Abstract
Diffusion imaging and brain connectivity analyses can reveal the underlying organizational patterns of the human brain, described as complex networks of densely interlinked regions. Here, we analyzed 1.5-Tesla whole-brain diffusion-weighted images from 64 participants – 15 patients with behavioral variant frontotemporal (bvFTD) dementia, 19 with early-onset Alzheimer’s disease (EOAD), and 30 healthy elderly controls. Based on whole-brain tractography, we reconstructed structural brain connectivity networks to map connections between cortical regions. We examined how bvFTD and EOAD disrupt the weighted ‘rich club’ – a network property where high-degree network nodes are more interconnected than expected by chance. bvFTD disrupts both the nodal and global organization of the network in both low- and high-degree regions of the brain. EOAD targets the global connectivity of the brain, mainly affecting the fiber density of high-degree (highly connected) regions that form the rich club network. These rich club analyses suggest distinct patterns of disruptions among different forms of dementia.
1. Introduction
Rapid advances in neuroimaging have revolutionized the study of brain connectivity, also known as ‘connectomics’ [1], revealing organizational principles in fiber connections and how these contribute to the functional and structural integrity of the brain. Structural and functional imaging can be used to create connectivity maps of the brain. To analyze these maps, advanced mathematical methods have been employed, such as graph theory, to better understand connectivity patterns in the healthy [2, 3] and diseased brain [4].
Diffusion weighted imaging (DWI) can be used in structural brain connectivity studies to assess the global and local breakdown of network integration in degenerative disease. Recent concepts that describe network properties - such as the “rich club” effect - can provide important information on the complexity and higher-order structure of the brain network. The rich club network is composed of densely interconnected components that are more heavily interconnected among themselves than would be expected by chance. Rich club components are highly central and interconnected regions of the brain [5] that have also been identified as “brain hubs” [2]. Studying the role and function of these hubs allows us to describe the brain in terms of a hierarchical ordering, specialization, and level of resilience [3] – identifying properties of brain networks in health and disease.
In this study we analyzed the nodal and global weighted rich club network in behavioral variant frontotemporal dementia (bvFTD) and early onset Alzheimer’s disease (EOAD), as compared to the healthy brain. Prior work suggests that if in particular the rich club organization is altered, it can cause damage to the cortical synchronization of the brain [3, 6]. Here, we hypothesize that the rich club network may be disrupted in both forms of dementia, perhaps leading to disrupted communication among cognitive systems of the brain. We expected frontal cortical regions to be disrupted in bvFTD [7], while in EOAD, we hypothesized differences in the posterior cingulate and precuneus regions [8]. Overall, we aimed to detect distinct patterns of disruption in the nodal and global organization of the rich club network. We found, for the first time, severely disrupted global connectivity in bvFTD participants with lower fiber density in both low- and high-degree cortical regions. This was accompanied by altered connectivity across more than 60% of the nodal connections of the brain. On the other hand, EOAD mainly affected the global connectivity of the network, and some of the high-degree cortical regions that form the rich-club. However, unlike in bvFTD, the overall organization of the brain network in EOAD was relatively preserved.
2. Methods
2.1 Participants and diffusion-weighted brain imaging
We analyzed diffusion-weighted images (DWI) from 30 healthy controls and 34 dementia patients – 15 bvFTD subjects and 19 age-matched EOAD subjects (Table 1). All 64 subjects underwent whole-brain MRI scanning on 1.5-Tesla Siemens Avanto scanners, at the MRI Center at UCLA. Standard anatomical T1-weighted sequences were collected (256×256 matrix; voxel size=1×1×1 mm3; TI=900, TR=2000 ms; TE =2.89 ms; flip angle=40 degrees), and diffusion-weighted images (DWI) using single-shot multisection spin-echo echo-planar pulse sequence (144×144 matrix; voxel size: 2×2×3 mm3; TR=9800 ms; TE=97 ms; flip angle=90; scan time=5 min 38 s). 31 separate images were acquired for each DTI scan: 1 T2-weighted images with no diffusion sensitization (b0 image) and 30 diffusion-weighted images (b = 1000 s/mm2). Image preprocessing was performed as described in [4]. This was not included here due to space limitations.
Table 1.
Demographic information for the 30 healthy controls, 15 bvFTD and 19 EOAD patients. The mean age and sex are listed for each diagnostic group.
| CTL | bvFTD | EOAD | Total | |
|---|---|---|---|---|
| Age | 59.5 ± 9.6 SD | 61.3 ± 10.8 SD | 57.9 ± 4.3 SD | 59.5 ± 8.7 SD |
| Sex | 13M/17F | 7M/8F | 7M/12F | 27M/37F |
2.2 NxN Connectivity Matrix Computation
We performed whole-brain tractography as described in [4]. We used a method based on the Hough transform to recover fibers, using a constant solid angle orientation distribution function to model the local diffusion propagator [9].
Each subject’s dataset contained ~10,000 useable fibers (3D curves) in total. 34 cortical labels per hemisphere, as listed in the Desikan-Killiany atlas [10], were automatically extracted from all aligned T1-weighted structural MRI scans with FreeSurfer (http://surfer.nmr.mgh.harvard.edu/).
For each subject, a 68×68 connectivity matrix was created whereby each element represented the total number of detected fibers, in that subject, that passed through each pair of ROIs. The connectivity matrices were normalized by the total number of fibers extracted for each brain.
2.3 Weighted Rich Club Networks
Graph theory metrics were used to examine the topology of the connectivity matrices. We used the Brain Connectivity Toolbox measures as described previously [11].
The weighted rich club coefficient is a function of the nodal degree, k – the number of edges that connect to a node. At a particular k level the nodal degree is computed as:
| (Eq. 1) |
where ki is the degree of a node i, and aij is a connections status between nodes i and j (aij = 1 if nodes i and j are connected and aij = 0 otherwise) [4, 12].
We computed the rich club coefficient for each subject’s anatomical network at a range of k value thresholds (i.e., k=1–22). To do this, we examined subnetworks, M, in the connectivity matrix, and computed the nodal degree by counting the links that interconnected each node i in the subnetwork with k other nodes. Nodes that had a nodal degree ≤ k were removed from the network. Then, we ranked all the connections in the network as a function of weight and stored them in a vector, Wranked. Within M, we selected the degrees larger than k; the number of links between the components of the subnetwork was counted, E>k, as well as the sum of their collective weight, W>k. Then, the weighted rich club, ϕw(k), was computed as the ratio between W>k and the sum of the ranked weights from Wranked (from the whole network) given by the top strongest connections in E>k [3].
| (Eq. 2) |
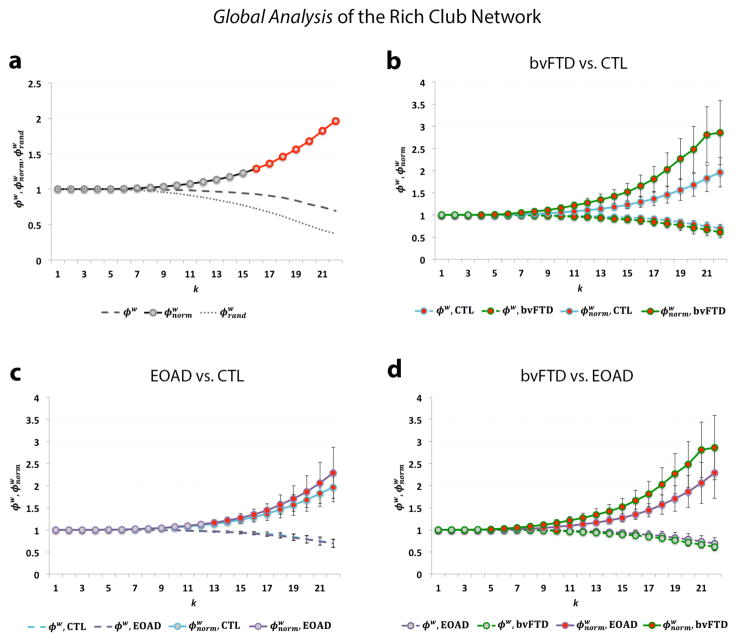
To normalize the measures, we compared the observed values to a rich club coefficient computed on an average calculated from 100 randomized networks of equal size and similar connectivity distribution. This is an important step in the analysis, as the absolute values provide limited information on network integration in the brain [2] (Fig. 2a).
Fig. 2.
(a) Shows the rich club curves, including the unnormalized (ϕw), normalized ( ) and randomized rich club ( ) as a function of nodal degree, k, for the weighted group average networks in healthy controls; a rich club is formed at k>15. (b) Shows significant differences (red) in the normalized (FDR critical p-value=0.016) and unnormalized rich club coefficient (FDR critical p-value=0.010) between bvFTD (green) and controls (blue) across most of the k-value regime. (c) Shows significant differences in the normalized (FDR critical p-value<0.016) rich club coefficient between EOAD (purple) and controls mostly in the high-level k-value regime. (d) Shows significant differences in the normalized (FDR critical p-value=0.016) and unnormalized rich club coefficient (FDR p-value=5×10−4) between bvFTD and EOAD participants. Gray dots on the curves indicate that no differences were detected. Error bars indicate standard error.
| (Eq. 3) |
Rich club subnetworks, as described throughout the study, were set at a high-degree k-levels (k>15), as previously reported [3]. As part of our nodal analysis, we investigated the rich club networks at k=16 by thresholding the connectivity matrices at nodal degree, k. We computed the nodal degree on the rich club networks at k=16 and compared it between bvFTD and controls, using a linear regression, with healthy coded at 0 and diseased coded as 1; we covaried for age, sex and brain volume. Similarly, we compared the EOAD group to controls and, separately, bvFTD to EOAD. For our global analyses, we tested how the unnormalized and separately, normalized, rich club coefficients at all 22 k-levels differed in bvFTD, relative to controls, using the same setup for the linear regression as described above. Then, we compared EOAD to controls and finally, bvFTD to EOAD. We used the false discovery rate procedure (FDR) to correct for the multiple tests performed at each cortical region and at each k level. In addition, we corrected for the 3 comparisons between diagnostic groups by adjusting the significance threshold to 0.05/3.
3. Results
3.1 Nodal Analysis
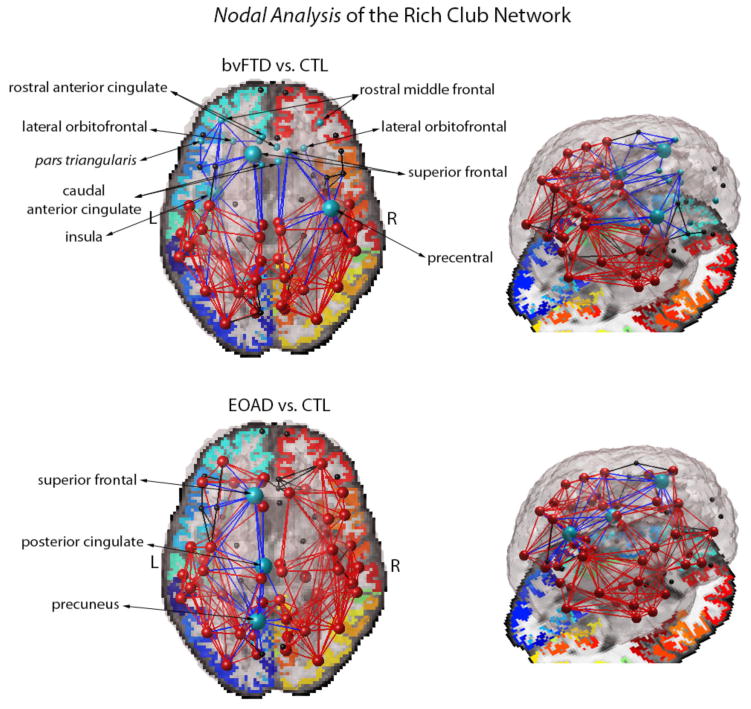
In our nodal analysis of the rich club network (at k=16), over 60% of the cortical regions in the bvFTD brain network were less interconnected (43 regions of 68), relative to healthy controls (FDR critical p-value=0.016). Among these, the most affected regions (p-value<10−10) were the left and right hemisphere caudal and rostral anterior cingulate, lateral orbitofrontal, rostral middle frontal and superior frontal regions. In addition, the left hemisphere insula and pars triangularis were also less interconnected in bvFTD, as was the precentral gyrus in the right hemisphere (p-value<10−10) (Fig. 1). Not all regions are listed due to space limitations. On the other hand, 20% of the cortical regions (14 regions of 68) in the rich club had a lower nodal degree in EOAD participants, relative to healthy controls (FDR critical p-value=0.010) with most affected regions found in the left hemisphere posterior cingulate, precuneus and superior frontal region (p-value<10−4) (Fig. 1).
Figure 1.
Average pattern of network connections and group differences in the nodal degree (at k=16) between bvFTD and controls (CTL) (top), and EOAD and CTL (bottom). Nodes and connections in red indicate the presence of rich club components (at k=16) averaged across all subjects for bvFTD and EOAD participants; components in black are in the low-degree k-level regime (k<16), not included in the rich club network. Most affected cortical regions in disease with a decrease in nodal degree are indicated in blue along with their connections to neighboring nodes; blue large spheres are part of the rich club network, but small spheres are not. The bvFTD network shows a visibly sparse organization, especially in the frontal lobe; EOAD targets the rich club components of the left hemisphere, but the overall organization of the rich club network is preserved.
When compared to each other, the brain network of bvFTD participants was significantly less interconnected than the EOAD brain (FDR critical p-value=0.013). The left and right hemisphere caudal and rostral anterior cingulate, lateral orbitofrontal, rostral middle frontal and superior frontal, pars triangularis had a lower nodal degree in bvFTD (p-value<10−5); furthermore, the left hemisphere lateral and medial orbitofrontal and insula were also more affected in bvFTD. Overall, the left hemisphere was most affected in both bvFTD and EOAD, indicating that it might be more vulnerable to network disruptions than the right.
3.2 Global analysis
The unnormalized rich club coefficient was lower in the diseased groups, relative to healthy controls (Fig. 2). The unnormalized rich club coefficient was lower in bvFTD participants, relative to controls (FDR critical p-value=0.010), but no difference was detected in EOAD. Furthermore, the bvFTD group had a lower unnormalized rich club coefficient than EOAD participants (FDR critical p-value=5×10−4).
In a separate analysis, the normalized weighted rich club coefficient was higher in the diseased groups, relative to healthy controls (Fig. 2). The normalized rich club was significantly higher in bvFTD subjects across most of the k-value regime (FDR critical p-value=0.016). Similarly, the normalized rich club coefficient was also higher in EOAD subjects, relative to healthy controls, but mostly in the high k-level network (k>13) (FDR critical p-value=0.016). When we compared the diseased global networks to each other, bvFTD participants had a significantly higher normalized rich club coefficient than EOAD across the low and high k-level regime (FDR critical p-value=0.016).
4. Discussion
Here we analyzed structural brain connectivity by examining the weighted rich club organization in cognitively healthy controls and participants with dementia (bvFTD and EOAD). The weighted rich club curves revealed distinct patterns of disruption in each disease group – bvFTD subjects showed severe nodal and global network disruptions in fiber density across the entire k-value regime, while EOAD participants showed disruptions mainly in the rich club network (i.e., high k-value regime), suggesting an overall more robust network than bvFTD.
The rich club phenomenon describes the hierarchical “assortative” organization of the human brain where high degree nodes are more likely to be interconnected among themselves than expected by chance [3, 13]. The human brain, in both health and disease, exhibits networks with high connectivity density indicating that the communication hubs of the brain operate collectively, and not as individual entities [3]. The findings from our nodal analysis are in line with prior reports that the central hubs of the human and non-human brain, often called the ‘central brain module’, include the superior frontal regions, precuneus, posterior cingulate and insula [2, 5, 14]. We found that these rich club regions are disrupted in disease: bvFTD participants showed severe alterations in the left and right hemisphere frontal regions and insula, while EOAD participants showed greatest disruptions in the left hemisphere precuneus, posterior cingulate and superior frontal region (Fig. 1).
Failure of the central brain module may severely affect global network efficiency, and the communication among network components [3, 15]. Meanwhile, EOAD targets the rich club cortical regions of the left hemisphere, which is in line with some prior work suggesting that the left hemisphere structural connectivity might be more affected in late-onset AD than the right [4].
The global analysis of the weighted right club coefficient takes into account the interconnectivity of the densest subnetworks of the brain, as a function of weight (i.e., fiber density), at each k-level, relative to the top k ranked weights across of the whole network. Hence, the unnormalized rich club coefficient decreased with increasing k-value thresholds as nodes are “peeled off” (Fig. 2). When normalized, using random networks of the same size and degree distribution, this pattern inverts. The bvFTD brain network is more vulnerable to the erosive decomposition method of the rich club showing disruptions in fiber density throughout most of the k-value regime (Fig. 2b). This is indicated by a lower unnormalized rich club coefficient and higher normalized rich club coefficient, relative to healthy elderly. In contrast, the early stages of AD showed a more robust network, unaffected in the low k-value regime, but fiber density was disrupted among the rich club nodes of the EOAD network, relative to controls (Fig. 2c). Similarly, this is indicated by a higher normalized rich club coefficient while no differences were detected in the unnormalized rich club coefficient, compared to controls. Finally, among the two disease groups, bvFTD seemed to have a more drastically altered global connectivity, with lower fiber density compared to EOAD (Fig. 2d).
There is room for possible speculation regarding the biological origins of the low and high-degree k-value regime. It may be that the low-degree k-value regime, where low degree nodes are eliminated from the rich-club subnetwork, may reflect a high level of specialization of these nodes [3]. Meanwhile, the high-degree k-value regime may indicate the absence of a densely interconnected connectome, where low-degree connections between cortical regions are missing; this may also reflect levels of differentiation between the densely connected hubs of the network [3]. In our analyses, bvFTD participants had a severely impaired fiber density across both the low- and high-degree k-value regime; the lower fiber density may have eliminated some of the potentially highly specialized low-degree nodes and reduced the connections among the high-degree nodes, leading to a more sparse rich club network. This may impair communication with neighboring nodes, and possibly function. Although EOAD affects the fiber density of major hubs in the network, the organizational integrity of the high-degree nodes in the rich club network is, however, relatively preserved.
One limitation of this study is the low spatial resolution of connectome – we represented the human brain as a network of 68 segmented cortical regions. This low network resolution may affect the topological properties of the recovered network [3]. In addition, the number of tractography fibers (~10,000) may also impact the detection of changes in complex structure and architecture of the white matter bundles. Overall, our analyses have successfully outlined distinct patterns of disruption in two different forms of dementia, providing insight into how damage to the human connectome may occur in degenerative brain disorders.
Acknowledgments
Algorithm development and image analysis for this study was funded, in part, by grants to PT from the NIBIB (R01 EB008281, R01 EB008432) and by the NIA, NIBIB, NIMH, the National Library of Medicine, and the National Center for Research Resources (AG016570, AG040060, EB01651, MH097268, LM05639, RR019771 to PT). Data collection and sharing for this project was funded by NIH Grant 5R01AG034499-05. This work was also supported in part by a Consortium grant (U54 EB020403) from the NIH Institutes contributing to the Big Data to Knowledge (BD2K) Initiative, including the NIBIB and NCI.
Contributor Information
Madelaine Daianu, Email: madelaine.daianu@ini.usc.edu, Imaging Genetics Center, Institute for Neuroimaging & Informatics, University of Southern California, Los Angeles, CA, USA.
Neda Jahanshad, Email: neda.jahanshad@ini.usc.edu, Imaging Genetics Center, Institute for Neuroimaging & Informatics, University of Southern California, Los Angeles, CA, USA.
Julio E. Villalon-Reina, Email: julio-villalon@ini.usc.edu, Imaging Genetics Center, Institute for Neuroimaging & Informatics, University of Southern California, Los Angeles, CA, USA
Mario F. Mendez, Email: mmendez@ucla.edu, Alzheimer’s Disease Research Center, Department of Neurology, UCLA School of Medicine, Los Angeles, CA, USA
George Bartzokis, Email: gbar@ucla.edu, Alzheimer’s Disease Research Center, Department of Neurology, UCLA School of Medicine, Los Angeles, CA, USA.
Elvira E. Jimenez, Email: elvira@ucla.edu, Alzheimer’s Disease Research Center, Department of Neurology, UCLA School of Medicine, Los Angeles, CA, USA
Simantini J. Karve, Email: karves@smccd.edu, Alzheimer’s Disease Research Center, Department of Neurology, UCLA School of Medicine, Los Angeles, CA, USA
Joseph Barsuglia, Email: joseph.barsuglia@va.gov, Alzheimer’s Disease Research Center, Department of Neurology, UCLA School of Medicine, Los Angeles, CA, USA.
Paul M. Thompson, Email: pthomp@usc.edu, Imaging Genetics Center, Institute for Neuroimaging & Informatics, University of Southern California, Los Angeles, CA, USA
References
- 1.Toga AW, Thompson PM. Connectomics sheds new light on Alzheimer’s disease. Biol Psychiatry. 2013;73(5):390–2. doi: 10.1016/j.biopsych.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sporns O, Honey CJ, Kötter R. Identification and classification of hubs in brain networks. PLoS ONE. 2007;2:e1049. doi: 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daianu M, Jahanshad N, Nir TM, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM the Alzheimer’s Disease Neuroimaging Initiative. Breakdown of Brain Connectivity between Normal Aging and Alzheimer’s Disease: A Structural k-core Network Analysis. Brain Connectivity. 2013;3(4):407–22. doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honey CJ, Sporns O. Dynamical consequences of lesions in cortical networks. Hum Brain Mapp. 2008;29:802–809. doi: 10.1002/hbm.20579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu PH, Lee GJ, Shapira J, Jimenez E, Mather MJ, Thompson PM, Bartzokis G, Mendez MF. Regional differences in white matter breakdown between frontotemporal dementia and early-onset Alzheimer’s disease. J Alzheimers Dis. 2014;39(2):261–269. doi: 10.3233/JAD-131481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer’s disease. J Neuroscience. 2003;23(3):994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aganj I, Lenglet C, Sapiro G, Yacoub E, Ugurbil K, Harel N. Reconstruction of the orientation distribution function in single and multiple shell Q-Ball imaging within constant solid angle. Magn Reson Med. 2010;64(2):554–466. doi: 10.1002/mrm.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–80. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–69. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 13.Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011;54:1262–1279. doi: 10.1016/j.neuroimage.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Zamora-López G, Zhou C, Kurths J. Cortical hubs form a module for multisensory integration on top of the hierarchy of cortical networks. Front Neuroinform. 2010;4:1. doi: 10.3389/neuro.11.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]