Abstract
The Pacific white shrimp Litopenaeus vannamei is a euryhaline penaeid species that shows ontogenetic adaptations to salinity, with its larvae inhabiting oceanic environments and postlarvae and juveniles inhabiting estuaries and lagoons. Ontogenetic adaptations to salinity manifest in L. vannamei through strong hyper-osmoregulatory and hypo-osmoregulatory patterns and an ability to tolerate extremely low salinity levels. To understand this adaptive mechanism to salinity stress, RNA-seq was used to compare the transcriptomic response of L. vannamei to changes in salinity from 30 (control) to 3 practical salinity units (psu) for 8 weeks. In total, 26,034 genes were obtained from the hepatopancreas tissue of L. vannamei using the Illumina HiSeq 2000 system, and 855 genes showed significant changes in expression under salinity stress. Eighteen top Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were significantly involved in physiological responses, particularly in lipid metabolism, including fatty-acid biosynthesis, arachidonic acid metabolism and glycosphingolipid and glycosaminoglycan metabolism. Lipids or fatty acids can reduce osmotic stress in L. vannamei by providing additional energy or changing the membrane structure to allow osmoregulation in relevant organs, such as the gills. Steroid hormone biosynthesis and the phosphonate and phosphinate metabolism pathways were also involved in the adaptation of L. vannamei to low salinity, and the differential expression patterns of 20 randomly selected genes were validated by quantitative real-time PCR (qPCR). This study is the first report on the long-term adaptive transcriptomic response of L. vannamei to low salinity, and the results will further our understanding of the mechanisms underlying osmoregulation in euryhaline crustaceans.
Introduction
Salinity is one of the main environmental factors that exert a selection pressure on aquatic organisms, and variations in ambient salinity can directly impact the composition and osmolality of body fluids in aquatic animals [1]. Aquatic crustaceans inhabit environments with varying salinities, such as freshwater to seawater, and a change of environment requires crustaceans to regulate hemolymph osmolytes via osmoregulation [2, 3]. Crustaceans display several patterns of osmoregulation, including osmoconformation, hyper-osmoregulation and hypo-osmoregulation [2, 4]. Studies have shown that salinity stress can reduce salt diffusion between hemolymph and the environment because water is absorbed from the medium, which leads to swollen cells [5]. When confronted with salinity stress, aquatic animals are forced to osmoregulate by altering the expression of various enzymes and transporters, and the physiological adaptations associated with such functional changes are energy intensive [6]. Therefore, it is important to determine the amount of energy that is required during adaptations to different salinities. In addition, although the biochemical osmoregulation mechanisms of crustaceans have been studied [1, 3, 4], the molecular adaptive mechanisms for energy mobilization are not known.
The Pacific white shrimp Litopenaeus vannamei is a typical euryhaline crustacean species that lives in coastal and oceanic environments, and its larvae develop in the ocean, whereas the postlarvae, juveniles and adults live in estuaries and lagoons [7]. A hyper-hypo-osmoregulation process exists in the life history of L. vannamei, and an adaptive mechanism must exist to cope with the environmental salinity fluctuation or long-term low salinity stress. Therefore, L. vannamei can serve as an animal model in the study of adaptive mechanisms in euryhaline crustacean to changes in salinity. Because L. vannamei is an important commercial penaeid species in inland aquaculture at low salinity, extensive research has been conducted on its osmoregulation capabilities. However, inconsistent results have been found in the literature regarding the iso-osmotic point for growth and survival [8–11], [12], immune ability [13], and stress resistance [12, 14].
To understand the molecular mechanism underlying salinity adaptation in L. vannamei, various genes have been cloned, including those from the gill for ion transport [15], sarco/endoplasmic reticulum Ca2+-ATPase [16], glutamate dehydratase [17,18], hyperglycemic hormone [19,20] and molt-inhibiting hormone [20]. Moreover, suppression-subtractive hybridization has been used to identify genes and pathways in juvenile L. vannamei that have been exposed to long-term low salinity. However, the most common genes in these libraries are immune-related proteins and enzymes [21], whereas genes or pathways related to energy metabolism have not been found.
Whole-transcriptome shotgun sequencing, which is known as RNA sequencing (RNA-seq) [22], has been employed to reveal a snapshot of the transcriptome [23] that can be used to capture and annotate the transcriptome [24], analyze digital gene expression in hemocytes to gain knowledge on the immune response [25] and discover novel transcribed regions in the genomes of aquatic animals [24,26,27]. To understand the complex molecular mechanism of a specific physiological process, RNA-seq is a practical and efficient method of determining nearly all of the genes and pathways involved in a corresponding physiological function [28–30]. In this study, we aimed to compare the transcriptomic response of L. vannamei to low-salinity stress and reveal the pathways and genes involved in the process of salinity adaption. The results of this study will provide a foundation for understanding the mechanism of osmoregulation in euryhaline crustacean species.
Results
Sequencing, de novo assembly and differential expression genes
We obtained 97.1 million reads and 98.1 billion nucleotides from the shrimp at both 3 and 30 practical salinity units (psu) (Table 1). After quality trimming and adapter clipping, 93.8 million reads accounting for 96.6% of the total reads were obtained. In addition, 26,034 genes and 38,237 unigenes with an average length of 1,610 bp were obtained by de novo assembly using Trinity software after splicing and removing redundancy. Among 26034 genes, 855 genes were significantly up or down regulated, and all the differential expression genes were show in S1 Table. A total of 2,341 unigenes were down-regulated, and 2,363 unigenes were up-regulated in low salinity with the absolute fold change >2. Among the unigenes, the largest and smallest unigenes were 24,554 bp and 351 bp, respectively (Table 2); the length distribution of unigenes is shown in S1 Fig. To assess the quality and coverage of the transcriptome data, we mapped the assembly unigenes by using Bowtie software, and the mapping data accounted for over 90% of the data, suggesting that the transcriptome dataset provided good gene coverage and enriched the transcriptome information of L. vannamei in the present study.
Table 1. Summary of Illumina-expressed short read production and filtered transcriptomic responses to salinity stress in Litopenaeus vannamei.
| Salinity | Reads | Nucleotides | Q20 (%) | Q30 (%) |
|---|---|---|---|---|
| 3 psu | 48,338,820 | 4,882,220,820 | 96.50 | 91.18 |
| 30 psu | 48,767,738 | 4,925,541,538 | 96.68 | 91.51 |
| Trimmed | ||||
| 3 psu | 46,640,504 | 4,599,887,662 | 99.07 | 94.49 |
| 30 psu | 47,118,402 | 4,653,045,098 | 99.09 | 94.64 |
Note: Q20 indicates that every 100 bp of sequencing reads will have an error, and Q30 indicates that every 1000 bp of sequencing reads will have an error.
Table 2. Summary of de novo assembly results of the transcriptomic responses to salinity stress in Litopenaeus vannamei.
| Type | Number |
|---|---|
| Total genes | 26034 |
| Total unigenes | 38237 |
| Total residues | 61573030 bp |
| Average length | 1610.3 bp |
| Largest unigene | 24554 bp |
| Smallest unigene | 351 bp |
Annotation of unigenes
The predicted sequences (predicted open reading frame nucleotide sequences) and unpredictable sequences (unpredictable nucleotide sequences) were annotated using BLASTp and BLASTx, respectively, and then blasted to protein databases, including the nonredundancy (NR), STRING, COG and KOG databases (BLAST 2.2.28+, E-value < 1e—5) (Table 3). Among the annotated predicted sequences, a total of 17,232 (76.83%), 6298 (28.08%), 3720 (16.58%) and 302 (1.35%) sequences were unambiguous alignments relative to the reference when BLASTx was used against the NR, STRING, KOG, and COG databases, respectively. However, among the unpredictable sequences, only 2,235 (14.14%), 746 (4.72%), 509 (3.22%), 313 (1.98%), 243 (1.54%), 128 (0.81%), and 56 (0.35%) of the 15,806 sequences were matched against the NR, gene ontology (GO), NT, STRING, KOG and COG databases, respectively.
Table 3. Summary of the annotations of Litopenaeus vannamei unigenes.
| Predicted sequences | Unpredictable sequences | |||
|---|---|---|---|---|
| Number | Ratio (%) | Number | Ratio (%) | |
| All genes | 22431 | 100 | 15806 | 100 |
| Annotated in NR | 17232 | 76.82 | 2235 | 14.14 |
| Annotated in NT | None | None | 509 | 3.22 |
| Annotated in GO | None | None | 746 | 4.72 |
| Annotated in string | 6298 | 28.08 | 313 | 1.98 |
| Annotated in COG | 3720 | 16.58 | 128 | 0.81 |
| Annotated in KOG | 5408 | 24.11 | 243 | 1.54 |
| Annotated in NOG | 302 | 1.35 | 56 | 0.35 |
After parsing the GO annotation output, a total of 8,779 unigenes were finally annotated with GO terms, with 50.02% annotated to biological processes, 28.93% annotated to cellular components and 21.05% annotated to molecular functions. The distribution of GO terms showed that cellular processes and metabolic processes were the well-represented terms among the biological processes. Cells and cell parts were significantly enriched in cellular components, and analytic activity and binding consisted of a large proportion of molecular functions (S2 Fig).
Clusters of orthologous groups (COGs) of proteins were determined to predict and classify the possible functions of unigenes. The COG annotation analysis showed that three types of function were obtained, including information storage and processing, cellular processes and signaling, and metabolic pathways. The hits from the COG analysis were functionally classified into 25 categories, and the most enriched terms were related to general functions and then transcription and signal transduction mechanisms (S3 Fig).
KEGG pathway analysis annotation and functional enrichment analysis of GO and KEGG pathways
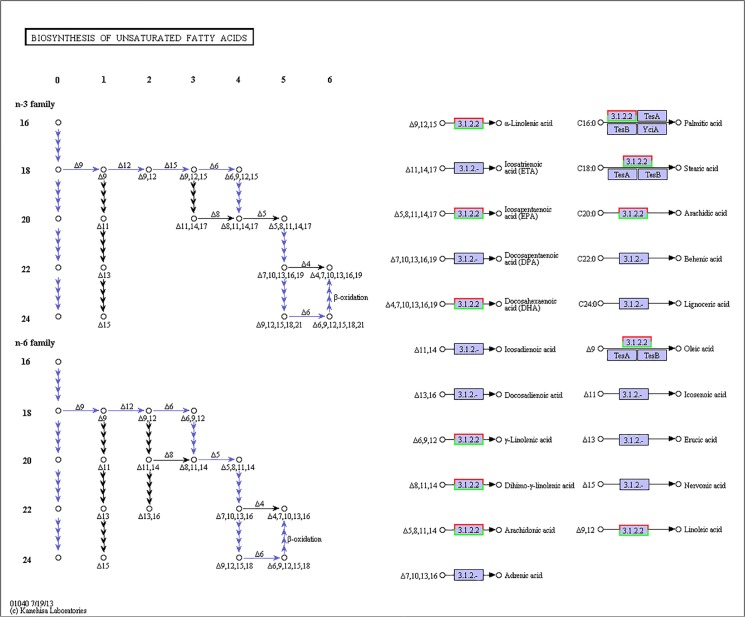
Various molecular pathways were obtained by Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation. A total of 9,621 unigenes were mapped onto 317 pathways, and the most enriched sequences were metabolic pathways, which was followed by the biosynthesis of secondary metabolites, spliceosome and RNA transport. The top 20 pathways with the greatest number of annotated sequences are shown in Table 4. A total of 47 significantly changed GO terms were obtained, and the most significant change was in molecular functions, which was followed by catalytic activity, histone H4 acetylation, structural constituents of cuticle and chitin binding. All of the significantly changed (P < 0.05) GO pathways are listed in Table 5. A KEGG pathway enrichment analysis was performed for the gene expression between salinity treatments to identify the number of significantly changed samples along the pathway that were relevant to the background number. The most significantly changed KEGG pathways were the glycosphingolipid biosynthesis, lysine degradation, glycosaminoglycan biosynthesis and malaria pathways. The gene recorded as B3GNT1,2 was significantly up-regulated both in the glycosphingolipid biosynthesis pathway and glycosaminoglycan biosynthesis pathway when shrimp were exposed to salinity at 3 psu (Figs 1 and 2). In addition, fatty-acid biosynthesis (Fig 3) was significantly enhanced, especially in short-carbon-chain fatty acids (C8-C18). In addition, low-salinity conditions enhanced polyunsaturated fatty-acid (PUFA) biosynthesis, especially that of highly unsaturated fatty acids such as ARA, EPA and DHA (Figs 4 and 5). All of the significantly changed (P < 0.05) KEGG pathways are listed in Table 6. These annotations provide valuable information for studying the specific biological and metabolic processes and functions and molecular mechanisms under salinity stress in L. vannamei.
Table 4. The top 20 pathways with the greatest number of annotated sequences.
| Pathway ID | Pathway definition | Number of sequences |
|---|---|---|
| path:ko01100 | Metabolic pathways | 1476 |
| path:ko01110 | Biosynthesis of secondary metabolites | 419 |
| path:ko03040 | Spliceosome | 278 |
| path:ko03013 | RNA transport | 260 |
| path:ko04144 | Endocytosis | 247 |
| path:ko05169 | Epstein-Barr virus infection | 245 |
| path:ko00230 | Purine metabolism | 236 |
| path:ko05205 | Proteoglycans in cancer | 232 |
| path:ko05200 | Pathways in cancer | 231 |
| path:ko01120 | Microbial metabolism in diverse environments | 227 |
| path:ko04142 | Lysosome | 219 |
| path:ko05166 | HTLV-I infection | 216 |
| path:ko04151 | PI3K-Akt signaling pathway | 216 |
| path:ko04510 | Focal adhesion | 215 |
| path:ko04141 | Protein processing in endoplasmic reticulum | 187 |
| path:ko05016 | Huntington's disease | 186 |
| path:ko04530 | Tight junction | 185 |
| path:ko05203 | Viral carcinogenesis | 180 |
| path:ko05168 | Herpes simplex infection | 180 |
| path:ko04120 | Ubiquitin mediated proteolysis | 172 |
Table 5. Significantly changed GO pathways of L. vannamei between the two salinities.
| GO ID | Pathway description | P value | Type |
|---|---|---|---|
| GO:0003674 | molecular function | 1.88E-06 | Molecular function |
| GO:0003824 | catalytic activity | 2.12E-06 | Molecular function |
| GO:0043967 | histone H4 acetylation | 0.000136 | Biological process |
| GO:0042302 | structural constituent of cuticle | 0.000223 | Molecular function |
| GO:0008061 | chitin binding | 0.000638 | Molecular function |
| GO:0097367 | carbohydrate derivative binding | 0.000884 | Molecular function |
| GO:0006030 | chitin metabolic process | 0.0016 | Biological process |
| GO:1901071 | glucosamine-containing compound metabolic process | 0.0021 | Biological process |
| GO:0006022 | aminoglycan metabolic process | 0.0041 | Biological process |
| GO:0016573 | histone acetylation | 0.0047 | Biological process |
| GO:0018393 | internal peptidyl-lysine acetylation | 0.0047 | Biological process |
| GO:0018394 | peptidyl-lysine acetylation | 0.0047 | Biological process |
| GO:0006475 | internal protein amino acid acetylation | 0.0061 | Biological process |
| GO:0000123 | histone acetyltransferase complex | 0.0075 | Cellular component |
| GO:0006473 | protein acetylation | 0.0079 | Biological process |
| GO:0043543 | protein acylation | 0.0129 | Biological process |
| GO:1901564 | organonitrogen compound metabolic process | 0.0139 | Biological process |
| GO:0018205 | peptidyl-lysine modification | 0.0139 | Biological process |
| GO:0006040 | amino sugar metabolic process | 0.0163 | Biological process |
| GO:0008152 | metabolic process | 0.0188 | Biological process |
| GO:0016491 | oxidoreductase activity | 0.0314 | Molecular function |
| GO:0005576 | extracellular region | 0.0476 | Cellular component |
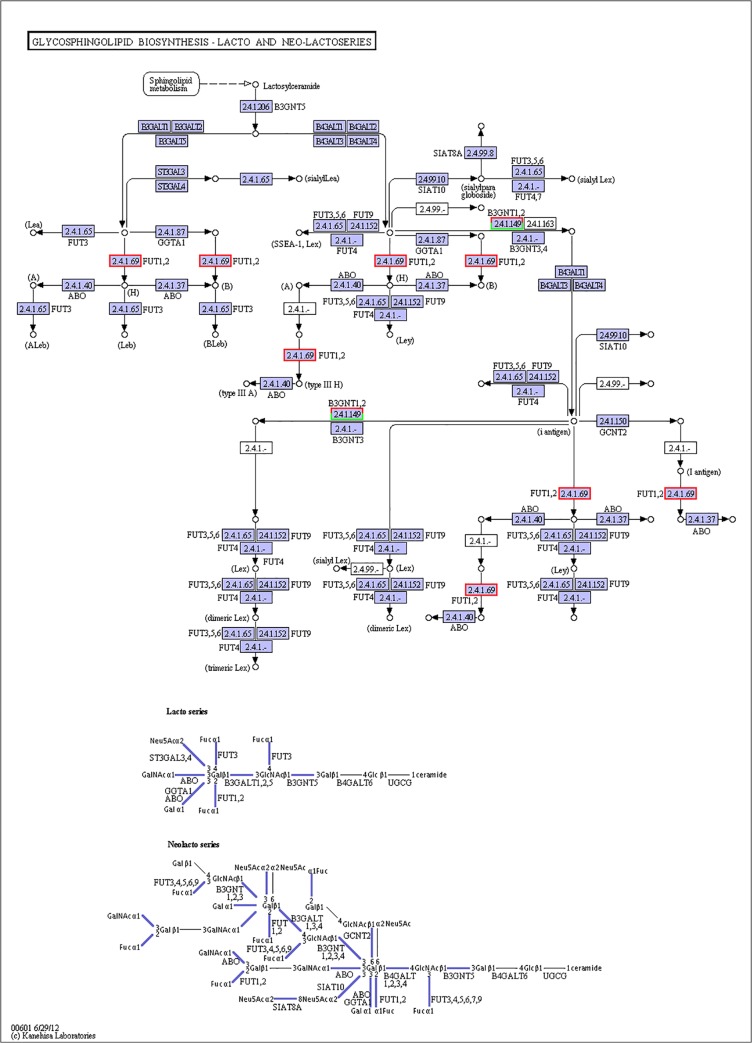
Fig 1. Pathways of glycosphingolipid biosynthesis: lacto and neolacto series (ko00601).
The red frames represent the genes were up-regulated, while the green frames represent that the genes were down-regulated. The frames with both red and green indicate that these genes have more than one unigenes, and some of them were up-regulated, but others were down-regulated.
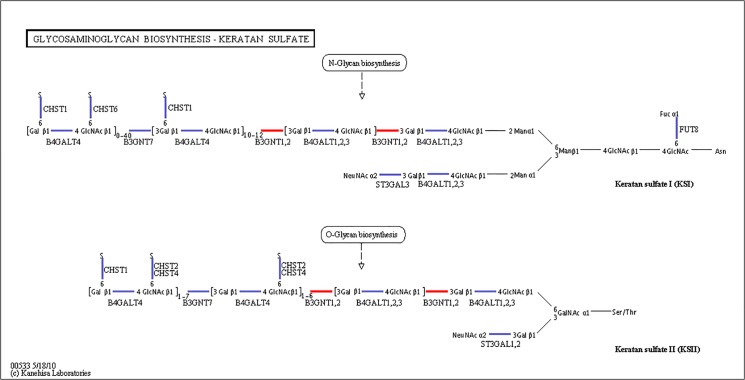
Fig 2. Pathways of glycosaminoglycan biosynthesis: keratan sulfate (ko00533).
The red line indicates that the genes were up-regulated.
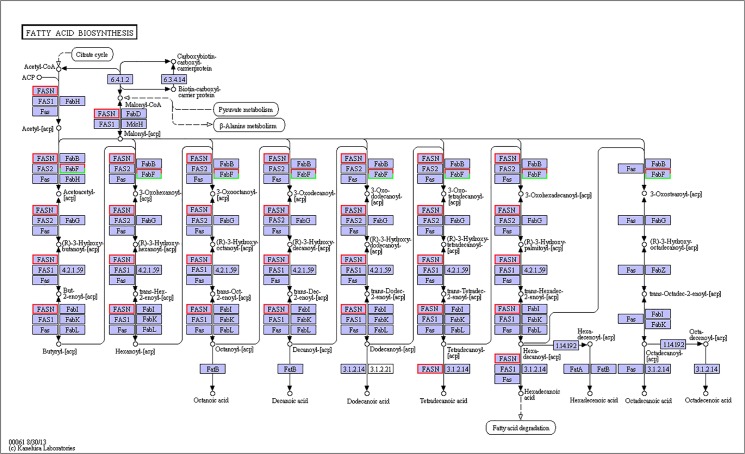
Fig 3. Pathways of fatty-acid biosynthesis (ko00061).
The red frames represent the genes were up-regulated, while the green frames represent that the genes were down-regulated. The frames with both red and green indicate that these genes have more than one unigenes, and some of them were up-regulated, but others were down-regulated.
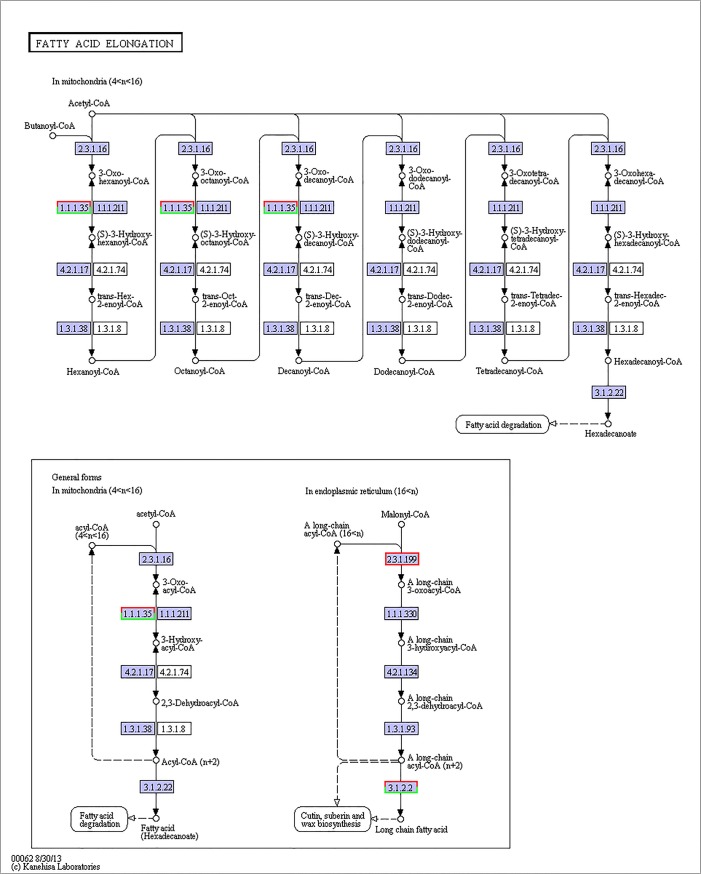
Fig 4. Pathways of fatty-acid elongation (ko00062).
The red frames represent the genes were up-regulated, while the green frames represent that the genes were down-regulated. The frames had both red and green indicated that these genes had more than one unigenes, and some of them were up-regulated, others were down-regulated.
Fig 5. Pathways of unsaturated fatty acid biosynthesis (ko01040).
The red frames represent the genes were up-regulated, while the green frames represent that the genes were down-regulated. The frames with both red and green indicate that these genes had more than one unigenes, and some of them were up-regulated, but others were down-regulated.
Table 6. Significantly changed KEGG pathways of L. vannamei between the two tested salinities.
| Pathway description | KEGG ID | Sample number | Background number | P-value |
|---|---|---|---|---|
| Glycosphingolipid biosynthesis—lacto and neolacto series | ko00601 | 7 | 16 | 0.001 |
| Lysine degradation | ko00310 | 23 | 102 | 0.002 |
| Glycosaminoglycan biosynthesis—keratan sulfate | ko00533 | 7 | 17 | 0.002 |
| Malaria | ko05144 | 11 | 41 | 0.007 |
| Phosphonate and phosphinate metabolism | ko00440 | 4 | 8 | 0.009 |
| Glycerophospholipid metabolism | ko00564 | 20 | 98 | 0.011 |
| Steroid hormone biosynthesis | ko00140 | 8 | 29 | 0.017 |
| Glycosaminoglycan degradation | ko00531 | 11 | 48 | 0.023 |
| Adipocytokine signaling pathway | ko04920 | 14 | 67 | 0.025 |
| Fatty-acid biosynthesis | ko00061 | 3 | 6 | 0.026 |
| Synthesis and degradation of ketone bodies | ko00072 | 8 | 32 | 0.031 |
| Ether lipid metabolism | ko00565 | 9 | 38 | 0.031 |
| Drug metabolism—cytochrome P450 | ko00982 | 13 | 63 | 0.033 |
| Drug metabolism—other enzymes | ko00983 | 15 | 76 | 0.033 |
| TGF-beta signaling pathway | ko04350 | 12 | 57 | 0.034 |
| Arachidonic acid metabolism | ko00590 | 14 | 70 | 0.035 |
| Metabolism of xenobiotics by cytochrome P450 | ko00980 | 14 | 71 | 0.039 |
| Cholinergic synapse | ko04725 | 14 | 71 | 0.039 |
Note: The sample number means differently expressed gene number, and the background number means the total gene number in the pathway.
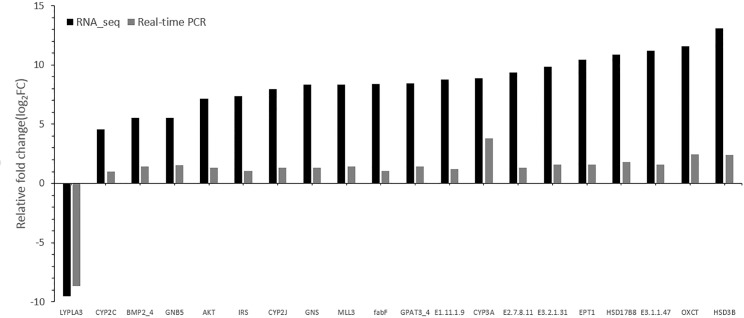
Validation of RNA-seq profile results by qPCR
Twenty randomly selected genes were measured in the same hepatopancreas RNA samples by qPCR, and the expression levels of these genes were significantly associated with the RNA-seq results (R = 0.91, Fig 6). These results confirm the reliability of RNA-seq and accuracy of the Trinity assembly.
Fig 6. Validation results of RNA-seq profiles by qPCR.
Discussion
Osmoregulation in crustaceans is a complex process because of the diverse range of salinities that they are exposed to in their natural habitat. High-throughput RNA-seq is a good method in determining the underlying molecular mechanisms of osmoregulation [31]. In addition, transcriptome analysis has been used in the discovery of single nucleotide polymorphisms (SNPs) [32] and identification of immune gene [33] and genes and pathways responsible for various pathogen challenges [34–37] in L. vannamei. In the present study, we used RNA-seq on L. vannamei exposed to low salinity to determine the metabolic pathways and perform expression profiling. We found that most of the metabolic activities that were significantly involved in adaptive responses were related to lipid metabolism. This study is the first of its type to report on the key energetic pathways relevant to osmoregulation in L. vannamei.
Relationship between lipid metabolism and response strategies to salinity stress
Shrimp gill is one of the main tissues for osmoregulation and icon exchange, and the polyunsaturated fatty acids synthesized in hepatopancreas are the main components of shrimp gill. Previous study has shown that under salinity stress, shrimp gill needs extra energy (most from saturated fatty acids) and polyunsaturated fatty acids to ensure the osmoregulation and icon exchange in L. vannamei [38]. In this study, a number of lipid metabolism pathways in the hepatopancreas of L. vannamei were influenced by salinity stress, including the fatty-acid biosynthesis pathway, arachidonic acid metabolism pathway (S4 Fig), adipocytokine signaling pathway (S5 Fig), glycerophospholipid metabolism pathway (S6 Fig), ether lipid metabolism pathway (S7 Fig) and ketone body synthesis and degradation pathway (S8 Fig). These pathways are found in the Chinese mitten crab Eriocheir sinensis under osmotic stress [31]. Lipids, including the fatty-acid structure, composition and metabolism, reduce osmotic shock in aquatic animals (Sui et al., 2007) by providing sufficient energy to maintain the ion balance and regulate the structure of biological membranes [6,39]. This process can directly or indirectly function in “compensatory processes” and “limiting processes," which are the two major osmoregulation strategies in crustaceans [4, 40].
In this study, most of the significant pathways related to salinity adaptations were associated with the above two major strategies. The limiting process is a strategy to adjust the permeability of the boundary structures to maintain hemolymph osmolality/ions in gill membranes, which is effective at reducing ion diffusion and water influx because the ion transport mechanism requires additional energy [4]. The ability to regulate gill permeability is an adaptive response that is crucial for decapod crustaceans because it facilitates long-term habitation in environments with variable salinity [4, 40]. Therefore, the cell membranes of the gill should play an important role in osmoregulation [41]. The “compensatory process” strategy is accomplished via the active exchange of solutes in hemolymph to counterbalance passive diffusion and maintain osmolality [4]. Because this process is energetically costly, a number of energy metabolism pathways and ion regulation pathways must be involved.
Energy from lipid metabolism to maintain the ion balance
Fatty-acid biosynthesis was significantly enhanced, especially for short-carbon-chain fatty acids (C8-C18), and arachidonic acid was also used to convert fatty acids or other metabolic products in the present study. When shrimp suffer from ambient salinity stress, additional energy is required through nutrient intake to maintain homeostasis by osmoregulation via the “compensatory process,” in which lipids play significant roles [42–45].
However, ketone bodies are indispensable for energy that is metabolized from fatty acids, which was revealed by the KEGG analysis in this study. Ketone bodies are produced by the liver from fatty acids during periods of low food intake or carbohydrate restriction. When carbohydrates are scarce, energy must be obtained from the breakdown of fatty acids from body tissue instead of glucose [46, 47]. Interestingly, in the adipocytokine signaling pathway, we found that the gene FACS functioned in long-chain fatty-acid biosynthesis and was significantly down-regulated under low-salinity stress. CPT-1 is another relevant gene for long-chain fatty-acid β-oxidation, and it was significantly down-regulated. Therefore, it is reasonable to assume that L. vannamei prefers to use shorter-chain fatty acids for energy supplementation and selectively retains longer-chain unsaturated fatty acids [39, 45]. The enhancement of saturated fatty acid biosynthesis in hepatopancreas (the main site for lipogenesis) would provide sufficient energy for osmoregulation in shrimp gill at low salinity, and this result is consistent with our previous findings in L. vannamei cultured at low salinity [38]. Penaeus monodon also prefers to use shorter-chain fatty acids in energy metabolism and selectively retains longer-chain unsaturated fatty acids [48].
Polyunsaturated fatty acids in osmoregulation of L. vannamei
In this study, fatty acid elongation (n > 18) was significantly enhanced, and the biosynthesis of α-linolenic acid, EPA, DHA, arachidic acid, arachidonic acid and other PUFAs was up-regulated at low salinity. Saturated fatty-acid biosynthesis was enhanced, and PUFA biosynthesis was also strengthened at low salinity. PUFAs can improve the resistance to osmotic shock in aquatic animals because PUFAs are mainly incorporated in cell membranes and can increase membrane permeability and fluidity [49, 50]. Free fatty acids, especially LC-PUFA, have the potential to modulate fatty-acid composition in the gill membrane and increase enzymatic efficiency [45, 51]. Modifications to the fatty-acid composition in the gills that increase the level of LC-PUFA can increase the gill area to enhance the osmoregulatory capacity in shrimp at low salinity, thereby increasing survival [39]. In addition, arachidonic acids in fish can enhance the branchial Na+/K+-ATPase activity and influence the ion balance [52]. Furthermore, arachidonic acid metabolites can influence the regulation of ion balances across the gill membrane [53].
Glycosphingolipid and glycosaminoglycan metabolism pathways in regulating membrane structure
In this study, the most influential pathways were associated with glycosphingolipid biosynthesis, glycosaminoglycan biosynthesis and glycosaminoglycan degradation, and they are all related to gill permeability. Glycosphingolipids (including lacto-/neolactoglycolipids and sphingolipids) function to protect the cell surface by maintaining the stability of the membrane or plasma membrane to protect against harmful environmental factors by forming a mechanically stable and chemically resistant outer leaflet for the plasma membrane lipid bilayer [54, 55]. Glycosaminoglycans are classified into four groups: keratan sulfate, heparin/heparan sulfate, chondroitin/dermatan sulfate and hyaluronic acid. One of the main functions of keratan sulfates is the maintenance of tissue hydration and implantation and migration of endothelial cell [56].
When shrimp were exposed to salinity of 3 psu, the B3GNT1,2 gene, which participates in both the glycosphingolipid biosynthesis pathway and glycosaminoglycan biosynthesis pathway, was significantly up-regulated. The FUT1_2 gene was also significantly up-regulated in the glycosphingolipid biosynthesis pathway. Thus, we believe that the genes B3GNT1,2 and FUT1_2 can promote glycolipid biosynthesis (Fig 1). Glycolipids constitute the lipid bilayer of the plasma membrane and play an important role in maintaining the physical state of the membrane [57]. Thus, glycolipid biosynthesis might promote the “limiting process” and provide resistance to low-salinity stress at 3 psu salinity. However, the effect of keratan sulfate and glycosaminoglycan degradation in low-salinity ambient osmoregulation is still not clear and requires further study.
Glycerophospholipids are glycerol-based phospholipids that are the main component of biological membranes [58], and their biosynthesis was significantly enhanced at low salinity (Fig 1). Although direct interactions were not observed between glycerophospholipid and osmoregulation in this study, glycerophospholipids could indirectly improve osmoregulation by affecting membrane permeability. Ether lipids are ubiquitous and constitute a major portion of the cell membranes in mammals [59]. In addition, these lipids play an important role in the generation of lipid second messenger systems, such as prostaglandins and arachidonic acid, which are important in signal transduction [60]. Ether lipids also act directly in cell signaling [61] and are involved in osmotic stress signal transduction. Another possible function of ether lipids is as an antioxidant against oxidative stress, which has been demonstrated in cell culture under salinity stress. Therefore, these lipids might play a role in serum lipoprotein metabolism [62] and lipid metabolism may play an important and indispensable role in osmoregulation [38].
Potential pathways in L. vannamei under chronic low salinity stress
Because of the complexity of the physiological response to low-salinity stress in L. vannamei, several pathways (in addition to lipid metabolism) show potential importance in the shrimp's ability to cope with salinity stress. However, clear evidence of the direct involvement of these pathways during salinity challenges has not been observed; thus, the putative functions of these pathways including lysine degradation, cholinergic synapse, drug metabolism pathway, steroid hormones metabolism pathway, phosphonate and phosphinate metabolism, are only briefly discussed.
Lysine is metabolized in eukaryotes to yield acetyl-CoA via lysine acetylation [63, 64]. Acetyl-CoA participates in osmoregulation as an intermediate metabolite commonly produced from energy metabolism, lipid metabolism and carbohydrate metabolism. In our results, the gene expression of 3-hydroxyacyl-CoA dehydrogenase, which can produce acetyl-CoA, was significantly up-regulated, resulting in more acetyl-CoA entering the citrate cycle for energy production. The above pathway could indirectly influence ion transfer or energy metabolism and promote “compensatory processes," which is consistent with reports on the Chinese mitten crab under salinity stress [31]. On the other hand, the choline in animal tissues is a primary component of neurotransmitter acetylcholine and functions with inositol as a basic constituent of lecithin [65]. Choline prevents the formation of fat deposits in the liver and facilitates the movement of fat into cells [66]. Thus, considering the result of our previous study [38], it seems that more fat may be used in supplying energy through β-oxidation.
Various physiological and pathological factors can affect drug metabolism, including age, individual variations, enterohepatic circulation, nutrition, intestinal flora and sex differences. Cytochrome P450 influences arachidonic acid metabolism [67, 68] and fatty-acid metabolism [69] and may have an indirect influence on osmoregulation by influencing the regulation of fatty-acid metabolism and other physiological and biochemical processes. However, the effect of drug metabolism on osmoregulation is still unknown and requires further study. Among the malaria pathways, we found that low-density lipoprotein receptor-related protein 1 (LRP1) was significantly changed when shrimp suffered from salinity stress. LRP1 might have an important role in low-density lipoproteins and thereby influence osmoregulation.
Steroid hormones help control metabolism, inflammation, immune functions and salt and water balance, influence sex characteristics and promote illness and injury prevention [70–72]. Moreover, Birukawa found that steroid hormones are involved in the osmoregulation of cetaceans [73]. Phosphonates are effective chelating agents that remain stable under harsh conditions. Phosphonates are also regularly used in reverse-osmosis systems [74]. However, the interaction or correlation between osmoregulation and phosphonates is still not clear and requires further study in aquatic animals.
Conclusion
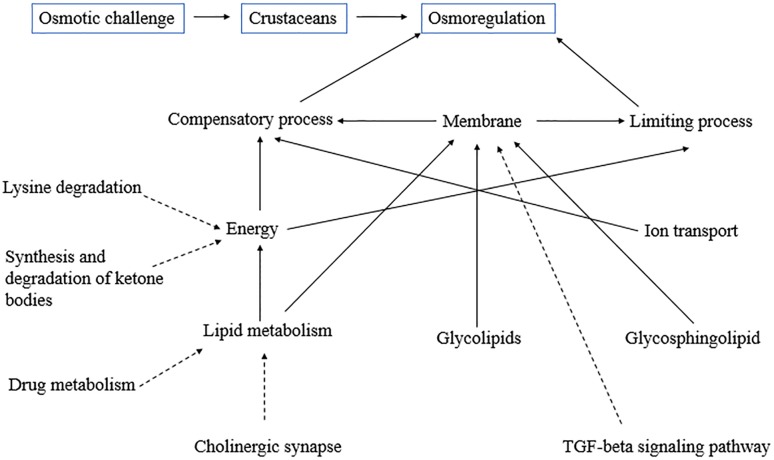
This study revealed that osmoregulation is a complex physiological adaptation that involves a number of pathways, especially the lipid metabolism pathway. When L. vannamei is subjected to an osmotic challenge at low salinity, shrimp can improve itself ability against osmotic stress by both “compensatory processes” and “limiting processes," which are the two major osmoregulatory strategies in crustaceans. These two osmoregulatory strategies may utilize various osmoregulatory mechanisms that are largely dependent on energy metabolism and cell membrane regulation (Fig 7). Not only will lipid metabolism supply sufficient energy to ensure the energy metabolism demand, but also many pathways (e.g. lysine degradation, cholinergic synapse, drug metabolism etc.) may indirect affect energy metabolism by generating some metabolic intermediates to indirectly influence the energy metabolism or affect lipid metabolism. On the other hand, other lipid metabolism pathways, including glycolipids and glycosphingolipid, are involved to improve osmoregulation capacity by increasing related enzymatic activity or changing gill permeability. This study provides some insights into the pathways involved in L. vannamei osmoregulation under low salinity. However, the mechanisms underlying osmoregulation in L. vannamei are complex and require further study, especially in relation to lipid metabolism and osmoregulation.
Fig 7. Relationship between the most influenced pathways and osmoregulation.
The dotted-line arrows are indirect effects, and solid-line arrows indicate direct influence.
Materials and Methods
Experimental animals, design and facilities
Juvenile white shrimp (1.98 ± 0.28 g) were obtained from the Shenzhen base of the South China Sea Fisheries Research Institute, Shenzhen, China, and they were stocked in six tanks at a density of 40 shrimp per tank (500 L) at 17 psu salinity for one week. The shrimp were acclimated to 3 psu and 30 psu in three tanks through daily increments of 2 psu prior to the start of the 8-week experiment. During the acclimation and experimental periods, the shrimp were fed a commercial diet (10% moisture, 40% crude protein, 8% crude lipid, 12% ash, 30% carbohydrates, 16.7 kJ g-1 digestible energy) three times daily at 08:00, 16:00 and 22:00 h. Based on the amount of feed left over from the previous day, daily rations were adjusted to a feeding level slightly over satiation. The unused feed was removed daily with a siphon tube. The photoperiod was 12 h light and 12 h dark. Seawater was pumped from the Dayawan Coast in Shenzhen and filtered through an activated carbon cartridge for at least 3 d before entering the culture system. The tap water was aerated before it was added to the tank to adjust the salinity level. During the experiment, water equal to 1/3 of the tank volume was exchanged once daily. The water quality parameters were monitored 2–3 times a week throughout the feeding trial and maintained at pH 7.5–7.9, temperature 26–28°C, dissolved oxygen 4.8–6.4 mg L-1, and total ammonia nitrogen < 0.02 mg L-1 during the trial.
At the end of the experiment, shrimp were fasted for 24 h before sampling. Five shrimp at the intermolt stage C in each tank were dissected to obtain the hepatopancreas tissue for the transcriptome analysis.
Eukaryote de novo transcriptome sequencing
Total RNA was extracted from the hepatopancreas using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and genomic DNA was removed using DNase I (TaKaRa). RNA quality was determined by a 2100 Bioanalyzer (Agilent), and the RNA was quantified using the NanoDrop 2000 spectrophotometer (ND-2000, Gene Company Ltd.). Only high-quality RNA samples (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, > 10 μg) were used to construct the sequencing library.
Library preparation and Illumina Hiseq2000 sequencing
The RNA-seq transcriptome library was prepared following the TruSeq RNA (Illumina, San Diego, CA) sample preparation instructions using 5 μg total RNA. Briefly, messenger RNA was isolated according to the polyA selection method by oligo beads [75] and then segmented (100 bp to 400 bp) by a fragmentation buffer. Next, double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) with random hexamer primers (Illumina). Then, the synthesized cDNA was subjected to end repair, phosphorylation and ‘A’ base addition according to Illumina’s library construction protocol. The libraries were size-selected for cDNA target fragments of 200–300 bp on 2% low-range ultra-agarose followed by PCR amplification using Phusion DNA polymerase (New England Biolabs) for 15 PCR cycles. After quantification by a TBS-380 fluorometer, the paired-end RNA-seq library was sequenced with the Illumina HiSeq 2000 system (2 × 100 bp read length). Raw reads were archived at the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accession No. SRP048814.
De novo assembly and annotation
The raw paired-end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Clean data from the samples were then used to perform RNA de novo assembly with the program Trinity (http://trinityrnaseq.sourceforge.net/) [76]. All of the assembled transcripts were searched against the NCBI protein NR, STRING and KEGG databases using BLASTx to identify the proteins that had the highest sequence similarity with the given transcripts to retrieve their function annotations, and a typical E-value cut-off was set at < 1.0×10−5. BLAST2GO (http://www.blast2 go.com/b2 ghome) [77] was used to obtain the GO annotations of uniquely assembled transcripts for describing biological processes, molecular functions and cellular components. A metabolic pathway analysis was performed using KEGG databases (http://www.genome.jp/kegg/) [78, 79].
Differential expression analysis and functional enrichment
To identify differentially expressed genes (DEGs) between two different samples, the expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FRKM) method. RSEM (http://deweylab.biostat.wisc.edu/rsem/) [80] was used to quantify gene and isoform abundances, and the R statistical package software EdgeR (empirical analysis of digital gene expression in R, http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) [81] was used for the differential expression analysis. A functional-enrichment analysis that KEGG was performed to identify the DEGs that were significantly enriched in GO terms and metabolic pathways (at a Bonferroni-corrected P-value 0.05) relative to the whole-transcriptome background. The GO functional enrichment and KEGG pathway analysis were conducted by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) [82].
Experimental validation of RNA-seq profiles by qPCR
Twenty randomly selected genes with significant expression in KEGG pathways were validated with quantitative real-time PCR (qPCR) and gene-specific primers designed using Primer Premier 6 (Table 7). Total RNA was extracted from the target hepatopancreas tissues using a TRIpure Reagent kit (Aidlab, RN01). Samples of polyadenylated RNA were reverse-transcribed using a TaKaRa kit (Cat. No. RR036A). The reactions were conducted in a total volume of 20 μl with the following reaction components: 2 μl 5X PrimeScript RT Master Mix (Perfect Real Time), 1 μg total RNA, and RNase-free dH2O up to 20 μl. The protocol for reverse transcription was 37°C for 15 min and 85°C for 5 s. The qPCR analysis was conducted in the CFX96 Real-Time PCR system (Bio-Rad Laboratories, Richmond, CA) using the Ultra SYBR Mixture (WCBIO, CW0957). The amplifications were performed in a 96-well plate in a 20 μl reaction volume containing 10 μl Ultra SYBR Mixture (WCBIO, CW0957), 0.4 μl each gene-specific forward and reverse primer, 8.4 μl RNase-free water and 0.8 μl cDNA. The thermal profile for the Ultra SYBR Mixture PCR analysis was 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The β-actin gene was used as the reference, and each gene had three replicated wells. Relative fold changes were calculated in the Relative Expression Software Tool version 2009 based on the cycle threshold values generated by qPCR [83].
Table 7. Primers used for the qPCR analysis.
| Gene name | Product size | Primer sequences (5’-3’) | |
|---|---|---|---|
| MLL3 | F | 90 | GACATCTCCTACCACATATACT |
| R | TTGACATACAGCACACCAT | ||
| LYPLA3 | F | 121 | TGGAACAGTCAACCTAAGAA |
| R | GTCAGAGTCACGCAAGAT | ||
| GPAT3_4 | F | 147 | TAGCAAGGAGATTACGAGAG |
| R | TGGCGACTGGATAGATGA | ||
| E2.7.8.11 | F | 132 | ATTCTCCGCATCTACTACAC |
| R | CCTCCAGAGTCCTATTCCA | ||
| EPT1 | F | 117 | ATGACCAAGAGCGAGATG |
| R | ACAGACACAATACAGGAGAT | ||
| HSD3B | F | 95 | CCAACACAATGCCTTCCT |
| R | CTTCCTCAGAGCCATGAC | ||
| HSD17B8 | F | 98 | AGAGAAGGAGCACGAGTG |
| R | CTTACCGCCAGATGATTATTG | ||
| CYP3A | F | 139 | TAGGCATCATAGGCAGGAA |
| R | TCTGGCAGGTTGTCTTCA | ||
| fabF | F | 120 | AGCCATCCTCACCATTCT |
| R | TATTCTTCTGTCCGCCATC | ||
| E3.1.1.47 | F | 145 | GAGCACAGAGACAGTTCC |
| R | CTGGCTGTTCCTGAGTTC | ||
| E1.11.1.9 | F | 119 | CTGAATGGCGTCCGTTAC |
| R | CGAAGTCTGTGTCTGTGTAT | ||
| CYP2J | F | 106 | TCCTACCAGCACAAGAGT |
| R | GCCAGGTAAGTGTCAGTC | ||
| IRS | F | 114 | AGAGGAGAGTGCCATATCA |
| R | ACCGCTGTTGTTAGTTGT | ||
| AKT | F | 118 | AGCACGAGACCTCCTTAG |
| R | CAGTTGATGGTGATGTAGAAG | ||
| GNS | F | 149 | GAGGACTCGTGGAACAAC |
| R | CGCTCTTCAGGTCATACAT | ||
| E3.2.1.31 | F | 129 | ATTCGCACTCTTGGATGG |
| R | CACTTGAGGAGGCTGAAG | ||
| OXCT | F | 126 | AACGGACGGAATTACATCA |
| R | GCACATTGGTAGGTTGAAG | ||
| CYP2C | F | 125 | CTGACGGCTCTGTATCTG |
| R | TGTGCTTGATGTGGTCTC | ||
| BMP2_4 | F | 122 | ACCAATACCTCGCTGATG |
| R | GATGTTCGTCACGTTGAAG | ||
| GNB5 | F | 106 | GCAGGATACAACGACTACA |
| R | GAGACATCTTCAGACAGGAG |
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by grants from the National Natural Science Foundation of China (No. 31472291, 31172422), the Special Fund for Agro-scientific Research in the Public Interest (No. 201003020, 201203065), National ‘Twelfth Five-Year’ Plan for Science & Technology Support (2012BAD25B03), the National Basic Research Program (973Program, No. 2014CB138803), Shanghai University Knowledge Service Platform Shanghai Ocean University aquatic animal breeding center (ZF1206), and partly by the E-Institute of Shanghai Municipal Education Commission (No. E03009) and ECNU innovation fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Charmantier G, Charmantier-Daures M (2001) Ontogeny of osmoregulation in crustaceans: the embryonic phase. American Zoologist 41: 1078–1089. [Google Scholar]
- 2. Charmantier G, Charmantier-Daures M, Towle D (2009) Osmotic and ionic regulation in aquatic arthropods Osmotic and Ionic Regulation Cells and Animals CRC press, Boca Raton, FL, New York, NY, Oxford, UK: 165–230. [Google Scholar]
- 3. Romano N, Zeng CS (2012) Osmoregulation in decapod crustaceans: implications to aquaculture productivity, methods for potential improvement and interactions with elevated ammonia exposure. Aquaculture 334: 12–23. [Google Scholar]
- 4. Pequeux A (1995) Osmotic regulation in crustaceans. Journal of Crustacean Biology: 1–60. [Google Scholar]
- 5. Davis DA, Saoud IP, McGraw WJ, Rouse DB (2002) Considerations for Litopenaeus vannamei reared in inland low salinity waters. Avances en Nutrición Acuícola VI Memorias del VI Simposium Internacional de Nutrición Acuícola 3. [Google Scholar]
- 6. Tseng YC, Hwang PP (2008) Some insights into energy metabolism for osmoregulation in fish. Comparative Biochemistry and Physiology C-Toxicology & Pharmacology 148: 419–429. [DOI] [PubMed] [Google Scholar]
- 7.FAO (2004) Manejo sanitario y mantenimiento de la bioseguridad de los laboratorios de postlarvas de camarón blanco (Penaeus vannamei) en América Latina. FAO Documento Técnico de Pesca 66.
- 8. Castille FL Jr, Lawrence AL (1981) The effect of salinity on the osmotic, sodium and chloride concentrations in the hemolymph of euryhaline shrimp of the genus Penaeus . Comparative Biochemistry and Physiology Part A: Physiology 68: 75–80. [Google Scholar]
- 9. Pan LQ, Zhang LJ, Liu HY (2007) Effects of salinity and pH on ion-transport enzyme activities, survival and growth of Litopenaeus vannamei postlarvae. Aquaculture 273: 711–720. [Google Scholar]
- 10. Diaz F, Farfan C, Sierra E, Re AD (2001) Effects of temperature and salinity fluctuation on the ammonium excretion and osmoregulation of juveniles of Penaeus vannamei, Boone. Marine & Freshwater Behaviour & Physiology 34: 93–104. [Google Scholar]
- 11. Buckle LF, Baron B, Hernandez M (2006) Osmoregulatory capacity of the shrimp Litopenaeus vannamei at different temperatures and salinities, and optimal culture environment. Revista de Biología Tropical 54: 745–753. [DOI] [PubMed] [Google Scholar]
- 12. Li EC, Chen LQ, Zeng C, Chen XM, Yu N, Lai QM, et al. (2007) Growth, body composition, respiration and ambient ammonia nitrogen tolerance of the juvenile white shrimp, Litopenaeus vannamei, at different salinities. Aquaculture 265: 385–390. [Google Scholar]
- 13. Ponce-Palafox J, Martinez-Palacios CA, Ross LG (1997) The effects of salinity and temperature on the growth and survival rates of juvenile white shrimp, Penaeus vannamei, Boone, 1931. Aquaculture 157: 107–115. [Google Scholar]
- 14. Li EC, Chen LQ, Zeng C, Yu N, Xiong ZQ, Chen XM, et al. (2008) Comparison of digestive and antioxidant enzymes activities, haemolymph oxyhemocyanin contents and hepatopancreas histology of white shrimp, Litopenaeus vannamei, at various salinities. Aquaculture 274: 80–86. [Google Scholar]
- 15. Pan LQ, Liu HY, Zhao Q (2014) Effect of salinity on the biosynthesis of amines in Litopenaeus vannamei and the expression of gill related ion transporter genes. Journal of Ocean University of China 13: 453–459. [Google Scholar]
- 16. Wang YH, Luo P, Zhang LP, Hu CQ, Ren CH, Xia JJ (2013) Cloning of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) gene from white shrimp, Litopenaeus vannamei and its expression level analysis under salinity stress. Molecular Biology Reports 40: 6213–6221. 10.1007/s11033-013-2733-x [DOI] [PubMed] [Google Scholar]
- 17. Li EC, Arena L, Lizama G, Gaxiola G, Cuzon G, Rosas C, et al. (2011) Glutamate dehydrogenase and Na+-K+ ATPase expression and growth response of Litopenaeus vannamei to different salinities and dietary protein levels. Chinese Journal of Oceanology and Limnology 29: 343–349. [Google Scholar]
- 18. Li EC, Arena L, Chen LQ, Qin JG, Van Wormhoudt A (2009) Characterization and Tissue-Specific Expression of the Two Glutamate Dehydrogenase Cdnas in Pacific White Shrimp, Litopenaeus Vannamei . Journal of Crustacean Biology 29: 379–386. [Google Scholar]
- 19. Shinji J, Kang BJ, Okutsu T, Banzai K, Ohira T, Tsutsui N, et al. (2012) Changes in crustacean hyperglycemic hormones in Pacific whiteleg shrimp Litopenaeus vannamei subjected to air-exposure and low-salinity stresses. Fisheries Science 78: 833–840. [Google Scholar]
- 20. Lago-Leston A, Ponce E, Munoz ME (2007) Cloning and expression of hyperglycemic (CHH) and molt-inhibiting (MIH) hormones mRNAs from the eyestalk of shrimps of Litopenaeus vannamei grown in different temperature and salinity conditions. Aquaculture 270: 343–357. [Google Scholar]
- 21. Gao WH, Tan BP, Mai KS, Chi SY, Liu HY, Dong XH, et al. (2012) Profiling of differentially expressed genes in hepatopancreas of white shrimp (Litopenaeus vannamei) exposed to long-term low salinity stress. Aquaculture 364: 186–191. [Google Scholar]
- 22. Morin RD, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh TJ, et al. (2008) Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques 45: 81–92. 10.2144/000112900 [DOI] [PubMed] [Google Scholar]
- 23. Chu YJ, Corey DR (2012) RNA Sequencing: Platform Selection, Experimental Design, and Data Interpretation. Nucleic Acid Therapeutics 22: 271–274. 10.1089/nat.2012.0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qian X, Ba Y, Zhuang QF, Zhong GF (2014) RNA-Seq Technology and Its Application in Fish Transcriptomics. Omics-a Journal of Integrative Biology 18: 98–110. 10.1089/omi.2013.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Q, Pan L, Ren Q, Hu D (2015) Digital gene expression analysis in hemocytes of the white shrimp Litopenaeus vannamei in response to low salinity stress. Fish Shellfish Immunology 42: 400–407. 10.1016/j.fsi.2014.11.020 [DOI] [PubMed] [Google Scholar]
- 26. Palstra AP, Beltran S, Burgerhout E, Brittijn SA, Magnoni LJ, Henkel CV, et al. (2013) Deep RNA Sequencing of the Skeletal Muscle Transcriptome in Swimming Fish. Plos One 8(1): e53171 10.1371/journal.pone.0053171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, et al. (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Research 22: 577–591. 10.1101/gr.133009.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith S, Bernatchez L, Beheregaray LB (2013) RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genomics 14(1). 375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xia JH, Liu P, Liu F, Lin G, Sun F, Tu RJ, et al. (2013) Analysis of Stress-Responsive Transcriptome in the Intestine of Asian Seabass (Lates calcarifer) using RNA-Seq. DNA Research 20: 449–460. 10.1093/dnares/dst022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu J, Ji PF, Wang BS, Zhao L, Wang J, Zhao ZX, et al. (2013) Transcriptome sequencing and analysis of wild amur ide (Leuciscus waleckii) inhabiting an extreme alkaline-saline lake reveals insights into stress adaptation. PLoS One 8(4): e59703 10.1371/journal.pone.0059703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li E, Wang S, Li C, Wang X, Chen K, Chen L (2014) Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis . Physiological genomics 46: 177–190. 10.1152/physiolgenomics.00191.2013 [DOI] [PubMed] [Google Scholar]
- 32. Yu Y, Wei JK, Zhang XJ, Liu JW, Liu CZ, Li FH, et al. (2014) SNP Discovery in the Transcriptome of White Pacific Shrimp Litopenaeus vannamei by Next Generation Sequencing. Plos One 9(1): e87218 10.1371/journal.pone.0087218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robalino J, Almeida JS, McKillen D, Colglazier J, Trent HF, Chen YA, et al. (2007) Insights into the immune transcriptome of the shrimp Litopenaeus vannamei: tissue-specific expression profiles and transcriptomic responses to immune challenge. Physiological Genomics 29: 44–56. [DOI] [PubMed] [Google Scholar]
- 34. Xue SX, Liu YC, Zhang YC, Sun Y, Geng XY, Sun JS, et al. (2013) Sequencing and de novo analysis of the hemocytes transcriptome in Litopenaeus vannamei response to white spot syndrome virus infection. Plos One 8(10): e76718 10.1371/journal.pone.0076718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen XH, Zeng DG, Chen XL, Xie DX, Zhao YZ, Yang CL, et al. (2013) Transcriptome analysis of Litopenaeus vannamei in response to white spot syndrome virus infection. Plos One 8(8): e73218 10.1371/journal.pone.0073218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li CZ, Weng SP, Chen YG, Yu XQ, Lu L, Zhang HQ, et al. (2012) Analysis of Litopenaeus vannamei transcriptome using the next-generation DNA sequencing technique. Plos One 7(10): e47442 10.1371/journal.pone.0047442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clavero-Salas A, Sotelo-Mundo RR, Gollas-Galvan T, Hernandez-Lopez J, Peregrino-Uriarte AB, Muhlia-Almazan A, et al. (2007) Transcriptome analysis of gills from the white shrimp Litopenaeus vannamei infected with white spot syndrome virus. Fish & Shellfish Immunology 23: 459–472. [DOI] [PubMed] [Google Scholar]
- 38. Chen K, Li E, Gan L, Wang X, Xu C, Lin HZ, et al. (2014) Growth and lipid metabolism of the Pacific White Shrimp Litopenaeus vannamei at different salinities. Journal of Shellfish Research 33: 825–832. [Google Scholar]
- 39. Palacios E, Bonilla A, Luna D, Racotta IS (2004) Survival, Na+/K+-ATPase and lipid responses to salinity challenge in fed and starved white pacific shrimp (Litopenaeus vannamei) postlarvae. Aquaculture 234: 497–511. [Google Scholar]
- 40. Rainbow PS, Black WH (2001) Effects of changes in salinity on the apparent water permeability of three crab species: Carcinus maenas, Eriocheir sinensis and Necora puber . Journal of Experimental Marine Biology and Ecology 264: 1–13. [Google Scholar]
- 41. Freire CA, Onken H, McNamara JC (2008) A structure-function analysis of ion transport in crustacean gills and excretory organs. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 151: 272–304. [DOI] [PubMed] [Google Scholar]
- 42. Lemos D, Phan VN, Alvarez G (2001) Growth, oxygen consumption, ammonia-N excretion, biochemical composition and energy content of Farfantepenaeus paulensis Perez-Farfante (Crustacea, Decapoda, Penaeidae) early postlarvae in different salinities. Journal of Experimental Marine Biology and Ecology 261: 55–74. [DOI] [PubMed] [Google Scholar]
- 43. Luvizotto-santos R, Bianchini A (2003) Lipids as energy source during salinity acclimation in the euryhaline crab Chasmagnathus granulata dana, 1851 (crustacea-grapsidae). Journal of Experimental Zoology Part A: Comparative Experimental Biology 295: 200–205. [DOI] [PubMed] [Google Scholar]
- 44. Minh Sang H, Fotedar R (2004) Growth, survival, haemolymph osmolality and organosomatic indices of the western king prawn (Penaeus latisulcatus Kishinouye, 1896) reared at different salinities. Aquaculture 234: 601–614. [Google Scholar]
- 45. Palacios E, Bonilla A, Pérez A, Racotta IS, Civera R (2004) Influence of highly unsaturated fatty acids on the responses of white shrimp (Litopenaeus vannamei) postlarvae to low salinity. Journal of Experimental Marine Biology and Ecology 299: 201–215. [Google Scholar]
- 46. Ranallo R, Rhodes E (1998) Lipid metabolism during exercise. Sports Medicine 26: 29–42. [DOI] [PubMed] [Google Scholar]
- 47. Bradbury MW (2006) Lipid metabolism and liver inflammation. I. Hepatic fatty acid uptake: possible role in steatosis. American Journal of Physiology-Gastrointestinal and Liver Physiology 290(2): G194–G198. [DOI] [PubMed] [Google Scholar]
- 48. Deering MJ, Fielder DR, Hewitt DR (1997) Growth and fatty acid composition of juvenile leader prawns, Penaeus monodon, fed different lipids. Aquaculture 151: 131–141. [Google Scholar]
- 49. Martins TG, Cavalli RO, Martino RC, Rezende CEM, Wasielesky W Jr (2006) Larviculture output and stress tolerance of Farfantepenaeus paulensis postlarvae fed Artemia containing different fatty acids. Aquaculture 252: 525–533. [Google Scholar]
- 50. Sui L, Wille M, Cheng Y, Sorgeloos P (2007) The effect of dietary n-3 HUFA levels and DHA/EPA ratios on growth, survival and osmotic stress tolerance of Chinese mitten crab Eriocheir sinensis larvae. Aquaculture 273: 139–150. [Google Scholar]
- 51. Hurtado MA, Racotta IS, Civera R, Ibarra L, Hernández-Rodríguez M, Palacios E (2007) Effect of hypo- and hypersaline conditions on osmolality and Na+/K+-ATPase activity in juvenile shrimp (Litopenaeus vannamei) fed low- and high-HUFA diets. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 147: 703–710. [DOI] [PubMed] [Google Scholar]
- 52. Van Anholt RD, Spanings FA, Koven WM, Nixon O, Wendelaar Bonga SE (2004) Arachidonic acid reduces the stress response of gilthead seabream Sparus aurata L. Journal of Experimental Biology 207: 3419–3430. [DOI] [PubMed] [Google Scholar]
- 53. Beckman B, Mustafa T (1992) Arachidonic acid metabolism in gill homogenate and isolated gill cells from rainbow trout, Oncorhynchus mykiss: the effect of osmolality, electrolytes and prolactin. Fish Physiology and Biochemistry 10: 213–222. 10.1007/BF00004515 [DOI] [PubMed] [Google Scholar]
- 54. Bartke N, Hannun YA (2009) Bioactive sphingolipids: metabolism and function. Journal of Lipid Research 50: S91–S96. 10.1194/jlr.R800080-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brown DA, London E (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. Journal of Biological Chemistry 275: 17221–17224. [DOI] [PubMed] [Google Scholar]
- 56. Trowbridge JM, Gallo RL (2002) Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology 12: 117R–125R. [DOI] [PubMed] [Google Scholar]
- 57. Bhoite S, Roy R (2013) Role of membrane lipid in osmoregulatory processes during salinity adaptation: a study with chloride cell of mud crab, Scylla serrata . Marine and Freshwater Behaviour and Physiology 46: 287–300. [Google Scholar]
- 58. Tocher DR (1995) Glycerophospholipid metabolism. Biochemistry and molecular biology of fishes 4: 119–157. [Google Scholar]
- 59. Paltauf F (1994) Ether lipids in biomembranes. Chemistry Physics Lipids 74(2): 101–139. [DOI] [PubMed] [Google Scholar]
- 60. Spector AA, Yorek MA (1985) Membrane lipid composition and cellular function. Journal of Lipid Research 26: 1015–1035. [PubMed] [Google Scholar]
- 61. Demopoulos C, Pinckard R, Hanahan DJ (1979) Platelet-activating factor. Evidence for 1-O-alkyl-2-acetyl-sn-glyceryl-3-phosphorylcholine as the active component (a new class of lipid chemical mediators). Journal of Biological Chemistry 254: 9355–9358. [PubMed] [Google Scholar]
- 62. Brosche T, Platt D (1998) The biological significance of plasmalogens in defense against oxidative damage. Experimental Gerontology 33: 363–369. [DOI] [PubMed] [Google Scholar]
- 63. Starai V, Celic I, Cole R, Boeke J, Escalante-Semerena J (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298: 2390–2392. [DOI] [PubMed] [Google Scholar]
- 64. Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proceedings of the National Academy of Sciences 103: 10224–10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wauben IPM, Wainwright PE (1999) The influence of neonatal nutrition on behavioral development: A critical appraisal. Nutrition Reviews 57: 35–44. [DOI] [PubMed] [Google Scholar]
- 66.Blum K, Downs BW, Waite RL, Heaney WJ (2012) Nutrigenomic methods to overcome carbohydrate bingeing and overeating. Google Patents.
- 67. Capdevila JH, Falck JR, Harris RC (2000) Cytochrome P450 and arachidonic acid bioactivation: molecular and functional properties of the arachidonate monooxygenase. Journal of Lipid Research 41: 163–181. [PubMed] [Google Scholar]
- 68. Kroetz DL, Zeldin DC (2002) Cytochrome P450 pathways of arachidonic acid metabolism. Current opinion in lipidology 13: 273–283. [DOI] [PubMed] [Google Scholar]
- 69. Hardwick JP (2008) Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochemical pharmacology 75: 2263–2275. 10.1016/j.bcp.2008.03.004 [DOI] [PubMed] [Google Scholar]
- 70. Funder JW, Krozowski Z, Myles K, Sato A, Sheppard KE, Young M (1996) Mineralocorticoid receptors, salt, and hypertension. Recent Progress in Hormone Research 52: 247–260; Discussion 261–242. [PubMed] [Google Scholar]
- 71. Gupta B, Lalchhandama K (2002) Molecular mechanisms of glucocorticoid action. Current Science-Bangalore 83: 1103–1111. [Google Scholar]
- 72. Frye C (2009) Steroids, reproductive endocrine function, and affect. A review. Minerva ginecologica 61: 541–562. [PubMed] [Google Scholar]
- 73. Birukawa N, Ando H, Goto M, Kanda N, Pastene LA, Nakatsuji H, et al. (2005) Plasma and urine levels of electrolytes, urea and steroid hormones involved in osmoregulation of cetaceans. Zoological Science 22: 1245–1257. [DOI] [PubMed] [Google Scholar]
- 74.Soderquist CA, Zeiher EK (1994) Method for inhibiting scale formation and/or dispersing iron in reverse osmosis systems. Google Patents.
- 75. Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–U130. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 78. Goto S, Kanehisa M (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39: W316–322. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.