Abstract
A topological model for transcription initiation by RNA polymerase II (RNAPII) has recently been proposed. This model stipulates that wrapping of the promoter DNA around RNAPII and the general initiation factors TBP, TFIIB, TFIIE, TFIIF and TFIIH induces a torsional strain in the DNA double helix that facilitates strand separation and open complex formation. In this report, we show that TFIIA, a factor previously shown to both stimulate basal transcription and have co-activator functions, is located near the cross-point of the DNA loop where it can interact with TBP, TFIIE56, TFIIE34, and the RNAPII-associated protein (RAP) 74. In addition, we demonstrate that TFIIA can stimulate basal transcription by stimulating the functions of both TFIIE34 and RAP74 during the initiation step of the transcription reaction. These results provide novel insights into mechanisms of TFIIA function.
Initiation of transcription by RNA polymerase II (RNAPII)1 proceeds through the formation of a preinitiation complex containing RNAPII and the general transcription factors (TF) TBP (the TATA box-binding protein of TFIID), TFIIB, TFIIE, TFIIF, and TFIIH on promoter DNA (reviewed in Refs. 1 and 2). The first step in preinitiation complex assembly is the recognition of the TATA element of the promoter by TBP. The binding of TBP to the TATA box induces a DNA bend of ~90° (3, 4). TFIIB can associate with the TBP-promoter complex (5). Mammalian TFIIF, which is composed of the subunits RAP74 and RAP30, directly binds to RNAPII and has been shown to participate in recruitment of the enzyme to the preinitiation complex (6, 7). TFIIE, which is also composed of two subunits called TFIIE56 and TFIIE34, is involved in the melting of promoter DNA at the transcription initiation site through a mechanism that is ATP-independent (8, 9). Finally, TFIIH, which has kinase and helicase activities, mediates the ATP-dependent melting of the promoter DNA in the region of the initiation site and is involved in the transition between the initiation and elongation states of the complex (10–15).
Recent results describing both the structure of the basal transcription machinery and the topological organization of the preinitiation complex have considerably improved our understanding of transcription initiation mechanisms. Determination of the atomic structure of yeast RNAPII at 2.8 angstroms resolution and that of elongating yeast RNAPII at 3.3 angstroms by Kornberg and co-workers (16, 17) has revealed key features of both the interaction between the enzyme and template DNA and the basis of its catalytic activity. Analysis of the molecular organization of the preinitiation complex using site-specific protein-DNA photo-cross-linking has provided insights on the topology of the preinitiation complex containing RNAPII and the general transcription factors (18–26). Recently, we have proposed a topological model, the DNA wrapping model, which describes transcription initiation by RNAPII (23, 26, 27). This model accounts for our photo-cross-linking data and several additional data obtained in various laboratories. The DNA wrapping model stipulates that a role for the general transcription factors is to help in the wrapping of the promoter DNA in the preinitiation complex in such a way that a torsional strain is progressively developed upstream of the transcription start site and results in the partial unwinding of the DNA helix. This region of unwound DNA is used as a substrate by the single-stranded DNA helicases of TFIIH that catalyze open complex formation (26).
First isolated as a general transcription factor, TFIIA has a rather controversial role in transcription initiation. Human TFIIA is composed of three subunits: α (35 kDa), β (19 kDa), and γ (12 kDa) (28–31). The α and β subunits are encoded by the same gene and are produced by posttranslational cleavage of a precursor (29, 30). In yeast, TFIIA is composed of only two subunits encoded by two different genes, TOA1 and TOA2 (32). The N-terminal part of the polypeptide produced from the TOA1 gene is homologous to the human α subunit, and the C-terminal part is homologous to the β subunit (29, 30). TOA2 encodes a polypeptide homologous to γ (28, 31). The posttranslational cleavage of human α/β has been demonstrated to be non-essential because wild type activity can be recovered with uncleaved recombinant α/β and γ renatured together (33). TFIIA is not essential for basal transcription in vitro, but it has been shown to stimulate basal transcription in a variety of systems (28, 33–37). TFIIA binds TBP and increases the affinity of TBP for the TATA box (36–38). TFIIA can displace certain repressors, including Dr1-DRAP1/NC2, topoisomerase 1, HMG1, and Mot-1, from the TFIID complex, indicating that TFIIA is involved in antirepression (39–43). Human TFIIA also plays a role in activated transcription, being required for the functioning of some activators (28, 29, 31, 33, 34, 44–47). For example, TFIIA binds to the activator Zta and mediates its stimulation of TFIID binding to the TATA box (28). Similarly, TFIIA enhances the activation of transcription by the activators Sp1, VP16, and NTF1 (34). The activators VP16 and Zta, which bind TFIIA, stimulate the assembly of a TFIIA-TFIID-promoter complex, consistent with the roles in vivo of these factors in activated transcription (46). The function of TFIIA in transcriptional antirepression and activation have been separated and is associated with distinct subunits of the factor (35). Subunits β and γ are essential for antirepression, whereas α is not. Conversely all three subunits are required for activation.
Previous photo-cross-linking experiments performed with a TBP-TFIIA-promoter complex have revealed that TFIIA makes promoter contacts both in the region of the TATA box and upstream of it (18, 20). We have now determined the position of TFIIA in a preinitiation complex assembled in the presence of TBP, TFIIB, TFIIE, TFIIF, and RNAPII. Our results indicate that TFIIA makes promoter contacts not only on the TATA box and upstream of it in the −40 region, as is observed in the TBP-TFIIA-promoter complex, but also in the +26 region. Given the two extreme promoter positions approached by TFIIA and the small size of TFIIA (67 kDa), these results suggest that TFIIA is located near the cross-point of the wrapped DNA structure, where it can simultaneously contact nucleotides −40 and +26. TFIIF, TFIIE, and RNAPII also cross-link to the −40 and +26 positions (23, 26), suggesting that TFIIA may directly interact with these factors. We report here that TFIIA directly interacts with RAP74, TFIIE56, and TFIIE34 in addition to the previously determined interaction with TBP. Furthermore, we use an abortive initiation assay to provide evidence that the stimulatory effect of TFIIA on basal transcription is exerted through a stimulation of the activity of RAP74 and TFIIE34 at the initiation stage of transcript formation.
EXPERIMENTAL PROCEDURES
Protein Factors
Recombinant yeast TBP (48), human TFIIB (49), human RAP30 (50), human RAP74 (wild type and deletion mutants) (50), human TFIIE56 and TFIIE34 (51–53), and calf thymus RNAPII (54) were prepared as previously described. Natural human TFIIA (nTFIIA) was partly purified using protein-affinity chromatography with immobilized TBP as we described (36). Recombinant human TFIIA (rTFIIA) was produced from uncleaved α/β and γ subunits that carried histidine tags (28). The two polypeptides were independently purified on Ni-NTA agarose columns (Qiagen) under denaturating conditions and were either used individually as TFIIAα/β and TFIIAγ or renatured together to produce rTFIIA.
Protein-DNA Photo-cross-linking
The synthesis of the photoreactive nucleotide N3R-dUMP, the preparation of the probes, and the conditions for binding reactions were as described (55). Two photo-probes containing the modified nucleotide at positions −39/−40 and +26 were used. For each probe, the concentration of poly(dI-dC) in the binding reactions was optimized to favor specific over nonspecific binding. A typical reaction with all the factors contained 200 ng each of TBP, TFIIB, RAP30, RAP74, TFIIE56, TFIIE34, rTFIIA, and RNAPII. UV irradiation, nuclease treatment, and SDS-PAGE analysis of radiolabeled photo-cross-linking products were performed as described previously (55).
Protein-Protein Interactions
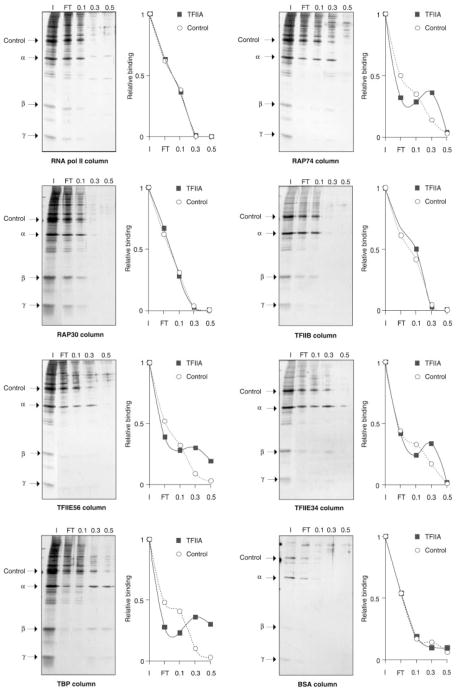
Protein-protein interactions were analyzed essentially as we previously described (22). RAP74 wt, RAP74 deletion mutants, RAP30, TFIIE34, TFIIE56, RNAPII, TFIIB, TBP, and bovine serum albumin (BSA) were immobilized on Affigel 10 (Bio-Rad) at a concentration of 1 to 5 mg/ml resin. Microcolumns were made with ~20 μl of this resin. A volume of 50 μl of nTFIIA (Fig. 2), recombinant TFIIAα/β, recombinant TFIIAγ (Fig. 3), or rTFIIA (Fig. 4), which contained 100 ng of nTFIIA or 200 ng of TFIIAα/β, TFIIAγ or rTFIIA, was then loaded on the different columns. The flowthrough was collected and the columns successively eluted with 50 μl of ACB buffer (10 mM Hepes, pH 7.9, 0.2 mM EDTA, 20% glycerol and 1 mM dithio-threitol) containing 0.1 M, 0.3 M, and 0.5 M NaCl. An aliquot of the input, the flowthrough, and the various salt elutions were analyzed on SDS-PAGE and revealed by silver (Fig. 2 and Fig. 4) or zinc staining (Fig. 3). The intensity of the bands was evaluated using the UN-SCAN-IT software. It was considered that TFIIA, or one of its subunits, was binding to a particular column when the intensity of the 0.3 M salt band was higher than the intensity of both the 0.1 M band and the flowthrough band. In contrast, when the intensity of the 0.3 M band was lower than the 0.1 M or the flowthrough band, we considered that TFIIA was not binding to the column. The specificity of TFIIA binding was assessed by comparing the binding of the TFIIA subunits to the binding of a contaminant polypeptide of the nTFIIA preparation (Fig. 2).
Fig. 2. Interactions of natural TFIIA with components of the basal transcription machinery.
Protein-affinity chromatography was performed using microcolumns containing immobilized RNAPII, RAP74, RAP30, TFIIE56, TFIIE34, TFIIB, TBP, and BSA. A volume of 50 μl of nTFIIA (100 ng of α) was chromatographed through each column. The flowthrough was collected in each case. The columns were eluted with 50 μl of buffer containing increasing amounts of NaCl (e.g. 0.1, 0.3, and 0.5 M). The fractions were analyzed using SDS-PAGE and compared with the input (I). The positions of the nTFIIA subunits and a contaminant polypeptide of the TFIIA preparation that served as negative control are indicated. In each case, a diagram showing the relative intensities of both TFIIAα and a contaminant band (negative control) in the various fractions is shown.
Fig. 3. Interactions of TFIIAα/β and TFIIAγ with RAP74, TFIIE56, and TFIIE34.
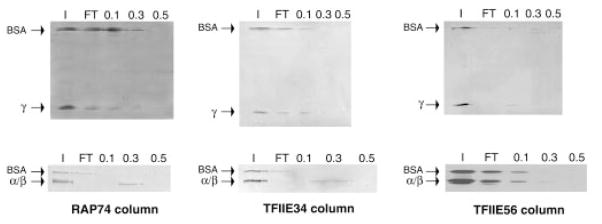
Protein-affinity chromatography was performed using microcolumns containing immobilized RAP74, TFIIE34, and TFIIE56. A volume of 50 μl containing 200 ng of TFIIAα/β and TFIIAγ was chromatographed through each column. Fractions were collected and analyzed as in Fig. 2. The positions of α/β, γ, and BSA (as an internal negative control) are indicated.
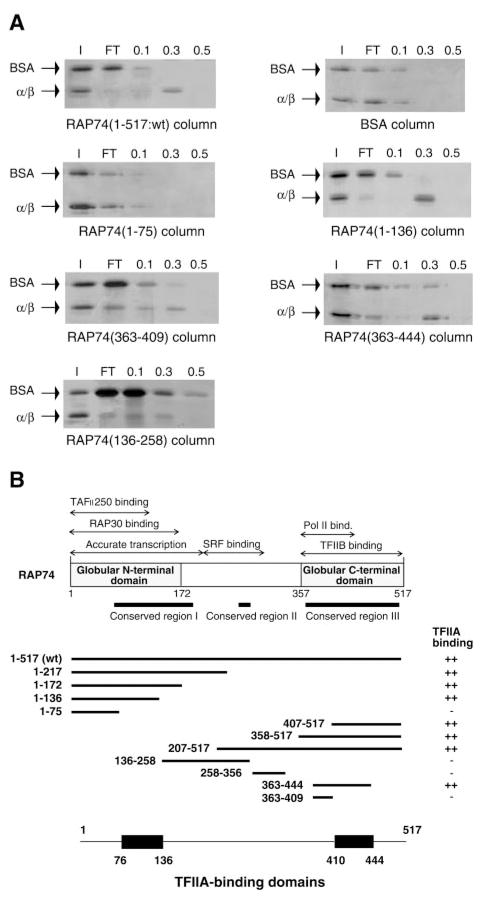
Fig. 4. Interactions of TFIIAα/β with RAP74 deletion mutants.
A, protein affinity chromatography was performed using columns containing immobilized RAP74 fragments (wild type or deletions mutants). A volume of 50 μl containing 200 ng of TFIIAα/β was chromatographed through each column. Fractions were collected and analyzed as in Fig. 2. The positions of TFIIAα/β and BSA (as an internal negative control) are indicated. B, summary of the interactions between the RAP74 deletion mutants and TFIIA. The various domains of RAP74 are indicated in the top part. The two TFIIA-binding domains of RAP74 are deduced from our analysis.
Gel Mobility Shift Assay
Plasmid DNA containing the adenovirus major late promoter (AdMLP) was digested with the restriction enzymes BamHI and DraI, and the 110-base pair fragment containing the promoter was filled-in using the Klenow fragment of DNA polymerase in the presence of [α-32P]dGTP. Gel mobility shift assays were performed as described previously (56). Complexes were assembled using highly purified TBP (20 ng), TFIIA (120 ng), and, when indicated, RAP74-(1–517) (640 ng),(1–136) (160 ng), (1–75) (91 ng), (363–409) (58 ng), and (363–444) (107 ng).
Transcription Assay
Transcription assays were performed as described previously (57). TBP (120 ng), TFIIB (120 ng), RAP30 (120 ng), RAP74 (260 ng), TFIIE34 (160 ng), TFIIE56 (240 ng), RNAPII (660 ng), and various amounts of TFIIA were incubated with 500 ng of the supercoiled DNA template containing the AdMLP from nucleotides −50 to +10 fused to a G-less cassette. Under these conditions a 391-nt run-off transcript is produced.
Abortive Initiation Assay
Templates were prepared by annealing two 80-base pair DNA oligonucleotides carrying the strands of the AdMLP from −45 to +35 as described (58). Typically, 12 ng of the double-stranded DNA template were incubated for 60 min at 30 °C with TBP (60 ng), TFIIB (30 ng), RAP30 (30 ng), RNAPII (165 ng), and, when indicated, RAP74 (65 ng), TFIIE34 (40 ng), TFIIE56 (60 ng), and various amounts of TFIIA in 20 μl of a reaction mixture containing 125 μM ATP, 125 μM CTP, 1.7 μM UTP, 2.5 μCi [α-32P]UTP, 1.25 mM MgCl2, 0.5 mM EGTA, and 125 units/ml RNase inhibitor. The synthesis was stopped by incubating at 68 °C for 3 min, and the reaction mixture then cooled on ice for 5 min. Calf intestine alkaline phosphatase (8 U, CIP) and 2 μl of 2× CIP buffer (500 mM Tris, pH 8.9, 1 mM EDTA) were added to 10 μl of the transcription reaction, and the resulting solution was incubated for 20 min at 37 °C to reduce the background caused by free radiolabeled nucleotides. The CIP reaction was stopped by adding 3 μl of loading buffer (50% glycerol, 200 mM EDTA, 0.05% bromphenol blue). Transcripts were analyzed on a 23% polyacrylamide denaturating gel containing 7 M urea and were quantitated using a PhosphorImager (Molecular Dynamics).
RESULTS AND DISCUSSION
TFIIA Is Located Near the Cross-point of the Wrapped DNA Structure in the Initiation Complex
We have previously shown that TFIIA assembled with TBP on the AdMLP (e.g. TBP-TFIIA-promoter complex) cross-linked to positions −31/−29, −25/−30, −39/−40, and −42 (18). We have now analyzed the position of TFIIA in a preinitiation complex composed of TBP, TFIIB, RAP74, RAP30, TFIIE56, TFIIE34, and RNAPII using site-specific protein-DNA photo-cross-linking. Both TFIIAα/β and TFIIAγ cross-linked in the region of the TATA box to photoprobe −31/−29 (data not shown), upstream of it to photoprobe −39/−40 (Fig. 1A, left panel) and downstream of it to photoprobe +26 (Fig. 1A, right panel). The cross-linking of TFIIA to positions −39/−40 required the presence of TBP but not that of RAP30 (Fig. 1A), TFIIB, or RNAPII (data not shown). The cross-linking of TFIIA to position +26, however, required the presence of TBP and RAP30 (Fig. 1A, right panel), as well as that of TFIIB and RNAPII (data not shown). These results indicate that promoter contacts by TFIIA in the −39/−40 region do not necessitate assembly of a preinitiation complex containing TFIIB, TFIIF, and RNAPII (e.g. a TBP-TFIIA-promoter complex is sufficient), whereas the promoter contact by TFIIA in the +26 region requires the assembly of a preinitiation complex (e.g. a TBP-TFIIA-TFIIB-TFIIF-RNAPII-TFIIE-promoter complex) in which promoter DNA adopts a wrapped structure (see Fig. 1B for a schematic representation).
Fig. 1. Photo-cross-linking of rTFIIA on the AdMLP.
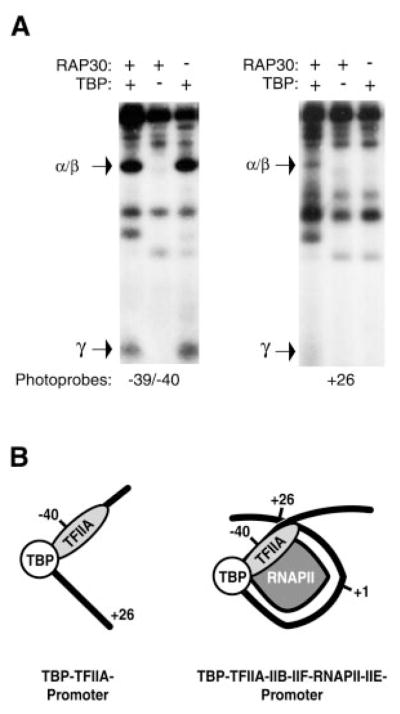
A, photo-cross-linking experiments with photoprobes −39/−40 and +26 were performed in the presence of rTFIIA (α/β and γ), TBP, TFIIB, TFIIF (RAP74 and RAP30), TFIIE (p56 and p34), and RNAPII. The specificity of the cross-linking signals was assessed by comparing reactions performed with all the factors to ones performed in the absence of TBP. Under our reaction conditions, the absence of TBP has the same effect as the use of a photoprobe with a mutation in the TATA element (TATAAAA to TAGAGAA) (55). The position of TFIIA α/β and TFIIA γ are indicated. B, schematic representation of promoter contacts by TFIIA in the context of the TBP-TFIIA-promoter complex and the TBP-TFIIA-TFIIB-TFIIF-RNAPII-TFIIE-promoter complex in which the DNA adopts a wrapped structure. Positions −39/−40, +26 and +1 are indicated. Only TBP, TFIIA, and RNAPII are represented to simplify the diagram.
The cross-linking of TFIIA to promoter regions downstream of the transcription initiation site (+10 to +30) was not unexpected. According to the DNA wrapping model, the DNA helices upstream of TATA (−40/−60 region) and downstream the initiation site (+10/+30 region) are juxtaposed in space. Our results provide additional support for the notion that promoter DNA is wrapped in the initiation complex and indicate that TFIIA is localized near the cross-point of the wrapped DNA structure.
TFIIA Directly Interacts with TBP, RAP74, TFIIE56, and TFIIE34
In the context of a preinitiation complex assembled with TFIIA, TBP, TFIIB, TFIIE, TFIIF, and RNAPII, we obtained cross-linking of TFIIA to photoprobes that are also cross-linked by other components of the complex. More specifically TFIIE34, RAP74, and RAP30 cross-link to photoprobe +26, while Rpb2, TFIIE34, RAP74, and RAP30 cross-link to photoprobe −39/−40 (23, 26). These observations indicate that TFIIA is in close proximity to these factors in the preinitiation complex, suggesting that TFIIA could directly interact with TFIIE, TFIIF, and RNAPII. To test this hypothesis, nTFIIA was chromatographed over different affinity columns containing immobilized RAP74, RAP30, TFIIE56, TFIIE34, TFIIB, and RNAPII. Columns containing TBP and BSA were used as positive and negative controls, respectively because TBP has been shown to interact with TFIIA (36, 37). The flowthrough was collected in each case and the columns were successively eluted with buffer containing 0.1 M, 0.3 M, and 0.5 M NaCl. The various fractions were analyzed by SDS-PAGE. Natural TFIIA (α, β, and γ subunits) was retained on the TFIIE56, TFIIE34, RAP74, and TBP columns but not on the TFIIB, RAP30, RNA-PII, and BSA columns (Fig. 2). Contaminant bands of the TFIIA fraction were visible in the flowthrough of all the columns. The binding of TFIIA to the affinity columns is most easily visualized by examining the elution of the β and γ subunits.
To further characterize these interactions, we next used the individual subunits of TFIIA in our affinity chromatography experiments. TFIIAα/β and TFIIAγ were individually chromatographed on affinity columns containing immobilized RAP74, TFIIE34 and TFIIE56. BSA was added to the input as an internal negative control. Fig. 3 shows that TFIIAα/β, but not TFIIAγ, interacts with RAP74 and TFIIE34. TFIIAα/β did not bind to the TFIIE56 column, whereas TFIIAγ was retarded on the TFIIE56 column, suggesting a weak interaction. This finding is surprising in view of the observation that nTFIIA binds strongly to the TFIIE56 column. Perhaps this is due to the association of TFIIAα/β and TFIIAγ resulting in a conformational change that favors binding to TFIIE56.
Two groups have previously reported interactions between TFIIA and TFIIE. Both used recombinant TFIIA, not the natural protein. In agreement with our results, Yamamoto et al. (59) obtained binding of TFIIE56 to TFIIAγ, and Yokomori et al. (60) obtained binding of TFIIE34 to TFIIAα/β. However, in contrast to both our data and that of Yokomori et al. (60), Yamamoto et al. (59) observed a weak binding of TFIIE34 to TFIIAγ but not to TFIIAα/β. The use of different binding assays may have caused this apparent discrepancy. Consistent with the interaction we observe, Yokomori et al. (60) have also shown that TFIIE interacts with the TFIIA-TBP complex on promoter DNA.
RAP74 Contains Two Distinct TFIIA-binding Domains
To determine the domain or domains of RAP74 responsible for the interaction with TFIIA, a series of RAP74 deletion mutants were immobilized on different affinity columns and rTFIIA (α/β and γ renatured together) chromatographed through each column. BSA was added to the input as an internal control, and a BSA column served as a negative control. Fig. 4A shows some representative data, and a summary is presented in Fig. 4B. All the N-terminal fragments of RAP74, except for RAP74-(1–75), which contains only the first 75 amino acids of the polypeptide, were bound by rTFIIA (Fig. 4A). Because rTFIIA did bind to RAP74-(1–136) but not to RAP74-(1–75), our results define a first domain of interaction between these two proteins that encompasses amino acids 76–136. We next used the C-terminal fragments of RAP74 in our affinity chromatography experiments. RAP74-(207–517), RAP74-(358–517), and RAP74-(407–517) were all observed to bind to RAP74, indicating the existence of a second interacting domain. To delineate this domain more precisely, two additional mutants, RAP74-(363–444) and RAP74-(363–409), were used. TFIIA bound well to RAP74-(363–444) but not to RAP74-(363–409) (Fig. 4A). These results define a second TFIIA-interacting domain of RAP74 located between amino acids 410 and 444. The existence of an additional putative TFIIA-binding domain in the central RAP74 region was ruled out by passing rTFIIA on columns containing RAP74–(136–258) and RAP74-(258–356). In each case, rTFIIA was not retained on the column. The two TFIIA-interacting domains of RAP74 that we identified are both localized in conserved regions of the protein, domain 76–136 being localized in conserved region I and domain 410–444 in conserved region III (see Fig. 4B for a schematic representation).
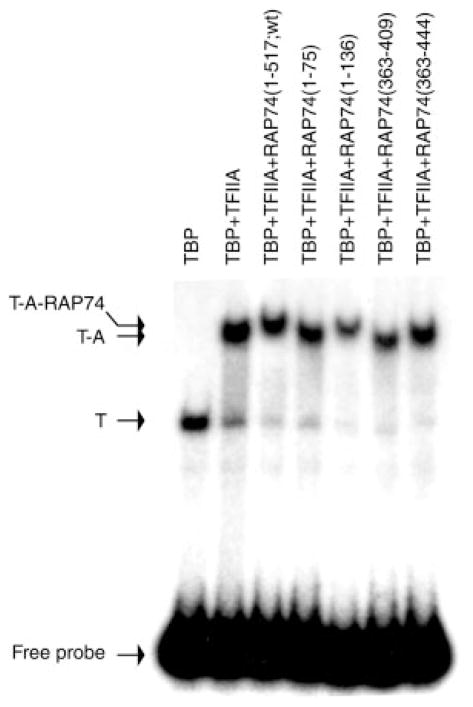
To assess whether or not RAP74 can associate with TFIIA on promoter DNA, we performed gel mobility shift experiments. RAP74 fragments carrying the TFIIA-binding domains (e.g. RAP74-(1–517;wt), RAP74-(1–136), and RAP74-(363–444)) but not fragments lacking the TFIIA-binding regions (e.g. RAP74-(1–75) and RAP74-(363–409)) gel shifted a TBP-TFIIA-promoter complex (Fig. 5). These results indicate that RAP74, through both its TFIIA-binding domains, can associate with TFIIA on promoter DNA.
Fig. 5. Association of RAP74 with a TBP-TFIIA-promoter complex.
Gel mobility shift assays were performed using a radiolabeled DNA fragment comprising the AdMLP in the presence of TBP alone, TBP, and TFIIA and TBP, TFIIA and various fragments of RAP74 (RAP74-(1–517;wt), RAP74-(1–75), RAP74-(1–136), RAP74-(363–409), and RAP74-(363–444)). The positions of the TBP (T), TBP-TFIIA (T-A), and TBP-TFIIA-RAP74 (T-A-RAP74) complexes and that of the free probe are indicated.
TFIIA Stimulates the Activity of TFIIE34 and RAP74 in Transcription Initiation
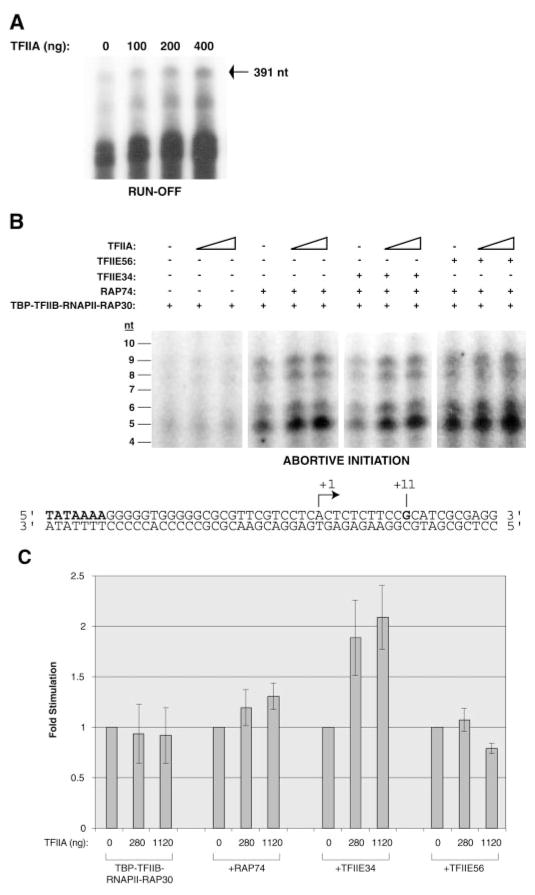
Several reports have shown that TFIIA can stimulate basal transcription by RNAPII in vitro, but is not essential for the basal transcription reaction (28, 33–37). For example, recombinant TFIIA increased the formation of a 391-nt run-off transcript from a supercoiled template carrying the AdMLP fused to a G-less cassette in the presence of TBP, TFIIB, TFIIE, TFIIF, and core RNAPII (Fig. 6A). To test whether or not TFIIA acts on the initiation step of the transcription reaction, we developed an abortive initiation assay in which an 80-base pair double-stranded oligonucleotide carrying the AdMLP was used to drive transcription initiation in the presence of RNAPII and the general transcription factors. In this assay only abortive transcripts of 2 to 10 nt in length can be synthesized because the transcription reaction is performed in the absence of GTP on a template that harbors a G at position +11 (see Fig. 6B). Abortive initiation was shown to be promoter-specific because the use of DNA templates with mutations in the AdMLP that impair transcription in run-off assays did not support synthesis of abortive transcripts.2 Under our reaction conditions, abortive initiation minimally requires the presence of TBP, TFIIB, RAP30, and RNAPII (Fig. 6). With this minimal set of factors, the addition of TFIIA does not stimulate the formation of abortive transcripts (Fig. 6, B and C). When either RAP74 alone, or RAP74 and TFIIE34 are added to the system, the formation of abortive transcripts is increased, indicating that these two factors are involved in transcriptional initiation (Fig. 6, data not shown, and Refs. 58, 61, 62). The addition of TFIIA to reactions containing either RAP74 alone, or RAP74 and TFIIE34, in addition to TBP, TFIIB, RAP30, and RNAPII, had a stimulatory effect (Fig. 6, B and C), indicating that TFIIA can stimulate the initiation stage of the transcription reaction. Because TFIIA stimulates transcription initiation only when RAP74 and TFIIE34 are present in the reaction, our results suggest that TFIIA enhances the functions of TFIIF and TFIIE to stimulate the basal transcription reaction.
Fig. 6. Stimulation of basal transcription by TFIIA.
A, run-off transcription assays were performed on a supercoiled template carrying the AdMLP using TBP, TFIIB, TFIIE, TFIIF, and RNAPII in either the absence or the presence of increasing amounts of TFIIA (100, 200, and 400 ng). The position of the accurately initiated transcript (391 nt) is indicated. B, abortive initiation assays were performed on a synthetic double-stranded oligonucleotide carrying the AdMLP using TBP, TFIIB, RAP30, and RNAPII in either the absence or the presence of RAP74 alone, RAP74 and TFIIE34, and RAP74 and TFIIE56. Increasing amounts of TFIIA were added to the reactions. The positions of the abortive transcripts (4–10 nt) are indicated. C, quantification of the stimulatory effect of TFIIA on abortive initiation. The intensity of the bands corresponding to the abortive transcripts from 4–6 experiments were quantitated using a PhosphorImager. The measured intensities were used to calculate the ratios of amount of transcript produced in the presence of TFIIA to that produced in its absence in each case (Fold Stimulation).
The role of TFIIA in RNAPII transcription is not completely resolved. However, several reports suggest that an important function of this factor is to act as a co-factor in transcriptional activation (28, 29, 33, 34, 44–47). In this paper, we establish the existence of structural and functional interactions between TFIIA and both TFIIE and TFIIF within the initiation complex. TFIIE and TFIIF have been shown to be involved in the melting of promoter DNA near the initiation site during open complex formation (9, 58). Considering that an activator such as Gal4-VP16, whose full activity requires the presence of TFIIA, can stimulate promoter melting near the initiation site (63), it is possible to envision an activation mechanism in which TFIIA functions as a bridge between the activator proteins and the general transcription factors TFIIE and TFIIF. This connection may help to explain how upstream activators influence and stimulate the biochemical events occurring at the transcription start site.
Acknowledgments
We thank the members of our laboratories for helpful discussions, Diane Bourque for artwork, and Will Home for critical reading of the manuscript.
Footnotes
This work was supported by grants from the Canadian Institutes for Health Research and the Cancer Research Society Inc. (to B. C.) and the National Institutes of Health (to Z. F. B.).
The abbreviations used are: RNAPII, RNA polymerase II; TF, transcription factor; TBP, TATA box-binding protein; RAP, RNA polymerase II-associated protein; nTFIIA, natural human TFIIA; rTFIIA, recombinant human TFIIA; PAGE, polyacrylamide gel electrophoresis; BSA, bovine serum albumin; AdMLP, adenovirus major late promoter; nt, nucleotide(s).
M. F. Langelier, Y. Porlier, and B. Coulombe, manuscript in preparation.
References
- 1.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Hampsey M. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y, Geiger JH, Hahn S, Sigler PB. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 4.Kim JL, Nikolov DB, Burley SK. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 5.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sopta M, Carthew RW, Greenblatt J. J Biol Chem. 1985;260:10353–10360. [PubMed] [Google Scholar]
- 7.Conaway RC, Garrett KP, Hanley JP, Conaway JW. Proc Natl Acad Sci U S A. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores O, Lu H, Reinberg D. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 9.Holstege FC, Tantin D, Carey M, van der Vliet PC, Timmers HT. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 11.Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 12.Lu H, Flores O, Weinmann R, Reinberg D. Proc Natl Acad Sci U S A. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serizawa H, Conaway RC, Conaway JW. Proc Natl Acad Sci U S A. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dvir A, Conaway RC, Conaway JW. Proc Natl Acad Sci U S A. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Q, Moreland RJ, Conaway JW, Conaway RC. J Biol Chem. 1999;274:35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- 16.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 17.Cramer P, Bushnell DA, Kornberg RD. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 18.Coulombe B, Li J, Greenblatt J. J Biol Chem. 1994;269:19962–19967. [PubMed] [Google Scholar]
- 19.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 20.Lagrange T, Kim TK, Orphanides G, Ebright YW, Ebright RH, Reinberg D. Proc Natl Acad Sci U S A. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forget D, Robert F, Grondin G, Burton ZF, Greenblatt J, Coulombe B. Proc Natl Acad Sci U S A. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TK, Lagrange T, Wang YH, Griffith JD, Reinberg D, Ebright RH. Proc Natl Acad Sci U S A. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert F, Douziech M, Forget D, Egly JM, Greenblatt J, Burton ZF, Coulombe B. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douziech M, Forget D, Greenblatt J, Coulombe B. J Biol Chem. 1999;274:19868–19873. doi: 10.1074/jbc.274.28.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim TK, Ebright RH, Reinberg D. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 26.Douziech M, Coin F, Chipoulet JM, Arai Y, Ohkuma Y, Egly JM, Coulombe B. Mol Cell Biol. 2000;20:8168–8177. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coulombe B, Burton ZF. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozer J, Moore PA, Bolden AH, Lee A, Rosen CA, Lieberman PM. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 29.Ma D, Watanabe H, Mermelstein F, Admon A, Oguri K, Sun X, Wada T, Imai T, Shiroya T, Reinberg D. Genes Dev. 1993;7:2246–2257. doi: 10.1101/gad.7.11.2246. [DOI] [PubMed] [Google Scholar]
- 30.DeJong J, Roeder RG. Genes Dev. 1993;7:2220–2234. doi: 10.1101/gad.7.11.2220. [DOI] [PubMed] [Google Scholar]
- 31.DeJong J, Bernstein R, Roeder RG. Proc Natl Acad Sci U S A. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranish JA, Lane WS, Hahn S. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 33.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 34.Yokomori K, Zeidler MP, Chen JL, Verrijzer CP, Mlodzik M, Tjian R. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 35.Ma D, Olave I, Merino A, Reinberg D. Proc Natl Acad Sci U S A. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coulombe B, Killeen M, Liljelund P, Honda B, Xiao H, Ingles CJ, Greenblatt J. Gene Expr. 1992;2:99–110. [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes P, Flores O, Reinberg D. Mol Cell Biol. 1992;12:413–421. doi: 10.1128/mcb.12.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buratowski S, Hahn S, Guarente L, Sharp PA. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 39.Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 40.Ge H, Roeder RG. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 41.Inostroza JA, Mermelstein FH, Ha I, Lane WS, Reinberg D. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 42.Meisterernst M, Roy AL, Lieu HM, Roeder RG. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 43.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Gralla JD, Carey M. Genes Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 45.Kang JJ, Auble DT, Ranish JA, Hahn S. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi N, Boyer TG, Berk AJ. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi T, Carey M. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 48.Ingles CJ, Shales M, Cress WD, Triezenberg SJ, Greenblatt J. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 49.Ha I, Lane WS, Reinberg D. Nature. 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 50.Finkelstein A, Kostrub CF, Li J, Chavez DP, Wang BQ, Fang SM, Greenblatt J, Burton ZF. Nature. 1992;355:464–467. doi: 10.1038/355464a0. [DOI] [PubMed] [Google Scholar]
- 51.Ohkuma Y, Sumimoto H, Hoffmann A, Shimasaki S, Horikoshi M, Roeder RG. Nature. 1991;354:398–401. doi: 10.1038/354398a0. [DOI] [PubMed] [Google Scholar]
- 52.Sumimoto H, Ohkuma Y, Sinn E, Kato H, Shimasaki S, Horikoshi M, Roeder RG. Nature. 354:401–404. doi: 10.1038/354401a0. [DOI] [PubMed] [Google Scholar]
- 53.Peterson MG, Inostroza J, Maxon ME, Flores O, Admon A, Reinberg D, Tjian R. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 54.Hodo HG, III, Blatti SP. Biochem. 1977;16:1334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- 55.Robert F, Coulombe B. Methods Mol Biol. 2001;148:383–393. doi: 10.1385/1-59259-208-2:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolner BS, Gralla JD. Mol Cell Biol. 2000;20:3608–3615. doi: 10.1128/mcb.20.10.3608-3615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burton ZF, Ortolan LG, Greenblatt J. EMBO J. 1986;5:2923–2930. doi: 10.1002/j.1460-2075.1986.tb04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan G, Greenblatt J. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 59.Yamamoto S, Watanabe Y, van der Speck PJ, Watanabe T, Fujimoto H, Hanaoka F, Ohkuma Y. Mol Cel Biol. 2001;21:1–15. doi: 10.1128/MCB.21.1.1-15.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yokomori K, Verrijzer PC, Tijan R. Proc Natl Acad Sci U S A. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lei L, Ren D, Burton ZFB. Mol Cel Biol. 1999;19:8372–8382. doi: 10.1128/mcb.19.12.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holstege FCP, van der Vliet PC, Timmers MHT. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W, Carey M, Gralla JD. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]