Abstract
Identifying and tracking cells in the brain is a challenging task, requiring advanced molecular biology techniques and precise anatomical information. We have applied the principle of RGB marking to the long-term marking and tracking of progenitor and mature cells in the adult brain. Briefly, multicolor RGB marking is based on the simultaneous, lentiviral vector-mediated expression of three genes encoding fluorescent proteins in the three basic colors (red, green, and blue). Here, we show the application of RGB marking to the stable multicolor marking of adult granule cells in the hippocampus. This technique provides each individual cell with a characteristic hue, which facilitates their anatomical identification and spatial tracking. Multicolor RBG marking of mature neurons can provide an effective approach to dissect the function of individual cells, as it allows combination with functional techniques, facilitating the understanding of complex neuronal populations such as that of the dentate gyrus.
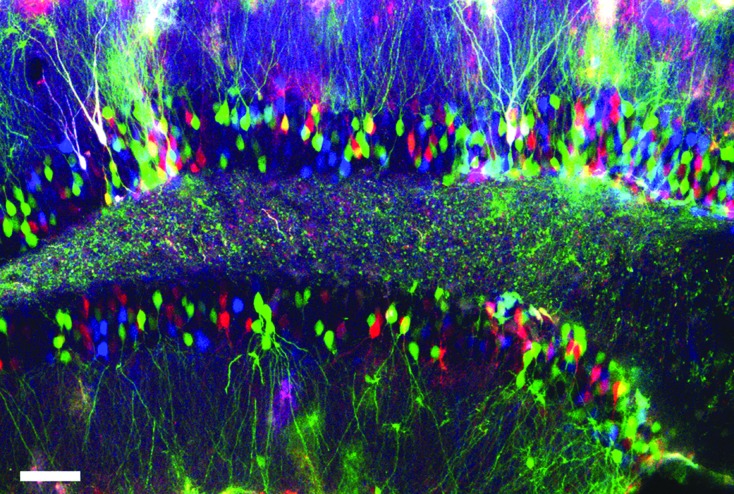
The analysis and genetic modification of complex neuronal populations at the single-cell level is a challenge in modern neuroscience. Lately, various transgenic mouse models have been introduced that facilitate spectacular color-coding of brain cells (for relevant references see Reference 1). To overcome the need for transgenic animals we have applied the principle of RGB marking2,3 to study complex neuronal populations in vivo.1 The simultaneous, lentiviral vector-mediated expression of three genes encoding fluorescent proteins in the three basic colors (red, green, and blue) provides multicolor labeling of different neural populations.1 In the image (Fig. 1), RGB marking of hippocampal mature granular neurons was achieved after intraparenchymal stereotaxic injection of lentiviral vectors,1 and analyzed 2 weeks later. The individual lentiviral vectors are incorporated at different copy numbers by each individual cell, generating the combinatorial and cell-specific expression of a color tag. These cell-specific color hues can be used for single-cell imaging, analysis of cell morphology, or cell development purposes, depending on the experimental setup and needs.1,4 When applied to the dentate gyrus, RGB marking permits the reconstruction of large numbers of neurons in a single experiment, which was not achievable previously without the use of alternative techniques allowing sparse labeling (i.e., retroviral tracing or Golgi staining).5 Given the stability of the color tags, RGB marking could be useful to investigate the long-term effects of the coexpression of relevant neurotrophic factors or key synaptic mediators in cell numbers or morphology. Also, given the photostability of fluorescent proteins, RGB-traced granule cells can be recorded by patch clamp,4 allowing the identification of the recorded single cells in subsequent imaging techniques, without the need for intracellular filling with tracers, or even allowing the precise pairing of neuron–synaptic boutons by complex morphometric analysis.6 RGB marking of granule cells, when combined with live imaging, would allow the understanding of the positioning and turnover of granule cells within the dentate gyrus in health and also in relevant models of disease.7 Identifying neurons at the single-cell level enables the precise quantification of region-dependent cell death in time-lapse experiments, critical to understand memory-dependent dynamics.
FIG. 1.
RGB marking of granular neurons after intraparenchymal administration of VSVG-CMV lentiviral vectors, analyzed 2 weeks after injection. Fluorescence is shown in red (mCherry), green (Venus), and blue (Cerulean). All images were analyzed by confocal microscopy. Scale bar, 25 μm.
This technique facilitates single-cell analysis and cell tracking over long periods of time and can be easily implemented in most neuroscience laboratories. Moreover, the use of lentiviral gene ontology (LeGO) vectors8 allows the coexpression of fluorescent proteins and genes of interest and/or short-hairpin RNAs (shRNAs), providing a flexible and potent toolbox for multiple and simultaneous gene modification. We have made RGB marking in the brain available in different formats: using the spleen focus-forming virus or cytomegalovirus promoter, which enables targeting of different cell types, and using lentiviral or gammaretroviral vectors, which enables targeting broad or selected (proliferating) cell populations.1 Multicolor RGB marking in the brain could be applied for time-lapse imaging, to develop gene therapy experiments and fate modification approaches, adapting well to the current needs of the research in neuroscience.
In the image (Fig. 1), fluorescence is shown in red (mCherry), green (Venus), and blue (Cerulean). The image was analyzed with an SP5 Leica confocal system. Scale bar, 25 μm.
Acknowledgments
The authors are indebted to many colleagues for kind support with various cells and constructs: R.Y. Tsien (Howard Hughes Medical Institute, USA) for mCherry cDNA, A. Miyawaki (RIKEN, Japan) and T. Schroeder (ETH Zürich, Switzerland) for Venus cDNA, D.W. Piston (Vanderbilt-Ingram Cancer Center, USA) for Cerulean cDNA, and Axel Schambach (MH Hannover, Germany) for the retroviral vector RSF91.GPF.pre*. This research was funded by the European Union Seventh Framework Programme under grant agreement IEF273243, by the Medical Research Council, Wessex Medical Research, and the DFG (SFB841, project SP2). K.R. was supported by a young investigator grant within the FFM Programme of the Univ. Med. Centre Hamburg-Eppendorf (NWF-12/09).
Author Disclosure Statement
The authors declare no competing financial interests exist.
References
- 1.Gomez-Nicola D, Riecken K, Fehse B, et al. . In-vivo RGB marking and multicolour single-cell tracking in the adult brain. Sci Rep 2014;4:7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber K, Thomaschewski M, Warlich M, et al. . RGB marking facilitates multicolor clonal cell tracking. Nat Med 2011;17:504–509 [DOI] [PubMed] [Google Scholar]
- 3.Weber K, Thomaschewski M, Benten D, et al. . RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat Protoc 2012;7:839–849 [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Nicola D, Suzzi S, Vargas-Caballero M, et al. . Temporal dynamics of hippocampal neurogenesis in chronic neurodegeneration. Brain 2014;137:2312–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahimi O, Claiborne BJ. Morphological development and maturation of granule neuron dendrites in the rat dentate gyrus. Prog Brain Res 2007;163:167–181 [DOI] [PubMed] [Google Scholar]
- 6.Wilke SA, Antonios JK, Bushong EA, et al. . Deconstructing complexity: serial block-face electron microscopic analysis of the hippocampal mossy fiber synapse. J Neurosci 2013;33:507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danzer SC. Depression, stress, epilepsy and adult neurogenesis. Exp Neurol 2012;233:22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber K, Bartsch U, Stocking C, et al. . A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol Ther 2008;16:698–706 [DOI] [PubMed] [Google Scholar]