ETP [34,39], the generation of which usually identifies with lymphoid lineage specification. Notably, the E2A transcription factor is also involved in lymphoid priming and shares many down-stream targets [10,43,44]. However, E2A may also acts upstream of Ikaros as it is required for the development of the LMPP from the multi-potent HSC [43]. These genetic studies are in line with biochemical studies that have identified E2A DNA binding motifs in the context of Ikaros chromatin enrichment sites in lymphoid specific enhancers[10].
While pro-B and NK are completely absent in Ikaros null mice, the ETP (DN1) in the thymus is still detected albeit at a highly reduced level [39]. Contribution of other Ikaros family members, such as Helios expressed in the HSC and through early stages of T cell but not B cell differentiation may provide some rescue in T cell differentiation. Ikaros dn mice show more profound defects with no T cell progenitors detected indicating interference with Helios function [6].
Priming of lymphoid specific gene expression in multi-potent progenitors
The establishment of lineage restriction can be observed at the molecular level by the induction and propagation of lineage-affiliated transcriptional programs that support lineage-specific differentiation. Comparative analyses of genome-wide transcriptional profiles of the HSC, LMPP, GMP and megakaryocyte-erythrocyte progenitor (MEP) [45] established nine lineage-affiliated signatures that fit into two transcriptional cascades that originate in the HSC [44]. One is propagated in the erythroid pathway, while the other, in the lympho-myeloid pathways (Fig. 2b, c). The front-runner in the lympho-myeloid cascade is a “stem-myeloid-lymphoid” (s-myly) signature, that is composed of genes primed in the HSC and further up-regulated in the LMPP. The second runner is a “restricted-myeloid-lymphoid” (r-myly) signature that comprises of genes primed in the LMPP and further induced in the downstream myeloid and lymphoid progenitors [44] (Fig. 2b, c).
Progenitor comparative transcriptional analysis has revealed that progenitor commitment into the lymphoid or myeloid lineages is gradual, whereas commitment into the erythroid lineage is rapid. Notably, the lymphoid and myeloid potential and their supporting gene expression programs are both maintained in the nominal myeloid progenitor (GMP) and B cell progenitor (CLP, pro-B) populations [44,46,47] The latent T cell activity of the GMP observed in vitro in mice and human [40,44] is consistent with the remaining expression of lymphoid genes in these cells (Fig. 2a, b). Although these progenitors may not support lymphoid differentiation in vivo, this study supports the idea of progressive lineage restriction along the lymphoid or myeloid pathways where either B cell or T cell potential is lost first followed by myeloid potential [48-50]. A similar approach to transcriptional profiling and computational analyses on human HSC and progenitors have likewise revealed transcriptional ‘landscapes’ that cross the lineage and population boundaries [42].
The multi-lineage (erythroid, myeloid and lymphoid) priming in the HSC observed by clustering of lineage specific gene expression profiles has been further confirmed at the single cell level. Erythroid and myeloid gene expression programs are primed together with lymphoid gene expression at a similar frequency, suggesting there is equal opportunity for all three fates that is further modulated by environmental inputs [44]. Lineage specific priming occurs in cells with an active stem cell gene expression program that is rapidly extinguished upon lineage restriction (Fig. 2b).
Transcriptional analyses of Ikaros null HSC and progenitors have revealed that up-regulation of the lymphoid genetic program and down-regulation of stem cell and myeloid programs controlled by Ikaros underlie this critical stage of lymphoid lineage specification at the LMPP stage [44] (Fig. 2c). A failure of such events results in a block of differentiation towards CLP and ETP and augmentation of the myeloid differentiation from the LMPP [34,44]. Examples of Ikaros target genes during this process are summarized in Fig. 2c. It will be important to determine whether maintenance of stem cell gene expression in mutant myeloid progenitors can contribute to aberrant expansion and whether this is linked to myeloid proliferative disorders in human. Thus the first major role of Ikaros in early multi-potent progenitors is to awaken their lymphoid potential by increasing local chromatin accessibility through the NuRD complex while suppressing alternate fates of myeloid differentiation and self-renewal.
Proliferative expansion of lymphocyte precursors is harnessed by Ikaros
After lymphoid lineage specification, Ikaros expression is again increased at the small pre-B cell stage in the bone marrow and in double positive (DP) T cell precursors in the thymus. These are equivalent stages of B and T cell differentiation where precursor cells come out of cycle to undergo the second antigen receptor rearrangement and selection (Fig. 2a). Notably, when Ikaros activity is reduced in human and mice, B and T cell leukemias arise from the preceding proliferative stages (Fig. 3a). Thus Ikaros plays an important role as a tumor suppressor at the lymphoid precursor stages where proliferation and Rag expression provide a high risk for leukemic transformation.
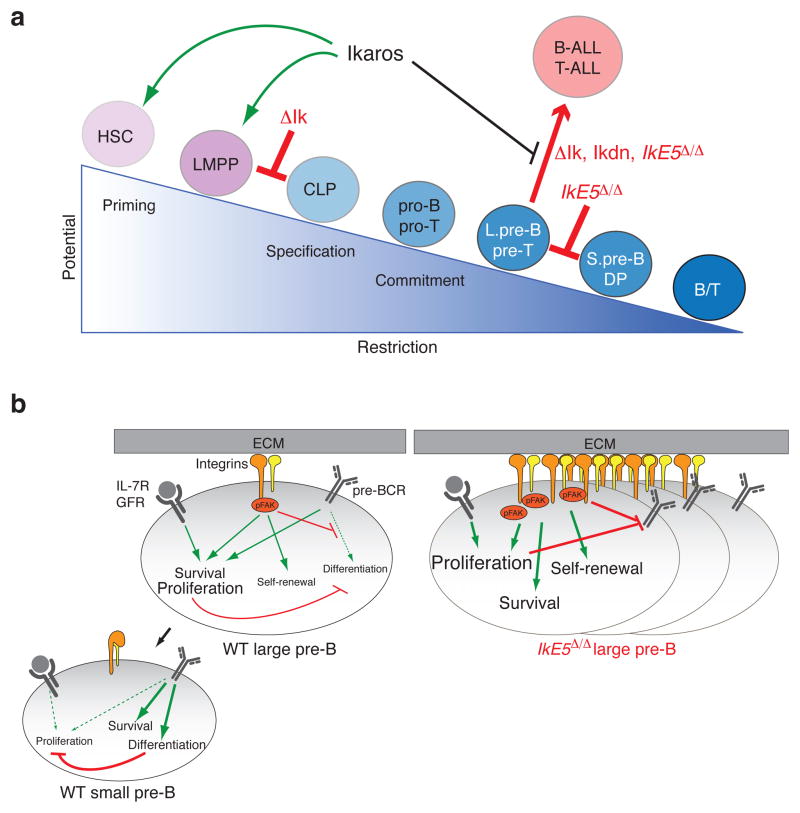
Figure 3. Effects of Ikaros mutations in early lymphoid development and homeostasis.
(a) A summary of Ikaros’ roles in early lymphocyte differentiation as revealed by various mouse genetic models. ΔIk, germline Ikaros null mutation; Ikdn. germline Ikaros dominant negative mutation; IkE5Δ/Δ, B cell specific conditional Ikaros dominant negative mutation. (b) A diagram of interactions between integrin, IL-7R and pre-BCR signaling in WT large pre-B cells and the effects of an Ikaros dominant negative mutation (IkE5Δ/Δ) on this signaling network. The strength of the effect of individual signaling pathways on cellular properties such as survival, self-renewal and proliferation is depicted by letter size. Green arrows and red bars indicate positive and negative interactions respectively. pFAK, phosphorylated FAK; IL-7R, Interleukin-7 receptor; GHR, growth factor and/or cytokine receptor.
Studies using germline knock-out mouse models have shown that homozygocity for Ikaros null and heterozygocity for Ikaros dn mutations result in a rapid development of T-ALL with a thymic origin. Leukemogenesis kinetics are faster in cells with the Ikaros dn isoforms due to interference with other Ikaros family members such as Helios and Aiolos that are also expressed in these cells [6,17]. One of the mechanisms of leukemic transformation in the thymus may relate to altered TCR signaling [51,52]. Recent studies have revealed that Ikaros suppresses Notch-dependent leukemia development by directly repressing transcription of Notch1 through upstream regulatory elements and promoters that include a cryptic intragenic promoter in DP thymocytes that supports expression of a ligand-independent Notch signaling [32,53,54]. Deletion of Ikaros in DP thymocytes unleashes de-regulated Notch signaling in these cells.
A new high-risk model of B-ALL supported by Ikaros mutations
Acute lymhoblastic leukemias (ALLs) are neoplasms of lymphoid precursors and are common among childhood malignancies [55]. With the advent of high-resolution genome-wide profiling approaches, a variety of genetic alterations, deletions and mutations have been newly identified on the ALL genomes that cooperate with previously characterized chromosomal alterations [56-58]. Notably, deletions and mutations in the genes encoding key transcription factors for early lymphoid development (e.g. PAX5, IKZF1, IKZF3, EBF, TCF3 and LEF) were identified in 40% of B-ALL cases [56]. Among these factors, deletion and mutation in the IKZF1 locus that encodes IKAROS is highly associated with BCR-ABL1-positive B-ALL that display poor prognosis [57]. These genetic alterations either give rise to functionally dominant negative IKAROS isoforms or result in haploinsufficiency. Both of these types of mutations can contribute to leukemia development.
Conditional inactivation of the Ikzf1 exon 5 (IkE5Δ/Δ), encoding N-terminal zinc fingers two and three, from the CLP or pro-B stages in mice resulted in production of dn isoforms and an arrest of B cell differentiation at the proliferative large pre-B stage [8] (Fig. 3a). Unlike WT counterparts, IkE5Δ/Δ large pre-B cells require BM stromal co-culture for survival and growth. Transcriptional profiling and functional analyses of IkE5Δ/Δ large pre-B cells revealed augmented integrin-mediated signaling that supports stable association with bone marrow stroma, survival and self-renewal [8] (Fig. 3b).
Inhibition of integrin-signaling by focal adhesion kinase (FAK) inhibitors induces the detachment of IkE5Δ/Δ large pre-B cells from BM stroma and apoptosis of mutant cells both in vitro and in vivo [8]. Immunodeficient mice transplanted with the IkE5Δ/Δ large pre-B cells develop oligoclonal pre-B-ALLs indicating selection of additional changes for malignant transformation [8]. These studies reveal a novel mechanism by which IKZF1 mutations contribute to poor prognosis pre-B-ALL that extends beyond the developmental block also generated by mutations in other lineage specific transcriptional regulators. Pre-B cell interactions with bone marrow niches are tightly controlled by Ikaros and provide a new target for therapeutic intervention in poor prognosis B-ALL associated with IKZF1 mutations.
A conditional Ikzf1 null mouse model that deletes exon eight gave a similar block in pre-B cell differentiation with accumulation of “aberrant pro-B” cells that are phenotypically similar to large pre-B cells [59]. Also in this mouse model, pre-BCR signaling is defective and cell adhesion programs of mutant pro-B cells are augmented. However, these mutant cells show a proliferation defect [59]. The differences between these two studies may be attributed to differences in the loss of Ikaros DNA binding activities; up-regulated Aiolos in the conditional Ikaros null mutant cells may compensate for the loss of Ikaros and prevent aberrant proliferative expansion that results upon loss of both Ikaros and Aiolos activities caused by the Ikaros dn isoforms generated by the IkE5Δ/Δ allele.
Transcriptional regulation of the Ikaros gene
The Ikzf1 (IKZF1) locus spans over 120kb on chromosome 11 in mice (Fig. 4a) and 7p in human in a highly conserved genomic locus. Deletions or loss of function mutations in the IKZF1 locus has been detected at a high frequency in a precursor B-ALL (20-30%), especially in over 80% of BCR-ABL+ B-ALL and nearly 70% of blast crisis of chronic myelogenous leukemia (CML) [56,57]. Large or smaller deletions at the 7p arm resulting in the loss of the entire IKZF1 allele, other null alleles or producing dn isoforms (IK-6), have been reported in B-ALL [18,60]. Mutations within the N-terminal Zn-finger domain that generate functional dn forms have been detected in human T-ALL and early T-cell precursor (ETP)-ALL [58,61] These studies show a strong correlation between Ikaros haploinsufficiency as well as expression of dn isoforms and ALL development that is strongly supported by past and recent mouse models.
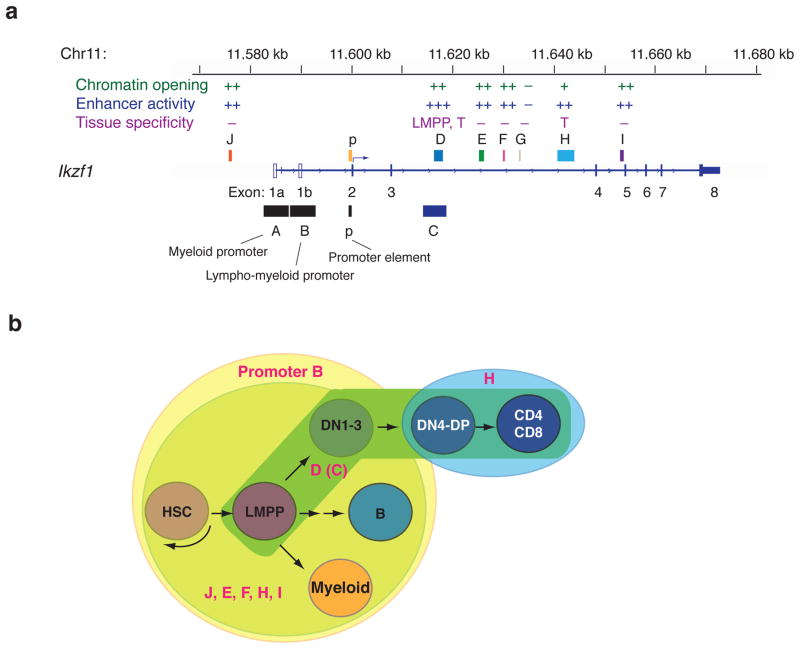
Figure 4. A summary of regulatory elements, their activities and stage specificity at the Ikaros gene (Ikzf1).
(a) A summary of the activities of the regions tested in transgenic mouse models (C, D, E, F, G, H, I, J) at the Ikzf1 locus [34,62,63]. Two promoter regions (A and B) and a promoter element (p) are shown. (b) A diagram of the activities of the lympho-myeloid promoter B and enhancers during lymphopoiesis. Promoter B (+p) provides Ikaros expression in the HSC, B cells, myeloid and DN1-3 thymocytes, however, enhancers J, E, F, H, I are required to maintain the expression in restricted chromatin environments. The enhancer C(D) is required for up-regulation of Ikaros from the HSC to the LMPP and it can further maintain the expression throughout T cell development. Although enhancer H cannot provide activity in the LMPP, it can support the expression past DN3 stages.
Thus, an understanding of the mechanisms that support Ikaros expression at stages of lymphocyte development that are dependent on Ikaros activity is critical. Our biochemical and genetic studies on Ikzf1 regulation have established the activities of hemo-lymphoid stage-specific promoters and enhancers at the Ikzf1 locus. Of the two promoters identified, one is the major promoter in lymphoid-myeloid cells (Fig. 4a, b). Of the six enhancers active in lymphoid and myeloid cells, only two enhancers are active in T cells and only one is responsible for up-regulation of Ikaros expression during the transition from the HSC to the LMPP [34,62,63](Fig. 4a, b). Several known transcription factors (TFs) important in the lymphoid system, such as E-box proteins, TCF-1, Runx1 and Ikaros itself, have been shown to bind these Ikaros locus regulatory regions in vivo [63]. Thus Ikzf1 expression is likely regulated by a complex regulatory network of cis-regulatory elements targeted by key lymphoid lineage promoting transcription factors. Further investigation on the functional contribution of these regulatory elements and TFs and the signaling events supporting their activation may provide a handle to understand and manipulate Ikaros expression during development and in hematopoietic malignancies such as multiple myeloma [64,65].
Recent studies revealed an unexpected role of IKAROS and AIOLOS in multiple myeloma (MM). Ikaros proteins in MM cells are targets of lenalidomide, a salidomide-like drug [66] that binds ubiquitin E3 ligase and alters its specificity, thereby promoting Ikaros protein degradation [64,65]. This suggests that in MM cells unlike B-ALL, the presence rather than the absence of Ikaros proteins promotes proliferation. Other mechanisms are also possible. Aiolos proteins are required for the generation of high affinity long-lived plasma cells, resident in the bone marrow and responsible for long-term humoral immunity. However, loss of Aiolos does not interfere with generation and affinity maturation of short-lived plasma cells suggesting an effect on the self-renewing potential of this long-lived effector cell population [67]. Importantly, these long-lived high affinity plasma cells are the normal counterparts from which MM is derived [68] [69].
Conclusions and future directions
The balanced production of lymphocytes is important for organismal health. Disruption of the mechanisms that support this process can cause disorders ranging from immune cell deficiencies to cancers of hematopoietic but also non-hematopoietic cell origin. Ikaros activity in the HSC–MPP identifies with lymphoid lineage potential, while Ikaros activity at later stages of lymphocyte differentiation regulates normal proliferative expansion and differentiation to a non-proliferative stage. Deletions and mutations in the IKZF1 locus in both coding and non-coding regions (i.e. highly conserved regulatory regions) may interfere with IKAROS activity and provide several independent ways to support leukemia development.
Past studies on Ikaros in lymphocyte differentiation are paving the road to new studies that seek to delineate Ikaros-based regulatory and signaling networks in both normal development and immune cell based disease. As such these studies bear important therapeutic implications for human health.
Acknowledgments
We thank to Drs. Taku Naito (Toho University Faculty of Medicine), John R. Seavitt (Baylor College of Medicine) and Bruce A. Morgan (Massachusetts General Hospital) for critical reading of the manuscript.
Footnotes
Conflict of interest: The author has no conflicts of interest to disclose.
References
- 1.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 2.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molnar A, Wu P, Largespada DA, Vortkamp A, Scherer S, Copeland NG, Jenkins NA, Bruns G, Georgopoulos K. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J Immunol. 1996;156:585–592. [PubMed] [Google Scholar]
- 5.Koipally J, Heller EJ, Seavitt JR, Georgopoulos K. Unconventional potentiation of gene expression by Ikaros. J Biol Chem. 2002;277:13007–13015. doi: 10.1074/jbc.M111371200. [DOI] [PubMed] [Google Scholar]
- 6.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 7.Papathanasiou P, Perkins AC, Cobb BS, Ferrini R, Sridharan R, Hoyne GF, Nelms KA, Smale ST, Goodnow CC. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity. 2003;19:131–144. doi: 10.1016/s1074-7613(03)00168-7. [DOI] [PubMed] [Google Scholar]
- 8.Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, Georgopoulos K. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol. 2014;15:294–304. doi: 10.1038/ni.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, Lawson GW, Bensinger SJ, Farnham PJ, Witte ON, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14:1073–1083. doi: 10.1038/ni.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2011;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. Embo J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. Embo J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, Morgan BA. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 16.Honma Y, Kiyosawa H, Mori T, Oguri A, Nikaido T, Kanazawa K, Tojo M, Takeda J, Tanno Y, Yokoya S, et al. Eos: a novel member of the Ikaros gene family expressed predominantly in the developing nervous system. FEBS Lett. 1999;447:76–80. doi: 10.1016/s0014-5793(99)00265-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 18.Kastner P, Dupuis A, Gaub MP, Herbrecht R, Lutz P, Chan S. Function of Ikaros as a tumor suppressor in B cell acute lymphoblastic leukemia. Am J Blood Res. 2013;3:1–13. [PMC free article] [PubMed] [Google Scholar]
- 19.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 20.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 21.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 22.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 27.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 28.Shimono Y, Murakami H, Kawai K, Wade PA, Shimokata K, Takahashi M. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- 29.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, Kee B, Vooijs M, Radtke F, Redondo JM, et al. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity. 2010;33:685–698. doi: 10.1016/j.immuni.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 37.Yokota T, Sudo T, Ishibashi T, Doi Y, Ichii M, Orirani K, Kanakura Y. Complementary regulation of early B-lymphoid differentiation by genetic and epigenetic mechanisms. Int J Hematol. 2013;98:382–389. doi: 10.1007/s12185-013-1424-7. [DOI] [PubMed] [Google Scholar]
- 38.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 39.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 40.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–594. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 41.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, April C, Fan JB, Dick JE. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14:756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide Lineage-Specific Transcriptional Networks Underscore Ikaros-Dependent Lymphoid Priming in Hematopoietic Stem Cells. Immunity. 2009 doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 46.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19-early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- 48.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 49.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 50.Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol Rev. 2010;238:23–36. doi: 10.1111/j.1600-065X.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 51.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 52.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190:1039–1048. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeannet R, Mastio J, Macias-Garcia A, Oravecz A, Ashworth T, Geimer Le Lay AS, Jost B, Le Gras S, Ghysdael J, Gridley T, et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood. 2010;116:5443–5454. doi: 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, Kelliher M, Kastner P, Chan S, Aster JC. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urayama KY, Chokkalingam AP, Manabe A, Mizutani S. Current evidence for an inherited genetic basis of childhood acute lymphoblastic leukemia. Int J Hematol. 2013;97:3–19. doi: 10.1007/s12185-012-1220-9. [DOI] [PubMed] [Google Scholar]
- 56.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 57.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupuis A, Gaub MP, Legrain M, Drenou B, Mauvieux L, Lutz P, Herbrecht R, Chan S, Kastner P. Biclonal and biallelic deletions occur in 20% of B-ALL cases with IKZF1 mutations. Leukemia. 2013;27:503–507. doi: 10.1038/leu.2012.204. [DOI] [PubMed] [Google Scholar]
- 61.Marcais A, Jeannet R, Hernandez L, Soulier J, Sigaux F, Chan S, Kastner P. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res. 2010;34:426–429. doi: 10.1016/j.leukres.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 62.Kaufmann C, Yoshida T, Perotti EA, Landhuis E, Wu P, Georgopoulos K. A complex network of regulatory elements in Ikaros and their activity during hemo-lymphopoiesis. Embo J. 2003;22:2211–2223. doi: 10.1093/emboj/cdg186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida T, Landhuis E, Dose M, Hazan I, Zhang J, Naito T, Jackson AF, Wu J, Perroti EA, Kaufmann C, et al. Transcriptional regulation of the Ikzf1 locus. Blood. 2013 doi: 10.1182/blood-2013-01-474916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, Wu L, Ito T, Ando H, Waldman MF, Thakurta A, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN. ) Br J Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong KK, Bradner JE, Kaelin WG., Jr The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe R, Tokuhira M, Kizaki M. Current approaches for the treatment of multiple myeloma. Int J Hematol. 2013;97:333–344. doi: 10.1007/s12185-013-1294-z. [DOI] [PubMed] [Google Scholar]
- 67.Cortes M, Georgopoulos K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J Exp Med. 2004;199:209–219. doi: 10.1084/jem.20031571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annu Rev Pathol. 2011;6:249–274. doi: 10.1146/annurev-pathol-011110-130249. [DOI] [PubMed] [Google Scholar]
- 69.Hosen N. Multiple myeloma-initiating cells. Int J Hematol. 2013;97:306–312. doi: 10.1007/s12185-013-1293-0. [DOI] [PubMed] [Google Scholar]