Abstract
Recent advances in the development of genome editing technologies based on programmable nucleases have significantly improved our ability to make precise changes in the genomes of eukaryotic cells. Genome editing is already broadening our ability to elucidate the contribution of genetics to disease by facilitating the creation of more accurate cellular and animal models of pathological processes. A particularly tantalizing application of programmable nucleases is the potential to directly correct genetic mutations in affected tissues and cells to treat diseases that are refractory to traditional therapies. Here we discuss current progress towards developing programmable nuclease-based therapies as well as future prospects and challenges.
Of the approximately 25,000 annotated genes in the human genome, mutations in over 3,000 genes have already been linked to disease phenotypes (www.omim.org/statistics/geneMap), and more disease-relevant genetic variations are being uncovered at a staggeringly rapid pace. Now, due to sharp drops in sequencing cost, the completion of the human genome project, and the exponential growth of human genome sequencing data from diseased individuals, the role of genetics in human health has become a major focus of research, clinical medicine and the development of targeted therapeutics1. These advances in our understanding of the genetic basis of disease have improved our understanding of disease mechanisms and pointed toward potential therapeutic strategies. However, despite valid therapeutic hypotheses and strong efforts in drug development, there have only been a limited number of successes using small molecules to treat diseases with strong genetic contributions2. Emerging therapeutic strategies that are able to modify nucleic acids within disease-affected cells and tissues have potential for treatment of monogenic, highly penetrant diseases, such as severe-combined immunodeficiency (SCID), haemophilia, and certain enzyme deficiencies due to their well-defined genetics and often lack of safe, effective alternative treatments.
Two of the most powerful genetic therapeutic technologies developed thus far are gene therapy, which enables restoration of missing gene function by viral transgene expression, and RNA interference (RNAi), which mediates targeted repression of defective genes by knockdown of the target mRNA (reviewed in 3,4). Gene therapy has been used to successfully treat monogenic recessive disorders affecting the hematopoietic system, such as SCID and Wiskott-Aldrich syndrome, by semi-randomly integrating functional genes into the genome of hematopoietic stem/progenitor cells 5-7. RNAi has been used to repress the function of genes implicated in cancer, age related macular degeneration and transthyretin (TTR)-amyloidosis among others, resulting in a therapeutic effect in clinical trials (www.clinicaltrials.gov, trial numbers: NCT00689065, NCT01961921 and NCT00259753). Despite promise and recent success, gene therapy and RNAi have limitations that prevent their utility for a large number of diseases. For example, viral gene therapy may cause mutagenesis at the insertion site and result in dysregulated transgene expression6. Alternatively, RNAi use is limited to targets where gene knockdown is beneficial. Also, RNAi often cannot fully repress gene expression, and is therefore unlikely to provide a benefit for diseases where complete ablation of gene function is necessary for therapy. RNAi may also have poor specificity, posing potential safety concerns and sometimes decreasing the effectiveness of treatment8-10.
Genome editing technologies based on programmable nucleases such as meganucleases (reviewed in 11), zinc finger nucleases (reviewed in 12), transcription activator-like effector nucleases (reviewed in 13,14), and clustered regularly interspaced short palindromic repeat (CRISPR)-associated nuclease Cas9 (reviewed in 15) are opening the possibility of achieving therapeutic genome editing in diseased cells and tissues, resulting in the removal or correction of deleterious mutations or the insertion of protective mutations.
In this Review, we will describe the different nuclease-based genome editing technologies, the mechanisms by which they produce genetic changes, considerations for their uses in therapeutic settings and major challenges that will need to be addressed to realize their clinical translation. Although a large number of genome editing therapeutic efforts have focused on treatment of monogenic, highly-penetrant disorders, we will also discuss intriguing treatment strategies to apply this class of therapy to diseases such as viral infections and cancer.
Genome Editing Technologies
Programmable nucleases enable precise genome editing by introducing DNA double strand breaks (DSBs) at specific genomic loci. DSBs subsequently recruit endogenous repair machinery for either non-homologous end-joining (NHEJ) or homology directed repair (HDR) to the DSB site to mediate genome editing.
To date, four major classes of nucleases, meganucleases and their derivatives16-19, zinc finger nucleases (ZFNs) 20-28, transcription activator like effector nucleases (TALENs) 29-34, and CRISPR-associated nuclease Cas9 35-43 have been developed to enable site-specific genome editing (Table 1). These nuclease systems can be broadly classified into two categories based on their mode of DNA recognition - ZFN, TALEN, and meganucleases achieve specific DNA binding via protein-DNA interactions, Cas9 is targeted to specific DNA sequences by a short RNA guide molecule that base-pairs directly with the target DNA,protein-DNA interactions also have a role in its targeting. Meganucleases are endonucleases with large (>14bp) recognition sites, the DNA DNA binding domains of which are also responsible for cleavage of target sequences18.
Table 1.
Comparison of Different Programmable Nuclease Platforms.
| Zinc Finger Nuclease | TALEN | Cas9 | Meganuclease | |
|---|---|---|---|---|
| Recognition site | Typically 9 to 18 bp per ZFN monomer, 18 to 36 bp per ZFN pair | Typically 14 to 20 bp per TALEN monomer, 28 to 40bp per TALEN pair | 22bp (20bp guide sequence + 2bp PAM sequence for S. pyognes Cas9); up to 44 bp for double nicking | Between 14 and 40 bp |
| Specificity | Small number of positional mismatches tolerated | Small number of positional mismatches tolerated | Positional and multiple consecutive mismatches tolerated | Small number of positional mismatches tolerated |
| Targeting constraints | Difficult to target non-G-rich sequences | 5’ targeted base must be a T for each TALEN monomer | Targeted sequence must precede a PAM [AU: please define] | Targeting novel sequences often results in low efficiency |
| Ease of engineering | Difficult, may require substantial protein engineering | Moderate, requires complex molecular cloning methods | Easily re-targeted using standard cloning procedures and oligo synthesis | Difficult, may require substantial protein engineering |
| Immunogenicity | Likely low, as ZFs are based on human protein scaffold. Fokl is derived from bacteria and may be immunogenic | Unknown, protein derived from Xanthamonas sp. | Unknown, protein derived from various bacterial species | Unknown, meganucleases may be derived from many organisms including eukaryotes |
| Ease of ex vivo delivery | Relatively easy through methods such as electroporation and viral transduction | Relatively easy through methods such as electroporation and viral transduction | Relatively easy through methods such as electroporation and viral transduction | Relatively easy through methods such as electroporation and viral transduction |
| Ease of in vivo delivery | Relatively easy due to small size of ZFN expression cassettes, allows use in a variety of viral vectors | Difficult due to the large size of each TALEN and repetitive nature of DNA encoding TALENs, leanding to unwanted recombination events when packaged into lentiviral vectors | Moderate: The commonly used Cas9 from S. pyogenes is large and may impose packaging problems for viral vectors such as AAV, but smaller orthologs exist. | Relatively easy due to small size of meganucleases, allows use in a variety of viral vectors. |
| Ease of multiplexing | Low | Low | High | Low |
ZFNs and TALENs are chimeric enzymes consisting of a DNA binding domain fused to the sequence agnostic nuclease domain, FokI20,31. Re-targeting of ZFNs and meganucleases requires protein engineering, whereas re-targeting of TALENs requires complex molecular cloning18,44,45. In contrast, the Cas9 protein is invariant and can be easily retargeted to new DNA sequences by changing a small portion of the sequence of an accompanying RNA guide that base-pairs directly with target DNA. A potential advantage of Cas9 is its ability to introduce multiple DSBs in the same cell (also referred to as multiplexing) via expression of distinct guide RNAs. All four types of nucleases have been demonstrated to achieve efficient genome editing in a wide range of model organisms and mammalian cells and efforts are now underway in both industry and academia to develop these tools as therapeutics 46-49.
Once the DSB has been made, the lesion may be repaired by either NHEJ or HDR depending on the cell state and the presence of a repair template (Fig. 1). NHEJ may repair the lesion by directly rejoining the two DSB ends in a process that does not require a repair template. Although NHEJ-mediated DSB repair can be accurate, repeated repair of the same DSB by NHEJ machinery eventually results in the formation of small insertion or deletion mutations (indels) bridging the break site. Indels introduced into the coding sequence of a gene can cause frameshift mutations that lead to mRNA degradation by nonsense-mediated decay, or result in the production of nonfunctional truncated proteins 50. Thus, NHEJ may be used to suppress gene function similar to RNAi, however, it may lead to permanent inactivation by introducing loss of function mutations to the gene in targeted cells.
Figure 1. Types of Therapeutic Genome Modifications.
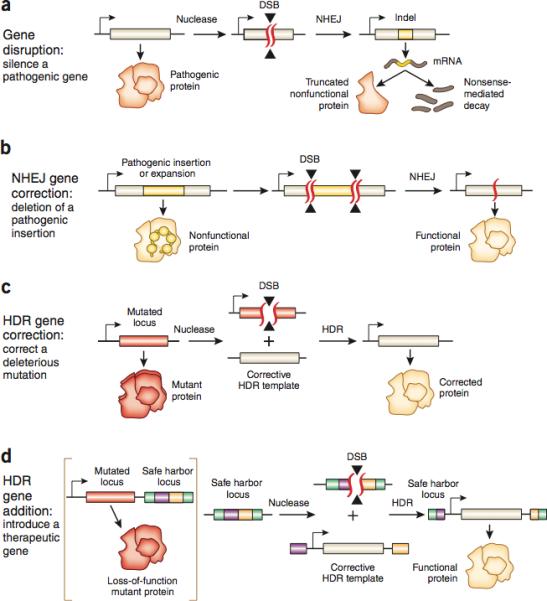
The specific type of genome editing therapy depends on the nature of the mutation causing disease. a, In gene disruption, the pathogenic function of a protein is silenced by targeting the locus with NHEJ. Formation of indels on the gene of interest often result in frameshift mutations that create premature stop codons and a non-functional protein product, or non-sense mediated decay of transcripts, suppressing gene function. c, HDR gene correction can be used to correct a deleterious mutation. A DSB is induced near the mutation site in the presence of an exogenously provided, corrective HDR template. HDR repair of the break site with the exogenous template corrects the mutation, restoring gene function. d, An alternative to gene correction is gene addition. This introduces a therapeutic transgene into either the native or a non-native locus in the genome. A DSB is induced at the desired locus and an HDR template containing homology to the break site, a promoter, a transgene and a polyadenylation (polyA) sequence is introduced to the nucleus. HDR repair recovers gene function in the target locus albeit without true physiological control over gene expression. b, In NHEJ gene correction two DSBs targeted to both sides of a pathogenic expansion or insertion may be resolved by NHEJ, causing a deletion of the intervening sequences to mediate therapy. This form of treatment would require multiplexed targeting of disease causing mutations.
In comparison, HDR allows researchers to use an exogenous DNA template to specify the outcome of the DSB repair 22,51-56. Upon introduction of a targeted DSB, HDR machinery may use exogenously provided single or double stranded DNA templates with sequence homology to the break site to synthesize DNA that is used to repair the lesion, incorporating any changes encoded in the template DNA. For example, HDR may be used along with an appropriately designed repair template to replace a mutated gene directly, thereby restoring gene function while preserving physiological regulation of gene expression.
Therapeutic Genome Editing Strategies
Genome editing based therapy can be achieved through a number of approaches including correction or inactivation of deleterious mutations, introduction of protective mutations, addition of therapeutic transgenes, or disruption of viral DNA.
Pathogenic mutations can be broadly classified as gain- or loss-of-function. A gain-of-function mutation, such as in the cases of the HTT gene for huntington disease (http://omim.org/entry/143100) and FGFR3 for Achondroplasia (http://omim.org/entry/100800), which result in the expression of a dominant negative mutant gene, may be corrected by using NHEJ-mediated induced mutations to specifically inactivate the mutant gene while leaving the wildtype copy intact on the other allele (Fig. 1a). Additionally, targeting of two nucleases to make two DSBs around and insertion may be useful for treatment of nucleotide expansion disorders such as spino-cerebellar ataxia (http://www.omim.org/entry/164400), huntington disease, and fredriech ataxia (http://omim.org/entry/229300) (Fig. 1B). A combination of DSBs may also be used to edit multiple loci to achieve a therapeutic effect.
However, some gain-of-function mutations, such as the SOD1 G93A mutation in Amyotrophic Lateral Sclerosis (ALS) (http://omim.org/entry/147450), are point mutations, which might not be sufficiently different from the functioning allele on the homologous chromosome to be distinguished by the current generation of programmable nucleases, potentially leading to an undesirable complete loss of function of the protein if it is corrected using NHEJ. In this case HDR could be used to change the gain-of-function allele to the wildtype sequence, recovering gene function and eliminating pathogenic activity while preserving physiological levels of gene expression (Figure 1C). Similarly, loss-of-function mutations, including those found in Tay-Sachs disease (http://omim.org/entry/272800) would necessitate precise sequence changes to eliminate pathogenicity, requiring HDR gene correction to revert the loss-of-function mutation to the wildtype sequence. This same logic can also be extended to mutations which protect against infectious or genetic disease which may be loss-of-function in the case of CCR5 for HIV46,57 (Figure 1a) or PCSK9 for hypercholesterolemia58 which require inactivation by NHEJ, or gain-of-function in the case of APP (p.A673T) in alzheimers disease59 which would require correction by HDR.
For deleterious loss of function mutations and protective gain of function mutations, a therapeutic effect may also be achieved by introducing a copy of the wildtype gene or gain-of-function mutant respectively (Figure 1D). The therapeutic transgene may be inserted into a new locus, including identified ‘safe harbor’ loci - a region of the genome which does not lead to discernable phenotypic effects when disrupted - to recover missing gene function60-62. Gene insertion may also be used to stably confer cells novel functions that protect against disease, such as the insertion of chimeric-antigen receptors (CAR) into T-cells to target certain leukemias63. Such gene insertion strategies are similar to viral-mediated gene therapy, but with the advantage of providing better control over transgene copy number and expression levels, which may be important for gene targets whose function is sensitive to expression levels.
Programmable nucleases may also be targeted to foreign DNA, such as viral genomes, which may be integrated proviruses or maintained extrachromosomally64-68. Targeting of extrachromosmal DNA may lead to depletion of viral genomes while mutagenesis of the provirus genome at important coding sequences or regulatory regions may inactivate viral replication. Additionally, the use of multiple DSBs might be used to excise proviral genomes64. As viral sequences may bear little sequence homology to the host genome, this class of treatments may exhibit less off target effects than editing therapies targeting endogenous loci.
Factors Influencing Therapeutic Efficacy
Genome editing has been successfully applied to a number of diseases at the preclinical level as well as in a phase I clinical trial (Table 2) 46,48,49,69,70. In evaluating the feasibility of a genome editing based therapy, the therapeutic effect of the desired genetic change should first be clearly established. Subsequently, the success of a given strategy will depend on the ease with which a therapeutic modification ‘threshold’ is achieved, a criteria that is governed by the fitness of edited cells, the DSB repair pathway utilized to edit the genome, and the efficiency of delivery of genome editing molecules to target cell types.
Table 2.
Examples of applications of genome editing to therapeutic models.
| Disease Type | Nuclease Platform Employed | Therapeutic Strategy | References |
|---|---|---|---|
| Hemophilia B | ZFN | HDR-mediated insertion of correct gene sequence | 48 |
| HIV | ZFN and CRISPR | NHEJ-mediated inactivation of CCR5 | 46,69,70,131 |
| DMD | CRISPR and TALEN | NHEJ-mediated removal of stop codon, and HDR-mediated gene correction | 132,133 |
| HBV | TALEN and CRISPR | NHEJ-mediated depletion of viral DNA | 65,66 |
| SCID | ZFN | HDR-mediated insertion of correct gene sequence | 47 |
| Cataract | CRISPR | HDR-mediated correction of mutation in mouse zygote | 134 |
| Cystic fibrosis | CRISPR | HDR-mediated correction of CFTR in intestinal stem cell organoid | 135 |
| Hereditary tyrosinemia | CRISPR | HDR-mediated correction of mutation in liver | 49 |
Fitness of Edited Cells
If edited cells have an increased fitness relative to unedited cells, this will result in a selective advantage for edited cells, reducing the number of cells that initially needs to be edited to reverse disease symptoms (Figure 2). For example SCID-X1 is caused by mutations in the IL2RG gene, the function of which is required for proper development of the hematopoietic lymphocyte lineage (http://www.omim.org/entry/300400). Hematopoietic progenitor cells with an active IL2RG gene selectively expand relative to their unedited counterparts. For example in people with SCID-X1 who received viral gene therapy for SCID-X171,72, as well in a rare affected individual who had a spontaneous correction of a SCID-X1 mutation in a T-cell progenitor73, corrected hematopoietic progenitor cells were able to overcome the hematopoietic development block and expand relative to their diseased counterparts to mediate a therapeutic effect.
Figure 2. Factors Influencing Therapeutic Efficacy.
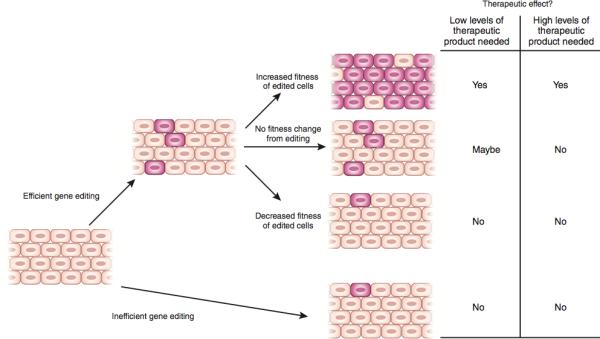
For a genome editing therapy to be efficacious, enough cells carrying the desired genome modification must exist in a tissue to reverse disease. If editing is efficient, treatment will create a population of cells carrying the desired genomic modification (depicted in pink). Depending on whether the editing event creates a fitness change in target cells, edited cells will proportionally increase, or decrease relative to unedited cells (depicted in brown) over time in tissues. Proportionally high levels of cells carrying therapeutic genome modifications in a disease-affected tissue are likely to result in a therapeutic effect. However, if low levels of a secreted gene product are needed to reverse disease, then successfully editing a small number of cells may be therapeutically efficacious.
In contrast, other diseases in which edited cells do not exhibit a change in fitness, the number of cells that must initially be modified to reach a therapeutic effect is higher, For example, chronic granulomatous disorder (CGD), is caused by mutations in genes encoding phagocytic oxidase proteins that are involved in the generation of reactive oxygen species by neutrophils to kill pathogens (http://www.omim.org/entry/306400). Dysfunction of phagocytic oxidases does not influence hematopoietic cell fitness or development thus there would probably be no preferential expansion of edited cells in this disease. Indeed, no selective advantage for gene corrected cells in CGD has been observed in gene therapy trials, leading to difficulties with long-term cell engraftment74,75.
Some diseases where edited cells do not confer a change in fitness can still have reversed diseases symptoms at low numbers of therapeutic modified cells. For example, genes that function in a non-cell-autonomous fashion may only require a small number of functioning alleles to produce enough gene product to treat disease. For instance, Haemophilia B, is caused by mutations in the gene encoding the secreted factor IX protein involved in the clotting cascade, severe disease is associated with less than 1% of normal activity, whereas restoration of at least 1% of factor IX activity prevents the most severe bleeding conditions, while higher levels of restoration will further improve other clinically relevant complications in human patients76,77. This suggests that small changes in the amount of factor IX activity through correction of alleles in even a small percentage of liver cells may be therapeutic. Indeed, a study using ZFNs to correct a mouse model of haemophilia B shortly after birth demonstrated that correction of 3-7% of mutated Factor IX alleles was sufficient to reverse disease symptoms, providing preclinical evidence for this hypothesis48.
In the case where editing imposes a fitness disadvantage, such as the correction of mutated tumor suppressor genes in cancer cells, modified cells would be outcompeted by their diseased counterparts, causing the benefit of treatment to be low The modification threshold of this final class of diseases would be extremely high requiring many cells to be directly modified and may not be suited for genome editing therapy. Therefore given the current state of technology genome editing therapies are most ideally suited for cases where editing confers a fitness advantage or where a small change in gene product levels can influence clinical outcomes.
Efficiency of Genome Editing
The efficiency of NHEJ and HDR mediated DSB repair varies significantly by cell type and cell state, NHEJ is most of the time more active than HDR. This difference in activity makes treating diseases that require gene correction or insertion of a gene more challenging than those requiring gene inactivation. NHEJ is thought to be active throughout the cell cycle and has been observed in a variety of cell types, including dividing and post-mitotic cells78,79. Therefore NHEJ may be used to facilitate high levels of gene disruption in target cell populations. In contrast, HDR acts primarily during S/G2 phase, and is therefore largely restricted to cells that are actively dividing, limiting treatments that require precise genome modifications to mitotic cells 80,81.
The efficiency of correction by HDR may be controlled by a number of factors. First, the nature of genome modification may influence editing rates as large HDR mediated insertions have been found to occur at a reduced rate relative to HDR mediated small deletions, insertions or substitutions61,82. Second, the exact sequence changes made through HDR may influence therapeutic efficacy, as editing events that do not destroy the nuclease recognition site may be subject to further mutagenesis by NHEJ, potentially reducing therapeutic editing rates. Third, increasing the extent of homology between the repair template and the DSB site may increase HDR, possibly by promoting the stability of D-loop intermediates formed during synthesis from a template82-84. Fourth, the topology of the HDR template may influence editing efficiency as single-stranded DNA oligonucleotides and viruses may yield higher HDR rates than double-stranded substrates85,86. Last, suppressing competing DNA repair pathways such as NHEJ has also been shown to increase HDR rates moderately87 although the safety of this strategy is not known and should be carefully assessed before implementation in a therapeutic context.
In addition to these approaches, further investigations aimed at improving HDR efficiency will be necessary to address a broader range of diseases with genome editing. Furthermore, many of these approaches may be synergistic and can be implemented in combination to increase the rate HDR past the therapeutic editing threshold needed to treat many diseases. Despite the challenges associated with HDR, proof-of-concept preclinical HDR treatments have now been described for mouse models of haemophilia B and hereditary tyrosinemia 48,49.
Modes of Delivery: Ex Vivo vs. In Vivo Editing
Achieving therapeutic editing requires delivery of programmable nucleases to target cells, which can be achieved either ex vivo, by modifying and autologously transplanting cells, or in vivo, by direct application of nucleases to diseased cells in the body (Figure 3). Given that nucleases can potentially be mutagenic, the ideal delivery system would permit transient nuclease activity. Currently, nucleases can be delivered as nucleic acids encoding the desired editing system, or directly as proteins themselves. While nucleic acids and protein are both capable of achieving transient expression in target cell types, protein is likely to provide the best control over nuclease dosage, since there is no signal amplification. Another important consideration is that DNA based nuclease expression systems pose risks of insertional mutagenesis by the vector itself. So far, a variety of delivery methods have been developed.
Figure 3. Ex Vivo vs. In Vivo Editing Therapy.
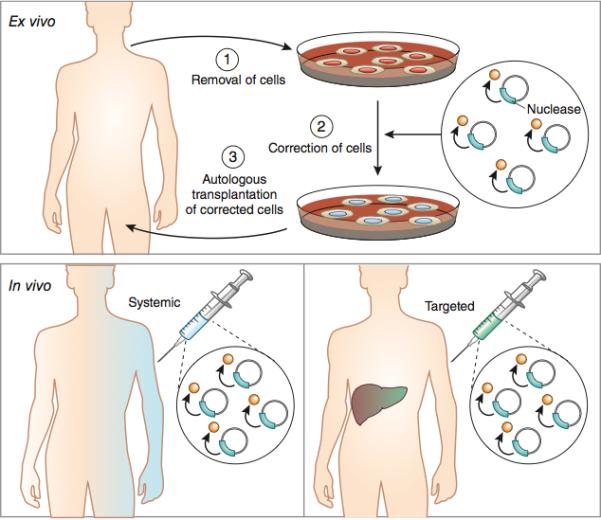
In ex vivo editing therapy cells are removed from a patient, edited and then re-engrafted (top panel). For this mode of therapy to be successful, target cells must be capable of survival outside the body and homing back to target tissues post-transplantation. In vivo therapy involves genome editing of cells in situ (bottom panels). For in vivo systemic therapy, delivery agents that are relatively agnostic to cell identity or state would be used to effect editing in a wide range of tissue types. For example systemic delivery of AAV serotype 8 vectors has been used in preclinical models to target liver tissue with high efficiency. Alternatively, in vivo therapy, may also be achieved through local injection of viral vectors to the affected tissue, such as the eye, brain, or muscle.
In ex vivo editing therapy the target cell population is removed from the body, modified with programmable nucleases and then transplanted back into the original host (Figure 3-top panel). This mode of therapy allows the target cell population to be manipulated with a wide range of delivery platforms such as, electroporation, cationic lipids, cell penetrating peptides, carbon nanowires and viral vectors. Ex vivo therapies are likely to achieve high editing rates, due to the extensive development of these delivery systems for research and gene therapy applications. Moreover, many ex vivo therapies afford control over the specific dosage of therapeutic molecules delivered to cells. This may be particularly important when off-target modifications are a concern, as titrating the amount of nuclease may decrease such mutations88.
However, there are two large drawbacks with ex vivo. First, target cells must be capable of surviving manipulation outside the body which is a challenge for many tissues because cells either fail to survive, or lose properties necessary for their function in vivo. Thus, ex vivo therapy is largely limited to tissues with adult stem cell populations amenable to culture and manipulation, such as the hematopoietic system. Second, cultured cells often engraft poorly upon re-introduction into a patient, decreasing the effectiveness of treatment. However, engraftment may be enhanced for hematopoietic cells by ablative conditioning regimens that deplete unedited cells prior to transplantation. These are clinically feasible but introduce significant risks to patients 89.
In vivo genome editing involves direct delivery of programmable nucleases to disease affected cells in their native tissues (Figure 3 bottom panels). There are two advantages of in vivo editing therapy over ex vivo approaches. First, in vivo editing can be applied to diseases where the affected cell population is not amenable to ex vivo manipulation. Second, in vivo delivery has the potential to target multiple tissue types, potentially allowing for the treatment of diseases that affect multiple organ systems. These properties will probably allow in vivo treatment to be applied to a wider range of diseases than ex vivo therapies.
To date, in vivo editing has largely been achieved through the use of viral vectors with defined, tissue-specific tropism. Such vectors are currently limited in terms of cargo carrying capacity and tropism, restricting this mode of therapy to organ systems where transduction with clinically useful vectors is efficient, such as the liver, muscle and eye 90-92. Another major potential barrier for in vivo delivery is the immune response that may be raised in response to the large amounts of virus necessary for treatment, this phenomenon is not unique to genome editing and is observed with other virus based gene therapies93. It is also possible that peptides from editing nucleases themselves will be presented on MHC Class I molecules to stimulate an immune response, although there is little evidence to support this happening at the preclinical level. Another major difficulty with this mode of therapy is controlling the distribution and consequently the dosage of genome editing nucleases in vivo, leading to off-target mutation profiles that may be difficult to predict. To address some of the concerns associated with viral vectors, non-viral delivery systems are under active development to reduce the potential risks currently associated with the use of viral vectors and expand the range of targetable tissues (reviewed in 94).
The potential clinical complications faced by therapeutic genome editing overlap significantly with those of gene therapy, which make use of similar delivery agents and result in expression of novel gene products in the host. For a more in-depth discussion of the safety concerns regarding transgene expression and viral vectors for therapy, the reader is referred to recent reviews and studies on gene therapy.95,96
Examples of Successful Genome Editing Therapeutic Strategies
Ex Vivo Editing Therapy
The long standing clinical expertise with the purification, culture and transplantation of hematopoietic cells has made diseases affecting the blood system such as SCID, Fanconi anemia, Wiskott-Aldrich syndrome and sickle cell anemia the focus of ex vivo genome editing therapy. Another reason to focus on hematopoietic cells is that, thanks to previous efforts to design gene therapy for blood disorders, delivery systems of relatively high efficiency already exist. Despite these advantages, the often low efficiency of cell engraftment upon transplantation necessitates that this mode of therapy be applied to diseases where edited cells possess a fitness advantage, so that a small number of engrafted, edited cells can expand and treat disease. One such disease is HIV, as HIV infection results in a fitness disadvantage to CD4+ T cells.
The rationale for genome editing for HIV treatment originates from the observation that individuals homozygous for loss of function mutations in CCR5, a cellular co-receptor for the virus, are highly resistant to infection and otherwise healthy, suggesting that mimicking this mutation with genome editing could be a safe and effective therapeutic strategy57. This idea was clinically validated when an HIV infected patient was given an allogeneic bone marrow transplant from a donor homozygous for a loss of function CCR5 mutation, resulting in undetectable levels of HIV and restoration of normal CD4+ T-cell counts97. Although bone marrow transplantation is not a realistic treatment strategy for most HIV patients, due to the limitied numbers of CCR5-null donors and potential graft vs. host disease, HIV therapies that convert a patient's own T-cells into CCR5 null cells are.
Early studies using ZFNs and NHEJ to knockout CCR5 in humanized mouse models of HIV showed that transplantation of CCR5 edited CD4+ T cells improved viral load and CD4+ T-cell counts 70. Importantly, these models also showed that HIV infection resulted in selection forcells not expressing CCR5. As a result of this promising study, genome editing therapy that knocks out CCR5 in patient T cells has now been tested in humans. In a recent phase I clinical trial, CD4+ T cells from patients with HIV were removed, edited with ZFNs designed to knockout the CCR5 gene, and autologously transplanted back into patients46. Early results from this trial suggest that genome editing through ZFNs of the CCR5 locus is safe, although the follow up time is too short to fully understand the risks and efficacy of treatment.
Gene correction strategies have also been successfully demonstrated in a recent study in which a mutated IL2RG gene was restored in hematopoietic stem cells (HSCs) obtained from a patient suffering from SCID-X147. First, HSCs were transduced using integration-deficient lentivirus containing an HDR template encoding a therapeutic cDNA for IL2RG. Following transduction, cells were electroporated with mRNA encoding ZFNs targeting a mutational hotspot in IL2RG to stimulate HDR based gene correction. To increase HDR rates, culture conditions were optimized with small molecules to encourage HSC division. This strategy resulted in gene corrected HSCs from the SCID-X1 patient being obtained in culture at therapeutically relevant rates. HSCs from unaffected individuals that underwent the same gene correction procedure could sustain long-term hematopoiesis in mice. HSCs are capable of giving rise to all hematopoietic cell types and can be autologously transplanted, making them an extremely valuable cell population for all hematopoietic genetic disorders 98. Gene corrected HSCs could, in principle, be used to treat a wide range of genetic blood disorders making this study an exciting breakthrough for therapeutic genome editing.
In Vivo Genome Editing Therapy
In vivo genome editing therapy faces similar challenges to ex vivo strategies and is also limited by the small number of efficient delivery systems. Inefficient modification of target loci will be compounded by any inefficiencies in delivery, making tissues lacking robust delivery platforms particularly difficult to treat with this mode of therapy. For organ systems where delivery is efficient, however, there have already been a number of exciting preclinical therapeutic successes.
The first example of successful in vivo editing therapy was demonstrated in a mouse model of haemophilia B48. Recovering Factor IX activity to above 1% of its levels in severely affected individuals can transform the disease into a milder form, as infusion of recombinant Factor IX into such patients prophylactically from a young age to achieve such levels largely ameliorates the most severe bleeding complications76. In addition, Factor IX is synthesized and secreted by the liver, an organ that can be transduced efficiently by viral vectors encoding editing systems.
Using hepatotropic adeno-associated viral (AAV) serotypes encoding ZFNs and a corrective HDR template, up to 7% gene correction of a mutated [AU: 7% of what?], humanized Factor IX gene in the murine liver was achieved48. This resulted in improvement of clot formation kinetics, a measure of the function of the clotting cascade, demonstrating for the first time that in vivo editing therapy is not only feasible, but also efficacious.
Building on this study, other groups have recently used in vivo genome editing of the liver with CRISPR-Cas9 to successfully treat a mouse model of hereditary tyrosinemia and to create mutations that provide protection against cardiovascular disease49,99. These two distinct applications demonstrate the versatility of this approach for disorders that involve hepatic dysfunction. Application of in vivo editing to other organ systems will be necessary to prove that this strategy is widely applicable. Currently, efforts to optimize both viral and non-viral vectors are underway to expand the range of disorders that can be treated with this mode of therapy90,94.
Challenges to Clinical Translation
Translating genome editing technologies to the clinic faces major challenges, primarily in terms of the safety and efficacy of these treatments. Due to the distinctly different molecular nature of these therapies compared to small molecule and biologic therapies, engineering developments in several areas will be needed to bring these tools to bear on clinical medicine.
Increasing Efficiency of Gene Correction
Although the amount of genome modification in a target cell population required to create a therapeutic effect differs depending on the disease, the efficacy of most editing treatments will be improved with increased editing rates. As previously noted, editing rates are controlled by the activity of DSB repair pathways.
Since NHEJ-mediated DSB repair is active in most cell types and are efficient, the primary challenge to date has been to increase the efficiency of HDR. So far, applications of HDR in genome editing have been primarily limited to dividing cells, due to the selective expression of HDR machinery during cell division and its down-regulation in slowly cycling or post-mitotic cells. Cell cycle regulation has now been somewhat by-passed for slowly cycling cell types through stimulation of mitosis with pharmacologic agents ex vivo47. However, truly post-mitotic cells are unlikely to be amenable to such manipulation, limiting the applicability of this strategy. Nevertheless, further work that will enable precise gene correction in post-mitotic cells such as neurons are critical to developing therapeutic strategies for a large number of untreatable neurological disorders. The solution to improved HDR in neurons will likely surface as we improve our understanding of DNA damage repair mechanism in the brain, and through harnessing of heterologous mechanisms. For example, the neurotrophic Herpes Simplex Virus (HSV), which depends on single-strand annealing (SSA), a form of HDR, to replicate, supplements exogenous viral proteins to facilitate viral replication, might provide answers to achieving efficient gene correction in post-mitotic cells100.
Additional, non-HDR-based strategies may also be successful for facilitating precise gene correction in post-mitotic cells. For example, attempts have been made to completely circumvent the need for HDR through direct ligation of DNA templates containing therapeutic transgenes into targeted DSBs. Such ligation events have been successfully demonstrated using ZFN101, but the rates are low for Cas9102,103. This difference might be due to cleavage patterns between ZFN and Cas9, whereas ZFN generates a predictable 4-bp overhang and Cas9 generates a blunt cut. Future structure-guided engineering may be able to alter the cleavage pattern of Cas9 to generate a sticky ends.
Understanding and Improving Specificity of Editing Nucleases
The specificity of genome editing tools is one of the main safety concerns for clinical application. Genetic modifications are permanent, and deleterious off-target mutations have the potential to create cells with oncogenic potential, reduced fitness, or functional impairment. Furthermore, oncogenic mutations resulting from off-target editing may lead to expansion of edited cells, thus, even low levels of off-target mutagenesis may have devastating consequences.
Two issues remain outstanding: evaluating and reducing off-target effects. A number of studies have attempted to evaluate the targeting specificity of ZFN, TALEN, and Cas9 nucleases. The limited number of studies characterizing ZFN104,105 and TALEN106 specificity have only highlighted the challenges of detecting ZFN and TALEN off-target activity. Of note, the two independent studies attempting to characterize the off-target profile of the same pair of CCR5-targeting ZFNs have returned distinct and non-overlapping off-target sites, which highlights the challenges associated with analysis of nuclease specificity.
Many studies have attempted to evaluate the specificity of Cas9, partly owing to the simplicity of the RNA-guided DNA targeting mechanism of Cas9, which makes it significantly easier to establish hypotheses regarding possible off-targeting mechanisms based on Watson-Crick base pairing rules. While initial bacterial39, biochemical40,41, and mammalian42 experiments have suggested that the 3’ 8-12bp seed region of the guide sequence can be sensitive to single base mismatches, further work have shown that this rule-of-thumb is not necessarily accurate, especially in situations where there is high concentration of Cas9 and guide RNA88,107-110. Many of these studies were carried out in cell lines and examined Cas9-mediated mutagenesis at genomic sites bearing high levels of homology to the on-target sequence and found that, unsurprisingly, subsets of highly homologous off-target sites were significantly mutated by the nuclease. However, the scope of possible off-target sites evaluated by these studies was limited to computationally predicted sites.
More recently, whole-genome sequencing of Cas9-edited cell lines revealed low incidence of off-target mutation, which suggests that Cas9-mediated genome editing may be specific 111-113. Despite these studies, unbiased assessment of genome wide off-targeting using more advanced methods like direct capture of DSBs 114, labeling of DSBs with oligo captures115, and techniques that can detect larger structural variations (i.e. translocations) potentially imposed by nuclease treatment116 will help us further understand the true risk of mutagenesis imposed by programmable nucleases. It is worth noting that off-target effects may be cell-type specific; for example off-target effects in transformed cell lines with dysregulated DSB repair pathways may overestimate off-target effects that would be observed in healthy primary cells.
In order to reduce the frequency of off-target effects, many groups are rapidly improving the targeting specificity of Cas9. For example, transformation of Cas9 into a single-strand DNA nickase that primarily generates DSBs by creating two separate single-strand breaks on opposite DNA strands via the expression of two separate guide RNAs, reduces off-target indel formation at computationally predicted off target sites102,109. Additionally, truncation of the guide RNA as well as RNA-guided FokI nuclease based on fusion between catalytically inactive Cas9 and the FokI nuclease domain are also able to achieve improved levels of targeting specificity117-119.
Alternative genome editing strategies not involving nucleases have also been explored, and may pose a lower mutagenic risk62. AAV genomes containing transgenes flanked by homology to target loci are capable of stimulating HDR in the absence of a nuclease, albeit at lower rates 48,86,120-122. Using this strategy one study targeted a factor IX cDNA to the highly expressed albumin locus, allowing for correction of the bleeding diathesis phenotype in factor IX deficient mice62. By targeting a highly expressed locus the authors could achieve 7-20% of factor IX levels, despite an HDR rate of 0.5%. Although this strategy may not be widely applicable due to the low absolute targeting rate, this and future, improved nuclease strategies should also be considered for therapeutic applications.
Delivery
Another major challenging facing clinical translation is delivery of editing systems to target cell types. A variety of nucleic acid or protein delivery methods may be used to introduce genome editing nucleases into target cells ex vivo or in vivo (Figure 3). Depending on the choice of delivery method, the nucleases may either be transiently or permanently expressed in the target cell. Given that nucleases may exhibit off-target cleavage activity or trigger immune responses, the delivery system should be carefully selected.
For ex vivo applications, such as editing of hematopoietic stem cells, electroporation may be used to achieve transient nuclease expression through delivery of DNA-based nuclease expression vectors, mRNA, or protein. Both integration-competent and deficient lentiviral vectors have also been successfully used to drive nuclease expression. However, integrating lentiviral vectors may be less desirable as they drive constitutive expression and may result in more off-target activity. In addition, all three nuclease platform have also been demonstrated to be amenable to modifications so that proteins can be directly delivered into cells either through engineered cell-penetrating peptides or chemical conjugation106,123,124.
For in vivo applications, the most promising delivery systems are viral vectors, particularly adeno-associated viral (AAV) vectors, which have recently been approved for clinical use125. AAV comes in many serotypes and have been shown to have high delivery efficacy for a variety of tissue types including the eye, brain, liver, and muscle126. However, the relatively small packaging capacity of AAV vectors post some challenges for nuclease delivery. Whereas ZFNs are relatively small and a dimeric ZFN pair can be packaged into a single AAV, a dimeric TALEN pair is much larger and will likely need to be packaged into two separate AAV vectors. For Cas9, short orthologs may be packaged along with guide RNAs into a single AAV. So far, AAV-mediated nuclease expression has been demonstrated to be successful in several tissue types, including liver and brain48,127. In the case of viral mediated Cas9 delivery, which may result in constitutive expression of nuclease proteins and cause genome instability and toxicity, self-cleaving mechanisms may be used to inactivate the nuclease transgene on the delivery vector128.
Despite the potential of AAV-mediated in vivo nuclease expression, there are several challenges that will require further development. First, AAV-mediated nuclease expression is often constitutive and it would be more desirable to be able to shut down nuclease expression after the genome editing event has successfully occurred in the target cell. Second, patients who have already been naturally exposed to AAV will likely have developed immunity against specific serotypes. Therefore AAV may not be an appropriate delivery vehicle for these patients.
To overcome these challenges faced by viral vectors, nanoparticle- and lipid-based in vivo mRNA or protein delivery systems may provide an attractive alternative123,129. Delivery of nuclease mRNA via nanoparticle conjugation, or nuclease proteins will permit more precise dosage control, which has been shown to affect the level of off-target mutation rate88,124. mRNA or protein delivery will also be transient, therefore minimizing any potentially undesirable nuclease-induced toxicity. Finally, for nuclease protein delivery, especially TALENs and Cas9, which are derived from microbial origins, exposure of the microbial proteins may stimulate immune reactions. Potential strategies for circumventing immunotoxicity resulting from protein delivery may include limiting dosage and protein humanization to reduce immunogenicity130.
Conclusion
The enormous excitement surrounding genome editing needs to be coupled with strategic planning and rigorous but enabling regulatory processes to ensure successful development of this class of potentially life-changing medicine. The technology will require a number of iterations to systematically optimize its efficacy, safety, and specificity. Despite being in its infancy, genome editing presents tantalizing opportunities for tackling a number of diseases that are beyond the reach of previous therapies. Given the accelerating pace of technological advances and broad range of basic science and clinical applications, the road ahead will undoubtedly be an exciting one.
Box: Using Genome Editing to Treat Non-monogenic Diseases.
Introduction of protective mutations for complex diseases treatment
The abundance of genetic information has made it possible to identify naturally occurring mutations that confer resistance to disease phenotypes. These mutations occur in both coding and non-coding regions of the genome and have received attention as therapeutic targets for complex, non-monogenic diseases such as cardiovascular disease58,136, HIV97, Alzheimer's disease59 and hemoglobinopathies137. Genome editing provides the possibility of introducing these protective mutations into patients to reverse illness.
Many known protective mutations involve loss of function alleles, which can be introduced via NHEJ-mediated gene disruption. This approach has rapidly gained traction due to the high efficiency of NHEJ in therapeutically accessible cells and treatment using this strategy is currently in clinical trials for HIV46. [AU: following paragraph removal ok to save space in the box. The HIV example is extensively discussed in the main text]
Mutations that protect against disease also lie hidden outside the coding region of the genome. GWAS and CHIP-seq were recently used together to identify non-coding mutations that may be important targets for genome editing therapy 138,137. A major factor controlling sickle cell disease severity is the expression level of fetal hemoglobin (HbF), increased HbF levels decreasing disease severity. A GWAS for regions controlling HbF expression identified variation within the BCL11A gene, the product of which is known to negatively regulate HbF expression 138,139. This variation promotes transcription factor binding within an intron that enhances BCL11A expression in the erythroid lineage, thereby decreasing HbF expression levels in red blood cells. TALENs were used to directly remove this intron from erythroid cells [AU:OK?] and showed that this resulted in an increase in HbF levels 137. However, this study was not carried out in HSCs, the most therapeutically relevant cell population for this disease. It will be exciting to see if this approach can be extended to these cells to provide a clinical benefit for patients. Furthermore, it is worth noting that non-coding regions will likely hold therapeutically important regions: 93% of GWAS hits, disease- and trait-associated are found within noncoding regions 140.
Programmable Nucleases as Antivirals
In addition, programmable nucleases may be developed as anti-viral strategies.
In principle, nucleases may be used to target viral sequences for cleavage and subsequent destruction. Additionally, NHEJ based mutagenesis of elements critical for viral fitness could render latent viruses incapable of propagating infection. Alternatively, multiplexed nucleases like Cas9 could be used to excise provirus from the genomes of infected cells, leading to their degradation by cellular nucleases.
Efforts to develop genome editing nuclease for antiviral therapy have largely focused on HIV, where large reservoirs of latent provirus can persist in the presence of anti-retroviral therapies and serve to re-activate infection once treatment is stopped. The long-terminal repeats (LTRs) of HIV drive viral gene expression and are critical for viral fitness. By targeting Cas9 to cleave LTR sequences, a study recently demonstrated the possibility of mutating the proviral LTR and significantly reduces expression of HIV genes in T cells 64. Although an exciting discovery, there are several additional challenges to translate this in vitro result to the clinic. Likely the biggest challenge will be delivering nucleases to all HIV carrying cells in an infected individual to eliminate all of the latent provirus. Currently, there are no therapeutic platforms capable of delivering genome editing nucleases to the majority of T cells. Similar strategies have been seen with HPV 141, and HBV 65,66 but most infectious diseases face the same problem as HIV: extremely efficient delivery of genome editing tools is likely required to achieve complete removal of viral infection.
References
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Thoene JG. Small molecule therapy for genetic disease. Cambridge University Press; Cambridge, UK ; New York: 2010. [Google Scholar]
- 3.Kay MA. State-of-the-art gene-based therapies: the road ahead. Nature reviews. Genetics. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 4.Vaishnaw AK, et al. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaspar HB, et al. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Science translational medicine. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 6.Howe SJ, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. The Journal of clinical investigation. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aiuti A, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO molecular medicine. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature reviews. Drug discovery. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 11.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature reviews. Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 13.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 14.Scharenberg AM, Duchateau P, Smith J. Genome engineering with TAL-effector nucleases and alternative modular nuclease technologies. Current gene therapy. 2013;13:291–303. doi: 10.2174/15665232113139990026. [DOI] [PubMed] [Google Scholar]
- 15.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPRCas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thierry A, Dujon B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: a new method for physical mapping of eukaryotic genomes. Nucleic acids research. 1992;20:5625–5631. doi: 10.1093/nar/20.21.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thierry A, et al. Cleavage of yeast and bacteriophage T7 genomes at a single site using the rare cutter endonuclease I-Sce I. Nucleic acids research. 1991;19:189–190. doi: 10.1093/nar/19.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic acids research. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boissel S, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic acids research. 2014;42:2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annual review of biophysics and biomolecular structure. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 22.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 23.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO journal. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature biotechnology. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 26.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 27.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 28.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic acids research. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 30.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 31.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nature biotechnology. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 33.Mahfouz MM, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic acids research. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 36.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 37.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 38.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic acids research. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isalan M. Zinc-finger nucleases: how to play two good hands. Nature methods. 2012;9:32–34. doi: 10.1038/nmeth.1805. [DOI] [PubMed] [Google Scholar]
- 45.Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnology and bioengineering. 2013;110:1811–1821. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 46.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. The New England journal of medicine. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genovese P, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature biotechnology. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 51.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Molecular and cellular biology. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Molecular and cellular biology. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic acids research. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plessis A, Perrin A, Haber JE, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Molecular and cellular biology. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudin N, Sugarman E, Haber JE. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 58.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nature genetics. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 59.Jonsson T, et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 60.Lombardo A, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nature methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 61.Moehle EA, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barzel A, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2014 doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. The New England journal of medicine. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin SR, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Molecular therapy. Nucleic acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:1889–1897. doi: 10.1038/mt.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Molecular therapy : the journal of the American Society of Gene Therapy. 2010;18:947–954. doi: 10.1038/mt.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber ND, et al. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PloS one. 2014;9:e97579. doi: 10.1371/journal.pone.0097579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holt N, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nature biotechnology. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nature biotechnology. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hacein-Bey-Abina S, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. The New England journal of medicine. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 72.Gaspar HB, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 73.Bousso P, et al. Diversity, functionality, and stability of the T cell repertoire derived in vivo from a single human T cell precursor. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:274–278. doi: 10.1073/pnas.97.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malech HL, et al. Prolonged production of NADPH oxidase-corrected granulocytes after gene therapy of chronic granulomatous disease. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12133–12138. doi: 10.1073/pnas.94.22.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang HJ, et al. Retroviral gene therapy for X-linked chronic granulomatous disease: results from phase I/II trial. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:2092–2101. doi: 10.1038/mt.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lofqvist T, Nilsson IM, Berntorp E, Pettersson H. Haemophilia prophylaxis in young patients--a long-term follow-up. Journal of internal medicine. 1997;241:395–400. doi: 10.1046/j.1365-2796.1997.130135000.x. [DOI] [PubMed] [Google Scholar]
- 77.Kaushansky K, Williams WJ. Williams hematology. McGraw-Hill Medical; New York: 2010. [Google Scholar]
- 78.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Molecular and cellular biology. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma S. Age-related nonhomologous end joining activity in rat neurons. Brain research bulletin. 2007;73:48–54. doi: 10.1016/j.brainresbull.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 80.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 82.Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene Targeting with Zinc-finger Nucleases. G3. 2013 doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews. Molecular cell biology. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Orlando SJ, et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic acids research. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nature methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller DG, et al. Gene targeting in vivo by adeno-associated virus vectors. Nature biotechnology. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- 87.Beumer KJ, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bunn HF, Aster J. Pathophysiology of blood disorders. McGraw-Hill; New York: 2011. [Google Scholar]
- 90.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nature reviews. Genetics. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nguyen TH, Ferry N. Liver gene therapy: advances and hurdles. Gene therapy. 2004;11(Suppl 1):S76–84. doi: 10.1038/sj.gt.3302373. [DOI] [PubMed] [Google Scholar]
- 92.Boye SE, Boye SL, Lewin AS, Hauswirth WW. A comprehensive review of retinal gene therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:509–519. doi: 10.1038/mt.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene therapy. 2004;11(Suppl 1):S10–17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 94.Yin H, et al. Non-viral vectors for gene-based therapy. Nature reviews. Genetics. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 95.Mingozzi F, High KA. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lamers CH, et al. Immune responses to transgene and retroviral vector in patients treated with ex vivo-engineered T cells. Blood. 2011;117:72–82. doi: 10.1182/blood-2010-07-294520. [DOI] [PubMed] [Google Scholar]
- 97.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. The New England journal of medicine. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 98.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ding Q, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circulation research. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weller SK, Sawitzke JA. Recombination promoted by DNA viruses: phage lambda to herpes simplex virus. Annual review of microbiology. 2014;68:237–258. doi: 10.1146/annurev-micro-091313-103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome research. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakade S, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nature communications. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nature methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature biotechnology. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 106.Guilinger JP, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nature methods. 2014;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mali P, et al. Barcoding cells using cell-surface programmable DNA-binding domains. Nature methods. 2013;10:403–406. doi: 10.1038/nmeth.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature biotechnology. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Veres A, et al. Low incidence of off-target mutations in individual CRISPR Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell stem cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan EP, Li Y, Del Castillo Velasco-Herrera M, Yusa K, Bradley A. Off-target assessment of CRISPR-Cas9 guiding RNAs in human iPS and mouse ES cells. Genesis. 2014 doi: 10.1002/dvg.22835. [DOI] [PubMed] [Google Scholar]
- 113.Yang L, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nature communications. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nature methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology. 2014 doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nature biotechnology. 2014 doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature biotechnology. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature biotechnology. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lisowski L, et al. Ribosomal DNA integrating rAAV-rDNA vectors allow for stable transgene expression. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20:1912–1923. doi: 10.1038/mt.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi Y, Falahati R, Zhang J, Flebbe-Rehwaldt L, Gaensler KM. Role of antigen-specific regulatory CD4+CD25+ T cells in tolerance induction after neonatal IP administration of AAV-hF.IX. Gene therapy. 2013;20:987–996. doi: 10.1038/gt.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Paulk NK, Loza LM, Finegold MJ, Grompe M. AAV-mediated gene targeting is significantly enhanced by transient inhibition of nonhomologous end joining or the proteasome in vivo. Human gene therapy. 2012;23:658–665. doi: 10.1089/hum.2012.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature biotechnology. 2014 doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nature methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wirth T, Parker N, Yla-Herttuala S. History of gene therapy. Gene. 2013;525:162–169. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- 126.Samulski RJ, Muzyczka N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annual Review of Virology. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 127.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nature biotechnology. 2014 doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moore R, et al. CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic acids research. 2014 doi: 10.1093/nar/gku1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kormann MS, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nature biotechnology. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 130.Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature. 1988;332:323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 131.Ye L, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ousterout DG, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:1718–1726. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Long C, et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell stem cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 135.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell stem cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 136.Tg, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. The New England journal of medicine. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Menzel S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nature genetics. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 139.Xu J, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pennisi E. The Biology of Genomes. Disease risk links to gene regulation. Science. 2011;332:1031. doi: 10.1126/science.332.6033.1031. [DOI] [PubMed] [Google Scholar]
- 141.Kennedy EM, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells using a bacterial CRISPR/Cas RNA-guided endonuclease. Journal of virology. 2014 doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


