Abstract
Molecular chaperones and their associated cofactors form a group of highly specialized proteins that orchestrate the folding and unfolding of other proteins and the assembly and disassembly of protein complexes. Chaperones are found in all cell types and organisms, and their activity must be tightly regulated to maintain normal cell function. Indeed, deregulation of protein folding and protein complex assembly is the cause of various human diseases. Here, we present the results of an extensive review of the literature revealing that the post-translational modification (PTM) of chaperones has been selected during evolution as an efficient mean to regulate the activity and specificity of these key proteins. Because the addition and reciprocal removal of chemical groups can be triggered very rapidly, this mechanism provides an efficient switch to precisely regulate the activity of chaperones on specific substrates. The large number of PTMs detected in chaperones suggests that a combinatory code is at play to regulate function, activity, localization, and substrate specificity for this group of biologically important proteins. This review surveys the core information currently available as a starting point toward the more ambitious endeavor of deciphering the “chaperone code”.
Keywords: Molecular chaperone, Post-translational modification, Hsp70, Hsp90, VCP
1. Introduction
Post-translational modifications (PTMs) encompass a number of specific chemical modifications that occur on proteins following their synthesis. PTMs can greatly affect the activity of a protein or its molecular interaction network, and constitute a tool for regulating protein function as well as one of the means through which proteome complexity is achieved. Although the identification of PTMs originally relied on shifts of the protein’s molecular weight and isoelectric properties, the techniques employed have since evolved tremendously with use of isotope labeling and mass spectrometry. Indeed, the development of powerful biochemical approaches such as the affinity purification of tagged proteins coupled with mass spectrometry analysis of the co-purified proteins, in conjunction with improved mass precision achieved by the new generation mass spectrometers, has played a pivotal role in the identification of post-translational-modifying enzymes, their targets and the PTMs themselves. Some groups of proteins are extensively targeted by PTMs due to the importance of their precise and rapid regulation. For example, PTMs of nucleosomal components were shown to orchestrate crucial functions like gene expression, DNA repair and replication. Patterns of modification were observed on histones, some with overlapping functions, some opposing and others appearing in a sequential manner. From this seemingly endless array of PTMs combinations, a “histone code” was nonetheless derived which opened up a new area of research: “epigenetics” or the study of heritable variations in gene expression not encoded directly in the DNA sequence.
Molecular chaperones are proteins involved in the folding or unfolding of other proteins as well as assembly or disassembly of larger macromolecular complexes. As the protocols and tools utilized in proteomics have improved, it has become apparent that chaperones are extensively targeted by PTMs, although the enzymes responsible and functional impacts of these modifications remain, for the most part, poorly understood. Given the overwhelming number of PTMs detected in chaperones, it is only logical to conclude that a code, similar to the one that has been proposed for histones, does exist although it will likely take years before its meaning is fully decrypted. This review aims at compiling the data available so far in the literature on this subject and the authors hope that this convergence of information may help others in finding recurring motifs in this new “chaperone code”.
Chaperones represent a wide and eclectic group of proteins and definition of what exactly constitutes a chaperone is somewhat inexplicit. For this reason we have decided to limit our reviewing effort to proteins for which there was the most abundant literature in regards to PTMs, namely, Hsp70, Hsp90 and VCP/p97. Although we have gone to great lengths to review all relevant studies, the task of compiling the collected data in a concise manner was an arduous one and we would therefore like to apologize to authors who may have published works on chaperone PTMs that are not included in this review.
2. Post-translational modification of Hsp70 chaperones
Hsp70 chaperones were first discovered as proteins that are overexpressed in Drosophila in response to heat exposure [1]. Hsp70 chaperones are not only expressed as a mean of surviving environmental stress, but have broader roles, notably in assisting de novo folding of nascent polypeptides by binding to exposed hydrophobic patches which promotes adoption of their native conformation and prevents their aggregation [2]. In addition to their function in protein folding, Hsp70 chaperones facilitate translocation of some cargo protein to their organellar subcompartments [3–5] and control the activity of a number of regulatory proteins including nuclear receptors, kinases and transcription factors [6]. The most prominent example is heat shock factor 1 (HSF-1). This transcription factor is maintained inactive through its association with Hsp chaperones in normal conditions, but is released during heat shock due to a greater affinity of the chaperones toward heat-denatured proteins, at which time HSF-1 can bind to Heat Shock promoter Elements (HSE) located in the genes of inducible forms of Hsp70 and Hsp90 and promote their expression [7].
The human genome encodes thirteen Hsp70 proteins (and four more distantly-related Hsp105/Hsp110 proteins), most of which localize predominantly in the cytoplasm like constitutive Hsc70 and inducible Hsp70, while others are more specialized and directed toward specific compartments, such as the Endoplasmic Reticulum (ER)-specific BiP/ GRP78 and mitochondrial GRP75 [8]. Hsp70 is a monomer divided in two functional domains: an N-terminal ATPase domain and a “client” protein binding domain. At the extreme C-terminal end of most cytoplasmic Hsp70 chaperones lies an EEVD motif that is known to interact with certain TetratricoPeptide Repeat (TPR)-containing proteins [9,10].
Many cochaperone of Hsp70 are believed to function by modulating the rate at which key steps occur during the chaperone’s ATP cycle. The diverse members of the Hsp40/DnaJ protein family bind to unfolded client proteins, thereby contributing to Hsp70 specificity, and have furthermore been shown to activate its ATPase activity, resulting in rapid hydrolysis of ATP from which the energy to create conformational changes in the chaperone is derived [11,12]. Bag-1 and Hsp105/Hsp110, on the other hand, have a nucleotide exchange activity which promotes ADP release and discharge of the Hsp70 substrate [13–15]. This effect is antagonized by yet another cochaperone, HIP, which stabilizes the ADP-bound state and prevents premature release of the client protein [14].
Hsp70 and Hsp90 cofactor HOP provides a platform upon which both chaperones can cooperate to facilitate proper folding. In this model, Hsp70 first interacts with the non-native protein and then passes it on to Hsp90 which would then more actively unwind the polypeptide and allow achievement of its native state [16]. CHIP is an E3 ubiquitin ligase that mediates ubiquitination and subsequent degradation of client proteins (for explanation see ubiquitin section of Hsp70 below) and could therefore act as a quality control regulator of the folding pathway [17]. Since both cochaperones are TPR-containing proteins, it is believed that they compete for chaperone binding through the EEVD motif, thereby generating an equilibrium between folding and degradation of Hsp cargo.
Expression levels of Hsp70 have been shown to be either up- or down-regulated in a variety of cancer types [18,19] and although some have questioned its validity as a prognostic marker [20] it would appear that depletion or inactivation of this chaperone sensitizes tumor cells to apoptosis [21–23]. Hsp70 can also provide a shielding effect against neurodegenerative aggregation disorders such as Huntington’s and Parkinson’s disease, either by directly promoting resolubilization of aggregates or by preventing apoptosis of the neurons [24,25]. However, Hsp70-mediated protection against cytotoxicity would appear to be lost with time as concentration of the chaperone decline in an age-dependant manner [26,27]. Interestingly, centenarians were shown to maintain elevated levels of the chaperone, which may be a factor accounting for longevity [28].
2.1. Phosphorylation
Phosphorylation is one of the best known and prevalent PTM in the cell. In most cases, a phosphate group is hydrolyzed from ATP and covalently linked to the hydroxyl group of serine, threonine or tyrosine of the substrate protein. Numerous wide-ranging studies have been conducted to identify variations in the phosphoproteome in response to diverse conditions. In many of these, phosphosites were identified within chaperones of the Hsp70 family. Unfortunately, given the overwhelming data stemming from such broad analyses, the role of these modifications on chaperone function has seldom been pursued further. All modifications encountered in the survey of the available scientific literature have been listed in a graphical format, starting with PTMs of Hsp70 in Fig. 1. More specific examples are listed below.
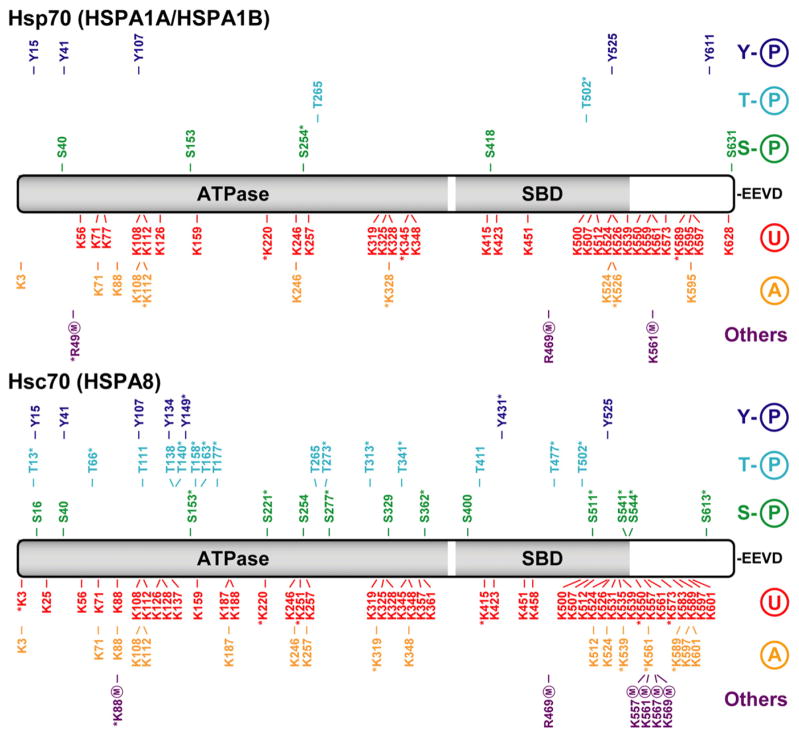
Fig. 1.
Hsp70 and Hsc70 post-translational modifications. Linear representation of the Hsp70 and Hsc70 isoforms of the Hsp70 family with ATPase domain, substrate binding domain (SBD) and C-terminal EEVD motif. All reported PTMs are marked according to a color code: tyrosine phosphorylation (Y-P) in dark blue, threonine phosphorylation (T-P) in light blue, serine phosphorylation (S-P) in green, ubiquitination (U) in red, acetylation (A) in orange and all others modifications in purple. Methylation (M) of arginine or lysine residues are the only infrequent modifications indexed in this instance. The majority of the data was provided by the public PTM database PhosphoSitePlus (http://www.phosphosite.org) and also from various sources in the scientific literature. PTMs identified on the basis of a single mass spectrum or reported in a single article are denoted with an asterisk.
It has been noted previously that Hsp70 translocates from the cytoplasm to the nucleus under environmental stress such as heat shock [29]. Since tyrosine phosphorylation was identified as a PTM that is significantly increased following heat shock [30], it appears possible that such a modification could account for Hsp70’s relocalization. Indeed, it was found that a specific tyrosine residue (Y525) is phosphorylated directly after heat shock and that phosphomimetic mutation (substitution of the tyrosine in question by a negatively charged residue like aspartic acid) increases nuclear accumulation and promotes cell survival following heat shock [31]. Conversely, one study reported that Hsp70 is hypophosphorylated overall following heat shock, although this effect could be due to the fact that only the cytosolic fraction was analyzed in that particular report [32].
Human Akt is a homologue of an oncogene of the murine leukemia virus AKT8 [33]. Unsurprisingly, the expression and activity of this kinase has been shown to be upregulated in a number of cancers [34]. Deregulation in Akt activity has also been noted in diabetes mellitus, and in a number of neurodegenerative diseases [35,36]. It has been reported that Akt could target a number of chaperones in vitro. Among these are Hsp70 and BiP/GRP78, although the specific phosphosites have not been determined [37]. Akt has also been shown to phosphorylate various Hsp90 isoforms including Hsp90α, Hsp90β and GRP94 (PTMs of Hsp90 isoforms will be discussed in greater details in the next section). The modification of ER-specific chaperones, like BiP/GRP78 and GRP94, by the cytosolic kinase Akt may appear dubious and could very well be the result of in vitro experimental design that does not take into account subcellular compartmentalization. On the other hand, there have been occurrences of resident chaperones of the endoplasmic reticulum being retrotranslocated to the cytoplasm, along with client proteins [38]. Researchers should however remain cautious when considering these phosphosites.
Most of the data collected so far on the phosphorylation of Hsp70 have remained somewhat anecdotal in that, although phosphosites were detected and sometimes effects of such modifications have been reported, the underlying molecular mechanisms were almost never clearly identified. However a late-breaking article on chaperone phosphorylation provides a better picture of the mechanistics of the chaperone code. It was shown that the C-terminus of both Hsp70 and Hsp90 could be phosphorylated by a number of kinases in vitro [39]. Due to its proximity to the EEVD motif of both chaperones, this modification resulted in a modulation in affinity toward their TPR-containing cofactors. Indeed, phosphorylated Hsp chaperones displayed an increased interaction with HOP and a concomitant reduced binding to CHIP. Phosphorylation was particularly strong in proliferating cancer cells, where the balance of chaperone activities is believed to be toppled toward assisted folding of the client proteins by the HOP/Hsp70/Hsp90 complex rather than ubiquitin-mediated degradation through the E3 ubiquitin ligase CHIP.
2.2. Acetylation
Acetylation is a process through which an acetyl group is removed from donor acetyl-coenzyme A and is covalently linked to a target protein with the help of a Histone AcetylTransferase (HAT). While co-translational N-terminal acetylation generally targets the protein for degradation [40], post-translational lysine acetylation can have a wide variety of effects. Acetylation is a reversible modification and the removal of the acetylation mark is catalyzed by Histone DeAcetylases Complexes (HDAC), a number of which were shown to co-immunoprecipitate with Hsp70 [41]. The Histone deacetylase inhibitor FK228 is a potent anticancer agent [42], but the exact mechanism by which it exerts its effect is still unknown. One group has shown that this compound promotes acetylation of Hsp70 [43]. Following treatment with FK228, Hsp70 presents increased binding to various oncogenic or potentially tumorigenic client proteins (Bcr-Abl, Raf-1, Cdk4, Akt, and survivin) over Hsp90 which usually ensures their stability [44,45]. These proteins are thus rapidly degraded and could theoretically account for the anticancer effect of FK228. Acetylation as a prominent protein modification has garnered more attention in recent years and a number of high-throughput analyses have uncovered a multitude of acetylated lysines in Hsp70 chaperones [46–48].
2.3. Ubiquitination
In ubiquitination, a small modifying peptide known as ubiquitin is adenylated and loaded onto a ubiquitin-activating enzyme (E1). Subsequently, the peptide is passed on to one of the 30–40 ubiquitin-conjugating enzymes (E2) before reaching its final target through the help of one of over 600 ubiquitin protein ligases (E3) that drive ubiquitination specificity [49]. This cascade can be repeated a number of times, creating a chain of covalently linked ubiquitins. Depending on which lysine residue is used in the preceding ubiquitin peptide to link to the following ubiquitin module, the significance of this modification can greatly vary. It is generally considered that K48-linked ubiquitin chains target the substrate protein to degradation by the 26S proteasome [50]. However, while other linkage types may still direct the target protein to proteosomal degradation [51], they may also affect the function of modified proteins by changing their conformation or altering their molecular interaction network. As mentioned previously, the Hsp70 and Hsp90 cofactor CHIP is an E3 ubiquitin ligase that ubiquitinates client proteins. Both Hsp70 and Hsp90 have also been shown to be ubiquitinated themselves by CHIP. The functional significance of this modification is a matter of debate as some authors have postulated that ubiquitinated Hsp70 is directed to the proteasome for degradation, while others observed no effect on the steady-state of the chaperone [52–55]. Parkin is yet another E3 ubiquitin ligase that was found to catalyze the addition of ubiquitin moieties to Hsp70 [56]. The enzyme mediates the mono-ubiquitination of Hsp70 at multiple sites with no apparent effect on its stability. As with acetylation, numerous high-throughput analyses have identified an array of ubiquitinated residues [47,57–59]. Interestingly, a lot of these sites overlap with acetylation targets (see Fig. 1) which may suggest a competition between the two modifications.
2.4. Methylation
Protein methylation consists in the enzymatic transfer of a methyl group from a methyl donor, usually S-adenosylmethionine, to a substrate protein. The residues typically targeted by this modification are lysine and arginine, although side chains of histidine, glutamine, asparagine, glutamic acid, aspartatic acid and cysteine have, on occasions, been shown to be methylated [60]. In addition, some proteins can also be methylated on their N- or C-terminal ends. This modification was initially believed to be more stable than phosphorylation or acetylation, for which reverse enzymes phosphatases and acetylases were known, but a number of demethylases have since been identified [61,62]. Hsp70 chaperones have been shown to be methylated on both lysine and arginine residues [63], and the pattern of methylation can also change during cellular proliferation and under various conditions, including transformation by the Rous sarcoma virus and arsenite treatment [64]. Recently, our group discovered an entirely new family of lysine methyltransferases that preferentially target molecular chaperones [65]. In the case of Hsp70 chaperones, one of these enzymes, METTL21A, was found to trimethylate multiple Hsp70 isoforms on a conserved lysine (K561). Another group confirmed that K561 of Hsp70 was indeed the target of methylation, although in this instance the residue appeared to be dimethylated and the enzyme responsible for this mark was proposed to be SETD1A [66]. It was further demonstrated that the modification resulted in translocation of the chaperone from the cytoplasm to the nucleus, where increased interaction with Aurora kinase B could be observed. Dimethylation of Hsp70 was found to stimulate Aurora kinase B activity in phophorylating histone H3 and thus promote mitosis, consistent with the observation that K561 dimethylation of Hsp70 is elevated in numerous cancer cells.
3. Post-translational modification of Hsp90 chaperones
Hsp90 participates in the post-translational folding and stabilization of a multiplicity of client proteins as well as the assembly of numerous protein complexes [67]. In fact it has been shown that in yeast, the Hsp90 homolog interacts with as much as 10% of the proteome [68]. The Hsp90 family consists of four different isoforms: cytosolic constitutive (beta) and inducible (alpha) forms, the latter of which has been shown to be secreted in certain conditions [69], a mitochondrial member (TRAP1) and an ER-resident protein (GRP94).
Hsp90 comprises an N-terminal ATP-binding domain, a middle (M) domain that accounts for much of Hsp90 affinity toward its various client proteins and a C-domain that mediates homodimerization of the chaperone [70]. As was the case for Hsp70, Hsp90 cytosolic isoforms have a C-terminal EEVD motif that targets the chaperone to various TPR-containing cochaperones [71]. Structural analyses have revealed that large conformational changes take place upon ATP binding, where the chaperone shifts from an “open” state where the various domain are free to move about via flexible linker regions to a “closed” or “tense” conformation [72–77]. The N-terminal domain, through a transient dimerization and association with the M-domain [78], becomes catalytically active and can thus hydrolyze ATP to return to the initial open state. The chaperoning function of Hsp90 is the direct result of these ATP-driven permutations and it is believed that cochaperones act primarily by affecting these transitions. Aha1 and Cpr6 stimulate the ATP cycle, while HOP, p23 and Cdc37 interfere with it [79–82]. Other molecules are known to affect the ATP cycle of Hsp90, including Geldanamycin and its analogs which hinder its chaperoning function by occupying the ATP-binding pocket [83].
Hsp90 has been implicated in a number of cancers, mostly due to the stabilization of overexpressed oncogenic proteins, both mutant and normal [84,85]. In this regard, Hsp90 inhibitors are now considered promising anti-cancer agents [86–88]. Despite its deleterious role in cancer, Hsp90 function is thought to be crucial in promoting evolution and guarding against possible disease-causing products of mutated genes by acting as a “capacitor” or “genetic buffer”. Based on this hypothesis, the chaperone binds normally occurring genetic variants of various proteins that might provide a selective trait in stress conditions or could otherwise be potentially harmful to homeostatic cells [89–91].
3.1. Phosphorylation
Hsp90 has long been known to be an important phosphoprotein (see [92] for a review and Fig. 2 for a list of reported Hsp90 modifications). Hsp90 and its ER isoform were initially believed to be capable of autophosphorylation [93,94]. However, it is more likely that this activity is rather the result of co-purified kinases. Phosphorylation of Hsp90 has been shown to be linked to its chaperoning function. In one report, unphosphorylated Hsp90 was found to bind to the reovirus protein σ1 and become phosphorylated in the process of releasing its client [95]. Moreover, it was demonstrated that geldanamycin, the Hsp90 inhibitor, interferes with phosphorylation of the chaperone [95].
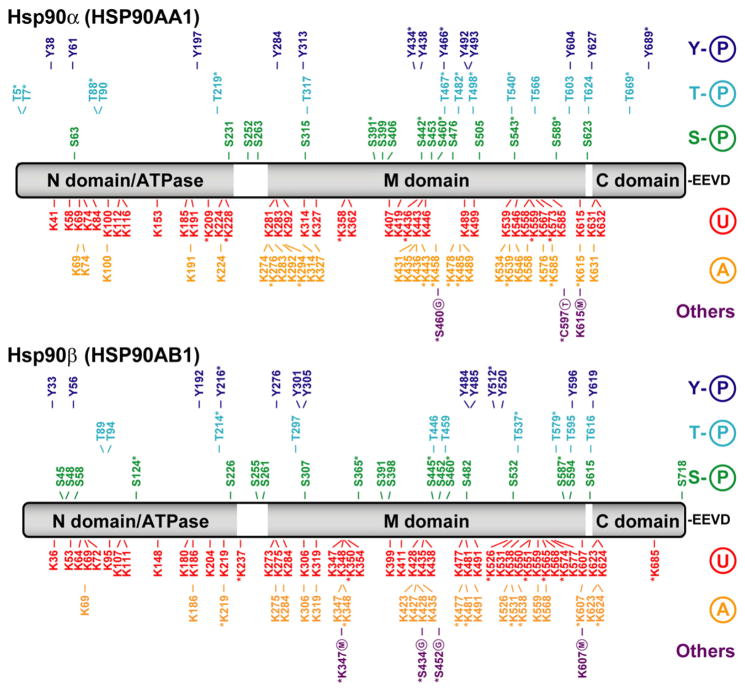
Fig. 2.
Hsp90α and Hsp90β post-translational modifications. Linear representation of the Hsp90α and Hsp90β isoforms of the Hsp90 family with N-terminal ATPase domain, middle (M) domain, C-terminal dimerization (C) domain and EEVD motif. All reported PTMs are marked according to a color code: tyrosine phosphorylation (Y-P) in dark blue, threonine phosphorylation (T-P) in light blue, serine phosphorylation (S-P) in green, ubiquitination (U) in red, acetylation (A) in orange and all others modifications in purple. Lysine methylation (M), glycosylation (G) and S-thionylation (T; a broad term which serves to describe a number of redox modifications that take place on the sulfur group of cysteine residues including disulfide oxidation, S-nitrosylation and S-glutathionylation) are among the infrequent modifications reported here. The majority of the data was provided by the public PTM database PhosphoSitePlus (http://www.phosphosite.org) and also from various sources in the scientific literature. PTMs identified on the basis of a single mass spectrum or reported in a single article are denoted with an asterisk.
Early characterization of Hsp90 showed that the pleiotropic kinase CK2 can phosphorylate Hsp90α at serine residues S231 and S263 as well as corresponding sites (S226 and S255) in the Hsp90β isoform [96]. The ER-specific isoform GRP94 is likewise phosphorylated by this kinase [97,98], raising again the logistical question of how an ER chaperone could possibly be modified by a cytosolic kinase. Although the exact biological function of these PTMs in human Hsp90 is still unclear, some authors have observed implications in apoptosome formation, arylhydrocarbon receptor-dependent transcriptional repression, B-Raf/MKK/ERK signaling and telomerase activation [99–102]. In another report, Hsp90 was shown to enhance phosphorylation of eukaryotic initiation factor 2α, but only when the chaperone itself had been phosphorylated beforehand by CK2 [103].
However CK2 is far from being the only serine/threonine kinase directed toward Hsp90. For example, Polo-like kinases PLK2 and PLK3 can phosphorylate Hsp90α, Hsp90β in vitro [104]. In the case of Hsp90β, the target residue of these kinases was shown to be S718, although the exact sites on other Hsp90 isoforms have yet to be identified. The alpha isoform of Hsp90 was also found to be phosphorylated at T5 and T7 by the DNA-dependent protein kinase (DNA-PK) and the beta isoform at T89, S391 and T616 in cells overexpressing the Pregnancy-upregulated Non-ubiquitous Calmodulin Kinase (PNCK) [105,106].
One of the most intensely-studied Hsp90 client proteins is the endothelial Nitric Oxide Synthase (eNOS). The chaperone has an activating effect on the enzyme resulting in enhanced Nitric Oxide (NO) production [107], which was shown to be a critical regulator of cardiovascular homeostasis [108] and to be lacking in patients with diabetes mellitus [109]. Reduced eNOS activity is believed to be the leading cause of diabetic vasculopathy. In this respect, it is interesting to note that high-glucose and diabetes mellitus both promote PKA-mediated Hsp90α phosphorylation at T90 and subsequent translocation to cell surface, which results in impaired eNOS activity and NO production [110].
Hsp90 isoforms were also found to be phosphorylated on tyrosines. For example, Y301 of Hsp90β is modified by c-Src in a Vascular Endothelial Growth Factor Receptor-2 (VEGFR-2)-dependent manner and has a direct implication in stimulation of eNOS activity and NO release. As such, tyrosine phosphorylation of Hsp90 could play a crucial role in mediating the proangiogenic effect of VEGF [111]. Y301 phosphorylation was furthermore shown to promote interaction with Aha1, which could account for the effect on eNOS activation as overexpression of the cochaperone itself yielded similar results on NO production [112].
In addition, Saccharomyces cerevisiae Wee1 phosphorylates both yeast and human Hsp90α on the conserved tyrosine Y38 [113]. Notably, this modification appears to occur in the nucleus during S-phase and leads to the subsequent ubiquitination and degradation of the chaperone. Non-phosphorylatable mutants showed decreased chaperoning function toward a subset of client proteins and limited interaction with cochaperones Aha1 and p23.
A prominent example of the impact of Hsp90 phosphorylation in defining a functional chaperone code was published recently and pertains to the role of three phosphosites on the association of various Hsp90 cofactors [114]. In vitro kinase reactions have shown that incubation of the chaperone with the kinase Yes results in phosphorylation of Y197, Y313 and Y627. An elegant model was proposed to integrate all interaction data from phosphomimetic mutants where Hsp90 first complexes with S13-phosphorylated Cdc37 and a client protein. TPR-containing phosphatase PP5 then joins the complex and dephosphorylates Cdc37. Next, the cochaperone is tyrosine phosphorylated by Yes, thus disrupting its association with the client protein which is not released from the complex as it still interacts with Hsp90. The chaperone itself is then phosphorylated at position Y197, leading to the release of Cdc37. Subsequent phosphorylation at Y313 promotes association with Aha1 and progression of the ATPase cycle. Once ATP has been hydrolyzed and the client protein has been chaperoned, final phosphorylation at Y627 antagonizes association with both Aha1 and PP5, with the final outcome being dis-aggregation of the complex. Little is known, however, about how the modified components, like Hsp90 and Cdc37, are recycled before another chaperoning cycle can take place. Nonetheless, this report was important because it points to an integral role of Hsp90 phosphorylation in the chaperone’s ATPase cycle, and therefore, its chaperoning ability. It also shed light on the equally essential modification of Hsp90 cofactors, such as phosphorylation of Cdc37.
Although the data collected on Hsp90 phosphorylation mostly pertains to the cytoplasmic alpha and beta isoforms, a few modifications have been attributed solely to the ER and mitochondrial counterparts. For instance, GRP94 is phosphorylated by the Golgi apparatus casein kinase (G-CK) at sites that are distinct from the ones that are targeted by CK2 [115]. Also, mitochondrial kinase PINK1 has been shown to phosphorylate TRAP1 [116]. Phosphorylation of TRAP1 is enhanced under oxidative stress and is crucial in protecting the cell against apoptosis. Interestingly, this effect was not observed for mutant forms of PINK that have been associated with autosomal forms of Parkinson’s disease.
3.2. Acetylation
Acetylation of Hsp90 was initially observed following treatment with the anticancer agent FK228 [117]. This molecule has the ability, as do many other histone deacetylase inhibitors, to inhibit growth and induce apoptosis in a variety of tumor cell lines [42,118]. It was found however that histone acetylation alone could not account for the molecule’s cytotoxic effect [119]. The molecule’s properties appear instead to be the result of increased acetylation of Hsp90, which interferes with binding and stabilization of oncoproteins p53, Raf-1, and ErbB2.
Hsp90 was later shown to form a complex with the cytoplasmic deacetylase HDAC6 in vivo, and that the enzyme could directly affect the acetylation status of Hsp90 [120,121]. HDAC6 had previously been implicated in clearance of misfolded protein aggregates [122], hinting at a possible role in regulating chaperone function. In cell depleted in HDAC6 by RNAi, activity of the Hsp90 client protein Glucocorticoid Receptor (GR) is compromised as a result of impaired formation of GR-Hsp90 heterocomplexes [120,123]. Furthermore, increased acetylation of Hsp90, either by siRNA-mediated knockdown of HDAC6 or treatment with the HDAC inhibitor trichostatin A, results in reduced affinity for ATP and cochaperone p23 [120,123].
The use of deacetylase inhibitors also lead to the discovery of various modified lysines within Hsp90. For example, acetylation of K294 was identified in Hsp90α purified from cells treated with trichostatin A, although the existence of additional sites was also noted [124]. Acetylation mimicking-mutants of Hsp90α at K294 displayed reduced affinity to a number of client proteins (ErbB2, p60v-Src, and androgen receptor) and cochaperones (p23, Cdc37, FKBP52, Aha1, Hsp70, HOP, and CHIP).
In cells treated with another HDAC inhibitor, LBH589, purified Hsp90α was shown to be acetylated at seven different lysine residues (K69, K100, K292, K327, K478, K546 and K558), although aforementioned K294 acetylation could not be confirmed [125]. In was also demonstrated that the histone acetyltransferase p300 can interact with and acetylate Hsp90α directly, but is not the sole chaperone-modifying HAT. Indeed, Hsp90 purified from p300-depleted cells still harbors acetylated lysines. Acetylation-mimetic mutants were once again employed to show that affinity of Hsp90 toward ATP, geldanamycin, Hsp70, cochaperone CHIP and client protein Raf-1 was either up- or down-regulated depending of which residue was mutated. Furthermore, hyperacetylation of Hsp90α through treatment with LBH589 as well as acetylation-mimetic mutants resulted in its extracellular export.
3.3. Methylation
Analysis of the protein interaction network of SET and MYND domain (SMYD) lysine methyltransferases revealed that Hsp90α interacts with both TPR-containing SMYD3 and SMYD2 [126]. The chaperone was methylated at K209 and K615 in vitro by SMYD2. These modifications are antagonized by the Hsp90 cofactor HOP, and methylation of K615 was further shown to be a reversible modification that can be removed by the demethylase LSD1. Yet another report demonstrated that SMYD2 was responsible for the monomethylation of K615 [127]. Highest level of SMYD2 expression and methylation of K615 were found in striated muscle tissue, where both proteins colocalize on the I-band region of myofibrils. SMYD2 and Hsp90 interact with the myofilament protein titin, a component of I-bands, although the formation of the complex is dependent upon methylation of the chaperone by its associated methyltransferase. This complex was shown to enhance titin stabilization and to be crucial in the formation of sarcomeric structures during muscle development.
3.4. S-Nitrosylation
S-Nitrosylation is a modification of cysteine residues where the thiol side chain is covalently linked to nitric oxide to form a thionitrite group [128]. No enzyme appears to be required for this reaction and only a subset of cellular proteins spontaneously nitrosylate at specific cysteine residues as a consequence of changes in the redox state of the cell. Hsp90α has been identified as a target of S-nitrosylation [129]. The positive regulatory effect of Hsp90 on eNOS function has already been mentioned in the preceding section on phosphorylation. Surprisingly, eNOS activation in endothelial cells promotes S-nitrosylation of Hsp90α at C597. The modified residue resides within a region proposed to be important for interaction with eNOS. This modification results in impairment of Hsp90α ATPase activity and reduced eNOS function. From these observations, an elegant model can be derived for an autoregulatory negative feedback loop of nitric oxide synthesis by eNOS.
3.5. Glycosylation
Glycosylation, or the enzymatic attachment of a sugar moiety to a protein, can involve a wide array of carbohydrates that can be covalently linked to the side chain of a number of amino acids. In the case of O-glycosylation, the reaction takes place on the hydroxyl group of serines, threonines and tyrosines, in a similar fashion as phosphorylation. While most protein glycosylation reactions occur within the secretory pathway, there is a plethora of well-known nuclear and cytoplasmic glycoconjugates. It has been reported that mouse Hsp90 can be O-glycosylated with N-acetyl-D-glucosamine (GlcNAc) in its middle domain on two serines for Hsp90β (S434 and S452) and a single serine for Hsp90α (S460) [130]. Although the exact function of this modification remains unknown, it is interesting to note that phosphorylation of Hsp90β S452 has also been observed, which may imply a competition between the two modifications in regulating Hsp90 function. Furthermore, it has been noted that the enzyme responsible for O-GlcNAcylation, O-linked β-N-acetylGlucosamine Transferase (OGT), shuttles between the nucleus and the cytoplasm [131] and contains a TPR domain which can interact directly with Hsp90. The chaperone appears to promote OGT stabilization, as inhibition of Hsp90 through the use of various inhibitors results in rapid proteasomal degradation of OGT [132]. It is thus tempting to speculate that a similar feedback mechanism could exist for O-GlcNAc modification as was shown for S-nitrosylation.
Most glycosylations occur within compartments of the secretory pathway and so it is unsurprising to note that ER-resident GRP94 is also glycosylated, although in this case the modification consists of N-glycosylation, more precisely, asparagine-linked oligosaccharides. In one report, it has been found that all GRP94 proteins contain basal glycosylation on N196 and that only a subset can be hyperglycosylated on various asparagine residues in the middle section of the protein (N424, N460 and N481) [133]. Contrasting with this all-or-none hyperglycosylation model, others have noted the presence of multiple, stable forms of GRP94 that differ only on the extent of their glycosylation [97,134]. Unfortunately, little is known about the functional significance of these modifications.
4. Post-translational modification of VCP
VCP/p97 is a member of the AAA (ATPases Associated with various cellular Activities) family of ATPases. The protein is composed of an approximately 200-amino acid N-terminal domain and two ATPase domains (D1 and D2), the former being critical for homohexamerization, while the latter is responsible for most of the protein’s ATPase activity and exhibits large conformational changes during ATP hydrolysis [135]. VCP, like Hsp70 and Hsp90 isoforms, is a ubiquitous and abundant protein that accounts for approximately 1% of all cell proteins and acts as a molecular chaperone in various cellular processes such as cell cycle progression [136], DNA repair [137–140], transcription factor control [141–143], autophagy [144,145], homotypic membrane fusion [146–148], and Endoplasmic Reticulum Associated protein Degradation (ERAD) [149–152]. It is believed that VCP achieves these various roles by recruiting many different cofactors.
Of these functions, degradation of misfolded proteins synthesized in the endoplasmic reticulum (ER) is the most extensively characterized. Newly synthesized proteins that fail to adopt their native conformation in the ER are quickly reassigned to a distinct compartment where they are ubiquitinated by membrane-bound Hrd1 or gp78 E3 ubiquitin ligase complexes [153] and retrotranslocated to the cytoplasm through a pore formed with Derlin-1 and/or Sec61 [154]. VCP, whether in complex with the Ufd1-Npl4 heterodimer or with other UBX-containing cofactors, interacts with the incorrectly folded protein as it emerges from the ER and provides the mechanical force needed to drive the extraction of the polypeptide into the cytoplasm using an ATP-driven “ratchet” or “milking” motion of reciprocally alternating conformations between the dual ATPase domains [155]. The chaperone also serves as a platform from which deglycosylation by Peptide N-Glycanase (PNGase) [156] and polyubiquitination by E4 multi-ubiquitination enzyme Ufd2 [157] can take place. Shuttle proteins, like Rad23 and Dsk2, then target the ubiquitinated substrate for degradation through direct interaction with both Ufd2 and the 26S proteasome complex [158]. Several deubiquitilating enzymes, such as Otu1, VCIP135, YOD1 and Ataxin-3 [148,159–161], have also been shown to interact with VCP, although their exact role in ERAD is still unknown.
Mutations in the VCP gene have been linked to the autosomal dominant disorder Inclusion Body Myopathy with Paget’s disease of bone and Fronto-temporal Dementia (IBMPFD) and, more recently, familial Amyotrophic Lateral Sclerosis (ALS) [162–168]. Most described VCP mutations reside within the N-terminal and D1 domains [169,170], which are domains proposed to be involved in cofactor association as well as interaction with ubiquitinated substrates [170–172]. Many studies were performed to define how these disease mutations affect the function of VCP. From a biochemical point of view, the most promising alteration concerned the increased ATPase activity, in conjunction with structural changes induced by ATP binding [173,174]. Histological manifestation of IBMPFD is similar to that of other IBMs, with muscle biopsies showing degenerating fibers, rimmed vacuoles, and sarcoplasmic inclusions of ubiquitin, VCP, and TAR DNA-binding protein 43 (TDP-43) [170]. Such VCP and ubiquitin inclusions are also observed in other neurodegenerative disorders as well as in Lewy bodies in Parkinson’s disease and dystrophic neurites in Alzheimer’s disease [175]. VCP has also been shown to interact with proteins containing polyglutamine expansions such as those responsible for Huntington’s disease and Machado-Joseph disease [176].
4.1. Phosphorylation and acetylation
Given VCP’s wide-ranging activities, it comes as no surprise that its post-translational modification would impact equally diverse cellular functions (see Fig. 3 for a list of reported VCP modifications). For example, it was demonstrated that the chaperone could be phosphorylated at S352, S745 and S747 by Akt under growth hormone activation or in response to hypoxia [177,178]. VCP phosphorylation has also been shown to take place during cold acclimation [179], sperm capacitation [180] and transitional ER assembly via Jak2 [181]. Furthermore, VCP was identified as an interactor of phosphatases PTPH1 [182] and PTPN22 [183].
Fig. 3.
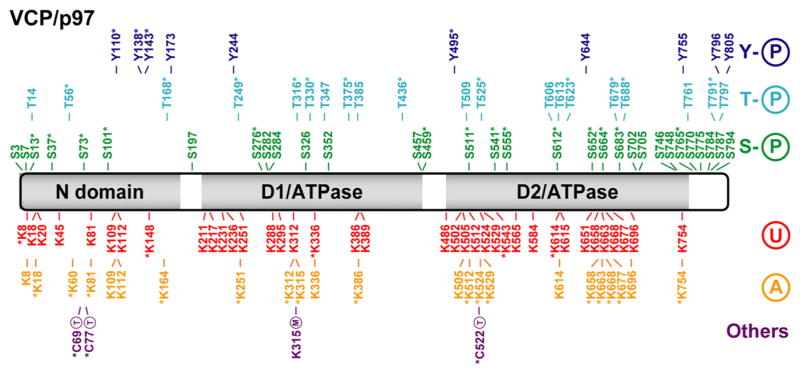
VCP/p97 post-translational modifications. Linear representation of the VCP/p97 protein with N-terminal (N) domain and dual ATPase domains D1 and D2. All reported PTMs are marked according to a color code: tyrosine phosphorylation (Y-P) in dark blue, threonine phosphorylation (T-P) in light blue, serine phosphorylation (S-P) in green, ubiquitination (U) in red, acetylation (A) in orange and all others modifications in purple. Lysine methylation (M) and S-thionylation (T; a broad term which serves to describe a number of redox modifications that take place on the sulfur group of cysteine residues including disulfide oxidation, S-nitrosylation and S-glutathionylation) are among the infrequent modifications reported here. The majority of the data was provided by the public PTM database PhosphoSitePlus (http://www.phosphosite.org) and also from various sources in the scientific literature. PTMs identified on the basis of a single mass spectrum or reported in a single article are denoted with an asterisk.
In addition to UBX-containing factors that bind the N-terminal domain of VCP, a number of VCP-interacting proteins have another structural element in common, the PUB domain [184]. PUB domain interaction with VCP is mediated by a short motif at the C-terminus of the chaperone [185]. This motif includes a tyrosine residue (Y805) that was previously identified as a target of phosphorylation in T cell receptor activation [186,187] and in centrosome association (or nuclear translocation in the case of yeast) during mitosis [188]. It was demonstrated that phosphorylation of this residue impedes interaction with PUB domain-containing PNGase and Ufd3, a factor that interacts with deubiquitinating enzyme Otu1 [160]. Given the fact that Ufd3 has been shown to compete with E4 multi-ubiquitination enzyme Ufd2 for binding with VCP [160], it is therefore possible that phosphorylation of T805 could promote Ufd2 recruitment and thus favor polyubiquitination and proteosomal degradation of client proteins instead of deubiquitination via Ufd3 and Otu1 and chaperoning.
The role of VCP in DNA repair is still obscure and has mostly been overshadowed by its function in ERAD. It has been demonstrated that a subpopulation of VCP resides in the nucleolus, where it interacts with the Werner syndrome protein [137,139], a helicase involved in the repair of double strand DNA breaks. In keeping with a possible role of the chaperone in the DNA damage response, VCP has been shown to be phosphorylated at S784 following UV irradiation or doxorubicin treatment, at which time it interacts more tightly with chromatin [138]. DNA-PK, which has been mentioned earlier in the Hsp90 section above, can phosphorylate this site in vitro, although data suggest that this particular kinase is not the only one responsible for this mark in vivo and that it could possibly be carried out by other PhosphatidylInositol-3 Kinase-related Kinases (PIKK), like the well-known transducers of DNA damage checkpoint ATM and ATR.
In cells where misfolded proteins aggregate, such as in polyglutamine disease and various other neurodegenerative disorders, it has been found that transcription is repressed through the active deacetylation of histones H3 and H4 [189]. It was reasoned that the decrease in production of new misfolded proteins through transcriptional repression might allow the cell to chaperone or degrade aggregated proteins. However, the link between accumulation of misfolded proteins and transcriptional suppression is still a matter of debate. VCP was proposed to play a role in this process. Indeed, in PC12 cells expressing an exogenous polyglutamine tract protein, VCP was found to translocate to the nucleus following phosphorylation of S612 and T613 and acetylation of K614 [190]. Phosphorylation and acetylation mimetic mutant of VCP induced deacetylation of histones H3 and H4 and transcriptional repression, indicating that the chaperone could in fact mediate transcriptional repression, although further study will be needed to elucidate the exact mechanisms through which VCP modifies chromatin.
4.2. Methylation
We and others have demonstrated that the previously un-characterized methyltransferase METTL21D, a homolog of Hsp70-targetting METTL21A, could methylate VCP on lysine residue K315 [65,191]. This residue had already been reported as a site of acetylation [192], but given the modest size discrepancy between lysine acetylation and trimethylation, it is possible that assignment of acetylation to this residue was rather the result of mis-annotation. The target lysine lies within VCP’s first ATPase domain, and it was shown that methylation of this residue results in abrogation of the individual domain’s ATPase activity in vitro. Our group went on to demonstrate that VCP methylation was concomitantly increased with the presence of UBX protein ASPSCR1, which also binds directly with METTL21D. Alternatively, ASPSCR1 had previously been ascribed a role in disassembly of the VCP hexamer [193] and since VCP appears to be more readily methylated as a monomer [191], it is therefore plausible that this cofactor may also promote methylation through this mechanism. Whatever the case may be, it appears as though IBMPFD and familial ALS-causing mutations can negatively impact on ASPSCR1-mediated methylation of the chaperone [65]. Methylation of VCP did not appear to result in a quantifiable effect on ubiquitin-mediated proteosomal degradation, autophagy or ATPase activity of the full chaperone in vivo [191]. METTL21D was shown, however, to promote tumor migration and invasiveness, consistent with its upregulation in mobilized macrophages and metastatic tumors [194].
4.3. S-Glutathionylation
S-Glutathionylation, like aforementioned S-nitrosylation, is a chemical modification that befalls on cysteine residues under oxidative stress. In the presence of oxidants, like diamide and peroxide, the overall ATPase activity of the chaperone is significantly decreased [195]. While C69, C77 and C522 were all shown to be S-glutathionylated in vitro, only the substitution of C522 shielded the ATPase activity of VCP against oxidative stress. Oxidation also strengthened the interaction with cofactors Npl4 and Ufd1 in vivo, although C522 did not appear to play a role in this phenomenon. C522 substitution did however protect cells against diamide-induced ER-derived cytoplasmic vacuolization and subsequent cell death. Although this particular cysteine residue is not conserved in S. cerevisiae, it was noted that substituting the corresponding residue for a cysteine sensitizes yeast cells to several oxidants. Oxidative stress has been shown to prevail in neurodegenerative disorder like Parkinson’s disease [196,197]. Furthermore, a correlation between aging and accumulation of oxidative damage has been observed [198]. It was thus posited that oxidation of VCP may play a role in the molecular physiopathology of neurodegenerative disorders, where the onset of symptoms typically comes at a later age.
5. Concluding remarks
Given the overwhelming number of PTMs that target chaperones, decrypting patterns of modifications into a unifying regulatory model may seem like a daunting task. However, surveying the data available so far allows us to identify common mechanisms by which modulation of chaperone function is enacted (Fig. 4). For example, alteration of the ATPase cycle at various steps has been proposed as a way by which cochaperones affect chaperone function and in some cases PTMs appear to work in the same way. Whether by direct modification of the nucleotide binding site or catalytic core, or by altering the overall structure of the protein through creation or destruction of more far-ranging intramolecular interactions, PTMs have been found to influence either binding, hydrolysis or release of ATP.
Fig. 4.
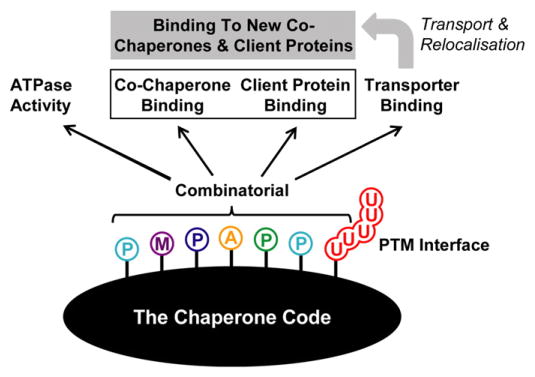
Schematic representation of the chaperone code. The post-translational modification (PTM) of molecular chaperones, through phosphorylation (P), acetylation (A), ubiquitination (U), methylation (M) and other chemical modifications, represents a widely used cellular strategy to regulate, with extreme rapidity and precision, the activity of these proteins that orchestrate protein folding/unfolding and protein complex assembly/disassembly. PTMs have been shown to modulate various aspects of chaperone function, including enzymatic activity (ATPase activity for Hsp90, Hsp70 and VCP), co-factor binding and protein client binding that together control chaperone specificity, and transporter binding that has the ability to relocalize chaperones in a cellular compartment where they can exert a distinct function by binding new cochaperones and client proteins. The notion of a chaperone code helps explaining how, by combining specific PTMs, a given molecular chaperone can discriminate between a multitude of substrates to respond efficiently to changing needs within the cell, both in a time and space controlled manner.
Altering cochaperone affinity is another way by which PTMs affect chaperone function. These cochaperones are rarely limited to their role in modulating the ATP cycle of their respective chaperone, but most often confer specificity to various substrates. Thus, modifications that guide the chaperone’s preference toward various cofactors may ultimately have critical consequences on which client proteins will be targeted as well as what fate will await these substrate proteins (i.e. silencing or activation though chaperoning; stability or degradation).
Translocation of chaperones is yet another recurring theme in chaperone regulation by PTMs. Amino acid modification can directly create or destroy interaction sites for carrier or anchor proteins; they can also create structural rearrangements, thereby revealing targeting sequences that would otherwise be embedded inside the chaperone. Those signaling peptides can direct the chaperone to various organelles and sub-compartments of the cell. Changing the localization of a chaperone may segregate it from its usual interactors, or it may simply put it in contact with a different set of cochaperones and client proteins.
Finally, while chaperones are often the effectors of protein turnover, PTMs may affect the stability of the chaperones themselves with obvious downstream effects for their respective client proteins.
While this additional layer of regulation represents further vulnerabilities that may be exploited during establishment of various disorders or infectious diseases, it is our hope that modulation of chaperones through PTMs may also provide novel diagnostic tools and, ultimately, therapeutic strategies to restore homeostasis in instances where chaperone function has gone awry.
Acknowledgments
We wish to thank members of our laboratory for helpful discussions. Our work relating to methyltransferases and chaperone methylation was supported by the Fonds de la recherche en santé du Québec (FRSQ) and the Canadian Institutes for Health Research (CIHR).
References
- 1.Tissieres A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84:389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- 2.Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 3.Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen TH, Law DT, Williams DB. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991;88:1565–1569. doi: 10.1073/pnas.88.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Thomas JO. The transport of proteins into the nucleus requires the 70-kilodalton heat shock protein or its cytosolic cognate. Mol Cell Biol. 1992;12:2186–2192. doi: 10.1128/mcb.12.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baler R, Zou J, Voellmy R. Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones. 1996;1:33–39. doi: 10.1379/1466-1268(1996)001<0033:efaroh>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–34432. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- 11.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 17.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 18.Ramp U, Mahotka C, Heikaus S, Shibata T, Grimm MO, Willers R, Gabbert HE. Expression of heat shock protein 70 in renal cell carcinoma and its relation to tumor progression and prognosis. Histol Histopathol. 2007;22:1099–1107. doi: 10.14670/HH-22.1099. [DOI] [PubMed] [Google Scholar]
- 19.Ricaniadis N, Kataki A, Agnantis N, Androulakis G, Karakousis CP. Long-term prognostic significance of HSP-70, c-myc and HLA-DR expression in patients with malignant melanoma. Eur J Surg Oncol. 2001;27:88–93. doi: 10.1053/ejso.1999.1018. [DOI] [PubMed] [Google Scholar]
- 20.Maehara Y, Oki E, Abe T, Tokunaga E, Shibahara K, Kakeji Y, Sugimachi K. Overexpression of the heat shock protein HSP70 family and p53 protein and prognosis for patients with gastric cancer. Oncology. 2000;58:144–151. doi: 10.1159/000012091. [DOI] [PubMed] [Google Scholar]
- 21.Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett. 2012;325:117–124. doi: 10.1016/j.canlet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nylandsted J, Rohde M, Brand K, Bastholm L, Elling F, Jaattela M. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc Natl Acad Sci U S A. 2000;97:7871–7876. doi: 10.1073/pnas.97.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonini NM. Chaperoning brain degeneration. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16407–16411. doi: 10.1073/pnas.152330499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 reduces alpha-synuclein aggregation and toxicity. J Biol Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 26.Gutsmann-Conrad A, Heydari AR, You S, Richardson A. The expression of heat shock protein 70 decreases with cellular senescence in vitro and in cells derived from young and old human subjects. Exp Cell Res. 1998;241:404–413. doi: 10.1006/excr.1998.4069. [DOI] [PubMed] [Google Scholar]
- 27.Njemini R, Abeele MV, Demanet C, Lambert M, Vandebosch S, Mets T. Age-related decrease in the inducibility of heat-shock protein 70 in human peripheral blood mononuclear cells. J Clin Immunol. 2002;22:195–205. doi: 10.1023/a:1016036724386. [DOI] [PubMed] [Google Scholar]
- 28.Ambra R, Mocchegiani E, Giacconi R, Canali R, Rinna A, Malavolta M, Virgili F. Characterization of the hsp70 response in lymphoblasts from aged and centenarian subjects and differential effects of in vitro zinc supplementation. Exp Gerontol. 2004;39:1475–1484. doi: 10.1016/j.exger.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Velazquez JM, Lindquist S. hsp70: nuclear concentration during environmental stress and cytoplasmic storage during recovery. Cell. 1984;36:655–662. doi: 10.1016/0092-8674(84)90345-3. [DOI] [PubMed] [Google Scholar]
- 30.Maher PA, Pasquale EB. Heat shock induces protein tyrosine phosphorylation in cultured cells. J Cell Biol. 1989;108:2029–2035. doi: 10.1083/jcb.108.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowlton AA, Grenier M, Kirchhoff SR, Salfity M. Phosphorylation at tyrosine-524 influences nuclear accumulation of HSP72 with heat stress. Am J Physiol. 2000;278:H2143–H2149. doi: 10.1152/ajpheart.2000.278.6.H2143. [DOI] [PubMed] [Google Scholar]
- 32.Cvoro A, Dundjerski J, Trajkovic D, Matic G. The level and phosphorylation of Hsp70 in the rat liver cytosol after adrenalectomy and hyperthermia. Cell Biol Int. 1999;23:313–320. doi: 10.1006/cbir.1998.0247. [DOI] [PubMed] [Google Scholar]
- 33.Staal SP. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc Natl Acad Sci U S A. 1987;84:5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 35.Rickle A, Bogdanovic N, Volkman I, Winblad B, Ravid R, Cowburn RF. Akt activity in Alzheimer’s disease and other neurodegenerative disorders. Neuroreport. 2004;15:955–959. doi: 10.1097/00001756-200404290-00005. [DOI] [PubMed] [Google Scholar]
- 36.Zdychova J, Komers R. Emerging role of Akt kinase/protein kinase B signaling in pathophysiology of diabetes and its complications. Physiol Res. 2005;54:1–16. doi: 10.33549/physiolres.930582. [DOI] [PubMed] [Google Scholar]
- 37.Barati MT, Rane MJ, Klein JB, McLeish KR. A proteomic screen identified stress-induced chaperone proteins as targets of Akt phosphorylation in mesangial cells. J Proteome Res. 2006;5:1636–1646. doi: 10.1021/pr0502469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duriez M, Rossignol JM, Sitterlin D. The hepatitis B virus precore protein is retrotransported from endoplasmic reticulum (ER) to cytosol through the ER-associated degradation pathway. J Biol Chem. 2008;283:32352–32360. doi: 10.1074/jbc.M807178200. [DOI] [PubMed] [Google Scholar]
- 39.Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2012 doi: 10.1038/onc.2012.314. (Advance online publication) [DOI] [PubMed] [Google Scholar]
- 40.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science (New York, NY) 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem. 2002;277:9590–9597. doi: 10.1074/jbc.M107942200. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res. 1998;241:126–133. doi: 10.1006/excr.1998.4027. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Wang SY, Zhang XH, Zhao M, Hou CM, Xu YJ, Du ZY, Yu XD. FK228 inhibits Hsp90 chaperone function in K562 cells via hyperacetylation of Hsp70. Biochem Biophys Res Commun. 2007;356:998–1003. doi: 10.1016/j.bbrc.2007.03.076. [DOI] [PubMed] [Google Scholar]
- 44.Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, Sessa WC, Altieri DC. Regulation of survivin function by Hsp90. Proc Natl Acad Sci U S A. 2003;100:13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neckers L. Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol Med. 2002;8:S55–S61. doi: 10.1016/s1471-4914(02)02316-x. [DOI] [PubMed] [Google Scholar]
- 46.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (New York, NY) 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 47.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science (New York, NY) 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 50.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 53.Kundrat L, Regan L. Identification of residues on Hsp70 and Hsp90 ubiquitinated by the cochaperone CHIP. J Mol Biol. 2010;395:587–594. doi: 10.1016/j.jmb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morales JL, Perdew GH. Carboxyl terminus of hsc70-interacting protein (CHIP) can remodel mature aryl hydrocarbon receptor (AhR) complexes and mediate ubiquitination of both the AhR and the 90 kDa heat-shock protein (hsp90) in vitro. Biochemistry. 2007;46:610–621. doi: 10.1021/bi062165b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008;105:1806–1819. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Mol Cell Proteomics. 2011;10:M110, 002089. doi: 10.1074/mcp.M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111, 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grillo MA, Colombatto S. S-adenosylmethionine and protein methylation. Amino Acids. 2005;28:357–362. doi: 10.1007/s00726-005-0197-6. [DOI] [PubMed] [Google Scholar]
- 61.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 63.Wang C, Lin JM, Lazarides E. Methylations of 70,000-Da heat shock proteins in 3T3 cells: alterations by arsenite treatment, by different stages of growth and by virus transformation. Arch Biochem Biophys. 1992;297:169–175. doi: 10.1016/0003-9861(92)90656-h. [DOI] [PubMed] [Google Scholar]
- 64.Wang C, Gomer RH, Lazarides E. Heat shock proteins are methylated in avian and mammalian cells. Proc Natl Acad Sci U S A. 1981;78:3531–3535. doi: 10.1073/pnas.78.6.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cloutier P, Lavallee-Adam M, Faubert D, Blanchette M, Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9:e1003210. doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho HS, Shimazu T, Toyokawa G, Daigo Y, Maehara Y, Hayami S, Ito A, Masuda K, Ikawa N, Field HI, Tsuchiya E, Ohnuma S, Ponder BA, Yoshida M, Nakamura Y, Hamamoto R. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat Commun. 2012;3:1072. doi: 10.1038/ncomms2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochim Biophys Acta. 2012;1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 69.Eustace BK, Sakurai T, Stewart JK, Yimlamai D, Unger C, Zehetmeier C, Lain B, Torella C, Henning SW, Beste G, Scroggins BT, Neckers L, Ilag LL, Jay DG. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 70.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 71.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 72.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 75.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 76.Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 78.Huai Q, Wang H, Liu Y, Kim HY, Toft D, Ke H. Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding. Structure. 2005;13:579–590. doi: 10.1016/j.str.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 79.Johnson JL, Halas A, Flom G. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol Cell Biol. 2007;27:768–776. doi: 10.1128/MCB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee P, Shabbir A, Cardozo C, Caplan AJ. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol Biol Cell. 2004;15:1785–1792. doi: 10.1091/mbc.E03-07-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McLaughlin SH, Sobott F, Yao ZP, Zhang W, Nielsen PR, Grossmann JG, Laue ED, Robinson CV, Jackson SE. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 82.Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 83.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 84.Neckers L. Heat shock protein 90: the cancer chaperone. J Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 85.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 86.Kim YS, Alarcon SV, Lee S, Lee MJ, Giaccone G, Neckers L, Trepel JB. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem. 2009;9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Messaoudi S, Peyrat JF, Brion JD, Alami M. Recent advances in Hsp90 inhibitors as antitumor agents. Anticancer Agents Med Chem. 2008;8:761–782. doi: 10.2174/187152008785914824. [DOI] [PubMed] [Google Scholar]
- 88.Neckers L. Chaperoning oncogenes: Hsp90 as a target of geldanamycin. Handbook of Experimental Pharmacology. 2006:259–277. doi: 10.1007/3-540-29717-0_11. [DOI] [PubMed] [Google Scholar]
- 89.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 90.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 91.Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 92.Mollapour M, Tsutsumi S, Neckers L. Hsp90 phosphorylation, Wee1 and the cell cycle. Cell cycle (Georgetown, Tex) 2010;9:2310–2316. doi: 10.4161/cc.9.12.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Csermely P, Kahn CR. The 90-kDa heat shock protein (hsp-90) possesses an ATP binding site and autophosphorylating activity. J Biol Chem. 1991;266:4943–4950. [PubMed] [Google Scholar]
- 94.Csermely P, Miyata Y, Schnaider T, Yahara I. Autophosphorylation of grp94 (endoplasmin) J Biol Chem. 1995;270:6381–6388. doi: 10.1074/jbc.270.11.6381. [DOI] [PubMed] [Google Scholar]
- 95.Zhao YG, Gilmore R, Leone G, Coffey MC, Weber B, Lee PW. Hsp90 phosphorylation is linked to its chaperoning function. Assembly of the reovirus cell attachment protein. J Biol Chem. 2001;276:32822–32827. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
- 96.Lees-Miller SP, Anderson CW. Two human 90-kDa heat shock proteins are phosphorylated in vivo at conserved serines that are phosphorylated in vitro by casein kinase II. J Biol Chem. 1989;264:2431–2437. [PubMed] [Google Scholar]
- 97.Cala SE. GRP94 hyperglycosylation and phosphorylation in Sf21 cells. Biochim Biophys Acta. 2000;1496:296–310. doi: 10.1016/s0167-4889(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 98.Cala SE, Jones LR. GRP94 resides within cardiac sarcoplasmic reticulum vesicles and is phosphorylated by casein kinase II. J Biol Chem. 1994;269:5926–5931. [PubMed] [Google Scholar]
- 99.Kurokawa M, Zhao C, Reya T, Kornbluth S. Inhibition of apoptosome formation by suppression of Hsp90beta phosphorylation in tyrosine kinase-induced leukemias. Mol Cell Biol. 2008;28:5494–5506. doi: 10.1128/MCB.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ogiso H, Kagi N, Matsumoto E, Nishimoto M, Arai R, Shirouzu M, Mimura J, Fujii-Kuriyama Y, Yokoyama S. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry. 2004;43:15510–15519. doi: 10.1021/bi048736m. [DOI] [PubMed] [Google Scholar]
- 101.Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, Litman ES, Croy CH, Meyer-Arendt K, Miranda JG, Brown RA, Witze ES, Schweppe RE, Resing KA, Ahn NG. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009;34:115–131. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Woo SH, An S, Lee HC, Jin HO, Seo SK, Yoo DH, Lee KH, Rhee CH, Choi EJ, Hong SI, Park IC. A truncated form of p23 down-regulates telomerase activity via disruption of Hsp90 function. J Biol Chem. 2009;284:30871–30880. doi: 10.1074/jbc.M109.052720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Szyszka R, Kramer G, Hardesty B. The phosphorylation state of the reticulocyte 90-kDa heat shock protein affects its ability to increase phosphorylation of peptide initiation factor 2 alpha subunit by the heme-sensitive kinase. Biochemistry. 1989;28:1435–1438. doi: 10.1021/bi00430a001. [DOI] [PubMed] [Google Scholar]
- 104.Salvi M, Trashi E, Cozza G, Franchin C, Arrigoni G, Pinna LA. Investigation on PLK2 and PLK3 substrate recognition. Biochim Biophys Acta. 2012;1824:1366–1373. doi: 10.1016/j.bbapap.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 105.Deb TB, Zuo AH, Wang Y, Barndt RJ, Cheema AK, Sengupta S, Coticchia CM, Johnson MD. Pnck induces ligand-independent EGFR degradation by probable perturbation of the Hsp90 chaperone complex. Am J Physiol Cell Physiol. 2011;300:C1139–C1154. doi: 10.1152/ajpcell.00167.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lees-Miller SP, Anderson CW. The human double-stranded DNA-activated protein kinase phosphorylates the 90-kDa heat-shock protein, hsp90 alpha at two NH2-terminal threonine residues. J Biol Chem. 1989;264:17275–17280. [PubMed] [Google Scholar]
- 107.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 108.Maxwell AJ. Mechanisms of dysfunction of the nitric oxide pathway in vascular diseases. Nitric Oxide. 2002;6:101–124. doi: 10.1006/niox.2001.0394. [DOI] [PubMed] [Google Scholar]
- 109.Stalker TJ, Gong Y, Scalia R. The calcium-dependent protease calpain causes endothelial dysfunction in type 2 diabetes. Diabetes. 2005;54:1132–1140. doi: 10.2337/diabetes.54.4.1132. [DOI] [PubMed] [Google Scholar]
- 110.Lei H, Venkatakrishnan A, Yu S, Kazlauskas A. Protein kinase A-dependent translocation of Hsp90 alpha impairs endothelial nitric-oxide synthase activity in high glucose and diabetes. J Biol Chem. 2007;282:9364–9371. doi: 10.1074/jbc.M608985200. [DOI] [PubMed] [Google Scholar]
- 111.Duval M, Le Boeuf F, Huot J, Gratton JP. Src-mediated phosphorylation of Hsp90 in response to vascular endothelial growth factor (VEGF) is required for VEGF receptor-2 signaling to endothelial NO synthase. Mol Biol Cell. 2007;18:4659–4668. doi: 10.1091/mbc.E07-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Desjardins F, Delisle C, Gratton JP. Modulation of the cochaperone AHA1 regulates heat-shock protein 90 and endothelial NO synthase activation by vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2012;32:2484–2492. doi: 10.1161/ATVBAHA.112.256008. [DOI] [PubMed] [Google Scholar]
- 113.Mollapour M, Tsutsumi S, Donnelly AC, Beebe K, Tokita MJ, Lee MJ, Lee S, Morra G, Bourboulia D, Scroggins BT, Colombo G, Blagg BS, Panaretou B, Stetler-Stevenson WG, Trepel JB, Piper PW, Prodromou C, Pearl LH, Neckers L. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol Cell. 2010;37:333–343. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu W, Mollapour M, Prodromou C, Wang S, Scroggins BT, Palchick Z, Beebe K, Siderius M, Lee MJ, Couvillon A, Trepel JB, Miyata Y, Matts R, Neckers L. Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Mol Cell. 2012;47:434–443. doi: 10.1016/j.molcel.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brunati AM, Contri A, Muenchbach M, James P, Marin O, Pinna LA. GRP94 (endoplasmin) co-purifies with and is phosphorylated by Golgi apparatus casein kinase. FEBS Lett. 2000;471:151–155. doi: 10.1016/s0014-5793(00)01378-8. [DOI] [PubMed] [Google Scholar]
- 116.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- 118.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 119.Brinkmann H, Dahler AL, Popa C, Serewko MM, Parsons PG, Gabrielli BG, Burgess AJ, Saunders NA. Histone hyperacetylation induced by histone deacetylase inhibitors is not sufficient to cause growth inhibition in human dermal fibroblasts. J Biol Chem. 2001;276:22491–22499. doi: 10.1074/jbc.M100206200. [DOI] [PubMed] [Google Scholar]
- 120.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 121.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 122.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 123.Murphy PJ, Morishima Y, Kovacs JJ, Yao TP, Pratt WB. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J Biol Chem. 2005;280:33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- 124.Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Y, Rao R, Shen J, Tang Y, Fiskus W, Nechtman J, Atadja P, Bhalla K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture JF. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- 127.Donlin LT, Andresen C, Just S, Rudensky E, Pappas CT, Kruger M, Jacobs EY, Unger A, Zieseniss A, Dobenecker MW, Voelkel T, Chait BT, Gregorio CC, Rottbauer W, Tarakhovsky A, Linke WA. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 2012;26:114–119. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stamler JS, Lamas S, Fang FC. Nitrosylation. The prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 129.Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Overath T, Kuckelkorn U, Henklein P, Strehl B, Bonar D, Kloss A, Siele D, Kloetzel PM, Janek K. Mapping of O-GlcNAc Sites of 20 S proteasome subunits and Hsp90 by a novel biotin-cystamine tag. Mol Cell Proteomics. 2012;11:467–477. doi: 10.1074/mcp.M111.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Whelan SA, Lane MD, Hart GW. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J Biol Chem. 2008;283:21411–21417. doi: 10.1074/jbc.M800677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang F, Snead CM, Catravas JD. Hsp90 regulates O-linked beta-N-acetylglucosamine transferase: a novel mechanism of modulation of protein O-linked beta-N-acetylglucosamine modification in endothelial cells. Am J Physiol Cell Physiol. 2012;302:C1786–C1796. doi: 10.1152/ajpcell.00004.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qu D, Mazzarella RA, Green M. Analysis of the structure and synthesis of GRP94, an abundant stress protein of the endoplasmic reticulum. DNA Cell Biol. 1994;13:117–124. doi: 10.1089/dna.1994.13.117. [DOI] [PubMed] [Google Scholar]
- 134.Feldweg AM, Srivastava PK. Molecular heterogeneity of tumor rejection antigen/heat shock protein GP96. Int J Cancer. 1995;63:310–314. doi: 10.1002/ijc.2910630227. [DOI] [PubMed] [Google Scholar]
- 135.Wang Q, Song C, Yang X, Li CC. D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J Biol Chem. 2003;278:32784–32793. doi: 10.1074/jbc.M303869200. [DOI] [PubMed] [Google Scholar]
- 136.Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Indig FE, Partridge JJ, von Kobbe C, Aladjem MI, Latterich M, Bohr VA. Werner syndrome protein directly binds to the AAA ATPase p97/VCP in an ATP-dependent fashion. J Struct Biol. 2004;146:251–259. doi: 10.1016/j.jsb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 138.Livingstone M, Ruan H, Weiner J, Clauser KR, Strack P, Jin S, Williams A, Greulich H, Gardner J, Venere M, Mochan TA, DiTullio RA, Jr, Moravcevic K, Gorgoulis VG, Burkhardt A, Halazonetis TD. Valosin-containing protein phosphorylation at Ser784 in response to DNA damage. Cancer Res. 2005;65:7533–7540. doi: 10.1158/0008-5472.CAN-04-3729. [DOI] [PubMed] [Google Scholar]
- 139.Partridge JJ, Lopreiato JO, Jr, Latterich M, Indig FE. DNA damage modulates nucleolar interaction of the Werner protein with the AAA ATPase p97/VCP. Mol Biol Cell. 2003;14:4221–4229. doi: 10.1091/mbc.E03-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang H, Wang Q, Kajino K, Greene MI. VCP, a weak ATPase involved in multiple cellular events, interacts physically with BRCA1 in the nucleus of living cells. DNA Cell Biol. 2000;19:253–263. doi: 10.1089/10445490050021168. [DOI] [PubMed] [Google Scholar]
- 141.Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin–proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 142.Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 143.Shcherbik N, Zoladek T, Nickels JT, Haines DS. Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr Biol. 2003;13:1227–1233. doi: 10.1016/s0960-9822(03)00457-3. [DOI] [PubMed] [Google Scholar]
- 144.Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vesa J, Su H, Watts GD, Krause S, Walter MC, Martin B, Smith C, Wallace DC, Kimonis VE. Valosin containing protein associated inclusion body myopathy: abnormal vacuolization, autophagy and cell fusion in myoblasts. Neuromuscul Disord. 2009;19:766–772. doi: 10.1016/j.nmd.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 147.Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- 148.Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 150.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 152.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochem Biophys Res Commun. 2009;390:758–762. doi: 10.1016/j.bbrc.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 155.Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 156.Allen MD, Buchberger A, Bycroft M. The PUB domain functions as a p97 binding module in human peptide N-glycanase. J Biol Chem. 2006;281:25502–25508. doi: 10.1074/jbc.M601173200. [DOI] [PubMed] [Google Scholar]
- 157.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 158.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 159.Ernst R, Mueller B, Ploegh HL, Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 161.Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- 162.Mehta SG, Khare M, Ramani R, Watts GD, Simon M, Osann KE, Donkervoort S, Dec E, Nalbandian A, Platt J, Pasquali M, Wang A, Mozaffar T, Smith CD, Kimonis VE. Genotype–phenotype studies of VCP-associated inclusion body myopathy with Paget disease of bone and/or frontotemporal dementia. Clin Genet. 2012 doi: 10.1111/cge.12000. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.DeJesus-Hernandez M, Desaro P, Johnston A, Ross OA, Wszolek ZK, Ertekin-Taner N, Graff-Radford NR, Rademakers R, Boylan K. Novel p.Ile151Val mutation in VCP in a patient of African American descent with sporadic ALS. Neurology. 2011;77:1102–1103. doi: 10.1212/WNL.0b013e31822e563c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tiloca C, Ratti A, Pensato V, Castucci A, Soraru G, Del Bo R, Corrado L, Cereda C, D’Ascenzo C, Comi GP, Mazzini L, Castellotti B, Ticozzi N, Gellera C, Silani V. Mutational analysis of VCP gene in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:630, e631–632. doi: 10.1016/j.neurobiolaging.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 166.Williams KL, Solski JA, Nicholson GA, Blair IP. Mutation analysis of VCP in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:1488, e1415–1486. doi: 10.1016/j.neurobiolaging.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 167.Abramzon Y, Johnson JO, Scholz SW, Taylor JP, Brunetti M, Calvo A, Mandrioli J, Benatar M, Mora G, Restagno G, Chio A, Traynor BJ. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2231.e1–2231.e6. doi: 10.1016/j.neurobiolaging.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Miller JW, Smith BN, Topp SD, Al-Chalabi A, Shaw CE, Vance C. Mutation analysis of VCP in British familial and sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging. 2012;33:2721, e2721–2722. doi: 10.1016/j.neurobiolaging.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 169.Djamshidian A, Schaefer J, Haubenberger D, Stogmann E, Zimprich F, Auff E, Zimprich A. A novel mutation in the VCP gene (G157R) in a German family with inclusion-body myopathy with Paget disease of bone and frontotemporal dementia. Muscle Nerve. 2009;39:389–391. doi: 10.1002/mus.21225. [DOI] [PubMed] [Google Scholar]
- 170.Weihl CC, Pestronk A, Kimonis VE. Valosin-containing protein disease: inclusion body myopathy with Paget’s disease of the bone and fronto-temporal dementia. Neuromuscul Disord. 2009;19:308–315. doi: 10.1016/j.nmd.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 173.Halawani D, LeBlanc AC, Rouiller I, Michnick SW, Servant MJ, Latterich M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol Cell Biol. 2009;29:4484–4494. doi: 10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Weihl CC, Dalal S, Pestronk A, Hanson PI. Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum Mol Genet. 2006;15:189–199. doi: 10.1093/hmg/ddi426. [DOI] [PubMed] [Google Scholar]
- 175.Mizuno Y, Hori S, Kakizuka A, Okamoto K. Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci Lett. 2003;343:77–80. doi: 10.1016/s0304-3940(03)00280-5. [DOI] [PubMed] [Google Scholar]
- 176.Hirabayashi M, Inoue K, Tanaka K, Nakadate K, Ohsawa Y, Kamei Y, Popiel AH, Sinohara A, Iwamatsu A, Kimura Y, Uchiyama Y, Hori S, Kakizuka A. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8:977–984. doi: 10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- 177.Klein JB, Barati MT, Wu R, Gozal D, Sachleben LR, Jr, Kausar H, Trent JO, Gozal E, Rane MJ. Akt-mediated valosin-containing protein 97 phosphorylation regulates its association with ubiquitinated proteins. J Biol Chem. 2005;280:31870–31881. doi: 10.1074/jbc.M501802200. [DOI] [PubMed] [Google Scholar]
- 178.Vandermoere F, El Yazidi-Belkoura I, Slomianny C, Demont Y, Bidaux G, Adriaenssens E, Lemoine J, Hondermarck H. The valosin-containing protein (VCP) is a target of Akt signaling required for cell survival. J Biol Chem. 2006;281:14307–14313. doi: 10.1074/jbc.M510003200. [DOI] [PubMed] [Google Scholar]
- 179.Imamura S, Ojima N, Yamashita M. Cold-inducible expression of the cell division cycle gene CDC48 and its promotion of cell proliferation during cold acclimation in zebrafishcells. FEBS Lett. 2003;549:14–20. doi: 10.1016/s0014-5793(03)00723-3. [DOI] [PubMed] [Google Scholar]
- 180.Ficarro S, Chertihin O, Westbrook VA, White F, Jayes F, Kalab P, Marto JA, Shabanowitz J, Herr JC, Hunt DF, Visconti PE. Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J Biol Chem. 2003;278:11579–11589. doi: 10.1074/jbc.M202325200. [DOI] [PubMed] [Google Scholar]
- 181.Lavoie C, Chevet E, Roy L, Tonks NK, Fazel A, Posner BI, Paiement J, Bergeron JJ. Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro. Proc Natl Acad Sci U S A. 2000;97:13637–13642. doi: 10.1073/pnas.240278097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang SH, Liu J, Kobayashi R, Tonks NK. Identification of the cell cycle regulator VCP (p97/CDC48) as a substrate of the band 4.1-related protein-tyrosine phosphatase PTPH1. J Biol Chem. 1999;274:17806–17812. doi: 10.1074/jbc.274.25.17806. [DOI] [PubMed] [Google Scholar]
- 183.Wu J, Katrekar A, Honigberg LA, Smith AM, Conn MT, Tang J, Jeffery D, Mortara K, Sampang J, Williams SR, Buggy J, Clark JM. Identification of substrates of human protein-tyrosine phosphatase PTPN22. J Biol Chem. 2006;281:11002–11010. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 184.Suzuki T, Park H, Till EA, Lennarz WJ. The PUB domain: a putative protein–protein interaction domain implicated in the ubiquitin–proteasome pathway. Biochem Biophys Res Commun. 2001;287:1083–1087. doi: 10.1006/bbrc.2001.5688. [DOI] [PubMed] [Google Scholar]
- 185.Zhao G, Zhou X, Wang L, Li G, Schindelin H, Lennarz WJ. Studies on peptide: N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation. Proc Natl Acad Sci U S A. 2007;104:8785–8790. doi: 10.1073/pnas.0702966104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Egerton M, Ashe OR, Chen D, Druker BJ, Burgess WH, Samelson LE. VCP, the mammalian homolog of cdc48, is tyrosine phosphorylated in response to T cell antigen receptor activation. EMBO J. 1992;11:3533–3540. doi: 10.1002/j.1460-2075.1992.tb05436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Egerton M, Samelson LE. Biochemical characterization of valosin-containing protein, a protein tyrosine kinase substrate in hematopoietic cells. J Biol Chem. 1994;269:11435–11441. [PubMed] [Google Scholar]
- 188.Madeo F, Schlauer J, Zischka H, Mecke D, Frohlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol Biol Cell. 1998;9:131–141. doi: 10.1091/mbc.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 190.Koike M, Fukushi J, Ichinohe Y, Higashimae N, Fujishiro M, Sasaki C, Yamaguchi M, Uchihara T, Yagishita S, Ohizumi H, Hori S, Kakizuka A. Valosin-containing protein (VCP) in novel feedback machinery between abnormal protein accumulation and transcriptional suppression. J Biol Chem. 2010;285:21736–21749. doi: 10.1074/jbc.M109.099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kernstock S, Davydova E, Jakobsson M, Moen A, Pettersen S, Maelandsmo GM, Egge-Jacobsen W, Falnes PO. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 192.Mori-Konya C, Kato N, Maeda R, Yasuda K, Higashimae N, Noguchi M, Koike M, Kimura Y, Ohizumi H, Hori S, Kakizuka A. p97/valosin-containing protein (VCP) is highly modulated by phosphorylation and acetylation. Genes Cells. 2009;14:483–497. doi: 10.1111/j.1365-2443.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 193.Orme CM, Bogan JS. The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-golgi intermediate compartment. J Biol Chem. 2012;287:6679–6692. doi: 10.1074/jbc.M111.284232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Thiele W, Novac N, Mink S, Schreiber C, Plaumann D, Fritzmann J, Cremers N, Rothley M, Schwager C, Regiert T, Huber PE, Stein U, Schlag P, Moll J, Abdollahi A, Sleeman JP. Discovery of a novel tumour metastasis-promoting gene, NVM-1. J Pathol. 2011;225:96–105. doi: 10.1002/path.2924. [DOI] [PubMed] [Google Scholar]
- 195.Noguchi M, Takata T, Kimura Y, Manno A, Murakami K, Koike M, Ohizumi H, Hori S, Kakizuka A. ATPase activity of p97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in Walker A motif. J Biol Chem. 2005;280:41332–41341. doi: 10.1074/jbc.M509700200. [DOI] [PubMed] [Google Scholar]
- 196.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 197.Koutsilieri E, Scheller C, Grunblatt E, Nara K, Li J, Riederer P. Free radicals in Parkinson’s disease. J Neurol. 2002;249(Suppl 2):II1–II5. doi: 10.1007/s00415-002-1201-7. [DOI] [PubMed] [Google Scholar]
- 198.Labunskyy VM, Gladyshev VN. Role of reactive oxygen species-mediated signaling in aging. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]