Abstract
Mitogen-activated dual-specificity MAPK phosphatases are important negative regulators in the MAPK signalling pathways responsible for many essential processes in plants. In a screen for mutants with reduced organ size we have identified a mutation in the active site of the dual-specificity MAPK phosphatase INDOLE-3-BUTYRIC ACID-RESPONSE5 (IBR5) that we named tinkerbell (tink) due to its small size. Analysis of the tink mutant indicates that IBR5 acts as a novel regulator of organ size that changes the rate of growth in petals and leaves. Organ size and shape regulation by IBR5 acts independently of the KLU growth-regulatory pathway. Microarray analysis of tink/ibr5-6 mutants identified a likely role for this phosphatase in male gametophyte development. We show that IBR5 may influence the size and shape of petals through auxin and TCP growth regulatory pathways.
Introduction
Leaf and floral organs grow by two basic cellular processes, cell proliferation and cell expansion to reach a given organ size [1]. Characterization of mutants with defects in proliferation and/or expansion has provided insight into how growth is regulated, with many of these regulatory factors appearing to act in independent pathways and having diverse predicted molecular functions [2]. A considerable number of genes have been identified that influence organ growth. These include genes involved in hormone signalling pathways, regulators of the timing and rate of proliferative or expansive growth and genes controlling identity and patterning of organs [3].
A number of auxin responsive genes have been identified in organ growth control. The auxin-induced ARGOS (AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE) gene contributes to regulating the timing of proliferation arrest [4]. ARGOS encodes a novel, plant specific protein which acts upstream of AINTEGUMENTA (ANT), encoding a member of the AP2/ERF transcription factor family. ARGOS promotes growth by stimulating ANT expression; ANT activity maintains the proliferative potential of cells in leaves and floral organs, with loss or gain of function leading to reduced or increased lateral organs, respectively [4–6]. Mitogen-activated protein kinases (MAPKs) have been implicated in auxin signalling and studies show a rapid MAPK activation in response to auxin in Arabidopsis seedling roots [7]. A screen for resistance to the inhibitory effects of the auxin precursor indole-3-butyric acid (IBA) on root growth identified a mutation in a MAPK phosphatase, INDOLE-3-BUTYRIC ACID-RESPONSE5 (IBR5) [8]. IBR5 is proposed to act as a positive regulator of auxin signalling in roots by inactivating MAPKs, including MPK12 [9]. MAPKs constitute a highly conserved family of enzymes in eukaryotes, and in plants MAPK-based signal transduction modules regulate a large number of physiological processes, including responses to environmental stresses and phytohormones [10].
Activation of MAPKs is regulated via dual phosphorylation of the conserved TXY motif located in the activation loop by upstream kinases (MAPKKs), and this activation can be reversed by dephosphorylation through protein phosphatases, including specialized dual-specificity MAPK phosphatases (MKPs) [11, 12]. There are five putative MKPs in Arabidopsis (AtMKP1, AtMKP2, DsPTP1, PHS1 and IBR5) which have the complete dual-specificty phosphatase active-site motif VxVHCx2GxSRSx5AYLM [8, 13–17]. Dephosyphorylation activitiy against Arabidopsis MAPKs has been shown for AtMKP2 (targeting MPK3 and MPK6), DsPTP1 (targeting MPK4) and IBR5 (targeting MPK12) in vitro [14, 15, 18]. IBR5 was confirmed to interact with MPK12 in vivo, and MPK12 acts as a negative regulator of auxin signalling [9]. It remains unclear how IBR5 modulates auxin responses in plants as it does not act through TIR1 or through destabilizing Aux/IAA repressor proteins [19]. Sensitivity to ABA also appears to act partially independently of auxin responses in ibr5 mutants as a suppressor that carries a mutation in PDR9/ABCG36, could restore ibr5 responses to a subset of auxins, but not to ABA [20].
Although the root phenotypes of ibr5 mutant plants have been described, above ground growth phenotypes have not been explored in detail. Mutant ibr5 plants have been described as having a shorter stature, epinastic leaves and defects in vascular patterning. A role for IBR5 in organ size control has not previously been characterized and in this paper we investigate which pathways are disrupted. A tinkerbell (tink/ibr5-6) mutant was identified in an EMS mutagenesis screen to identify novel regulators of growth control. tink/ibr5-6 plants have reduced petal size and a smaller stature compared to wild-type plants. Mapping revealed that the tink1 mutation disrupts the dual-specificity phosphatase active-site of IBR5. The growth dynamics of tink/ibr5-6 identified a role in regulating the rate of proliferative growth to maintain correct size and shape of organs. Microarray analysis suggests IBR5 plays a role in male gametophyte development, auxin and TCP growth regulatory pathways.
Results
The tink mutation affects organ size and shape by altering the rate of proliferative growth
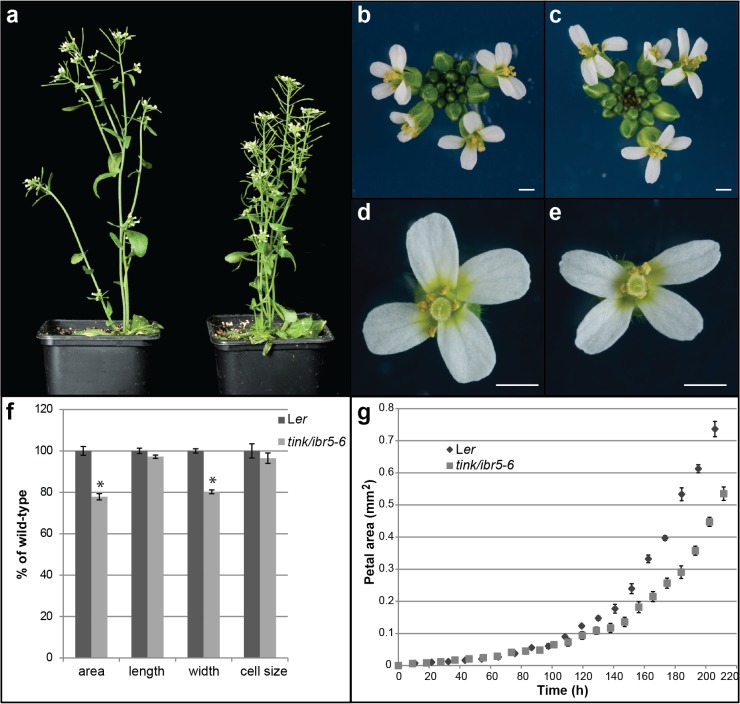
EMS mutagenesis of the klu-2 mutant in the Arabidopsis thaliana Landsberg erecta background was undertaken and approximately 2500 individual M2 lines were screened for reduced plant height and reduced petal size. Petals were used for screening as growth of floral organs is more consistent than leaf growth [21]. A mutant line that displayed significantly reduced petal and leaf size compared to klu-2 single mutants was selected for further investigation and backcrossed three times to wild type to separate the novel mutation and the klu-2 mutation. The novel mutation was named tinkerbell (tink) for its small stature (Fig 1A–1E and S1A Fig). Homozygous tink single-mutant plants show a 20% reduction in petal size compared to wild-type and this was due to a reduction in petal width (Fig 1F). Size of petals in heterozygous TINK/tink plants was indistinguishable from wild-type indicating this is a recessive mutation. To investigate the cause of the reduced petal size we measured cell size in mature petals of tink and wild-type plants. Cell size is not altered in tink plants compared to wild-type indicating that the reduced size of tink petals results from fewer cells (Fig 1F).
Fig 1. Above ground phenotypes of tink/ibr5-6 mutants.
a. Mutants in tink1/ibr5-6 (right) display reduced plant height and bushier phenotypes compared to Ler plants (left). An increased number of flowers in the inflorescence (c) and narrow petals (e) are observed in tink1/ibr5-6 compared to Ler (b, d) flowers. f. Measurement of tink1/ibr5-6 and Ler petal size showing statistically significant (shown by *) reduction in petal area (p value ≤ 4.5e-14) and petal width (p value ≤ 2e-37) in two tailed t-tests assuming unequal variance. g. Kinematic analysis of tink1/ibr5-6 and Ler petal size during development. Scale bar is 1 mm. Values are shown as mean ± SEM where n = 20.
To determine how TINK regulates organ size, we followed the growth dynamics of petal and leaf primordia in tink mutant and wild-type plants (Fig 1 and S1 Fig). Similar to petals, rosette area was significantly reduced from day 8 in tink plants compared to wild-type (illustrated by thick red line in S1B Fig). Mutant tink plants were shown to have a slightly decreased plastochron compared to wild-type which was visible by the increased number of flowers in the tink inflorescence (Fig 1C). Kinematic analysis of petal and leaf growth shows that tink plants have a reduced rate of organ growth compared to wild-type (Fig 1G and S1B Fig). This is particularly interesting as previous studies suggest that most regulators of organ size affect the transition between cell division and expansion rather than the rate of cell division itself [3].
The dual-specificity phosphatase indole-3-butyric acid-reponse5 (IBR5) is mutated in tink mutants
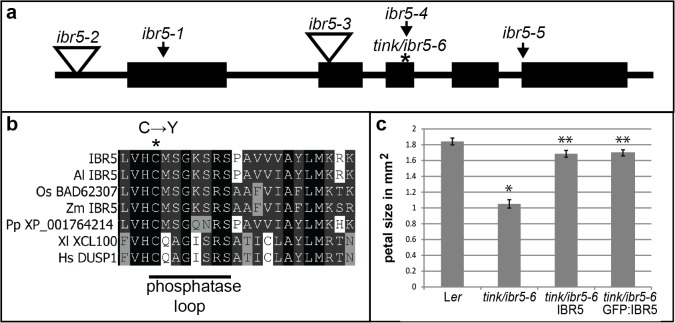
An F2 population of a backcross of tink mutants to Col-0 wild type was used for whole genome sequencing to identify the causal mutation. Rough mapping and analysis of SNPs distribution (S1C Fig) indicated that the mutation was located on the short arm of Chromosome 2. Further analysis of SNPs within this region revealed a G-to-A transition typical of EMS mutagenesis within the coding region of At2g04550 that was associated with the tink mutant (Fig 2A). At2g04550 corresponds to the previously characterised IBR5 gene that encodes a dual specificity protein phosphatase 1E [8]. Therefore tink represents a new mutant allele of IBR5 that will also be referred to as ibr5-6. Dual specificity protein phosphatases are characterized by a highly conserved active site motif VxVHCx2GxSRSx5AYLM, with the cysteine and arginine residues participating with the conserved aspartate in catalysis [18, 22]. The cysteine of this signature begins the dephosphorylation process with a nucleophilic attack on the phosphorus atom of the phosphotyrosine or phosphothreonine substrate. Disruption of this conserved cysteine has been shown to result in catalytic inactivity [19]. The G-to-A transition in tink/ibr5-6 changes the active cysteine residue to a tyrosine (Fig 2B). Complementation of the tink/ibr5-6 mutant with both p35S::IBR5 and p35S::GFP:IBR5 construct partially recovered wild-type petal size, indicating that loss of phosphatase activity of IBR5 contributes to the tink/ibr5-6 phenotype (Fig 2C).
Fig 2. Identification of the tink/ibr5-6 causal mutation.
a. Schematic representation of the IBR5 gene showing point mutation sites of mutants generated by EMS (indicated by arrows), ibr5-1 (causing a premature stop codon), ibr5-4 (G727 to A mutation in the third exon that changes G132 to E), ibr5-5 (G to A mutation in the intron of the last intron-exon junction) and tink/ibr5-6 in the third exon (asterix, G721 to A transition in tink/ibr5-6 changes C129 to Y in the conserved dual-specificity phosphatase catalytic domain), and T-DNA insertion sites of ibr5-2 and ibr5-3 in the promoter and second exon (open triangles). b. Alignment of IBR5 with plant homologues from Arabidopsis lyrata (Al), rice (Os), maize (Zm), Physcomitrella patens (Pp) and animal homologs from Xenopus laevis (Xl) and humans (Hs) showing the conserved phosphatase loop region. Identical residues are shaded in black and similar residues in at least two and four sequences are shaded in light and dark grey respectively. The G to A transition in tink/ibr5-6 that changes the active-site Cysteine residue to a Tyrosine is indicated with an asterisk. c. Petal size measurements of Ler, tink/ibr5-6, and tink/ibr5-6 plants complemented with p35S::GFP:IBR5 or p35S::IBR5 constructs. The significant reduction in size of tink/ibr5-6 petals compared to Ler (shown by *, p value ≤ 2.6e-16, two tailed t-test) is partially rescued in tink1/ibr5-6 GFP:IBR5 and tink1/ibr5-6 IBR5 petals. Petal size of tink1/ibr5-6 GFP:IBR5 and tink1/ibr5-6 IBR5 is significantly larger than that of tink/ibr5-6 (shown by **; p value ≤ 6e-12, IBR5:GFP and p ≤ 1.3e-11, IBR5) in two tailed t-tests assuming unequal variance. Values are shown as mean ± SEM, with n = 20.
To investigate if tink/ibr5-6 shows the characteristic reduced auxin sensitivity of other ibr5 mutants we performed root growth assays with or without the auxin IAA (Fig 3). Unlike ibr5-3, tink/ibr5-6 shows a very weak auxin insensitivity phenotype in the presence of 100 nm IAA (Fig 3B). The tink/ibr5-6 mutants show reduced root growth on standard growth media and a slight inhibition of root growth in the presence of auxin compared to the wild-type (Fig 3B and 3C). Previous complementation studies of IBR5 with a C-to-S phosphatase mutation in the ibr5-1 mutant background and the ibr5-4 allele which has a G-E transition in the catalytic domain show a weaker response than full loss of function ibr5 alleles [19, 23]. Additionally, ibr5-1 IBR5C-S lines fail to fully rescue 2,4-D resistance of ibr5-1 alleles [19]. These results suggest loss of IBR5 phosphatase activity has pleiotropic effects on root growth.
Fig 3. Root phenotype of ibr5 alleles compared to wild-type.
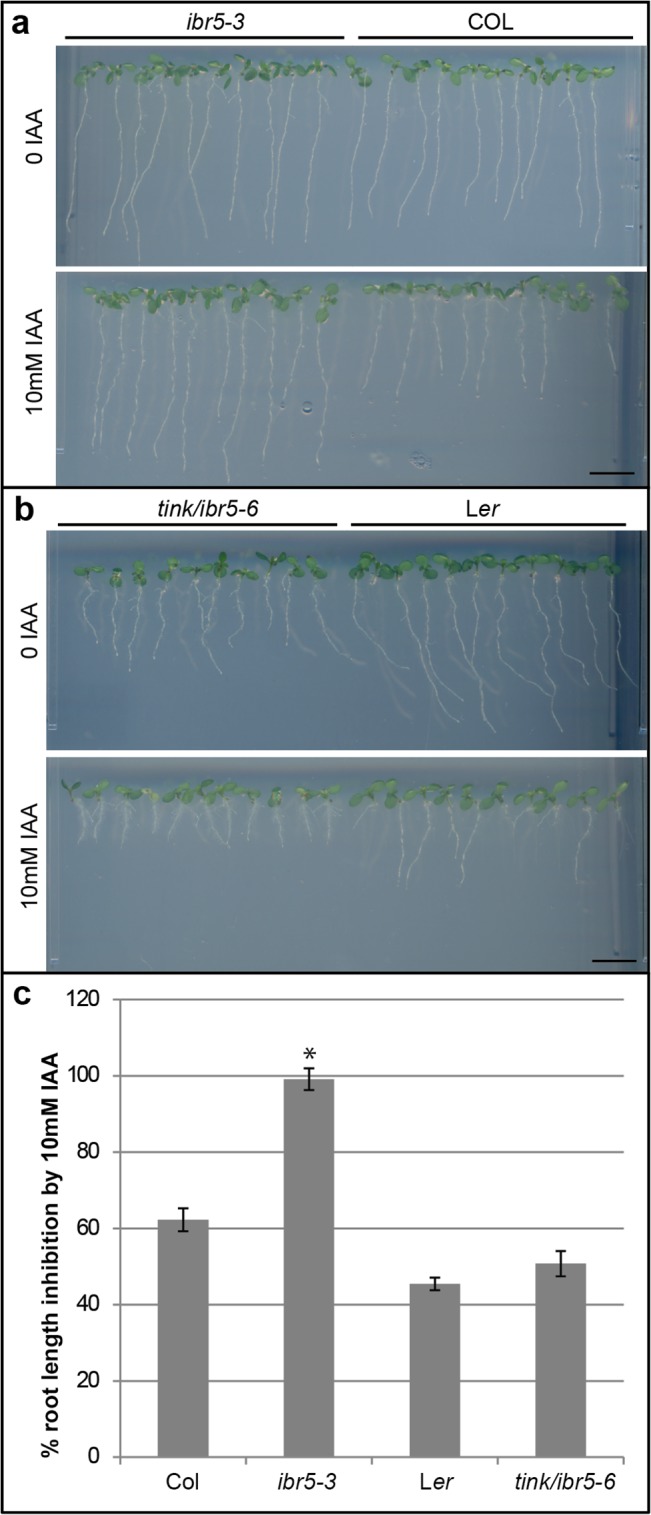
a. On standard growth medium (top panel) the ibr5-3 allele is indistinguishable from the wild-type (Col) whereas in medium containing 10 mM IAA, the ibr5-3 allele is insensitive to the inhibition of root growth seen in the wild-type (bottom panel). b. The tink/ibr5-6 allele shows reduced root growth compared to Ler on standard growth medium (upper panel) and medium containing 10 mM IAA (bottom panel). c. Inhibition of root length of Col, ibr5-3, Ler and tink/ibr5-6 plants grown on 10 mM IAA compared to un-supplemented medium. Col plants show a 38% reduction in root growth, compared to ibr5-3 mutants which are insensitive to the root growth inhibition (shown by *, p value ≤ 5.7e-14). Ler roots show a 55% decrease in root length when grown on 10 mM IAA compared to un-supplemented medium and tink/ibr5-6 plants show a similar root inhibition phenotype (p value ≤ 0.3). Scale is 1 cm. Values are shown as mean ± SEM, with n = 20.
Investigation of ibr5-3 mutant flowers revealed a narrow petal phenotype and tink/ibr5-6 displays vascular defects in petals similar to that observed in ibr5-3 plants (S1D–S1F Fig; [19]). A reduced size of adult ibr5 mutant plants compared to wild-type has previously been observed [8,19] and here we show that both petals and leaves are significantly smaller than wild-type. The above ground phenotypes of ibr5 mutant alleles varies. Presumed loss of function alleles ibr5-1 and ibr5-3, as well as an allele with altered transcript length, ibr5-5 show reduced plant height [23]. Alleles with altered IBR5 catalytic activity, ibr5-4 and ibr5-1 IBR5C-S do not affect plant height whereas the tink/ibr5-6 allele presented here appears to be consistent with the loss of function alleles.
Previous studies of the IBR5 promoter using GUS reporters showed IBR5 is expressed in roots, leaves and flowers and did not reveal any GUS expression in mature petals [8]. Other resources such as AtGenExpress microarray data suggest IBR5 is expressed in petals of young flowers (http://www.weigelworld.org/resources/microarray/AtGenExpress; [24]. It is possible pIBR5:GUS was not detected in petals in previous studies due to regulatory elements being missed in the promoter fragment (-2005 to -30) used, rather than the 3850bp upstream to the next upstream gene. Alternatively, IBR5 could be expressed earlier in petal development than the mature flower investigated or IBR5 acts non-cell autonomously to affect petal size. Identification of different splice variants of IBR5 that have some distinct functions may also play a role in post-transcriptional regulation of IBR5 in the peta ([23]). Previous studies show p35S:YFP:IBR5 fusion proteins located to the nucleus in root epidermal cells [9, 23]. Similarly our studies of the complementing p35S::GFP:IBR5 construct also show nuclear localization in the petal (S2 Fig).
Organ size and shape regulation by TINK/IBR5 acts independently of KLU regulatory pathways
The tink/ibr5-6 mutant was identified in a mutagenesis screen as an enhancer of the klu-2 mutant phenotype. The cytochrome P450 KLUH (KLU)/CYP78A5 is presumed to generate a growth-promoting signal that acts in a regulatory mechanism to coordinate the growth of individual organs [25]. Increased activity of KLU causes organ overgrowth, while klu mutants form smaller aerial organs consisting of fewer cells. Detailed investigation of double mutant tink/ibr5-6 klu-2 plants shows an additive effect to decrease petal size (Fig 4). This suggests that IBR5 acts independently of KLU regulatory pathways.
Fig 4. Double mutant analysis of tink/ibr5-6 klu-2 mutants compared to single mutants and wild-type.
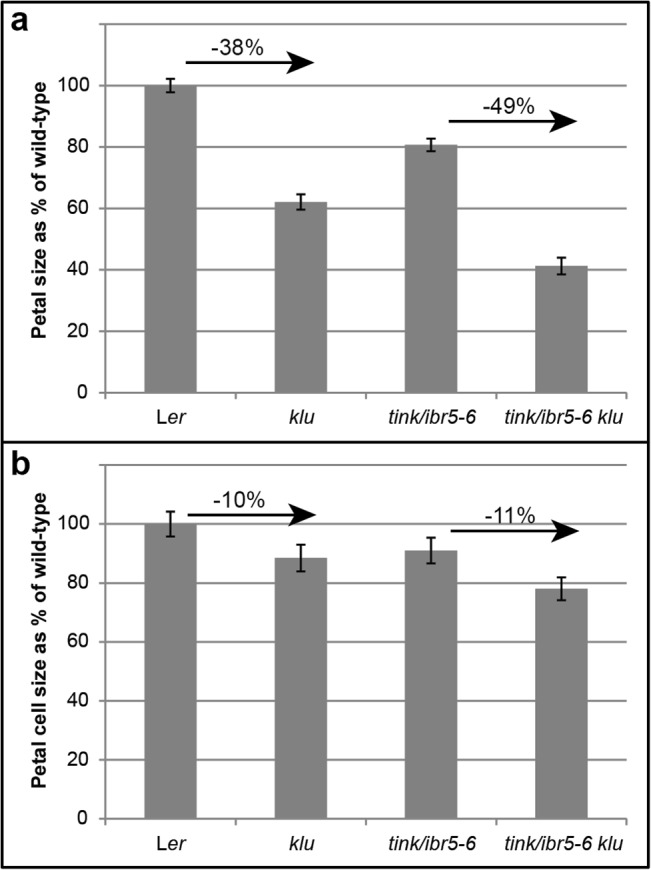
a. A significant reduction in petal area (p value ≤ 2.4e-7) is seen in klu-2 mutants compared to wild-type and the same relative decrease in petal size occurs in double mutants of tink/ibr5-6 with klu-2. b. Cell size in klu-2 mutants is significantly reduced (p value ≤ 0.02) compared to Ler and the same relative decrease in cell size occurs in tink/ibr5-6 klu-2 mutants. Values are shown as mean ± SEM, with n = 10.
Microarray analysis of tink/ibr5-6 mutants identifies a role for IBR5 in male gametophyte development
To gain further insight into the function of IBR5, transcriptional profiling of tink/ibr5-6 closed flowers was undertaken. A vast number of genes are mis-regulated in this mutant with an absolute log2 fold change above 1 compared to the wild-type (S1 Table). Gene ontology (GO) analysis, which categorises genes based of the gene product properties, such as cellular component, molecular function and biological process, shows an over-representation of genes involved in reproduction and more specifically male gametophyte development (Table 1). Expression of a subset of genes predicted to be involved in pollen development were confirmed as mis-regulated in the tink/ibr5-6 mutant using quantitative real-time (Q)-PCR (S3 Fig). To test for a defect in pollen function, pollen morphology of tink/ibr5-6 was observed, and the transmission efficiency of tink/ibr5-6 pollen was investigated by crossing tink/+ heterozygotes as male to wild-type female parents. Anthers and pollen of tink/ibr5-6 mutants did not show any gross morphological defects compared to wild-type. Also, no significant difference in transmission efficiency through the tink/ibr5-6 male gametophyte is evident (S3 Fig). Thus, despite the expression signature in tink/ibr5-6 mutant flowers, mutant pollen does not seem to be impaired in its function under standard laboratory conditions, and more specific conditions may be required to reveal a pollen phenotype.
Table 1. List of GO terms enriched in tink/ibr5-6 vs Ler microarray dataset.
| GOID | GOterm | Annotated | Significant | Expected | Fisher.p.value |
|---|---|---|---|---|---|
| GO:0090406 | pollen tube | 39 | 20 | 1.74 | 2.46E-17 |
| GO:0042995 | cell projection | 40 | 20 | 1.79 | 4.72E-17 |
| GO:0048610 | cellular process involved in reproduction | 164 | 31 | 8.53 | 3.48E-10 |
| GO:0009860 | pollen tube growth | 79 | 21 | 4.11 | 3.71E-10 |
| GO:0035295 | tube development | 102 | 23 | 5.31 | 1.88E-09 |
| GO:0048868 | pollen tube development | 102 | 23 | 5.31 | 1.88E-09 |
| GO:0004857 | enzyme inhibitor activity | 138 | 26 | 7.29 | 1.35E-08 |
| GO:0009932 | cell tip growth | 99 | 21 | 5.15 | 2.98E-08 |
| GO:0009856 | pollination | 158 | 27 | 8.22 | 4.44E-08 |
| GO:0046910 | pectinesterase inhibitor activity | 43 | 13 | 2.27 | 1.93E-07 |
| GO:0031225 | anchored to membrane | 219 | 28 | 9.79 | 5.49E-07 |
| GO:0030234 | enzyme regulator activity | 297 | 37 | 15.69 | 1.15E-06 |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | 396 | 44 | 20.92 | 2.69E-06 |
| GO:0032989 | cellular component morphogenesis | 271 | 33 | 14.1 | 5.03E-06 |
| GO:0030154 | cell differentiation | 350 | 39 | 18.2 | 6.36E-06 |
| GO:0048869 | cellular developmental process | 489 | 49 | 25.43 | 8.76E-06 |
| GO:0022414 | reproductive process | 1012 | 84 | 52.64 | 1.40E-05 |
| GO:0000003 | reproduction | 1030 | 84 | 53.57 | 2.66E-05 |
| GO:0032501 | multicellular organismal process | 1867 | 135 | 97.11 | 4.41E-05 |
| GO:0048235 | pollen sperm cell differentiation | 27 | 8 | 1.4 | 4.77E-05 |
| GO:0031224 | intrinsic to membrane | 699 | 53 | 31.26 | 1.20E-04 |
| GO:0009653 | anatomical structure morphogenesis | 559 | 50 | 29.07 | 1.36E-04 |
| GO:0004091 | carboxylesterase activity | 290 | 31 | 15.32 | 1.58E-04 |
| GO:0048232 | male gamete generation | 34 | 8 | 1.77 | 2.82E-04 |
| GO:0055046 | microgametogenesis | 38 | 8 | 1.98 | 6.31E-04 |
| GO:0007276 | gamete generation | 40 | 8 | 2.08 | 9.05E-04 |
| GO:0005623 | cell | 11775 | 556 | 526.61 | 7.00E-03 |
| GO:0044464 | cell part | 11775 | 556 | 526.61 | 7.00E-03 |
| GO:0016787 | hydrolase activity | 2578 | 163 | 136.22 | 7.44E-03 |
| GO:0019953 | sexual reproduction | 80 | 10 | 4.16 | 8.38E-03 |
| GO:0048609 | multicellular organismal reproductive process | 88 | 9 | 4.58 | 3.91E-02 |
IBR5 likely regulates growth through auxin and TCP growth regulatory pathways
Given the previous association of IBR5 with regulation of auxin pathways and results showing reduced expression of pDR5:GUS in the ibr5-1 mutant [8], we were interested to know if auxin responsive genes are mis-regulated in tink/ibr5-6. A set of genes identified in microarray studies as being significantly up- or down-regulated in tink/ibr5-6 compared to wild-type were validated by Q-PCR. These genes are annotated as being involved in auxin synthesis, transport, regulation and responses (S2 Table, Fig 5A). Intriguingly this list included a number of genes involved in auxin efflux (PINs). This supports previous studies that IBR5 is involved in auxin signalling pathways. Auxin transport assays performed on basal stem segments of ibr5-3 and tink/ibr5-6 mutants compared to the respective wild-types showed no significant differences (Fig 5B). To determine if changes in auxin distribution are responsible for the altered petal phenotype, the pDR5:GFP reporter was introduced into tink/ibr5-6 and ibr5-3 mutants [9]. In wild-type petals pDR5:GFP is localized at the tip of developing petals and in the vasculature [26]. No difference to wild-type pDR5:GFP pattern of expression was observed in the ibr5 mutant lines (S4 Fig). A reduction in the level of pDR5:GUS in roots and leaves has previously been observed in ibr5-1 mutants, even in tissues where pIBR5:GUS was not observed [8, 19]. From our investigation of pDR5:GFP expression in petals of ibr5 mutants, either no change in auxin levels occurs or our methods are not sensitive enough to detect potentially subtle changes in pDR5:GFP expression.
Fig 5. Investigation of auxin pathways in tink/ibr5-6 mutants.
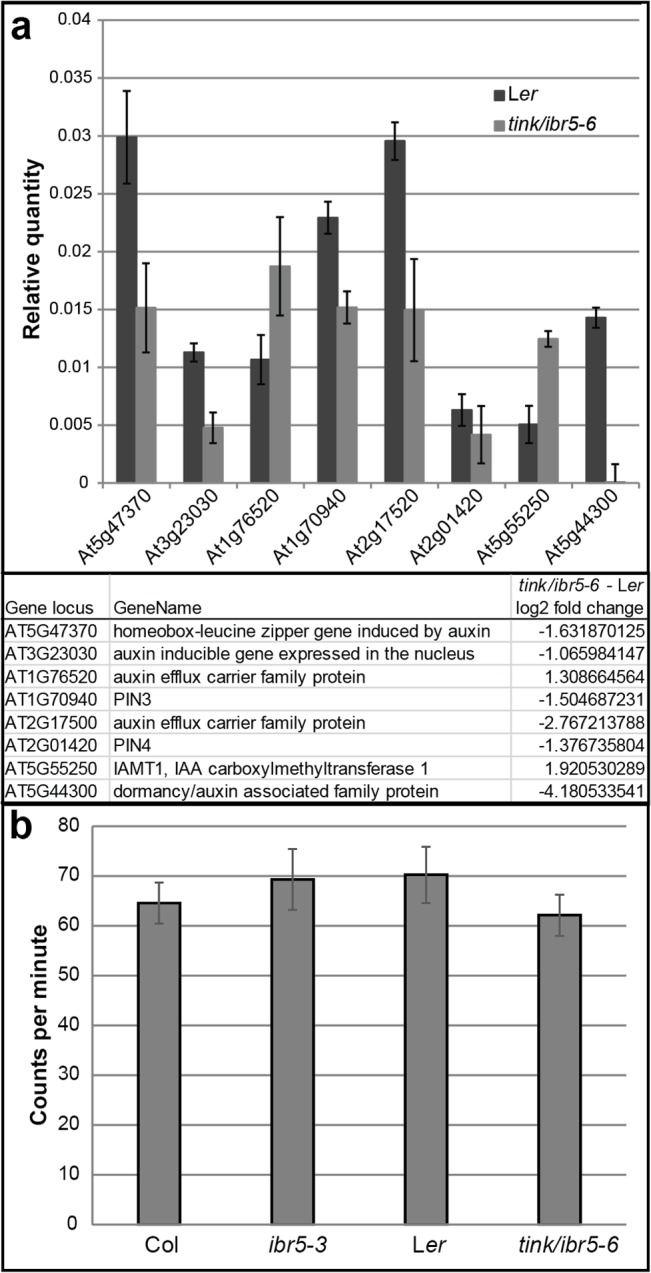
a. Real time quantitative (Q)-PCR validation (upper panel) of genes implicated in auxin biogenesis or transport identified in microarray analysis (lower panel) as being altered in tink/ibr5-6 mutants compared to wild-type flowers. Values in Q-PCR analysis are shown as mean ± SEM with expression levels normalized to that of the TUB6 gene for 3 biological and 3 technical replicates. b. Quantitation of bulk polar IAA transport in stem segments of the indicated genotypes. Values are represented as mean ± SEM of radiolabel transported (in cpm). There is no significant difference between wild-type and the respective ibr5 mutant alleles. At least 18 stem segments were assayed per genotype.
As cell proliferation is altered in tink/ibr5-6 mutants, the microarray data was searched for mis-regulated cyclin and related genes. A cyclin-dependent protein kinase, CYCP3;1 (AT2G45080), is significantly upregulated and a number of F-box family proteins altered in the tink/ibr5-6 mutants compared to wild-type (S3 Table). To identify other pathways through which IBR5 could be acting, MASTA analysis was used to compare the tink/ibr5-6 transcriptional profile to other available microarray data. This analysis identified a large overlap with the transcriptional profile of tcp14 and tcp15 mutants (S4 Table and S5 Table, S5 Fig). TCP transcription factors constitute a small family of plant-specific bHLH-containing, DNA-binding proteins that have been implicated in the control of cell proliferation in plants. To investigate if IBR5 overlaps with other TCP pathways, the tink/ibr5-6 dataset was also compared to a jaw-D microarray (S5 Fig). The JAW microRNA targets a number of class II TCPs, in particular TCP3 and TCP4, and dominant jaw-D alleles result in plants with uneven leaf shape and curvature [27]. Unlike tcp14 tcp15, no obvious overlap with tink/ibr5-6 and jaw-D microarray datasets occurs (S5 Fig). Reminiscent of the tink/ibr5-6 mutant, tcp14 and tcp15 mutants have a reduced plant height phenotype that results from a reduction in internode length. Leaf shape is also subtly altered in tcp14 tcp15 mutants indicating a role in regulating organ size and shape via changes in cell proliferation [28]. TCP15 is also implicated in regulating auxin homeostasis as the expression of an auxin-responsive promoter is induced by TCP15-EAR [29]. Further investigation to determine if IBR5 and TCP14 and TCP15 act in the same pathway to regulate organ size is an avenue for future research.
Discussion
tink/ibr5-6 alters the rate of proliferative growth
We have identified a novel role for the dual-specificity protein phosphatase IBR5 in regulating the shape and size of Arabidopsis organs. Loss of IBR5 function leads to narrow petals and leaves and its effect appears to be due to an altered rate of proliferative growth rather than changes in cell size. The reduced growth rate in tink/ibr5-6 in petals leads to 80% of the wild-type cell number. The plastochron of tink/ibr5-6 mutants is also shorter leading to an increased number of flowers in the inflorescence (Fig 1). The transition between cell proliferation and cell expansion is known to be a critical decision point during primordium growth. Unlike many other regulators of cell proliferative growth, IBR5 does not alter the length of time over which growth occurs and appears to be a novel regulator of growth. Consistent with an effect primarily on the rate of growth, rather than its duration, the tink/ibr5-6 mutation did not show any interaction with the klu mutation, which affects the timing of growth arrest, but not the growth rate.
Microarray analysis of tink/ibr5-6 revealed potentially novel roles in male gametogenesis and TCP regulatory pathways
Microarray analysis of tink/ibr5-6 mutant flowers identified a large number of mis-regulated genes compared to wild-type (S1 Table). An obvious pathway for organ size regulation was not identified whereas GO analysis of the tink/ibr5-6 microarray data suggests a significant overrepresentation of genes expressed in male gametophyte development and function. Transmission efficiency of tink/ibr5-6 gametes is not affected and the mechanism resulting in the change in gene expression remains somewhat elusive. Interestingly, a study of the phosphoproteome of mature Arabidopsis pollen identified an overrepresentation of mitogen-activated protein kinases [30]. The dual phosphorylation of MPK8 and MPK15 was confirmed yet no role in pollen development has been described to date. It is possible IBR5 plays a role in the male gametophyte through modulating activity of these or other MPKs. Although IBR5 did not interact with MPK8 or MPK15 in yeast-2-hybrid studies, an in vivo interaction cannot be ruled out [9].
Microarray comparison (MASTA) analysis revealed an interesting overlap of the tink/ibr5-6 profile with that of tcp14 tcp15 [31]. TCP transcription factors are key regulators of cell proliferation in growing organs and the balance between the growth-promoting class I factors and the negatively acting class II TCPs has been proposed to regulate the arrest of proliferative growth [32, 33]. Class I TCP factors are proposed to stimulate division required to produce the correct number of cells in young lateral primordia followed by a suppression of cell growth and division by class II TCP genes as cells exit the proliferative zone [34]. This view has been challenged in recent times as the class I TCPs, TCP14 and TCP15, can act to either promote or repress cell proliferation depending on the developmental context [28].
TCP14 and TCP15 are closely related class I TCP genes that modulate cell proliferation in the developing leaf blade and floral tissues and promote cell division in young internodes [28]. Overexpression of TCP14 (pAS1:TCP14) resulted in inhibition of internode elongation, inhibition of petal growth, reduced fertility and promotion of trichome development on sepals [35]. In tcp14 tcp15 double mutants and TCP14 overexpression lines the activity of the promoter of the mitotic factor CYCB1;2 is reduced or increased in stems, respectively [28, 35].
It is possible the ibr5 mutant growth phenotypes are mediated in part through altered activity of TCP14 and TCP15 transcription factors. It is tempting to suggest that this may occur through changes in phosphorylation status of these proteins. Recently a recombinant TCP8 was shown to be phosphorylated at Ser211 when expressed in Hi5 insect cells [36]. Characterisation of TCP14 and TCP15 in the ibr5 mutant background and generation of triple mutant tink/ibr5-6 tcp14 tcp15 mutants are interesting avenues for further investigation. Interestingly, TCP15 has been implicated in auxin homeostasis as it has been shown to bind the promoter regions of IAA3/SHY2 and the auxin responsive gene At1g29460 [29]. IBR5 is known to play a role in regulating auxin pathways yet the mechanism has remained cryptic.
IBR5 likely mediates growth through auxin pathways
The IBR5 loss of function mutant alleles ibr5-1 and ibr5-3 show less sensitivity to inhibitory concentrations of auxin in root growth assays. Expression of pDR5:GUS is reduced in these ibr5 alleles suggesting that IBR5 normally acts to promote auxin responses [8, 19, 20]. The role of IBR5 in auxin pathways remains unclear as IBR5 transcript levels do not alter in response to auxin, IBR5 acts independently of the TIR1 and AXR1 and AUX1 pathways and Aux/IAA proteins are not stabilized in ibr5 [19].
MPK12 has been identified as interacting with IBR5 and shown to be a negative regulator of auxin signalling in roots [9]. MPK12 activity is activated by auxin in vivo and reduced levels of MPK12 result in hypersensitivity to auxin [9]. The association of MPK12 with IBR5 has reinforced their roles in regulation of auxin signalling pathways. Unlike other ibr5 alleles, tink/ibr5-6 does not show strong reduced sensitivity to the inhibitory effects of auxin on root growth. The ibr5-1 mutation causes a premature stop codon that would result in a truncated product lacking the conserved phosphatase domain and the C-terminal region responsible for binding MAPK12 [8, 9]. As the tink/ibr5-6 mutation results in a full length protein with likely loss of phosphatase activity it is possible that it is able to sequester MPK12 and dampen its normal activity. This is likely given previous complementation studies of the ibr5-1 mutant using a mutated IBR5c129s only resulted in partial rescue of ibr5 defects [19]. Studies suggest that IBR5 may have some phosphatase independent functions and may bind and sequester its substrate(s) [19, 23]. This suggestion would fit with the root phenotypes of tink/ibr5-6 mutants as they display reduced root growth in the absence of auxin. The tink/ibr5-6 mutation could potentially cause the IBR5C-Y to act as a dominant negative in roots. This could occur through sequestration of its substrate(s) such as MPK12. Plants with reduced levels of MPK12 show hypersensitivity to auxin; however, they do not show root phenotypes in the absence of auxin. The tink/ibr5-6 root phenotype on standard media may reflect the IBR5C-T more completely suppressing MPK12 and potentially other MAPK substrates.
MPK12 is not expressed in the petals which may explain why ibr5 mutant phenotypes are more consistent in this organ. Complementation with a p35S::GFP:IBR5 construct largely recovered the tink/ibr5-6 petal size phenotype suggesting that the IBR5 phosphatase is indeed altered in the tink/ibr5-6 mutant (Fig 2). IBR5 could have a different MAPK substrate in petals as well as having both phosphate dependent and phosphate independent functions.
A role for IBR5 in modifying auxin transport could be inferred from the identification of pleiotropic drug resistance9 (pdr9) as a suppressor of ibr5 [20]. PDR9 encodes an ATP-binding cassette transporter implicated in cellular efflux of the synthetic auxin 2,4-D. Microarrays of tink/ibr5-6 identified a number of the auxin efflux PIN genes with altered expression profiles. A number of auxin responsive genes have been identified in organ growth control. The auxin-induced ARGOS (AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE) gene contributes to regulating the timing of proliferation arrest [4]. ARGOS encodes a novel, plant specific protein which acts upstream of AINTEGUMENTA (ANT), encoding a member of the AP2/ERF transcription factor family. ARGOS promotes growth by stimulating ANT expression; ANT activity maintains the proliferative potential of cells in leaves and floral organs, with loss or gain of function leading to reduced or increased lateral organs, respectively [4–6]. Genetic interaction studies with ant could potentially uncover an interaction of IBR5 and ANT pathways although given ANT and ANT-like genes act to regulate the length of proliferative growth, it seems likely that IBR5 acts independently of this pathway.
Auxin transport assays in ibr5-3 and tink/ibr5-6 stems did not reveal altered dynamics and localization of a pDR5:GFP reporter did not show any difference in levels or pattern of expression in ibr5-3 or tink/ibr5-6 mutant petals. It is possible that subtle changes in auxin signalling at the cellular level are responsible for the ibr5 above ground phenotypes and examination of PIN protein localization in ibr5 mutants would be an interesting avenue to further investigate these effects. Alteration of auxin patterning would explain the narrow petal phenotype as auxin is thought to act as a negative polarity organizer [26].
In this paper we have described a new allele of ibr5 and detail the above ground floral phenotypes. The pathways via which IBR5 regulates growth through auxin and TCP pathways open up further areas of research for this regulator of signalling pathways.
Experimental Procedures
Plant Material and growth conditions
The tink/ibr5-6 mutant was isolated from an EMS-mutagenesized population in the klu-2 background and backcrossed three times to Ler plants to isolate tink/ibr5-6 single mutants. klu-2 mutants are described in Anastisou et al., (2007). Plant growth conditions were as described in [37].
Identification of the TINK causal mutation
The tink/ibr5-6 mutation was rough mapped in a tink/ibr5-6 x Col F2 population by using described markers (http://carnegiedpb.stanford.edu/publications/methods/pps.uppl.html). Nuclei from 250 tink/ibr5-6 x Col F2 plants showing the mutant phenotype were extracted using the CellLytic PN kit semi-pure method (Sigma) to identify SNPs using whole genome sequencing. Library preparation, sequencing and analysis of SNP data are outlined in [38]. Genotyping of the tink/ibr5-6 causal mutation in At2g04550 was performed using KASP genotyping (LGCgenomics) with primers At2g04550_SNP12_FOR, 5’ GGCACGTGTTCTTGTGCATT 3’, At2g04550_SNP12_fam, 5’ GAAGGTGACCAAGTTCATGCTCTCACCTACTTTTCCCAGACATGC 3’, At2g04550_SNP12_VIC, 5’ GAAGGTCGGAGTCAACGGATTCTCACCTACTTTTCCCAGACATGT 3’.
Phenotypic Analysis
Organ and cell sizes, as well as growth dynamics of petals, were measured as described by Disch et al., (2006). Values are represented as mean ± SEM throughout. Each value corresponds to at least twenty petals from at least ten plants. Petals were imaged either on a Zeiss SteREO Lumar dissecting microscope or a Leica DM 6000 compound microscope and petal measurements were taken from images using Fiji (http://fiji.sc). For root elongation inhibition assays plants were grown aseptically on half MS media (Sigma) with 0.5%[w/v] sucrose solidified with 0.8% (w/v) agar, either alone or supplemented with 100nm IAA. Plants were grown for 10d at 22°C under standard long day conditions and the length of the primary root was measured. A minimum of 20 roots were measured for each genotype and values are represented as mean ± SEM throughout.
Generation of GFP-IBR5 constructs
The coding sequence corresponding to IBR5 was amplified from Arabidopsis leaf cDNA (Ler ecotype), using Phusion high fidelity DNA polymerase (New England Biolabs) and primers, At2g04550_Xba_Spe_F, 5’ TCTAGACAAACTAGTATGAGGAAGAGAGAAAGAGAG 3’ and At2g04550_R_Sal, 5’ ACGGTCGACCTAAGAGCCATCCATTGCA 3’ according to the manufacturer’s instructions. The IBR5 sequence was cloned into the FP101 vector using XbaI and Sal restriction sites and GFP inserted into the SpeI site at the N-terminus [39]. This binary vector was transformed into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis using the floral dip method [40].
Microarray Analysis
RNA was extracted from inflorescences with young flower buds from individual tink/ibr5-6 mutant or Ler plants with three replicates using the RNEasy kit (Qiagen). Hybridization of Affymetrix ATH1 arrays was performed by NASC Affymetrix service, Nottingham, UK. Arrays were normalized using gcrma with differentially expressed genes identified using the R/Bioconductor packages affy, limma and gcrma. Genes with absolute log2 fold change above 1 and BH corrected p-values below 0.05 were considered differentially expressed. Arrays were also analyzed using R/RankProd to extract the top 200 up- and down-regulated genes for comparison with the MASTA dataset [31]. Normalized signal intensities for jaw-d microarrays ([27]; PMID: 12931144) were downloaded from NCBI GEA (accession number GSE518) and analyzed the same way to be added to the comparison. Raw data for our microarrays are available at NCBI GEO under accession number GSE66419.
Quantitative real-time (Q)-PCR
Q-PCR was used to verify microarray results for a subset of genes involved in male gametogenesis and auxin regulation. RNA was extracted as above and cDNA synthesized from 1μg of RNA using the Superscript III (Invitrogen) reverse transcriptase according to the manufacturer’s instruction. Q-PCR was performed with SYBR-Green PCR Mastermix (Invitrogen) on a Bio-Rad DNA Engine with Chromo4 RT-PCR Detector. Three independent RNA samples for each genotype were assayed in triplicate. Expression levels were normalized to those of the constitutively expressed TUBULIN (At5g12250, [41] gene in each sample and are shown as mean ± SEM. Oligonucleotide sequences are given in S6 Table.
Auxin transport Assay
Auxin transport assays were performed as described [42] with the following slight modifications. Stem segments (18 mm long) excised from the most basal cauline internodes were incubated for 18 h in 1 × Arabidopsis salts medium (ATS) containing 1% sucrose and radiolabeled IAA (1 μM). The amount of transported radiolabel was quantified by scintillation counting (Top CountNXT; Packard Biosciences). Plants were cultivated in the greenhouse under long-day conditions with additional artificial light when needed. Six-week-old plants were used for analysis.
Supporting Information
a. Representative Ler (left) and tink/ibr5-6 mutant (right) plants show the rosette leaves of tink/ibr5-6 mutants are narrower than wild-type. b. Kinematic analysis of tink1/ibr5-6 and Ler rosette size during development show tink1/ibr5-6 has significantly smaller rosette area (shown by red bar, P-values below 0.05 using Wilcoxon Rank Sum tests) c. SNP distribution on chromosome 2 in a mapping population of tink/ibr5-6 (Ler) crossed to Columbia. The ratio of homozygous SNPs to heterozygous SNPs was plotted (y-axis). d. Measurement of ibr5-3 and Col petal size shows a statistically significant (shown by *) reduction in petal area (p value ≤ 3e-7) and petal width (p value ≤ 1e-14) in ibr5-3 mutants using two tailed t-tests assuming unequal variance. Petals of Ler (e) and tink/ibr5-6 (f) mutants show defects in vein patterning in tink/ibr5-6 compared to wild-type. Scale bar is 1 cm in (a) and 0.1 mm in (e,f).
(TIF)
GFP is located in the nucleus, DIC (a), GFP fluorescence signal (b), and merge (c). Scale bar is 10μm.
(TIF)
Phenotype of wild-type (Ler) (a) and tink/ibr5-6 (b) anthers show no obvious differences. c. Q-PCR (upper panel) and microarray (lower panel) analysis of genes specifically expressed in male gametophyte development, with altered expression in tink/ibr5-6 mutants compared to wild-type (Ler). d. Transmission efficiency of the tink/ibr5-6 mutation through male gametes. Scale in (a) and (b) is 0.1 mm. Values in (c) are shown as mean ± SEM from 3 biological and 3 technical replicates with expression levels normalized to that of the TUB6 gene.
(TIF)
Bright-field image is shown in a, d and g, GFP fluorescence is shown in b, e and h and merged image is shown in c, f and i. Scale bar is 100 μm.
(TIF)
The number of overlapping genes (A, C, E and G) and the inverse overlap (B, D, F, H) significantly up (red) or down (blue) regulated is shown for comparison of tink/ibr5-6 with tcp13 (A, B), tcp14 (C, D), tcp13/+ tcp14/+ (E, F) and jaw-D (G, H).
(TIF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to Susanna Sauret-Gueto for providing the pDR5:GFP line and to François Parcy for the FP101 plasmid. The assistance of members of the Lenhard and Bevan groups in providing comments on the manuscript and helpful discussions is gratefully acknowledged. This work was financially supported by a Biotechnology and Biological Sciences Research Council grant to fund K.J.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported in part by the Biotechnology and Biological Sciences Research Council grant and the Australian Research Council, Centre of Excellence in Plant Cell Walls (to KJ).
References
- 1. Mizukami Y. A matter of size: developmental control of organ size in plants. Current opinion in plant biology. 2001;4(6):533–9. 10.1016/S1369-5266(00)00212-0 . [DOI] [PubMed] [Google Scholar]
- 2. Hepworth J, Lenhard M. Regulation of plant lateral-organ growth by modulating cell number and size. Current opinion in plant biology. 2014;17:36–42. Epub 2014/02/11. 10.1016/j.pbi.2013.11.005 . [DOI] [PubMed] [Google Scholar]
- 3. Johnson K, Lenhard M. Genetic control of plant organ growth. The New phytologist. 2011;191(2):319–33. Epub 2011/04/27. 10.1111/j.1469-8137.2011.03737.x . [DOI] [PubMed] [Google Scholar]
- 4. Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. The Plant cell. 2003;15(9):1951–61. Epub 2003/09/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krizek BA. AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic acids research. 2003;31(7):1859–68. Epub 2003/03/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):942–7. Epub 2000/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mockaitis K, Howell SH. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant Journal. 2000;24(6):785–96. 10.1046/j.1365-313x.2000.00921.x . [DOI] [PubMed] [Google Scholar]
- 8. Monroe-Augustus M, Zolman BK, Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. The Plant cell. 2003;15(12):2979–91. Epub 2003/11/25. 10.1105/tpc.017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JS, Wang S, Sritubtim S, Chen JG, Ellis BE. Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. The Plant journal: for cell and molecular biology. 2009;57(6):975–85. Epub 2008/11/13. 10.1111/j.1365-313X.2008.03741.x . [DOI] [PubMed] [Google Scholar]
- 10.Smekalova V, Doskocilova A, Komis G, Samaj J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnology advances. 2013. Epub 2013/08/06. 10.1016/j.biotechadv.2013.07.009 . [DOI] [PubMed]
- 11. Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26(22):3203–13. Epub 2007/05/15. 10.1038/sj.onc.1210412 . [DOI] [PubMed] [Google Scholar]
- 12. Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends in plant science. 2010;15(2):106–13. Epub 2010/01/06. 10.1016/j.tplants.2009.12.001 . [DOI] [PubMed] [Google Scholar]
- 13. Gonzalez Besteiro MA, Ulm R. ATR and MKP1 play distinct roles in response to UV-B stress in Arabidopsis. The Plant journal: for cell and molecular biology. 2013;73(6):1034–43. Epub 2012/12/15. 10.1111/tpj.12095 . [DOI] [PubMed] [Google Scholar]
- 14. Lee JS, Ellis BE. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. The Journal of biological chemistry. 2007;282(34):25020–9. Epub 2007/06/26. 10.1074/jbc.M701888200 . [DOI] [PubMed] [Google Scholar]
- 15. Lumbreras V, Vilela B, Irar S, Sole M, Capellades M, Valls M, et al. MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. The Plant journal: for cell and molecular biology. 2010;63(6):1017–30. Epub 2010/07/16. 10.1111/j.1365-313X.2010.04297.x . [DOI] [PubMed] [Google Scholar]
- 16. Yoo JH, Cheong MS, Park CY, Moon BC, Kim MC, Kang YH, et al. Regulation of the dual specificity protein phosphatase, DsPTP1, through interactions with calmodulin. The Journal of biological chemistry. 2004;279(2):848–58. Epub 2003/10/23. 10.1074/jbc.M310709200 . [DOI] [PubMed] [Google Scholar]
- 17. Pytela J, Kato T, Hashimoto T. Mitogen-activated protein kinase phosphatase PHS1 is retained in the cytoplasm by nuclear extrusion signal-dependent and independent mechanisms. Planta. 2010;231(6):1311–22. Epub 2010/03/13. 10.1007/s00425-010-1135-8 . [DOI] [PubMed] [Google Scholar]
- 18. Gupta R, Huang YF, Kieber J, Luan S. Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant Journal. 1998;16(5):581–9. 10.1046/j.1365-313x.1998.00327.x . [DOI] [PubMed] [Google Scholar]
- 19. Strader LC, Monroe-Augustus M, Bartel B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC plant biology. 2008;8:41 Epub 2008/04/22. 10.1186/1471-2229-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strader LC, Monroe-Augustus M, Rogers KC, Lin GL, Bartel B. Arabidopsis iba response5 suppressors separate responses to various hormones. Genetics. 2008;180(4):2019–31. Epub 2008/10/04. 10.1534/genetics.108.091512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szecsi J, Wippermann B, Bendahmane M. Genetic and phenotypic analyses of petal development in Arabidopsis. Methods Mol Biol. 2014;1110:191–202. Epub 2014/01/08. 10.1007/978-1-4614-9408-9_8 . [DOI] [PubMed] [Google Scholar]
- 22. Sun H, Tonks NK. The Coordinated Action of Protein-Tyrosine Phosphatases and Kinases in Cell Signaling. Trends Biochem Sci. 1994;19(11):480–5. 10.1016/0968-0004(94)90134-1 . [DOI] [PubMed] [Google Scholar]
- 23. Jayaweera T, Siriwardana C, Dharmasiri S, Quint M, Gray WM, Dharmasiri N. Alternative splicing of Arabidopsis IBR5 pre-mRNA generates two IBR5 isoforms with distinct and overlapping functions. PloS one. 2014;9(8):e102301 Epub 2014/08/22. 10.1371/journal.pone.0102301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–6. 10.1038/Ng1543 . [DOI] [PubMed] [Google Scholar]
- 25. Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Developmental cell. 2007;13(6):843–56. Epub 2007/12/07. 10.1016/j.devcel.2007.10.001 . [DOI] [PubMed] [Google Scholar]
- 26. Sauret-Gueto S, Schiessl K, Bangham A, Sablowski R, Coen E. JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS biology. 2013;11(4):e1001550 Epub 2013/05/09. 10.1371/journal.pbio.1001550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425(6955):257–63. Epub 2003/08/22. 10.1038/nature01958 . [DOI] [PubMed] [Google Scholar]
- 28. Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. The Plant journal: for cell and molecular biology. 2011;68(1):147–58. Epub 2011/06/15. 10.1111/j.1365-313X.2011.04674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. Journal of experimental botany. 2012;63(2):809–23. Epub 2011/10/22. 10.1093/jxb/err305 . [DOI] [PubMed] [Google Scholar]
- 30. Mayank P, Grossman J, Wuest S, Boisson-Dernier A, Roschitzki B, Nanni P, et al. Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant Journal. 2012;72(1):89–101. 10.1111/j.1365-313X.2012.05061.x . [DOI] [PubMed] [Google Scholar]
- 31. Reina-Pinto JJ, Voisin D, Teodor R, Yephremov A. Probing differentially expressed genes against a microarray database for in silico suppressor/enhancer and inhibitor/activator screens. The Plant journal: for cell and molecular biology. 2010;61(1):166–75. Epub 2009/10/09. 10.1111/j.1365-313X.2009.04043.x . [DOI] [PubMed] [Google Scholar]
- 32. Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(36):12978–83. Epub 2005/08/27. 10.1073/pnas.0504039102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Viola IL, Reinheimer R, Ripoll R, Manassero NG, Gonzalez DH. Determinants of the DNA binding specificity of class I and class II TCP transcription factors. The Journal of biological chemistry. 2012;287(1):347–56. Epub 2011/11/15. 10.1074/jbc.M111.256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aguilar-Martinez JA, Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Frontiers in plant science. 2013;4:406 Epub 2013/10/19. 10.3389/fpls.2013.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, et al. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. The Plant cell. 2012;24(1):96–108. Epub 2012/01/24. 10.1105/tpc.111.093518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valsecchi I, Guittard-Crilat E, Maldiney R, Habricot Y, Lignon S, Lebrun R, et al. The intrinsically disordered C-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Molecular bioSystems. 2013;9(9):2282–95. Epub 2013/06/14. 10.1039/c3mb70128j . [DOI] [PubMed] [Google Scholar]
- 37. Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. The E3 ubiquitin ligase BIG BROTHER controls arabidopsis organ size in a dosage-dependent manner. Current biology: CB. 2006;16(3):272–9. Epub 2006/02/08. 10.1016/j.cub.2005.12.026 . [DOI] [PubMed] [Google Scholar]
- 38. Uchida N, Sakamoto T, Kurata T, Tasaka M. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant & cell physiology. 2011;52(4):716–22. Epub 2011/03/15. 10.1093/pcp/pcr029 . [DOI] [PubMed] [Google Scholar]
- 39. Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS letters. 2004;561(1–3):127–31. 10.1016/S0014-5793(04)00148-6 . [DOI] [PubMed] [Google Scholar]
- 40. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16(6):735–43. 10.1046/j.1365-313x.1998.00343.x . [DOI] [PubMed] [Google Scholar]
- 41. Guenin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of experimental botany. 2009;60(2):487–93. 10.1093/Jxb/Ern305 . [DOI] [PubMed] [Google Scholar]
- 42. Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology. 2006;16(6):553–63. 10.1016/j.cub.2006.01.058 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a. Representative Ler (left) and tink/ibr5-6 mutant (right) plants show the rosette leaves of tink/ibr5-6 mutants are narrower than wild-type. b. Kinematic analysis of tink1/ibr5-6 and Ler rosette size during development show tink1/ibr5-6 has significantly smaller rosette area (shown by red bar, P-values below 0.05 using Wilcoxon Rank Sum tests) c. SNP distribution on chromosome 2 in a mapping population of tink/ibr5-6 (Ler) crossed to Columbia. The ratio of homozygous SNPs to heterozygous SNPs was plotted (y-axis). d. Measurement of ibr5-3 and Col petal size shows a statistically significant (shown by *) reduction in petal area (p value ≤ 3e-7) and petal width (p value ≤ 1e-14) in ibr5-3 mutants using two tailed t-tests assuming unequal variance. Petals of Ler (e) and tink/ibr5-6 (f) mutants show defects in vein patterning in tink/ibr5-6 compared to wild-type. Scale bar is 1 cm in (a) and 0.1 mm in (e,f).
(TIF)
GFP is located in the nucleus, DIC (a), GFP fluorescence signal (b), and merge (c). Scale bar is 10μm.
(TIF)
Phenotype of wild-type (Ler) (a) and tink/ibr5-6 (b) anthers show no obvious differences. c. Q-PCR (upper panel) and microarray (lower panel) analysis of genes specifically expressed in male gametophyte development, with altered expression in tink/ibr5-6 mutants compared to wild-type (Ler). d. Transmission efficiency of the tink/ibr5-6 mutation through male gametes. Scale in (a) and (b) is 0.1 mm. Values in (c) are shown as mean ± SEM from 3 biological and 3 technical replicates with expression levels normalized to that of the TUB6 gene.
(TIF)
Bright-field image is shown in a, d and g, GFP fluorescence is shown in b, e and h and merged image is shown in c, f and i. Scale bar is 100 μm.
(TIF)
The number of overlapping genes (A, C, E and G) and the inverse overlap (B, D, F, H) significantly up (red) or down (blue) regulated is shown for comparison of tink/ibr5-6 with tcp13 (A, B), tcp14 (C, D), tcp13/+ tcp14/+ (E, F) and jaw-D (G, H).
(TIF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.