Abstract
The comprehensive study of protein structure and function, or proteomics, depends on the obtainability of full-length cDNAs in species-specific expression vectors and subsequent functional analysis of the expressed protein. Recombinational cloning is a universal cloning technique based on site-specific recombination that is independent of the insert DNA sequence of interest, which differentiates this method from the classical restriction enzyme-based cloning methods. Recombinational cloning enables rapid and efficient parallel transfer of DNA inserts into multiple expression systems. This unit summarizes strategies for generating expression-ready clones using the most popular recombinational cloning technologies, including the commercially available Gateway® (Life Technologies) and In-Fusion® (Clontech) cloning technologies.
Keywords: Recombinational, Cloning, Gateway®, In-Fusion®, High-throughput, Cre-Lox
INTRODUCTION
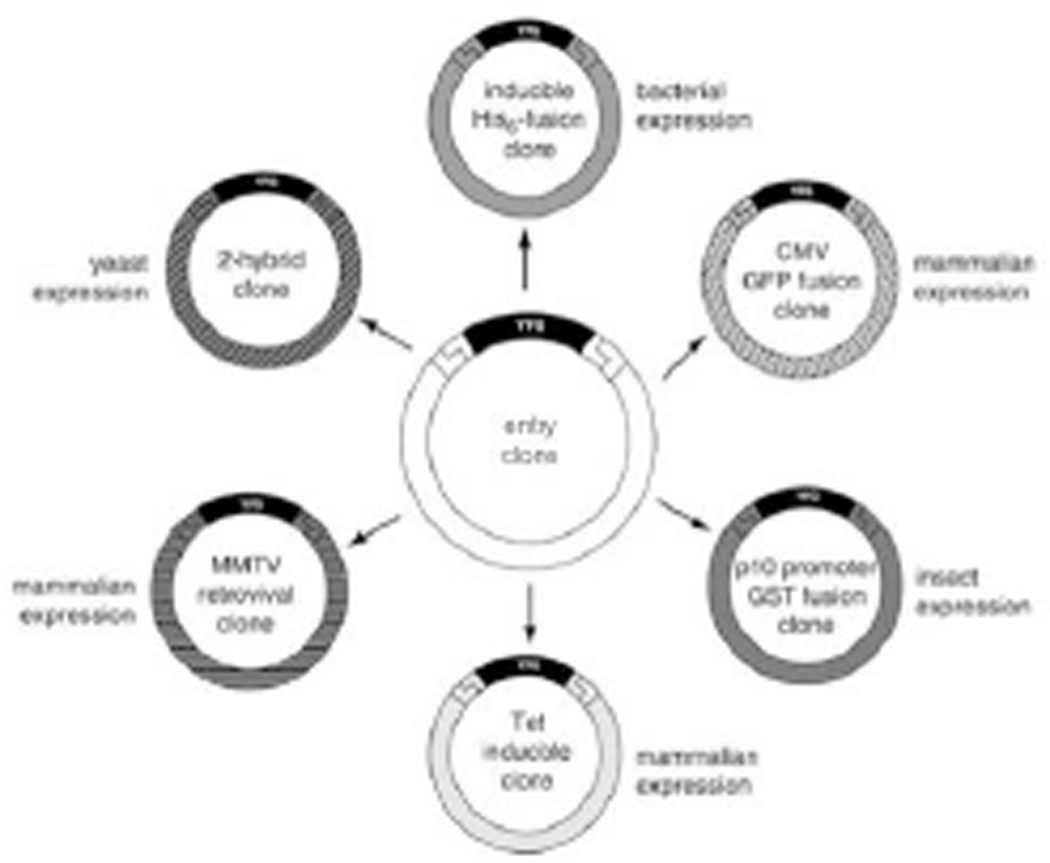
Large-scale experiments requiring protein expression from thousands of genes require an efficient method for cloning the genes into protein expression vectors. Traditional cloning methods based on restriction enzyme digestion and ligation are not practical due to the individual requirements and specifications for each construct. In contrast, recombinational cloning, or the transfer of DNA from one vector to another based on sequence homology, allows high-throughput cloning of genes into protein expression vectors under largely universal conditions without the use of restriction digests or ligation reactions. Recombinational cloning uses site-specific recombination, where both the insert and vector contain the required nucleotide sequences recognized by the cloning enzymes necessary for the recombinational event to occur. These nucleotide-specific sequences are referred to as the recombination sequences. Another advantage of recombinational cloning is the construction of an entry clone, an intermediate clone that functions as a holding or storage clone that allows the flexibility of transferring a single insert into multiple expression vectors (Fig. 3.20.1). In addition, multiple entry clones of varied sequences can be simultaneously transferred into a single expression vector to make a specific kind of library.
Figure 3.20.1.
Advantages of universal cloning technology. Recombination-based universal cloning technology enables efficient parallel transfer of your favorite gene (YFG) from an entry clone into various different expression systems for protein production and functional analysis.
The methods described in this unit are designed for use with two commercially available recombinational cloning systems, Gateway® (Life Technologies) and In-Fusion® (Clontech). Basic Protocol 1 describes the amplification of target genes and addition of the required recombination sites by PCR, Basic Protocols 2 and 3 describe the generation of entry (master) clones, and Basic Protocol 4 illustrates the construction of expression clones through the Gateway® system. Lastly, we provide a supplemental protocol describing the construction of expression clones utilizing the Cre-Lox system in Alternate Protocol 1. Although the Cre-Lox system is no longer commercially sold as a kit, the protocol is intended for those still currently using or have reagents for this system. In all cloning schemes described here, they all share the similar molecular mechanism of recombinational cloning. The differences between the systems can be observed in the sequence and length of the recombination sequences, enzymes required for the homologous recombination event to occur, and the need for an intermediate construction of an entry clone. The similarities and differences between the systems will be described in further detail below.
BASIC PROTOCOL 1
AMPLIFICATION OF TARGET GENES BY PCR
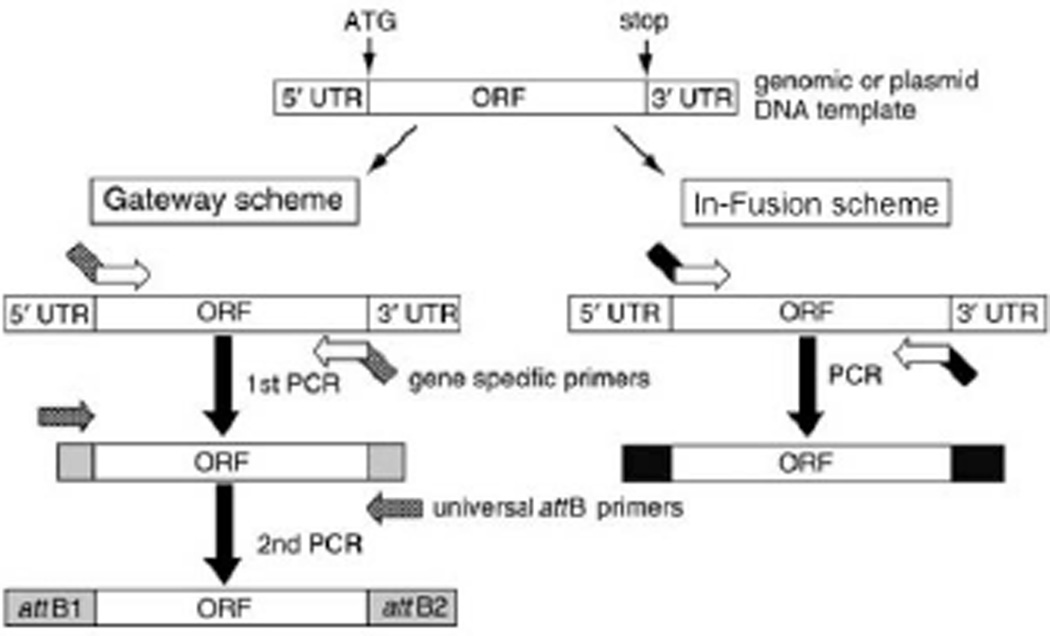
The PCR primers used to amplify a gene are designed to contain a 3’ gene-specific portion along with a common 5’ tail sequence for adding flanking recombination sites. If the required flanking recombination sequences are long (minimum of 35 base pair), as is often the case in the Gateway® scheme, some of the single stranded DNA in the primer synthesis may contain errors. In this case, the gene-specific primers can be designed to contain a 5’ tail sequence including only part of the recombination site. The rest of the recombination site can then be added efficiently by a secondary PCR employing a universal primer set. Applying a second universal set of primers also aids in lowering the cost of the PCR scheme by avoiding lengthier gene specific primers (Gateway® scheme; Fig. 3.20.2). For convenience, cost, and lower error rates, it is ideal to employ just one PCR step to build a recombination-competent fragment, which is usually possible with the In-Fusion® cloning scheme where only 15 base pair (bp) are needed for successful homologous recombination (In-Fusion® scheme; Fig. 3.20.2).
Figure 3.20.2.
Schematic view of primer design and PCR amplification. Only the open reading frame (ORF) will be amplified from ATG to the stop codon, using gene-specific primers. Note that in the Gateway® scheme the gene of interest will be amplified first with a gene-specific primer set (containing partial att sequences) and then reamplified with a universal primer set to create the entire attB sequence.
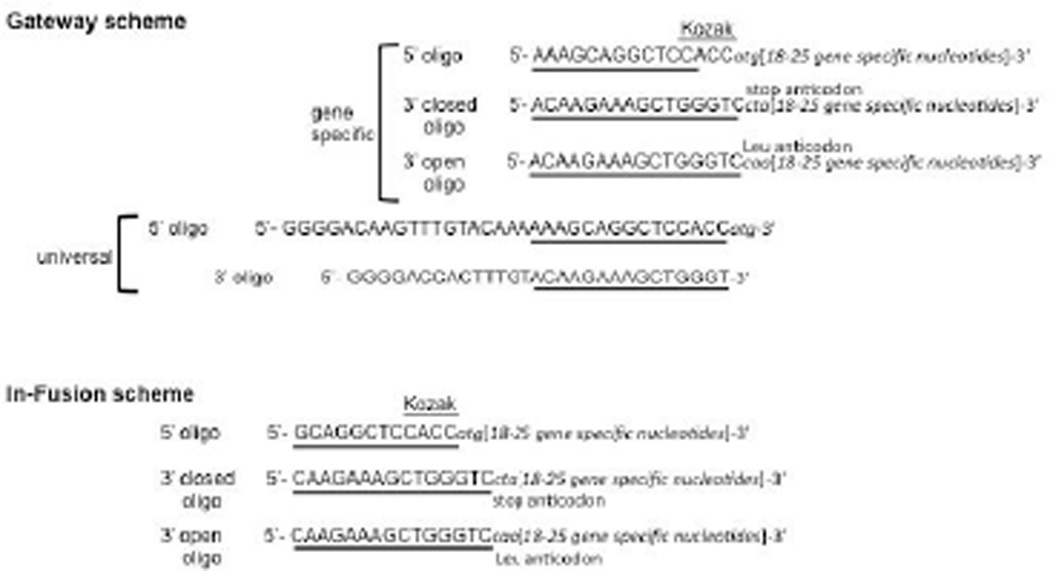
All primers must have a similar melting temperature and must be anchored in start or stop positions of the open reading frame. The gene-specific 5′ primer must start with ATG, and the 3′ primer must start at the end of the position of the common stop codon. On a practical level, such primers are designed using bases from the relevant end (5′ or 3′) of the target sequence and adding additional bases until the desired melting temperature (often 60°C) is reached. We recommend utilizing the nearest-neighbor algorithm to assistant in the primer design for optimal melting temperatures (SantaLucia, 1998). For high-throughput primer design, the nearest-neighbor thermodynamics Biopython module can be utilized (http://biopython.org/DIST/docs/api/Bio.SeqUtils.MeltingTemp-module.html). The recombination sequence can be appended once the design of the gene-specific portion is complete (Fig. 3.20.3). The primers place the coding sequences in-frame with the recombination sites. The Kozak sequence is added in front of ATG in all 5′ oligonucleotides for better protein expression. However, this sequence can be omitted when designing primers for cloning into a vector that results in a N-terminal tagged gene product. In the case of the 3′-open oligonucleotide, the common stop codon can be changed to leucine (Leu) to generate C-terminal fusion proteins.
Figure 3.20.3.
Sequence information for primers. Kozak sequences (CACC) were placed just in front of ATG sequences. In the Gateway® scheme, the partial att sequences are underlined in the case of gene-specific primers. In the In-Fusion® scheme, underline sequence highlight the 15 bases of homology to the entry vector. Shown is homology required to clone into Gateway® -compatible entry vector. To obtain C-terminal fusion proteins in the case of open primers, the common stop anticodon (CTA) is changed to the leucine anticodon (CAA). For the Gateway® scheme only, the universal primers contain complete att sequences and the matching sequences to gene-specific primers are underlined.
Materials
10 mM 4dNTPs
100% DMSO
2.0 U/µl Phusion® High-Fidelity DNA polymerase (New England BioLabs (NEB)) (Most high-fidelity polymerases should be sufficient)
5× Phusion® HF Buffer (NEB)
2× CloneAmp™ HiFi PCR Premix (Clontech)
DNA template: 10 ng/µl first-strand cDNA mix or 1–50 ng/µl plasmid DNA
10 µM gene specific 5′-oligonucleotide primer in sterile H2O (for primer design see Basic Protocol 1 introduction)
10 µM gene specific 3′-oligonucleotide primer in sterile H2O (for primer design see Basic Protocol 1 introduction)
100 µM universal 5′-oligonucleotide primer (Gateway® only)
100 µM universal 3′-oligonucleotide primer (Gateway® only)
PCR tubes (thin-walled) or 96-well plates
Multichannel pipettor (optional)
PCR Clean-Up kit (e.g., Qiagen or Macherey Nagel for high-throughput)
Centrifuge
Thermal cycler
Additional reagents and equipment for gel electrophoresis (see unit 2.5A) and DNA preparation (see unit 1.6 or commercial DNA prep kit).
Perform first-round Gateway® PCR
-
1Prepare the master mix for the first-round 50 µl PCR (multiply the volume for each component by the number of reactions plus two or three to allow for losses during transfer) as follows:
- 10 µl 5× Phusion® HF Reaction Buffer (1× final)
- 1 µl 10 mM 4dNTPs (0.2 mM final)
- 1.5 µl 100% DMSO (3% final)
- 0.5 µl 2.0 U/µl Phusion® High-Fidelity DNA polymerase (0.02 U final)
- 31 µl PCR-quality H2O.
-
2Dispense 44 µl master mix into PCR tubes for individual reactions or into each well of a 96-well plate for a large-scale format.For 96-well plate format a multichannel pipettor should be used.
-
3Add 2 µl 10 µM gene-specific 5′-oligonucleotide primer and 2 µl 10 µM gene-specific 3′-oligonucleotide primer to each reaction (final concentration 0.4 µM each primer).When primer stocks are delivered, they must be diluted or reconstituted to the working concentration of 10 µM.
-
4Add 2 µl DNA template (2ng final) to each well and mix well by pipetting up and down.Prepare plasmid DNA using any commercially available miniprep kit or standard miniprep method (e.g., unit 1.6), but note that quantifying the DNA (APPENDIX 3D) is important. A total of 2ng of plasmid DNA is optimal but as little as 0.1ng of DNA has also been shown to work.For cloning human genes, a first-strand cDNA mix of brain plus placenta (1:4) works well as template. First-strand cDNA, 100 ng per reaction, can be added to the master mix. To clone genes from specific tissue sources or other organisms, genomic DNA or an appropriate first-strand cDNA sample can be used (see unit 5.5 & 15.5 for making first-strand cDNA).
-
5aFor amplification with plasmid DNA as a template: Carry out PCR using the following amplification cycles:
Initial step: 30 sec 98°C (denaturation) 15 cycles: 10 sec 98°C (denaturation) 30 sec 60°C (annealing) 30 sec/kb 72°C (extension) Final Extension: 10 min 72°C (a.k.a polishing) Hold: 4°C (storage). Annealing temperature can vary based upon the melting temperature of the primers; 5°C below melting temperature is recommended. Given the dependence of this program on the length of the amplicon, it is helpful to organize the genes in batches of similar size. -
5bFor amplification with first-strand cDNA as a template: Carry out PCR using the following amplification cycles:
Initial step: 30 sec 98°C (denaturation) 20 cycles: 10 sec 98°C (denaturation) 30 sec 55°C (annealing) 30 sec/kb 72°C (extension) Final step: 10 min 72°C (polishing) Hold: 4°C (storage). For high throughput cloning, we recommend a general extension time of 1 to 2 min with overall target ORF sequences averaging <2 kb. If the overall ORF average is larger (2 to 4 kb), extension times of up to 6 to min are recommended. For larger ORF averages (>4 kb), we recommend using In-Fusion® cloning scheme (see In-fusion PCR below). Given the dependence of this program on the length of the amplicon, it is helpful to organize the genes in batches of similar size.
Perform second-round Gateway® PCR
-
6Prepare the master mix for the second-round 50 µl PCR (multiply the volume for each component by the number of reactions plus two or three to allow for losses during transfer) as follows:
- 0.0625 µl 100 µM universal 5′ primer (0.125 µM final)
- 0.0625 µl 100 µM universal 3′ primer (0.125 µM final)
- 10 µl 5× Phusion® HF Reaction Buffer (1× final)
- 1 µl 10 mM 4dNTPs (0.2 mM final)
- 1.5 µl 100% DMSO (3% final)
- 0.5 µl 2.0 U/µl Phusion® High-Fidelity DNA polymerase (0.02 U final)
- 36.9 µl (when using plasmid template) or 18.9 µl (when using cDNA template) PCR-quality H2O.
-
7
Aliquot 30 µl (for cDNA) or 48 µl (for plasmid DNA) of the master mix into fresh PCR tubes or 96-well plates.
-
8
Add 2 µl (5a product; plasmid template) or 20 µl (5b product; cDNA template) amplified DNA to each tube or well containing master mix.
-
9Carry out PCR using the following amplification cycles:
Initial step: 30 sec 98°C (denaturation) 20 cycles 10 sec 98°C (denaturation) 30 sec 58°C (annealing) 30 sec/kb 72°C (extension) Final step: 10 min 72°C (polishing) Hold: 4°C (storage). Annealing temperature can vary based upon the melting temperature of the primers; 5°C below melting temperature is recommended. Given the dependence of this program on the length of the amplicon, it is helpful to organize the genes in batches of similar size -
10
Store amplified DNA up to 6 months at −20°C.
-
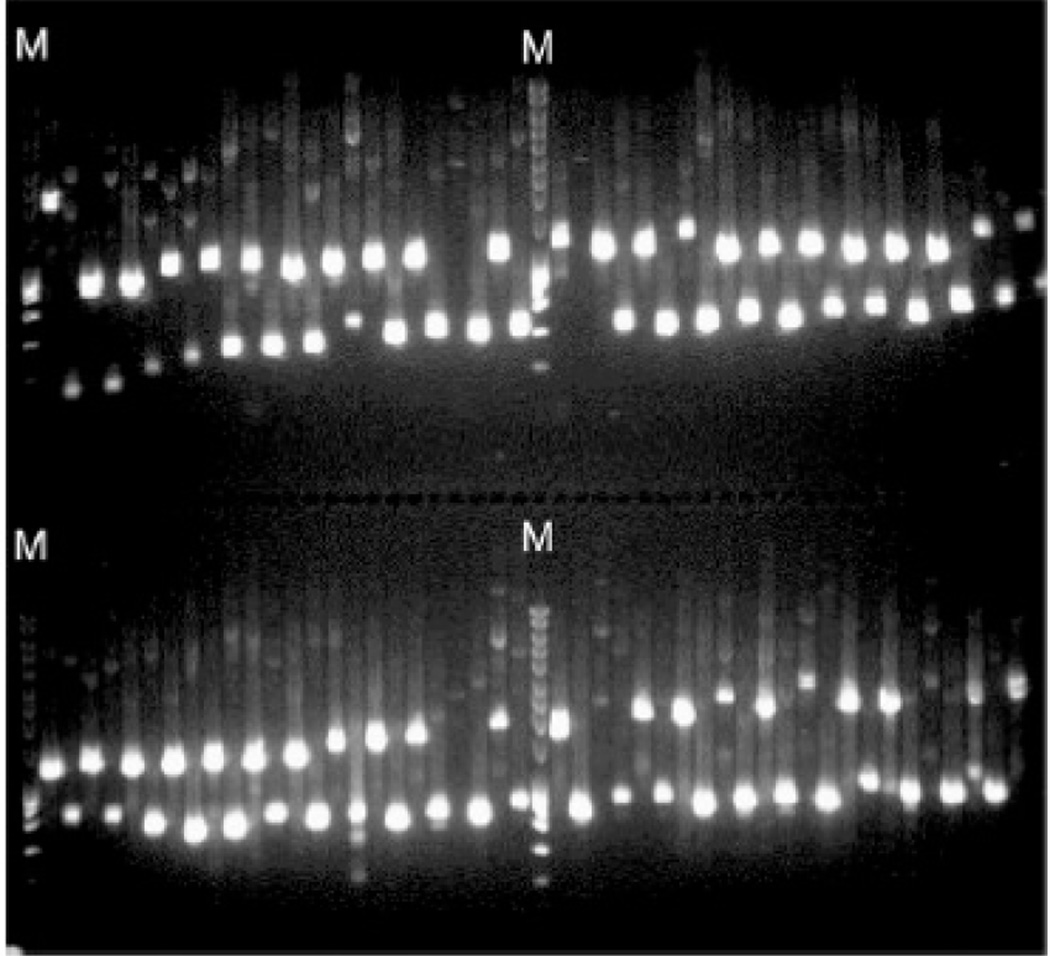
11Prepare a 1% agarose gel containing nucleic acid stain (unit 2.5A), load 2–5 µl of the PCR samples, load 1 kb DNA ladder in the first wells of each row, and run the gel to determine correct product size (see unit 2.5A).Amplification of a gene of interest from known plasmid DNA template using PCR is almost 100% efficient. However, to ensure successful downstream steps, validation of the PCR product size is performed by gel electrophoresis.In large cloning projects, a saw-tooth well arrangement will reduce the chance of contamination from neighboring bands (see Critical Parameters and Troubleshooting) (Fig. 3.20.8).
-
12Purify remaining PCR product from step 6 using single or high-throughput PCR cleanup kit according to manufacturer’s instructions.Additional minor and small products (e.g. primer dimers)can also be present in the final PCR reaction. These products will recombine with higher efficiency than the larger expected product, resulting in vectors containing aberrant sequences instead of the desired product.
-
13
Elute DNA from the DNA binding columns with 30 µl of PCR-grade H20 and determine DNA concentration (see APPENDIX 3D).
-
14
Store products at −20°C for up to six months.
Figure 3.20.8.
Examples of a saw-tooth pattern in agarose gel analysis of PCR product. PCR mixtures (96 samples) were run on a 1% agarose gel. A primer-ordering algorithm generates a map of oligonucleotide primers in 96-well format with alternating expected product size, so that the expected PCR bands would appear in a saw-tooth fashion. The letter M indicates DNA size markers.
Perform In-Fusion® PCR
-
1In-Fusion® cloning PCR is done with only one round of PCR with one set of primers. Prepare the master mix calculated for a 25 µl PCR reaction volume (multiply the volume for each component by the number of reactions plus two or three to allow for losses during transfer) as follows:
- 12.5 µl CloneAmp™ HiFi PCR Premix (1× final) (Clontech)
- 10.25 µl PCR-quality H2O.
-
2Dispense 22.75 µl master mix into PCR tubes for individual reactions or into each well of a 96-well plate for a large-scale format.For 96-well plate format a multichannel pipettor should be used.
-
3
Add 0.5 µl 10 µM gene-specific 5′-oligonucleotide primer and 0.5 µl 10 µM gene-specific 3′-oligonucleotide primer to each reaction (final concentration 0.2 µM each primer).
-
4
Add 1.25 µl of 2 µg DNA template (0.1 µg final) to each well and mix well by pipetting up and down.
-
5aPerform PCR using the following amplification cycles:
35 cycles: 10 sec 98°C (denaturation) 5–15 sec 55°C (annealing) 30 sec/kb 72°C (extension) Final step: 7 min 72°C (a.k.a. polishing) Hold: 4°C (storage). Annealing temperature can vary based upon the melting temperature of the primers; 5°C below melting temperature is recommended. Given the dependence of this program on the length of the amplicon, it is helpful to organize the genes in batches of similar size. -
6
Store amplified DNA up to 6 months at −20°C.
-
7
Validate and purify PCR products (see Gateway® PCR steps 11–14).
BASIC PROTOCOL 2
CAPTURE OF ORFS TO MAKE ENTRY CLONES FOR THE GATEWAY® SYSTEM: BP REACTION
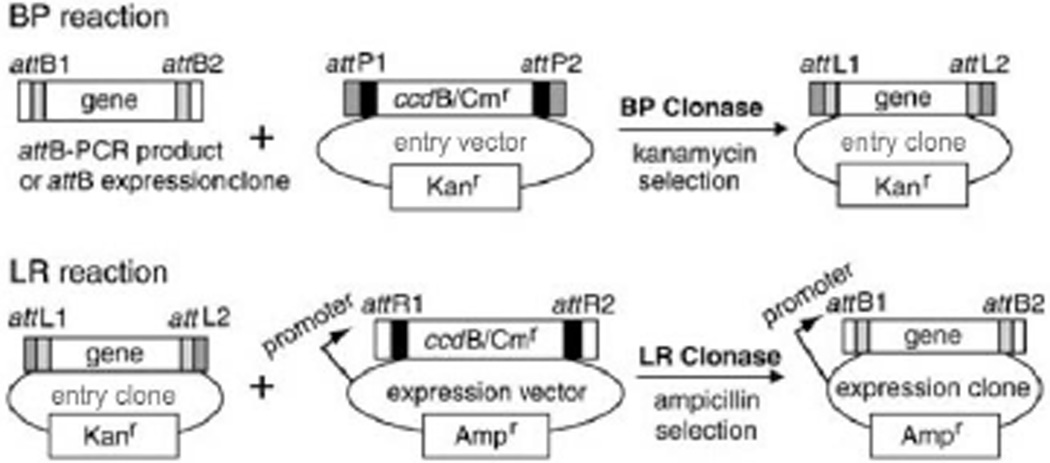
The amplified PCR product of a gene of interest must first be transferred into an entry vector to generate an entry clone. The transfer process requires recombination between the PCR product and the entry vector. This protocol describes the BP recombination reaction, so called because it mediates transfer of a gene of interest from an attB-PCR product or attB-expression clone to an attP-containing entry vector to create an entry clone (Fig. 3.20.4). For an efficient BP reaction, the attB substrate should be in linear form (PCR fragment or restriction-digested linearized plasmid), and the attP-containing entry vector should be supercoiled. Because the entry vector has the ccdB gene, which is toxic to common E. coli strains, a ccdB-resistant E. coli strain (One Shot® ccdB Survival™ 2 T1R; Life Technologies) is used for propagation of any ccdB-containing vectors. M13 forward (5’GTAAAACGACGGCCAGT) and reverse (5’CAGGAAACAGCTATGACC) primers can be used for sequence verification of constructed entry clone DNA.
Figure 3.20.4.
Gateway® BP and LR reactions. BP and LR Clonase® facilitate the recombination between attB and attP and between attL and attR, generating attL and attB sequences, respectively.
Materials
150 ng/µl entry vector (e.g., pDONR™221; Life Technologies)
Gateway® BP Clonase® II enzyme mix (Life Technologies)
10 to 50 ng/µl attB-PCR product prepared according to the Gateway® scheme (Basic Protocol 1)
PCR-grade H2O
LB plates supplemented with 50 µg/ml kanamycin (or other appropriate antibiotic; unit 1.1)
LB medium supplemented with 50 µg/ml kanamycin (or other appropriate antibiotic; unit 1.1)
50% (v/v) glycerol, sterile
Plasmid DNA isolation kit (e.g., Qiagen; optional)
96-well plate or 1.5 ml microcentrifuge tubes
Multichannel pipettor
25°C incubator
Additional reagents and equipment for growth of T1 phage resistant (T1R) E. coli competent cells (e.g. One Shot® MAX Efficiency® DH5α™ competent cells; Life Technologies) (unit 1.8), DNA miniprep (unit 1.6 or commercial DNA prep kit), and DNA sequencing (Chapter 7).
-
1Prepare the BP reaction master mix on ice for a 10 µl BP reaction (multiply the volume for each component by the number of reactions plus two or three to allow for losses during transfer) as follows:
- 1 µl 150 ng/µl pDONR™221 (15 ng/µl final)
- 2 µl BP Clonase® II enzyme mix.
- 2 µl PCR-grade H20.
-
2Dispense 5 µl master reaction mix into 1.5 ml microcentrifuge tubes or each well of a 96-well plate placed in ice.For 96-well plate format, a multichannel pipettor should be used.
-
3Add 5 µl of 10 to 20 ng/µl PCR product (50 to 100 ng) to the tubes or wells on ice. Incubate 1 hr at 25°C and place on ice or store at −20°C.The BP reaction mixture may be stored up to 6 months at −20°C.
-
4Add 2–3 µl BP reaction mixture to 50 µl E. coli competent cell and perform transformation using competent cell-specific transformation protocol.An efficient BP recombination transformation efficiency will yield >500 colonies from transforming the entire BP reaction. Larger genes (>2kb) will typically yield lower transformation efficiencies, however this can be potentially overcome by using cells with higher transformation efficiencies or In-Fusion® cloning (see Basic Protocol 3). Any lab made or commercially available competent cells with a transformation efficiency of greater than 1 × 108 cfu/ug will be sufficient.Refer to unit 1.8 for more information about bacterial transformation.
-
5Plate the transformation mix on an LB plate supplemented with 50 µg/ml kanamycin.Antibiotics should be carefully chosen to be appropriate to the entry vector (in this case, pDONR™221).
-
6
Incubate the plate overnight at 37°C.
-
7
Pick several (one to four) colonies and inoculate each into 1 ml LB medium containing 50 µg/ml kanamycin. Incubate overnight at 37°C.
-
8
Combine 0.7 ml overnight culture with 0.3 ml 50% (v/v) glycerol and store at −80°C.
-
9
Isolate plasmid DNA from the remainder of the culture using standard miniprep methods (e.g., unit 1.6) or a commercial kit (e.g., Qiagen or Macherey Nagel).
-
10Sequence the entry clone plasmid DNA covering the entire ORF and recombinational sequence region (see Chapter 7). Verify the sequence against the targeted DNA sequence information using bioinformatics tools (e.g., BLAST at http://www.ncbi.nlm.nih.gov/blast or Sequencher at http://www.genecodes.com/sequencher).The BP reactions are not perfect. Smaller fragments can be cloned more efficiently than the desired fragment and occasionally aberrant recombination products can be observed.It is highly recommended to fully sequence the insert and recombination sites to confirm that the correct, high-quality clone is captured in the entry vector.The authors usually pick four isolates per gene, fully sequence one clone and keep the others as backup reserves. The isolate will be selected if sequence is high-quality, if not, a reserve clone will be sequenced.
-
11
Select the clone with the highest quality gene sequence.
BASIC PROTOCOL 3
CAPTURE OF ORFS TO MAKE ENTRY CLONES UTILIZING IN-FUSION® CLONING
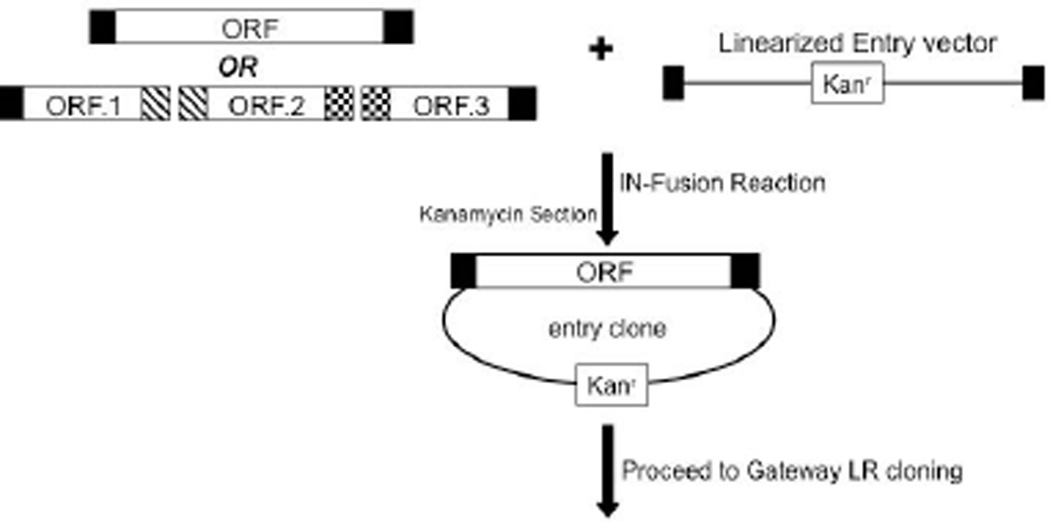
Capturing the coding sequences into an entry vector using the In-Fusion® system is mediated by homologous recombination between the end sequences of PCR products and the end sequences of linearized entry vector (Fig. 3.20.5). There is a 15 bp sequence constraint on the flanking sequences of the PCR product and the ends of the linearized vector. This shorter recombination sequence permits the overall primer length to nearly always be less than 40 bases, which in turn allows for only one PCR reaction to accomplish cloning using this system. In addition, the In-Fusion® kit will efficiently and consistently clone large DNA fragments (<15 kb) without requiring the cloning of multiple DNA products simultaneously. Lastly, the ORF of interest can be directly transferred to the final expression vector of interest without an intermediate entry clone step as in the Gateway® cloning scheme. However, for the ability to shuttle the ORF to multiple expression vectors in a lower cost and more efficient manner, it would be beneficial to transfer the ORF to an entry vector first as described below.
Figure 3.20.5.
Schematic diagram of In-Fusion® HD cloning reaction. In-Fusion® enzymes facilitate the homologous sequence recombination between PCR product(s) and linearized entry vector. Entry clones can then be taken through the Gateway® cloning scheme to generate expression clones.
Preparation of Linearized Vector
In order to achieve a successful In-Fusion® reaction, the entry vector must be linearized. To linearize the vector, a restriction digest is performed that results in the homologous recombination sites flanking the linearized vector. A modified pDONR™221 entry vector, pGWNcoEco (DNASU.org) can be linearized with a double digest that results in functional att sites flanking the linearized product, allowing for efficient In-Fusion® cloning.
A linearized vector can also be obtained through PCR by designing primers that initiate at the homologous recombination site and amplify around the vector.
Materials
50 ng/µl entry vector DNA (e.g., pGWNcoEco; DNASU.org)
Appropriate restriction enzymes (e.g. NcoI and Eco0109i for pGWNcoEco vector; NEB)
Appropriate restriction enzyme buffers (e.g., NEB)
Razor blades
Low-frequency UV light table
Eye protection for UV light
GelStar™ nucleic acid stain (Lonza)
Gel extraction kit (e.g., Qiagen)
5X In-Fusion® HD Enzyme Premix (Clontech)
150 ng/µl linearized entry vector
500 ng/µl PCR product (Basic Protocol 1: In-Fusion® PCR)
E. coli high-efficiency T1R competent cell culture (e.g., One Shot® MAX Efficiency® DH5α™ cells; Life Technologies)
LB plates supplemented with 50 µg/ml kanamycin (or appropriate antibiotic) (unit 1.1)
LB medium supplemented with 50 µg/ml kanamycin (or appropriate antibiotic) (unit 1.1)
50% (v/v) glycerol, sterile
PCR-grade H2O
Plasmid DNA isolation kit (e.g., Qiagen; optional)
96 well plates or 1.5 ml microcentrifuge tubes
PCR tubes (thin-walled)
Multichannel pipettor
Thermo cycler
Additional reagents and equipment for growth of E. coli competent cells (unit 1.8), DNA miniprep methods (unit 1.6), linearizing vectors using restriction enzymes (unit 3.1), DNA sequencing (see Chapter 7), agarose gel electrophoresis (unit 2.5A), determination of DNA concentration (APPENDIX 3D) and analyzing plasmid DNA using restriction enzymes (unit 3.1).
Linearization of entry vector using restriction enzyme digestion
-
1Linearize the entry vector (e.g., pGWNcoEco; DNASU.org) at the homologous recombination site using restriction enzymes (e.g. NcoI and Eco0109i for pGWNcoEco vector) (unit 3.1).pGWNcoEco vector contains the ccdB death cassette and needs to be propagated in ccdB resistant competent cells (e.g. One Shot® ccdB Survival™ 2T1R E. coli; Life Technologies) before purification of the vector.
-
2Prepare a 1% agarose gel containing 0.001% (v/v) GelStar™ nucleic acid stain, load entire restriction digest, load uncut entry vector, load 1kb DNA ladder in the first wells of each row, and run the gel (see unit 2.5A).In large cloning projects, a saw-tooth well arrangement will reduce the chance of contamination from neighboring bands (see Critical Parameters and Troubleshooting).Use the uncut vector as a negative control size maker for the digestion.
-
3
Excise the bands using a razor blade while visualizing the bands with GelStar™ nucleic acid stain on a low-frequency UV light box. Collect the gel fragment into 1.5 ml microcentrifuge tube and purify the DNA from the agarose using a gel extraction kit (e.g., QIAquick Gel Extraction kit; Qiagen).
-
4
Determine DNA concentration (see APPENDIX 3D).
-
5
Store linearized DNA at −20°C for up to six months.
In-Fusion® Cloning Reaction
-
6Prepare the master reaction mix on ice for the 10 µl In-Fusion® reaction (multiply the volume for each component for the number of reactions plus two or three to allow for losses during transfer) as follows:
- 2 µl 5× In-Fusion® HD Enzyme Premix (1× final)
- 1 µl 10× In-Fusion® reaction buffer (1× final)
- 1 µl of 150 ng/µl linearized entry vector (50 ng final).
- 3 µl of PCR-grade H20
-
7Dispense 7 µl master reaction mix into thin walled PCR tubes or 96-well plate.For 96-well plate format a multichannel pipettor should be used
-
8
Add 3 µl 500 ng/µl PCR product (100 ng final) (product generated in Basic Protocol 1: In-Fusion® PCR).
-
9Incubate reactions in thermo cycler for 15 min at 50°C, then place on ice.Reactions can be stored at −20°C until ready to proceed to next step or up to six months.
-
10Add 2.5 µl of In-Fusion® reaction mixture to 50 µl E. coli high-efficiency competent cell culture and perform transformation.Similar as in Gateway® cloning, any T1R competent cells with a transformation efficiency of greater than 1 × 108 cfu/ug is sufficient (e.g. One Shot® MAX Efficiency® DH5α-T1R competent cells; Life Technologies). Do not use more than 5 µl of diluted reaction mix for 50 µl competent cells.Refer to unit 1.8 for more information about bacterial transformation.
-
11
Plate the transformation mix on an LB plate supplemented with 50 µg/ml kanamycin (or appropriate antibiotic).
-
12
Incubate the plate overnight at 37°C.
-
13
Pick several (one to four) and inoculate each into 1 ml LB medium containing 50 µg/ml kanamycin (or appropriate antibiotic). Incubate overnight at 37°C.
-
14
Proceed as in Protocol 2, steps 8 through 10, to create glycerol stock cultures and sequence plasmid DNA covering the ORF and recombinational sequence region.
-
15
If a validated Gateway® compatible entry clone was generated at this step, proceed to Basic Protocols 4 to transfer the entry clone to an expression vector using the Gateway® cloning scheme.
BASIC PROTOCOL 4
GENERATING GATEWAY® EXPRESSION CLONES
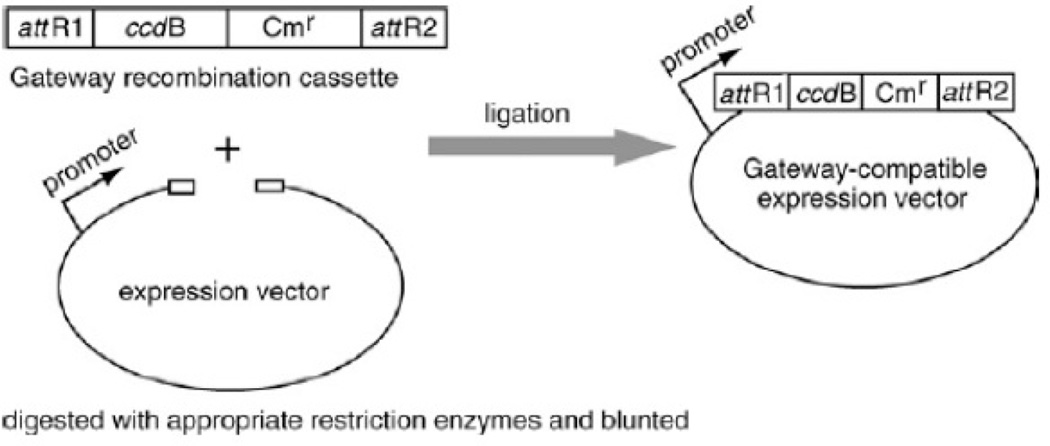
Expression clones are generated by the LR reaction between the attL-containing entry clone and attR-containing Gateway®-compatible expression vectors (see Fig. 3.20.4). Either the commercial Gateway® destination vectors or Gateway®-compatible destination vectors generated in-house can be used. Conversion to a Gateway®-compatible vector involves a simple insertion of a blunt-ended conversion cassette by ligation into the blunt-ended expression vector that is to be adapted to the Gateway® system. The conversion cassette, commercially available for the Gateway® system, includes a ccdB death gene, a GyrA inhibitor that kills the E. coli competent cells when the ccdB gene is expressed (Bernard P., Couturier M. (1992) J. Mol. Biol. 226:735–745.), and a chloramphenicol resistance (Cmr) gene flanked by attR sites (Fig.3.20.6). The new recombination sites created in the expression vector from this reaction will be attB sites, which are the shortest of the att sites.
Figure 3.20.6.
Conversion of an expression vector into a Gateway®-compatible vector. An expression vector of choice is linearized and blunted at favorable restriction sites. It is then ligated with an available blunt-ended cassette containing ccdB and Cmr genes. The Gateway®-compatible expression vector sequence should be analyzed to confirm the correct orientation.
Materials
Expression vector with a different selectable marker than the entry clone
Appropriate restriction enzymes (e.g., NEB)
T4 DNA polymerase or Klenow fragment (e.g., NEB)
TE buffer, pH 8.0 (APPENDIX 2)
T4 DNA ligase and buffer (e.g., NEB)
Gateway® recombination cassette (rfA, rfB, or rfC; Life Technologies), for making in-house Gateway®- compatible destination vectors
Sterile H2O
E. coli competent cell cultures (e.g., One Shot® ccdB Survival™ 2 T1R and DH5α; Life Technologies)
LB plates supplemented with 30 µg/ml chloramphenicol (or other antibiotic appropriate for the expression vector; unit 1.1)
LB medium supplemented with 30 µg/ml chloramphenicol (or other antibiotic appropriate for the expression vector; unit 1.1)
50% (v/v) glycerol, sterile
LR Clonase® II enzyme mix (Life Technologies)
150 ng/µl commercial Gateway® (e.g., pDEST™27; Life Technologies) or Gateway®-converted destination (expression) vector generated below
150 ng/µl entry clone plasmid DNA (Basic Protocol 2 or from plasmid resources (e.g., DNASU.org))
LB plates supplemented with 100 µg/ml ampicillin (or other appropriate antibiotic; unit 1.1)
LB medium supplemented with 100 µg/ml ampicillin (or other appropriate antibiotic; unit 1.1)
96-well plates or 1.5-ml microcentrifuge tubes
Multichannel pipettor
25°C incubator
Additional reagents and equipment for growth of E. coli competent cells (unit 1.8), linearizing the expression vector at the multiple cloning sites using restriction enzymes (unit 3.1), generating blunt ends using T4 DNA polymerase or Klenow fragment (units 3.5 & 3.16), and analyzing plasmid DNA using restriction enzymes (unit 3.1)
Prepare a Gateway®-compatible destination vector (optional)
-
1
Linearize the expression vector at the multiple cloning sites or the relevant position where the cDNA should be located using restriction enzymes (unit 3.1), and generate blunt ends using T4 DNA polymerase or Klenow fragment (units 3.5 & 3.16).
-
2
Adjust the linearized expression vector to a final concentration of 20 to 50 ng/µl in TE buffer, pH 8.0.
-
3Prepare the ligation reaction mix in a microcentrifuge tube as follows:
- 1 to 5 µl of 20 to 50 ng/µl dephosphorylated linearized vector
- 1 µl 10× T4 DNA ligase buffer
- 2 µl 5 ng/µl Gateway® recombination cassette
- 1 µl 1 U/µl T4 DNA ligase
- Sterile H2O to a final volume of 10 µl.
If a polypeptide fusion is to be added at either end of the gene, ensure that the chosen Gateway® recombination cassette is in the appropriate reading frame. -
4
Incubate 1 hr at room temperature.
-
5Add 1 to 2 µl reaction mixture to 50 µl ccdB Survival™ competent cells and perform transformation.One Shot® ccdB Survival™ 2 T1R competent cells are modified to allow growth of plasmids containing ccdB. Cells without ccdB resistance will be killed by the new destination vector that will carry a copy of the ccdB death gene.Transformation efficiency of >1 × 108 cfu/µg DNA is recommended.Refer to unit 1.8 for more information about bacterial transformation.
-
6Plate the transformation mix on an LB plate supplemented with 30 µg/ml chloramphenicol. Incubate overnight at 37°C.Antibiotics should be carefully chosen to be appropriate to the recombination cassette (in this case chloramphenicol).
-
7
Pick several (one to four) colonies and inoculate each into 1 ml LB supplemented with 30 mg/ml chloramphenicol. Incubate overnight at 37°C.
-
8
Combine 0.7 ml overnight culture with 0.3 ml 50% glycerol and store up to 6 months at −80°C.
-
9Isolate plasmid DNA from the remainder of the culture using standard miniprep methods (e.g., unit 1.6) or a commercial kit (e.g., Qiagen). Analyze plasmid DNA using appropriate restriction enzymes (unit 3.1) or by sequencing to identify clones with the proper orientation. Quantify plasmid DNA from a selected clone (APPENDIX 3D).Tranformants need to be screened for orientation because both orientations will be generated after blunt-end ligation. Appropriate restriction enzymes will cut the vector and cassette in an asymmetric fashion, yielding fragment lengths that depend on orientation.
Transfer an ORF into the expression vector using the Gateway® LR reaction
-
10Prepare the master reaction mix on ice for the 10 µl LR reaction (multiply the volume for each component for the number of reactions plus two or three to allow for losses during transfer) as follows:
- 1 µl 150 ng/µl commercial Gateway® (e.g., pDEST™27) or Gateway®-compatible (from step 9) destination vector
- 2 µl Gateway® LR Clonase® II Enzyme mix
- 4 µl PCR-grade H20.
-
11Mix well and dispense 7 µl master reaction mix into each well of a 96-well plate or 1.5 ml microcentrifuge tube on ice.For 96-well plate format a multichannel pipettor should be used.
-
12
Add and mix 3 µl entry clone plasmid DNA (Basic Protocols 2 or 3 or available resources, e.g., DNASU.org) to the 7 µl master reaction mix.
-
13Incubate the reaction for 1 hr at 25°C.The reaction mixture can be stored up to 6 months at −20°C after this step.
-
14Add 3 to 5 µl of reaction mixture to 50 µl E. coli competent cell culture and perform transformation.Any standard cloning strain that is ccdB-sensitive, such as T1R DH5α™, is sufficient. Competent cells can be made in lab or supplied from a vendor.Transformation efficiency of >1 × 108 cfu/µg DNA is recommended.Refer to unit 1.8 for more information about bacterial transformation.
-
15Plate the transformation mix on an LB plate supplemented with 100 µg/ml ampicillin. Incubate overnight at 37°C.Antibiotics should be carefully chosen to be appropriate to the expression vector, in this case, ampicillin for the commercial vector pDEST™27.
-
16Pick 1 to 2 colonies and analyze the plasmid DNA using restriction enzymes or DNA sequencing.Typically, the transfer reaction is very efficient (90% to 95%).
-
17
Choose the best clone and prepare for the desired use and store as a 15% glycerol stock at −80°C.
Alternate Protocol 1
GENERATING EXPRESSION CLONES UTILZING A CRE-LOX SYSTEM
Cre-Lox cloning strategy is another type of recombinational cloning system previously sold by Clontech (formally known as BD Clontech) as a kit called the Creator™ System. Although the Creator™ system is no longer marketed by Clontech, many researchers and plasmid repositories (e.g., DNASU.org) still possess or distribute Creator™-compatible master clones, so the knowledge of the transfer of entry (master) to expression vectors utilizing the Cre-Lox strategy is still worthy of discussion. Furthermore, the individual reagents and enzymes are still commercially available and/or purified in lab (Cantor and Chong, 2001), so this method can still be carried out by researchers if so desired. For more detailed information about Creator™ cloning system, see published reviews (Festa et al., 2013; Marsischky and LaBaer, 2004).
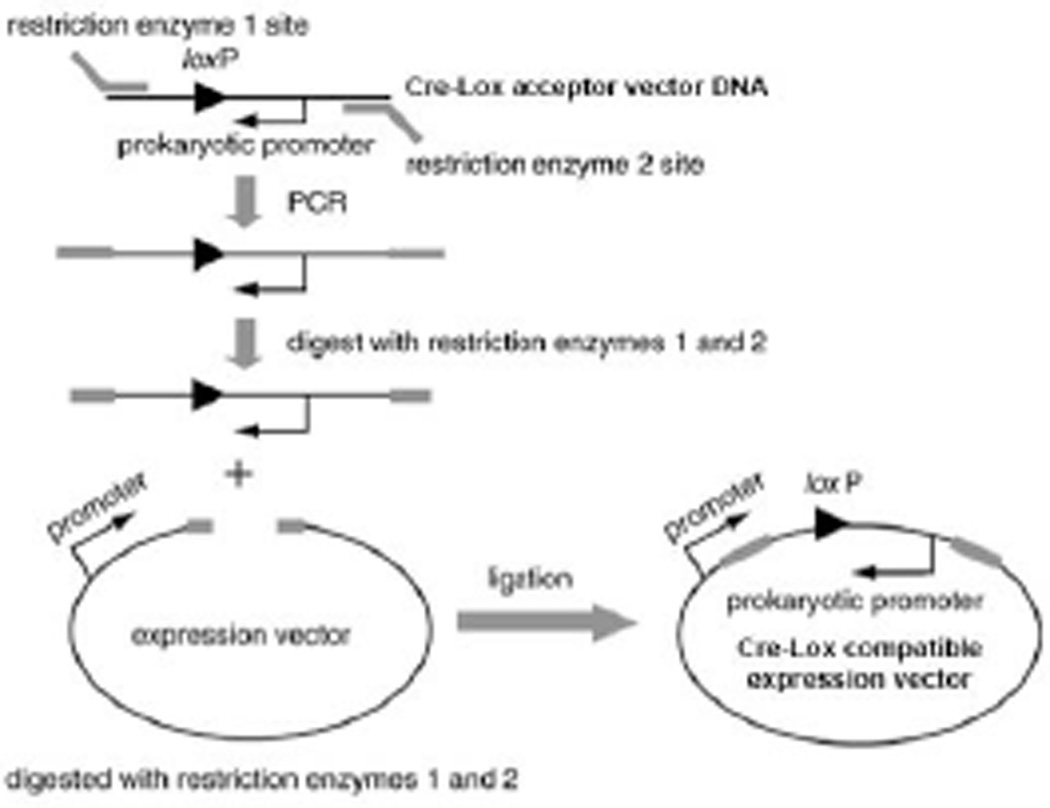
The Cre-Lox cloning strategy uses the same overall recombination principles as the Gateway® and In-Fusion® cloning systems, however different enzymes and recombination sites are utilized in the Cre-Lox system. LoxP recombination sites are incorporated in the entry vector and Cre recombinase is utilized for the generation of the final expression clone. Cre recombinase facilitates the gene or insert transfer from the entry clone to a Cre-Lox-compatible expression vector. The process can be performed using the commercially available vectors or Cre-Lox compatible vectors generated in-house. The generation of Cre-Lox compatible expression vectors involves (1) the amplification of the Cre-Lox conversion cassette with primers that contain restriction sites, (2) restriction of expression vector, and (3) ligation of the prepared cassette into the target expression vector (Fig. 3.20.7).
Figure 3.20.7.
Conversion of an expression vector into a Cre-Lox-compatible vector. The region flanking the loxP site and prokaryotic promoter is amplified with specific PCR primers containing specific restriction enzyme sites. Then, the PCR products and an expression vector are digested with appropriate restriction enzyme. A ligation reaction generates a Cre-Lox compatible expression vector.
Materials
Cre-Lox expression vector (e.g., pLP-CMVneo; DNASU.org) or other expression vector to make Cre-Lox compatible vector
LoxP primers
LoxP template (e.g., Addgene)
Restriction enzymes and buffers (NEB)
T4 DNA ligase and buffer (NEB)
E. coli competent cell culture (e.g., DH5α™; Life Technologies)
LB plates supplemented with 100 µg/ml kanamycin (or other appropriate antibiotic; unit 1.1)
LB plates supplemented with 30 µg/ml chloramphenicol (or other appropriate antibiotic; unit 1.1) and 7% (w/v) sucrose
Cre recombinase (NEB or lab purified (Cantor and Chong, 2001))
10× Cre reaction buffer (NEB)
20 ng/µl master or entry clone plasmid DNA
96-well plate or 1.5 ml microcentrifuge tubes
Multichannel pipettor
Additional reagents and equipment for PCR (Basic Protocol 1), agarose gel electrophoresis (unit 2.5A), growth of E. coli competent cells (unit 1.8), DNA plasmid preparation (unit 1.6 or commercial DNA prep kit), and analyzing the plasmid DNA using restriction enzymes (unit 3.1).
Prepare Cre-Lox-compatible expression vector (optional)
-
1Design PCR primers to amplify the loxP template as follows:
- 5′ primer: 5′-RS-ATA ACT TCG TAT AGC ATA CAT TAT-3′
- 3′ primer: 5′-RS-CAC GTC AGG TGG CAC TTT TCG-3′.
Restriction sites (RS) should be added, allowing the ligation of the cassette into the target vector. The target vector should have restriction enzyme sites that are not present in the loxP cassette. -
2
Amplify the loxP cassette by PCR (see Basic Protocol 1, steps 1 to 5a modified for 35 cycles).
-
3
Digest the PCR loxP fragment and the expression vector with the selected restriction enzymes.
-
4
Purify the digested PCR fragment (161 bp) by agarose gel electrophoresis (unit 2.5A).
-
5Ligate (see Basic Protocol 4, step 3) the PCR-amplified loxP cassette and the linearized expression vector.Use of the appropriate matched restriction enzymes for preparing the loxP cassette and the linearized expression vector are necessary for the ligation reaction to occur.
-
6Add 2.5 µl of ligation mixture to 50 µl E. coli competent cell culture, e.g., DH5α™, and perform transformation.Transformation efficiency of >1 × 108 cfu/µg DNA is recommended.Refer to unit 1.8 for more information about bacterial transformation.
-
7Plate the transformation mix on an LB plate supplemented with 100 µg/ml kanamycin. Incubate overnight at 37°C.Antibiotics should be carefully chosen to be appropriate to the expression vector.
-
8
Pick colonies and prepare overnight cultures and glycerol stocks (see, Basic Protocol 4, steps 7 and 8). Prepare plasmid DNA (unit 1.6 or commercial DNA prep kit) and quantify (APPENDIX 3D).
Transfer an ORF into the expression vector using Cre reaction
-
9Prepare the master reaction mix on ice for the 20 µl reaction (multiply the volume for each component for the number of reactions plus two or three to allow for losses during transfer) as follows:
- 200 ng commercial Cre-Lox (e.g., pLP-CMVneo; DNASU.org) or Cre-Lox-compatible (step 8) expression vector
- 1 µl Cre recombinase
- 2 µl 10× Cre reaction buffer
- Bring volume to 10 µl with H2O.
-
10Dispense 10 µl reaction mix into each well of a 96-well plate.For a 96-well plate format, a multichannel pipettor should be used.
-
11Add 10 µl 20 ng/µl entry clone plasmid DNA to the 10 µl reaction mix.Master clone plasmid DNA (150 to 200 ng) per reaction works well.
-
12
Incubate 30 min at 37°C.
-
13
Heat inactivate the reaction 15 min at 70°C.
-
14Add 2 to 3 µl reaction mixture to 50 µl E. coli competent cell culture, e.g., DH5α™, and perform transformation.Transformation efficiency of >2 × 108 cfu/µg DNA is recommended.More than 5 µl of reaction mix can be toxic to bacterial competent cells.Refer to unit 1.8 for more information about bacterial transformation.
-
15Plate the transformation mix on an LB plate supplemented with 30 µg/ml chloramphenicol and 7% (w/v) sucrose. Incubate the plates overnight at 37°C.Antibiotics should be carefully chosen to be appropriate to the expression vector (in this case, pLP-CMVneo).
-
16Pick good-sized colonies.It is important to pick good-sized colonies rather than small ones. It has been reported that the small colonies could be background and may harbor undesirable recombinants.
-
17Analyze the plasmid DNA of picked colonies using restriction enzymes (unit 3.1) or single read sequencing.Typically, the transfer reaction is very efficient (90% to 95%). For large-scale projects, in many cases, it is not necessary to pick several colonies and analyze them.
-
18
Choose the best clone and prepare for the desired use or store as a 15% glycerol stock up to 6 months at −80°C.
COMMENTARY
Background Information
Overview of universal cloning strategy
As molecular biology has matured, the number of experimental approaches that can be conceived and executed with a cloned cDNA copy of a gene has grown greatly. Proteins can be expressed in vitro or in vivo in a wide variety of organisms, ranging from bacteria to human cells; their expression can drive the production of purified protein for structural studies, or their regulated expression can be used for complementing genetic phenotypes; epitope tags (e.g., green fluorescent protein, GST, or HA) can be added as markers for localization, for use in purification, or as part of two-hybrid assay systems. Each of the applications that utilize protein expression demands the subcloning of the relevant cDNA into an appropriate plasmid vector that adds the relevant selectable markers, transcriptional and/or translational signals, any needed polypeptide tags, and other relevant sequences. Using traditional subcloning methods based on restriction enzymes and ligation, the effort required to transfer cDNAs from vector to vector can be substantial and time consuming. Large-scale experiments involving the examination of thousands of genes simultaneously using automated and parallel approaches, would be impossible using standard methods.
Recombinational cloning is based on site-specific recombination to transfer DNA sequences into multiple expression systems (Walhout et al., 2000; Reboul et al., 2001, 2003; Brizuela et al., 2002; Yamada et al., 2003; Marsischky and LaBaer, 2004; Saul et al., 2014). This cloning strategy is independent of the insert DNA sequence to be cloned, separating it from the limitations of classical restriction enzyme–based cloning where desired digestion sites are often located within the gene, in turn limiting the potential cloning strategies (Festa et al., 2013). Recombinational cloning enables rapid and efficient transfer of single inserts into multiple different expression systems for protein expression and functional analysis. It also allows the simultaneous transfer of a many different DNA sequences into an expression vector in parallel, enabling high throughput applications (Fig. 3.20.1). Presently, the Gateway® system (Life Technologies) and the In-Fusion® system (Clontech) are widely used and commercially available.
Our preferred recombinational approach employs the use of a set of entry clones from which numerous different expression clones can be easily produced. The entry clones contain the coding sequences (also called open reading frames, ORFs) situated in inert (non-protein-expressing) vectors, which are endowed with recombinational sequences that are recognized by the recombinational enzymes that drive the recombining of the insert into the desired vector. The use of inert vectors for the entry clones avoids inadvertent protein expression and consequent negative selection of clones that contain inserts toxic to the propagating E. coli strain. Using recombination, the coding sequences can then be transferred to any compatible protein expression vector that has the recombination sequences situated in the appropriate position and reading frame. Virtually any existing expression vector can be easily adapted to contain recombinational sequences for either of the existing commercial systems. Based upon the design of the original entry clones, the expression clones can either produce native protein (when entry clones contain a naturally positioned stop codon) or carboxyl-terminal tagged proteins (when the stop codon is omitted from the entry clone). The transfer process is a highly conservative molecular reaction with a very low nucleotide error rate. In turn, once the sequence of the entry clones has been verified, most all daughter clones contain the desired sequence, hence limiting the time and effort of expression clone validation.
The steps involved in making expression-ready entry clones include (1) identifying the genes that will be cloned, (2) obtaining sequence information regarding the coding sequences, (3) determining whether to include or exclude the stop codon, (4) designing gene-targeted oligos for PCR, (5) constructing entry clones using high-fidelity PCR and capture into the entry vector, (6) bacterial transformation and picking colonies, (7) DNA sequencing of the insert and important flanking sequences, and (8) sequence analysis and selection of entry clones that meet the acceptance criteria. Once the entry clones are constructed and their sequences verified, they can be transferred to appropriately constructed expression vectors to generate expression-ready clones.
The recombination reactions themselves are equilibrium reactions that result in a mixture of end products, starting substrates, and intermediates. The desired product may only be present as a small fraction of this mix, but by using the appropriate positive and negative selections, it is the only product that is recovered. This results in an overall efficiency that exceeds 95%, which can enable high throughput automation. Experiments can be easily expanded to genome scale once a complete high-quality collection has been constructed in a recombinational cloning system (Yu et al., 2014; Ceroni et al., 2010; Rolfs et al., 2008; Thanawastien et al., 2009; Murthy et al., 2007; Rolf et al., 2008). It should be noted that many of these collections of clones are publically available. Before producing new clones, researchers should check some of these open resources to see if they clones they desire are already available (DNASU.org, Addgene.org, Plasmid.med.harvard.edu, ATCC.org, Beiresources.org).
The Gateway® system
Gateway® technology is based on the bacteriophage lambda (λ) site-specific recombination system, which facilitates the integration of λ genome into the Escherichia coli (E. coli) chromosome, and thus the ability to grow lytically after integration (Ptashne, 1992; Hartley et al., 2000; Walhout et al., 2000). Recombination occurs between specific attachment (att) sites on the interacting DNA molecules. The DNA segments flanking the recombination sites are changed such that after recombination the att sites become hybrid sequences from the att sequences of each parental vector. For example, attL sites comprise sequences from attB and attP sites. Strand exchange occurs within a core region that is common to all att sites. For more detailed information about λ recombination, see published references and reviews (Landy, 1989; Ptashne, 1992; http://www.lifetechnologies.com/us/en/home/life-science/cloning/gateway-cloning.html).
The specific recombinational att sites are the first critical factor in the λ site-specific recombination system. The att sites serve as the binding site for recombination proteins and have been well characterized (Weisberg and Landy, 1983). Upon λ integration, recombination occurs between attB (bacteria) and attP (phage) sites to give rise to attL (left) and attR (right) sites. The attB and attP sites are different sizes, and because the attL and attR sites are hybrids, their sizes fall in between. The actual crossover occurs between homologous core regions (15 bp) on the two sites, but surrounding sequences are required as they contain the binding sites for the recombination proteins (Landy, 1989). In developing the Gateway® system, mutations were introduced into the core of the att sites to create different sets that are incompatible with each other, generating attB1 and attP1 sites and attB2 and attP2 sites. The attB1 recombine only with attP1, not with attP2. This allows a gene to be flanked by attL1 and attL2 sites on either side that can recombine with the corresponding attR sites and maintain the correct orientation. The different species of recombination sites (attB, attP, attL, and attR) are found in different vector constructs. The system is designed so that the attB site, the smallest of the four, appears in the final expression clone introducing an additional 9 amino acids into any fusion proteins.
The second major factor in in the λ site-specific recombination is the enzymes that recognize and drive the recombination events. The Lambda recombination is catalyzed by a mixture of enzymes that bind to specific att sequences described above. The recombination proteins differ depending upon which direction the reaction is run. When the reaction is run to mix attB and attP sites, such as when making entry clones, it is catalyzed by the bacteriophage λ integrase (Int) and E. coli integration host factor (IHF) proteins (BP Clonase® enzyme mix), whereas, if the reaction is between attL and attR sites it is catalyzed by the bacteriophage λ Int and excisionase (Xis) proteins, and the E. coli IHF protein (LR Clonase® enzyme mix). For more information about the recombination enzymes, see published references and reviews (Nash, 1977; Nash and Robertson, 1981; Landy, 1989; Ptashne, 1992). Two recombination reactions constitute the basis of the Gateway® technology. BP reaction, catalyzed by BP Clonase® enzyme mix, facilitates recombination of an attB substrate (attB-PCR product or a linearized attB-containing expression clone) with an attP substrate (entry vector) to create an attL-containing entry clone (Fig. 3.20.4). The BP cloning efficiency tends to drop rapidly with increasing insert DNA size larger than 2 kb (Marsischky and LaBaer, 2004). On the other hand, LR Reaction, catalyzed by LR Clonase® enzyme mix, facilitates recombination of an attL substrate (entry clone) with an attR substrate (expression vector) to create an attB-containing expression clone (Fig. 3.20.4).
The recombination reaction just described is an equilibrium reaction; therefore tight selection is required to ensure that only the desired product is obtained. In the Gateway® system, an antibiotic resistance marker mediates positive selection for the correct entry clone. To avoid contamination from starting vectors, there is counter-selection by the ccdB gene, which is toxic to general cloning bacteria due to the ability to inhibit bacterial DNA gyrase (Bernard and Couturier, 1992). Thus, only bacteria that contain entry clones where the death gene has been successfully replaced by the desired insert will survive selection. Vectors carrying the ccdB gene can be propagated using a special E. coli strain which expresses ccdA which forms a one-to-one complex with the ccdB protein and thus keeps it from killing the bacterium (One Shot® ccdB Survival™ 2 T1R). The combined positive and negative selection provides an overall BP or LR reaction efficiency of 95%. For more detail refer to the Gateway® manual from the manufacturer. Many Gateway® compatible clones and collections already exist and are publically available (DNASU.org).
The In-Fusion® system
The Clontech In-Fusion® system relies on homologous recombination to generate the desired constructs in a one-step isothermal in vitro recombination event. Unlike with the Gateway® BP with a maximum cloning efficiency for sequence lengths <2 kb (Marsischky and LaBaer, 2004), the In-Fusion® reaction has been shown to successfully capture insert sizes as large as 15 kb with efficiency of greater than 95% (http://www.clontech.com/US/Products/Cloning_and_Competent_Cells/Cloning_Kits/Cloning_Kits-HD-Liquid). The In-Fusion® system also allows for the cloning of up to four different 1 kb fragments simultaneously in a single reaction. The protocol for cloning multiple fragments into a single vector is the same as described above, however the PCR fragments must have complementary ends to the neighboring fragments and/or the entry vector (Fig. 3.20.5). Although, the cloning of multiple inserts utilizing the In-Fusion® system does lower the reaction efficiency from >95% to 70–80%. The last major advantage to the system, and unlike with Gateway® cloning where att sites must be inserted into the cloning vectors, the In-Fusion® system vectors do not have to be modified.
The In-Fusion® system of combining a linearized vector with one or more PCR products with overlapping ends to generate clones is based on the simple principle of homologous recombination. This system utilizes the recombinational functional properties of vaccinia virus DNA polymerase (VVpol), in addition to a proprietary buffer (Irwin et al. 2012, Hamilton et al. 2006, Zhu et al. 2007). The molecular mechanisms of this ligation-independent cloning system starts with the incubation of the In-Fusion® enzymes (optimized VVpol), PCR product(s), and linearized vector. The VVpol 3’-to-5’ exonuclease activity removes nucleotides from the ends of the double-stranded DNA samples (Irwin et al. 2012, Hamilton et al. 2006, Zhu et al. 2007). The exposed complementary single stranded ends of 15 bp homologous regions of the PCR fragments and linearized vector anneal, in which can be incited with the addition of the vaccinia virus I3 single-strand DNA binding protein. A DNA polymerase is present to insert bases into the gaps of DNA, and a DNA ligase functions to seal in the nicks, resulting in a seamless double stranded circular construct of a vector and the sequence of interest (Zhu et al. 2007, Irwin et al. 2012). Similar to the Gateway® system, positive selection for the correct entry clone is mediated by an antibiotic resistance marker. To avoid contamination from starting non-linearized vectors, there is counter-selection by the ccdB gene that when expressed is toxic to general cloning bacteria due to the ability to inhibit bacterial DNA gyrase (Bernard and Couturier, 1992). As previously stated, the commercially available In-Fusion® cloning kit also contains proprietary optimized reagents and buffers that generate more efficient and consistent reactions (Irwin et al. 2012).
New England Biolabs (NEB) also has a cloning kit for recombinational cloning that is also based on the mechanism of homologous recombination called Gibson Assembly® Cloning Kit (UNIT 3.22). This kit ensures the efficient joining of overlapping DNA fragments in a single isothermal reaction with the use of three different enzymes: an exonuclease, proprietary DNA polymerase, and a DNA ligase. The fundamentals of Gibson Assembly® were originally described in 2009 in a research publication, however the reagents have been optimized by NEB to obtain greater transformation efficiency of larger inserts up to 20kb. Although, one could use this publication as a framework for an in-house cost effective, albeit lower efficiency, cloning protocol for generating large DNA constructs. In theory, any proofreading thermostable DNA polymerase can be adapted with enhancer agents (i.e., DMSO, polyethyleneglycol (PEG), E. coli single-stranded DNA-binding protein (SSB), Gene 32 protein) in combination with commercially available ligase and exonucleoase.
Single step recombinational cloning directly into destination vectors
Gateway® and In-Fusion® cloning provide a useful system to shuttle DNA sequences between an intermediate entry clone and a wide variety of destination (expression) vectors. The entry clones serve as "storage constructs" that can be utilized to generate a new destination vector in a short experimental timeframe. However, several applications and/or researchers do not find a need for this intermediate cloning step, or the flexibility and advantages offered by the entry clone. For these circumstances, and in the effort to save time and money, Life Technologies has published a protocol for a single-step reaction for generating expression clones (Liang, 2013
In the single-step Gateway® reaction scheme, the PCR product is combined with an entry vector, expression vector, and the LR Clonase® II enzyme mix. The conditions are carried out in the same manner as in the LR reaction described in Basic Protocol 4. The efficiency of the single-step Gateway® reaction scheme has been shown to be similar to the efficiency of the standard dual-step Gateway® cloning pipeline (Fig. 3.20.4). The single-step protocol can also generate entry clones through the partitioning of the Gateway® transformation between two selection media appropriate for the entry and expression clones. The single-step strategy is, however, not as efficient at generating entry clones as the traditional BP cloning scheme (Liang, 2013).
The In-Fusion® system is advertised to allow a seamless, directional construction of any PCR fragment(s) into your desired destination vector in a single reaction. Clontech’s manual and cloning scheme does not include an entry clone generation step, and the only requirements is a 15 bp homologous region between your DNA sequence and a linearized destination vector. Clever strategies have been devised to insert genes into many expression vectors in parallel using the In-Fusion® system (Bird, 2011). However, with the entry clone step being bypassed, to move the DNA sequence subsequently into a new expression vector requires cloning from the beginning of the pipeline starting at the generation of the PCR product.
Modification for use with other vectors
The Gateway® recombinational cloning technology platforms provide for various forms of expression systems (e.g., bacterial, mammalian, insect, and in vitro transcription/translation). In many cases, however, users may want to modify their own expression vectors into recombination-compatible systems (Figs. 3.20.6). For this purpose, Life Technologies retails a vector conversion system (https://www.lifetechnologies.com/order/catalog/product/11828029) to provide a way to convert other expression vectors into recombination-compatible ones by simply inserting the recombination cassette using PCR and restriction-mediated cloning steps. Refer to standard cloning manuals or to the Gateway® conversion manuals for more details. For the In-Fusion® and Gibson® cloning schemes, the cloning vectors do not have to be modified or converted for recombinational cloning purposes.
Critical Parameters and Troubleshooting
Amplification of target DNA
Primer mapping
When performing agarose gel electrophoresis in handling multiple genes or large cloning projects using a 96-well plate format, it is helpful if the resulting gel bands form a saw tooth pattern in which the expected fragments progressively increase in size across the gel lanes, but alternate in size to avoid lane-to-lane contamination (Fig. 3.20.8). This effect is most easily planned at the stage of primer design, so that the genes are amplified in an order that will result in this pattern.
DNA polymerase
Fidelity and PCR success rate (% of genes with positive PCR product) are crucial parameters in high-throughput PCR cloning. The choice of polymerase has a dramatic effect on the success rate. In the authors' experience, Phusion® (NEB) and Pfx (Life Technologies) DNA polymerases were comparable (UNIT 3.5) and gave the highest success rates in cloning up to 4 kbp from human brain and placenta first-strand cDNA mix. In addition, Herculase II Fusion (Aligent Technologies), GC-rich polymerase (Roche), and Elongase® (Life Technologies) were successfully used in other systems (Walhout et al., 2000; Reboul et al., 2001). Running pilot experiments on small subsets of clones is recommended before executing a large-scale experiment in order to adjust reaction parameters, e.g., the amount of polymerase, melting temperature, cycle number, and GC content of the primers.
Template
PCR reactions using plasmid template are highly efficient. As little as 200 ng of template plasmid DNA is enough to reduce the number of PCR cycles and the rate of mutations. When the plasmid form for the target gene is not available, the cDNAs must be amplified from cDNA libraries, first-strand cDNA, or genomic DNA. Genomic DNA is an excellent source for a simple organism (i.e., one that lacks RNA splicing) because it is essentially a normalized library where each gene is represented equally. For eukaryotes, the authors have found increased success rates from first-strand cDNA with highest rates in the brain and placenta first-strand cDNA mixture (1:4) for general human gene cloning purposes; however, for cloning certain cell- or tissue-type-specific genes, use of template DNA generated from those cells or tissues is recommended.
Generation of entry clones
Pilot experiments
The concentration of purified PCR product can vary from gene to gene, especially in large-scale projects, and can influence the success rate of generating entry clones, even though all were successfully amplified. Consequently, running a pilot experiment with a small number of genes is strongly recommended, using positive and negative controls for estimating the range of PCR product concentration against the fixed entry vector concentration.
Transformation
The efficiency of transformation into competent E. coli cells is critical, especially in dealing with large-scale projects in a 96-well format. Transformation efficiencies of >1 × 108 cfu/µg DNA are recommended. For high-throughput projects LB agar can be prepared in 48-well grid square bioassay trays (Genetix), and the bacterial colonies can be automatically picked using robotics.
Selection
Because the recombination reaction does not go to completion, a mixture containing the starting plasmids, one or more intermediates, the desired product, and one or more byproducts is obtained. Thus, stringent antibiotic selection conditions are required to select only the desired recombinant containing the target gene in its correct orientation.
Verification
Enzymatic amplification using PCR can introduce unwanted random mutations in the DNA sequence. In addition, primer mutations can be introduced during synthesis of the oligonucleotide primers. The enzymatic contribution to this mutation rate can be decreased by reducing the number of amplification cycles. Nevertheless, each entry clone should be sequenced in its full length to verify the correct sequence of the target gene and the recombination site has been obtained. Once the entry clone is verified, we recommend a single sequence read for confirmation of subsequent expression clones.
Generation of expression clones
Transformation
As in generation of the entry clone, the efficiency of transformation into competent E. coli cells is critical in dealing with large-scale projects in a 96-well format. Bacterial competent cells with transformation efficiencies of ≥1 × 108 cfu/µg DNA are recommended for the Gateway® and In-Fusion® systems pipelines.
Pilot experiments
Unusual recombination events occasionally occur, especially in retroviral expression vectors. It is advisable to confirm that the expression clones harbor correct inserts in proper orientation by analyzing plasmid DNA of some expression clones using specific restriction enzymes in a pilot experiment.
Anticipated Results
Amplification of target DNA
Utilizing either plasmid DNA or genomic DNA (especially prokaryotic cells) as a template, PCR should amplify only the gene of interest, and the overall success rate is >95%. In the case of first-strand cDNA, the PCR success rate is ~70% because of the complexity and relatively low representation of each gene. Upon electrophoresis of first-strand PCR product the authors frequently observe a few bands in addition to those corresponding to the desired products, most likely because of the complexity of the mixture. Purification of the desired products alleviates that problem except in cases where the product bands are smears. In those cases PCR conditions should be optimized again.
Generation of entry clones
In the authors' experience, a >95% success rate for generating entry clones from plasmid DNA can be expected (with the screening of four selected colonies; 80% success with screening one colony), but with first-strand cDNA the situation is more complicated. Generally, upon electrophoresis, the PCR product band is faint compared to those from plasmid DNA template because of a lower DNA concentration. This is a result of the lower success rate in PCR steps (~70%) for first-strand DNA templates. Moreover, after DNA sequencing and analysis, more clones may be eliminated for the first-strand cDNA template because of more random mutations introduced during higher numbers of PCR cycles required as a result of the more complex and heterogeneous DNA template. Overall, based on the authors' experience, a >50% success rate is expected from a first-strand cDNA cloning scheme, compared to >95% overall success rate using plasmid DNA as template.
Generation of expression clones
The transfer reaction is robust, and a ~95% success rate is expected. The few ineffective transfers should be easily rescuable after an additional round of transfer reaction. Their failure is most often due to random losses during plasmid DNA preparation in the 96-well format. Using highly competent cells (2 × 108 cfu/µg DNA competency), at least fifty colonies in a 10-cm petri dish are normally obtained from a standard bacterial transformation protocol.
Because the entire process of transferring inserts into expression vectors is very robust, it is easily automated. In addition, because the reaction does not involve any amplification or rearrangements, if the insert DNA sequence has been fully verified and is in-frame with the recombination sites, only a conformational single read resequence is needed after constructing expression clones.
Any functioning expression vector can be converted to accept inserts in the Gateway® or In-Fusion® systems. Attention must be paid when designing primers to ensure that all fusion proteins will be in-frame. The expression vector should not use the same antibiotic marker as that of the entry vector.
Time Considerations
The basic procedures can be completed in ~1 week with activities broken down as follows: 1 day for PCR and DNA purification, 1 day for capture reactions and transformation, 1 day for colony picking and DNA minipreps, 1 to 2 days for sequencing, 1 day for transfer of ORF into expression vector, and 1 to 2 days for DNA preparation and analysis.
Acknowledgements
This work was funded by the National Institute of General Medical Sciences (NIGMS; U01 GM098912 to J.L.).
Footnotes
Internet Resources
http://www.lifetechnologies.com/us/en/home/life-science/cloning/gateway-cloning.html Highly recommended website for more information and troubleshooting related to Life Technologies Gateway® Technology.
http://www.clontech.com/US/Products/Cloning_and_Competent_Cells/Cloning_Kits/Cloning_Kits-HD-Liquid
Highly recommended website for more information and troubleshooting related to Clontech In-Fusion® Technology.
https://www.neb.com/products/e5510-gibson-assembly-cloning-kit
Highly recommended website for more information related to NEB Gipson Assembly Cloning kit.
Literature Cited
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- Bird LE. High throughput construction and small scale expression screening of multi-tag vectors in Escherichia coli. Methods. 2011;55:29–37. doi: 10.1016/j.ymeth.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Brizuela L, Richardson A, Marsischky G, LaBaer J. The FLEXGene repository: Exploiting the fruits of the genome projects by creating a needed resource to face the challenges of the post-genomic era. Arch. Med. Res. 2002;33:318–324. doi: 10.1016/s0188-4409(02)00372-7. [DOI] [PubMed] [Google Scholar]
- Cantor EJ, Chong S. Intein-mediated rapid purification of cre recombinase. Protein Expression and Purification. 2001;22:135–140. doi: 10.1006/prep.2001.1428. [DOI] [PubMed] [Google Scholar]
- Ceroni A, Sibani S, Baiker A, Pothineni VR, Bailer SM, LaBaer J, Hass J, Campbell CJ. Systematic analysis of the IgG antibody immune response against varicella zoster virus (VSV) using a self-assembled protein microarray. Mol. BioSyst. 2010;6:1604–1610. doi: 10.1039/c003798b. [DOI] [PubMed] [Google Scholar]
- Festa F, Steel J, Bian X, LaBaer J. High-throughput cloning and expression library creation for functional proteomics. Proteomics. 2013;13:1381–1399. doi: 10.1002/pmic.201200456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson DG, Young L, Chuang R, Venter JC, Hutchison CA, Hamilton OS. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hamilton MD, Nuara AA, Gammon DB, Buller RM, Evans DH. Duplex strand joining reactions catalyzed by vaccinia virus DNA polymerase. Nuc. Acid. Res. 2006;35:143–151. doi: 10.1093/nar/gkl1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin CR, Farmer A, Willer DO, Evans DH. In-Fusion® cloning with vaccinia virus DNA polyermase. In: Isaacs SN, editor. Vaccinia Virus and Poxvirology: Methods and Protocols, Vol. 890: Methods in Molecular Biology. New York, New York: 2012. pp. 23–35. [DOI] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Liang X, Peng L, Baek C, Katzen R. Single Step BP/LR combined Gateway® Reaction. Biotechniques. 2013;5:265–268. doi: 10.2144/000114101. [DOI] [PubMed] [Google Scholar]
- Marsischky G, LaBaer J. Many paths to many clones: A comparative look at high-throughput cloning methods. Genome Res. 2004;14:2020–2028. doi: 10.1101/gr.2528804. [DOI] [PubMed] [Google Scholar]
- Murthy T, Rolfs A, Hu Y, Shi, Zhenwei, Raphael J, Moreira D, Kelley F, McCarron S, Jepson D, Taycher E, Zuo D, Mohr SE, Fernadez M, Brizuela L, LaBaer J. A full-genomic sequence-verified protein-coding collection for Francisella tularensis. PLoS One. 2007;2(6):e577. doi: 10.1371/journal.pone.0000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA. Integration and excision of bacteriophage lambda. Curr. Top. Microbiol. Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Nash HA, Robertson CA. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J. Biol. Chem. 1981;256:9246–9253. [PubMed] [Google Scholar]
- Ptashne M. A Genetic Switch: Phage (Lamda) and Higher Organisms. Cambridge, MA: Cell Press; 1992. [Google Scholar]
- Reboul J, Vaglio P, Tzellas N, Thierry-Mieg N, Moore T, Jackson C, Shin-I T, Kohara Y, Thierry-Mieg D, Thierry-Mieg J, Lee H, Hitti J, Doucette-Stamm L, Hartley JL, Temple GF, Brasch MA, Vandenhaute J, Lamesch PE, Hill DE, Vidal M. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, Moore T, Hudson JR, Jr, Hartley JL, Brasch MA, Vandenhaute J, Boulton S, Endress GA, Jenna S, Chevet E, Papasotiropoulos V, Tolias PP, Ptacek J, Snyder M, Huang R, Chance MR, Lee H, Doucette-Stamm L, Hill DE, Vidal M. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Rolfs A, Montor WR, Yoon SS, Hu Y, Bhullar B, Kelley F, McCarron S, Jepson DA, Shen B, Taycher E, Mohr SE, Zuo D, Williamson J, Mekalanos J, LaBaer J. Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae. PNAS. 2008;105:4364–4369. doi: 10.1073/pnas.0712049105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfs A, Hu Y, Ebert L, Hoffman D, Zuo D, Ramachandran N, Raphael J, Kelley F, McCarron S, Jepson DA, Shen B, Baqui MMA, Pearlberg J, Taycher E, DeLoughery C, Hoerlein A, Korn B, LaBaer J. A biomedically enriched collection of 7000 human ORF clones. PLoS One. 2008 doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. PNAS. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul J, Petritis B, Sau S, Rauf F, Gaskin M, Ober-Reynolds B, Mineyev I, Magee M, Chaput J, Qiu j, LaBaer J. Development of full-length human protein production pipeline. Protein Sci. 2014 doi: 10.1002/pro.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawastien A, Montor WR, LaBaer J, Mekalanos JJ, Yoon SS. Vibrio cholera proteome-wide screen for immunostimulatory proteins identifies phosphatidylserine decarboxylase as a novel toll-like receptor 4 agonist. PLoS Path. 2009 doi: 10.1371/journal.ppat.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Weisberg RA, Landy A. Site-Specific Recombination in Phage Lamda. In: Weisberg RA, editor. Lamda II. Cold Spring Harbor, NY.: Cold Spring Harbor Press; 1983. pp. 211–250. [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, Pham P, Cheuk R, Karlin-Newmann G, Liu SX, Lam B, Sakano H, Wu T, Yu G, Miranda M, Quach HL, Tripp M, Chang CH, Lee JM, Toriumi M, Chan MM, Tang CC, Onodera CS, Deng JM, Akiyama K, Ansari Y, Arakawa T, Banh J, Banno F, Bowser L, Brooks S, Carninci P, Chao Q, Choy N, Enju A, Goldsmith AD, Gurjal M, Hansen NF, Hayashizaki Y, Johnson-Hopson C, Hsuan VW, Iida K, Karnes M, Khan S, Koesema E, Ishida J, Jiang PX, Jones T, Kawai J, Kamiya A, Meyers C, Nakajima M, Narusaka M, Seki M, Sakurai T, Satou M, Tamse R, Vaysberg M, Wallender EK, Wong C, Yamamura Y, Yuan S, Shinozaki K, Davis RW, Theologis A, Ecker JR. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yu X, Bian X, Throop A, Song L, Del Moral L, Park J, Seiler C, Fiacco M, Steel J, Hunter P, Saul J, Wang J, Qiu J, Pipas JM, LaBaer J. Exploration of Panviral proteome: high-throughput cloning and functional implications in virus-host interactions. Theranostics. 2014;8:808–822. doi: 10.7150/thno.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Cai G, Hall EO, Freeman GJ. In-Fusion® ™ assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. BioTechniques. 2007;43:34–359. doi: 10.2144/000112536. [DOI] [PubMed] [Google Scholar]