Abstract
Phagocytic clearance of the spent photoreceptor outer segments (OS) by RPE cells is regulated by circadian rhythm cycle and is essential for photoreceptor integrity and function. Mertk regulates RPE phagocytosis and a deficiency in Mertk causes photoreceptor degeneration and visual loss. This study aimed to investigate Mertk regulation of the microRNAs (miRNA), potentially regulating expression of their target genes, which affect phagocytosis. The differentially expressed miRNAs were identified using miRCURY™ microRNA Arrays from total RNA isolated at 0900hr and 1900hr from the mechanically dissociated RPE sheets of the WT and Mertk−/− mice, which were housed in a 12-hour light-dark cycle with the lighting onset at 0700hr (7:00am). Validation of the differentially expressed miRNAs and assessment of the putative miRNA target gene expression were performed by real-time PCR. Among the differentially expressed miRNAs in the Mertk−/− RPE, seven miRNAs were up-regulated and thirteen were down-regulated in the morning groups. Similarly, twenty four miRNAs were found to be up-regulated and thirteen were down-regulated in the evening groups. To search for those that may participate in regulating expression of cytoskeletal proteins, we examined the predicted target genes that might participate in phagocytosis were examined by real-time PCR. Of nine potential altered targets, four deregulated genes were myosin subunits. Notably, multiple members of the 21 up-regulated miRNAs can theoretically recognize these down-regulated mRNAs, particularly MyH14 and Myl3. This study shows that loss of Mertk alters miRNA expression, which in turn affects expression of the downstream target genes, potentially affecting phagocytosis.
Introduction
The retinal pigment epithelium (RPE) cells are highly polarized and tightly associated with retinal photoreceptor cells. Once the retina becomes functional, the RPE is essential for phagocytic clearance of the spent photoreceptor outer segment (OS) tips [1–3]. Defects in phagocytic clearance of the OS by RPE lead to photoreceptor death and retinitis pigmentosa (RP) disease. The RPE functions in the rod OS turnover have been extensively studied in the Royal College of Surgeons (RCS) rat. [4, 5]. The RPE cells in these rats carry an inherited defect in rod OS phagocytosis and the mutant gene is the Mertk receptor [6, 7]. Mertk null mutation in causing photoreceptor degeneration has also been found in Mertk knockout mice [8, 9] and human RP patients [10–13]. Mertk mutation causes photoreceptor death due to an impaired phagocytosis of the shedding OS by the RPE, which normally expresses the Mertk receptor.
Many functions and molecular events of RPE cells display a unique circadian pattern. Phagocytic uptake of OS exhibits a robust light-driven and circadian burst of activity within the first few hours after exposure to light [14, 15]. Some molecules, especially those participating in the RPE phagocytosis, display diurnal regulation of their expression [16–19].
Gene expression is regulated at multiple stages, including mRNA stability and translation processes. MicroRNA plays several important roles in regulation of the gene expression by binding to complementary sequences within the 3’ untranslated region (3’UTR) of target mRNAs and causing subsequent translational repression or degradation of these mRNAs [20, 21]. There is evidence that shows miRNAs function in a variety of biological processes, including embryonic patterning and developments, cell lineage determination, and tumorigenesis. miRNA expression is tissue-specific and has been detected at high levels in the mouse eye, including the lens, cornea, and retina [22, 23]. Circadian regulation of the eye-specific miRNAs and of the relevant target genes has been shown to play important roles in regulation of circadian rhythm [24–26]. The expression profiles and functional roles of the miRNAs have been studied in retinitis pigmentosa [27].
Despite efforts to elucidate the expression profile of miRNAs in the mammalian eye, little is known about the specific functions of miRNAs in the mouse RPE cells. To identify the miRNAs that are regulated by Mertk and in turn regulate the expression of the target genes which potentially affect RPE phagocytosis, we performed a comprehensive analysis of the miRNA expression profiles. In this analysis, we compared the differential expressed species in the Mertk−/− RPE with the WT controls, using miRCURY LNA microRNA arrays. Since RPE phagocytosis is governed by circadian-regulation, we also examined expression profiles, especially those that were altered during the diurnal lighting cycle. Differentially expressed miRNAs identified by microarray was further confirmed by real-time qPCR. This study showed that several miRNAs were altered in the Mertk−/− RPE relative to the control, and some of them exhibited a diurnal expression pattern under circadian regulation.
Methods
Animal
All animals were housed in a pathogen-free facility and handled according to the regulations of the Institutional Animal Care and Use Committee (IACUC #10131) of University of Louisville and all procedures adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
RPE cell preparation
Mice were housed in 12-h light/12-h dark cycles with the light onset at 0700 hr and the offset at 1900 hr. For total RNA isolation, the RPE cells were quickly prepared at 0900 hr (2 hr after the onset of light) and 1900 hr (12 hr after lighting) from the wild type and Mertk−/− mice at age of 4 weeks. After the eyes were enucleated and rinsed with 1xPBS, the eyecups were prepared by removing the anterior eye tissues (cornea, iris epithelium, lens) and the neural retina. The RPE cells were isolated by mechanically scraping out of the RPE sheet from the eyecups.
Total RNA isolation
Each sample of the total RNA was isolated and pooled from 10 eyes using TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions. The purity and concentration of total RNA were measured on a spectrophotometer (NanoDrop ND-1000) by the absorbance (260/280 nm). The RNA integrity was confirmed by electrophoresis on a 1.5% agarose gel.
LNA-based miRNA microarray analysis
The miRNA profiling was performed in duplicates on two independent preparations of RPE RNAs using LNA mercury™ microarray (Exiqon, Denmark). 1 μg total RNA from each group of the Mertk−/− or WT RPE cells were labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array Labeling kit (Exiqon, Denmark). Equal amount of fluorescence-labeled sample and control was mixed and hybridized to the miRCURY™ LNA Array version 11.0 (Exiqon), which contains 1769 capture probes, complementary to all human, mouse, rat, and their related viral sequences from the v11.0 release of miRBase (http://microrna.sanger.ac.uk/sequences/). The hybridization was performed according to the miRCURY™ LNA Array manual and the array images were obtained by an Axon GenePix 4000B microarray scanner and normalized by GenePix pro V6.0 (Axon Instruments).
Quantitative real-time PCR
The real-time qPCR validation of miRNA expression was performed using Mir-X miRNA first-strand synthesis and quantification kit (Clontech), according to the manufacturer's instructions. The real-time qPCR quantification of specific miRNA sequences was performed on a 60-fold dilution of the cDNA samples using the SYBR Advantage qPCR premix and mRQ 3’ Primer (Clontech). The PCR reaction was carried out on a Stratagene Mx3005P instrument using the conditions of 1-cycle denaturation at 95°C, 10 sec; 40-cycle amplification at 95°C, 5 sec and 60°C, 20 sec; and dissociation at 95°C, 60 sec; 55°C, 30 sec; 95°C, 30 sec. All reactions were performed in triplicate and included with no template control for each gene. U6 snRNA was included in each sample as an endogenous control to determine the Ct for determination of the relative level using delta-delta Ct (ddCt) calculation method [23], ΔCt=Ct miRNA−Ct U6; ΔΔCt=Ct test group−ΔCtcontrol group; relative miRNA expression=2−ΔΔCT.
The level of putative target mRNA was quantified by real-time RT-PCR as described previously [28]. Expression levels of the genes were normalized to 18S rRNA. Each sample was compared in 3 independent RT-PCR amplifications.
Statistical analysis
For calculation of differentially expressed species, the statistical significance of differentially expressed miRNA was analyzed by student’s t-test after normalization. For each probe, an arithmetic mean of four replicates from two independent hybridizations was calculated. The probes with the mean values <50 were filtered as undetectable.
Results
Mertk mutation causes aberrant expression of microRNAs
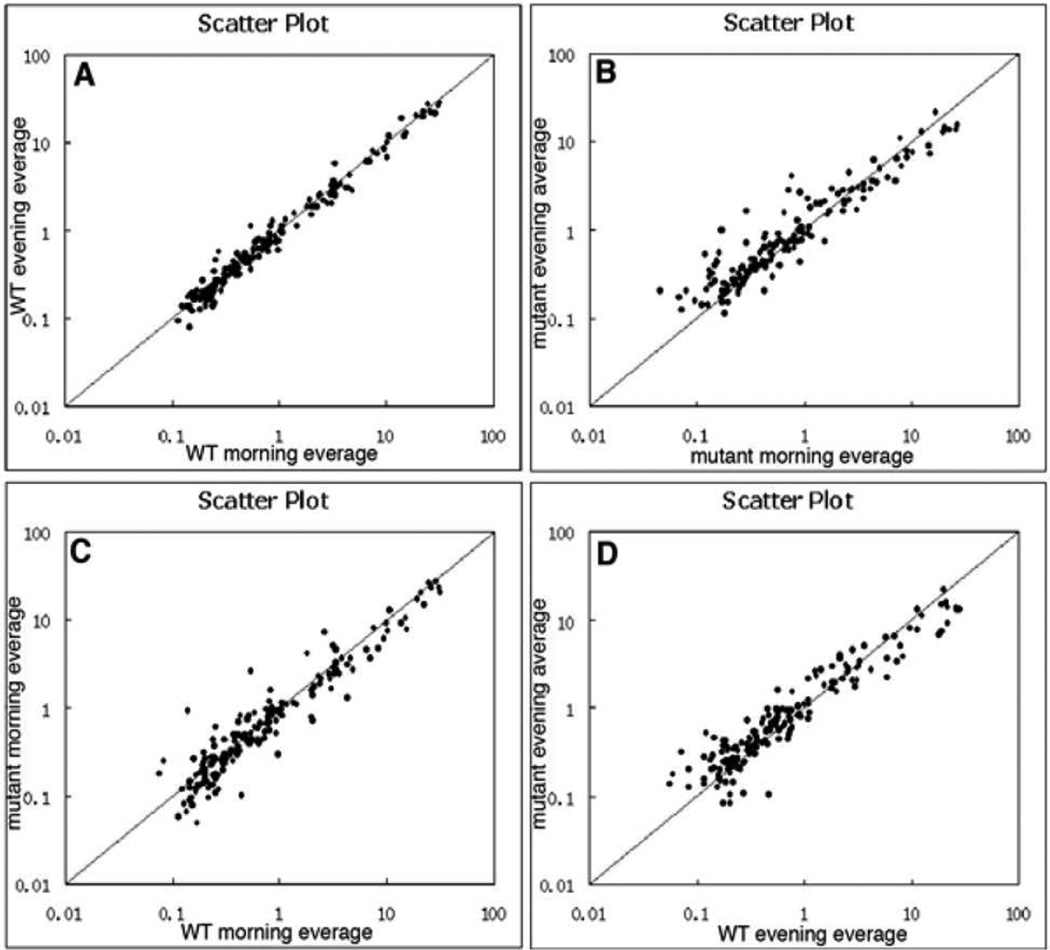
To search for the microRNAs that were altered in the Mertk−/− RPE during diurnal cycle and might participate in regulation of RPE phagocytosis, we performed a comprehensive analysis of the microRNA expression profile in the WT and Mertk−/− RPE cells. To compare the changes in miRNA expression at the phagocytic peak and off-peak, we collected samples at 2 and 12 hours after the onset of lighting. Cluster analysis was performed on the miRNA expression profiles between the morning and evening groups of WT or Mertk−/− cells, independently. The Scatter Plot revealed a greater variation between morning and evening in the Mertk−/− groups compared to a milder regulation in the WT cells (Fig. 1B vs 1A). A similar large alteration was also observed on the Scatter Plots of the comparison between the Mertk−/− and WT groups in either the morning or the evening experimental sets (Fig. 1C, 1D). Those comparisons indicate that Mertk mutation causes a greater alteration in miRNA expression.
Figure 1. MiRNA expression profiling in the Mertk mutant and wild-type control RPE cells during diurnal lighting cycle.
Scatter plots show a comparison of the miRNA expression profiles between the morning and evening groups in the WT (A) and Mertk−/− (B) RPE cells. The miRNA expression profiles were also compared between the Mertk−/− and WT RPE cells isolated in the morning (C) and evening (D).
Diurnal regulation of miRNA in the WT and Mertk−/− RPE cells
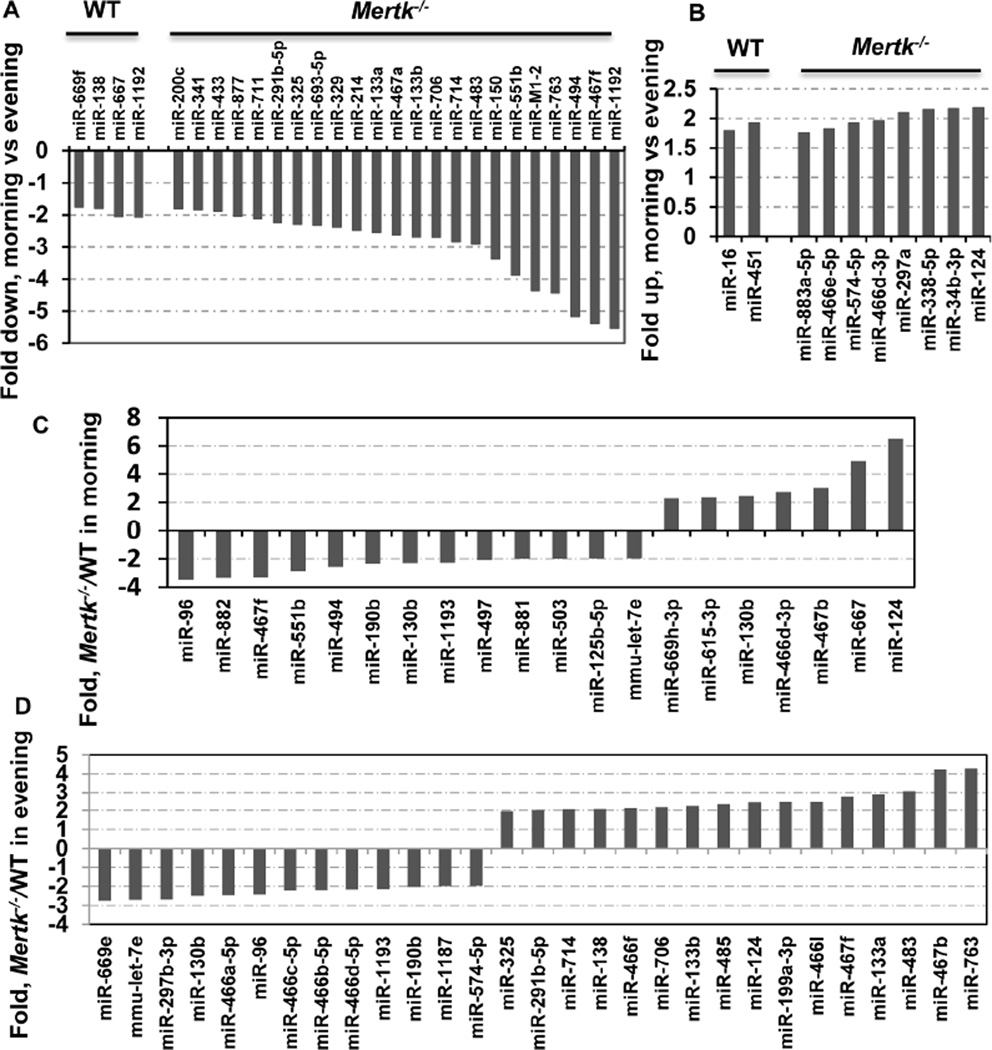
The phagocytic uptake of OS debris in mice exhibits a robust, light-driven diurnal peak 2 hours after light exposure and diminishes gradually thereafter [14, 29]. To detect miRNAs regulated by the circadian cycle, we compared the expression profiles in the morning versus the evening groups. The expression levels of 6 in WT and 32 in Mertk−/− RPE cells significantly differed between the cells from morning and evening time points. Four miRNAs in WT morning group showed a decrease of more than 1.8-fold when compared with the evening group; and 2 were upregulated by factors of 1.5 to 2.0 (Fig. 2A, 2B). Differential expression between two Mertk−/− groups was more noteworthy. There were 23 down- and 8 up-regulated genes with changes equal or more then 1.8-fold in the morning Mertk−/− RPE compared with the evening mutant cells (Fig. 2A, B). These data suggests that Mertk deficiency causes more significantly altered miRNA expression during the diurnal lighting cycle.
Figure 2. MiRNAs regulation during the diurnal lighting cycle. (A and B).
Comparison of miRNA expression between the morning and evening groups. MiRNAs expression levels are normalized to U6 internal control and plotted as fold changes in the miRNA level expressed in the morning as compared to the level in the evening. More miRNAs in the Mertk−/− RPE was differentially expressed between the morning and evening groups (A, B, right). Data are expressed as the average of two independent experiments. Only changes ≥ ±1.8-fold are shown. (C and D) Comparison of miRNA expression between the Mertk−/− and WT groups. Expressions of miRNAs are normalized to U6 internal controls and plotted as fold-changes for Mertk−/− over the WT controls. The comparison was performed between the morning (C) and the evening (D) groups. Data are expressed as the average of two independent experiments. Fold changes with ≥ ±2.0-fold are shown.
Mertk deficiency alters miRNA expression profiles in the RPE cell
To identify microRNAs that are aberrantly expressed in the Mertk−/− RPE cells, and may in turn regulate expression of their target genes potentially participating in phagocytosis, we compared miRNA expression levels in the mutant cells with controls using student’s t-test (p-value ≤0.05). We identified 7 up- and 13 down-significantly regulated miRNAs in the Mertk−/− RPE compared with the WT cells, in the morning groups (Fig. 2C). When similar comparison was done on the evening groups, we found that 16 miRNAs were significantly up-regulated and 13 were down-regulated (Fig. 2D). Of the 13 down-regulated miRNAs in the Mertk−/− morning group, five also showed a ≥2-fold decrease and one revealed an 1.5-fold decrease in the Mertk−/− evening group, while six showed no change or changes with ≤1.5 fold (Table 1). In addition, 8 miRNA down-regulated by ≥2-fold were found only in the Mertk−/− evening group but not in the morning group (Table 1). On the other hand, of 7 up-regulated miRNAs in the Mertk−/− morning group, 3 were also increased by ≥2-fold and 4 exhibited either no change or changes by ≤2-fold in the evening group (Table 1); in addition, 14 miRNAs were up-regulated by ≥2-fold only found in the evening group (Table 1). Notably, the miR467f and miR483* exhibited a diverse regulation pattern with down-regulation in the morning and up-regulation in the evening (Table 1).
Table 1. Differentially expressed miRNAs in the Mertk−/− RPE.
Table shows down-regulated (A) and up-regulated (B) miRNAs in the Mertk−/− RPE. The columns “Morning” and “Evening” represent fold changes in the Mertk−/− RPE over the WT RPE in the comparisons obtained from morning or evening groups, respectively. “-” indicates changes of <1.5-fold.
| (A) Down-regulated | ||
|---|---|---|
| miRNA | Morning | Evening |
| miR-96 | −3.5 | −2.4 |
| miR-190b | −2.3 | −2.0 |
| miR-130b | −2.3 | −2.5 |
| miR-1193 | −2.3 | −2.2 |
| mmu-let-7e | −2.0 | −2.7 |
| miR-882 | −3.3 | −1.5 |
| miR-467f | −3.3 | 2.7 |
| miR-551b | −2.9 | - |
| miR-494 | −2.6 | - |
| miR-497 | −2.1 | - |
| miR-881 | −2.0 | - |
| miR-503 | −2.0 | - |
| miR-125b-5p | −2.0 | - |
| miR-669e | - | −2.8 |
| miR-297b-3p | - | −2.7 |
| miR-466a-5p | - | −2.5 |
| miR-466c | - | −2.2 |
| miR-466b-5p | - | −2.2 |
| miR-466d-5p | - | −2.2 |
| miR-1187 | - | −2.0 |
| miR-574-5p | - | −2.0 |
| (B) Up-regulated | ||
|---|---|---|
| miRNA | Morning | Evening |
| miR-467b | 3.04 | 4.22 |
| miR-667 | 4.92 | 1.94 |
| miR-124 | 6.61 | 2.42 |
| miR-669h-3p | 2.30 | 1.59 |
| miR-615-3p | 2.34 | 1.50 |
| miR-130b | 2.44 | - |
| miR-466d-3p | 2.73 | - |
| miR-466f | 1.86 | 2.1 |
| miR-485 | 1.67 | 2.3 |
| miR-291b-5p | - | 2.0 |
| miR-325 | - | 2.0 |
| miR-714 | - | 2.1 |
| miR-138 | - | 2.1 |
| miR-706 | - | 2.2 |
| miR-133b | - | 2.2 |
| miR-199a-3p | - | 2.4 |
| miR-466l | - | 2.5 |
| miR-467f | −3.30 | 2.7 |
| miR-133a | - | 2.9 |
| miR-483 | −1.62 | 3.1 |
| miR-763 | - | 4.3 |
Real time qPCR confirmation of microRNA changes in the Mertk−/− RPE cells
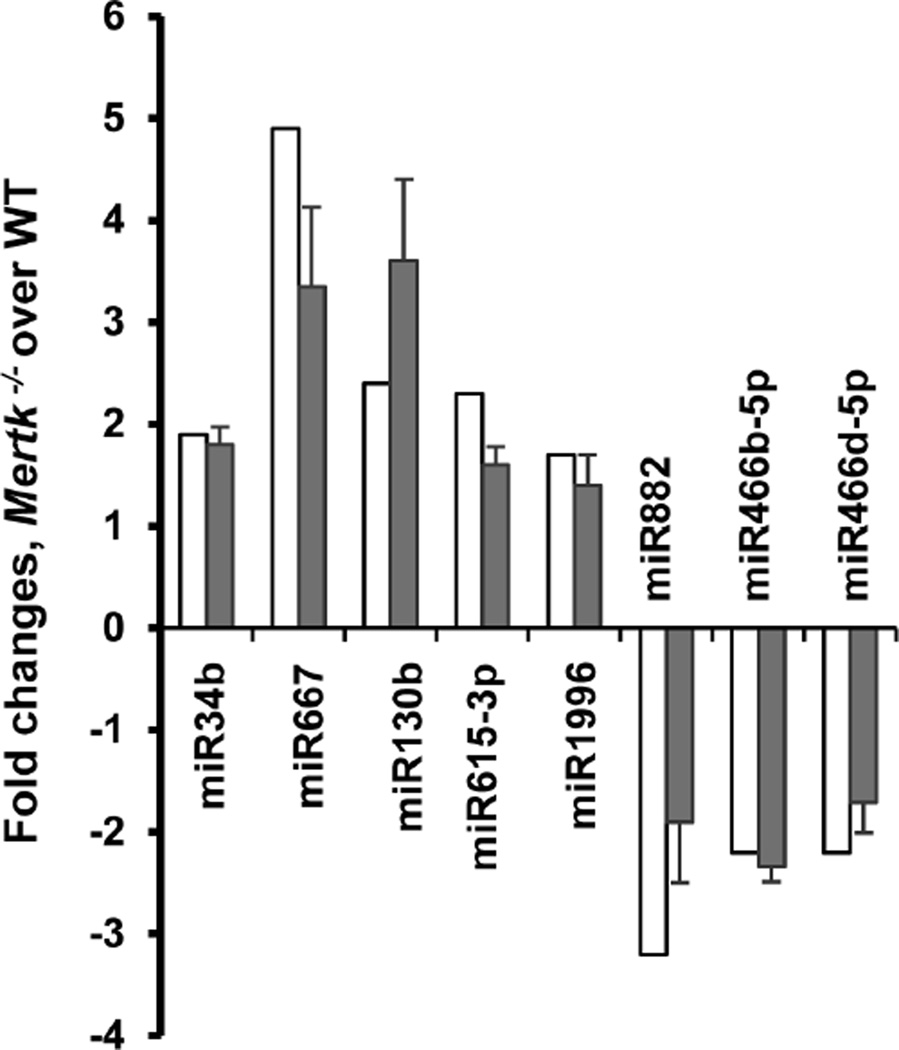
Of these total 58 miRNAs differentially expressed according to the microarray data through comparison between the Mertk−/− and WT cells or between the morning and evening groups of the Mertk−/− cells (shown in figure 2), 13 were selected for validation of the changes by real time quantitative PCR, based on their potential targets that may involve in phagocytosis. Eight of the 13 selected miRNAs showed concordance between microarray and real-time PCR results (Fig. 3). Five of those (miR-485, miR-290-3p, miR-466d-3p, miR-467b, and miR-669h-3p) showed discordance between microarray and real-time PCR results. All five of those were increased in the Mertk−/− RPE according to microarray, but showed either no significant change (miR-485) or a decrease by real-time PCR. The discordance of these data may be explained by different sensitivity of the methods used or by the low abundance of miRNA expression in the cells.
Figure 3. Comparison of miRNA expression profile in Mertk−/− RPE cells by microarray (open bars) and real-time PCR (solid bars) methods.
Hybridization signal and qPCR level was normalized to small non-coding RNA U6b and expressed as fold changes of miRNA level in the Mertk−/− RPE compared with controls. Mean of triplicate experiments ±SD is shown. The qPCR data represent one of three independent experiments.
Real time qPCR examination of the selected target genes
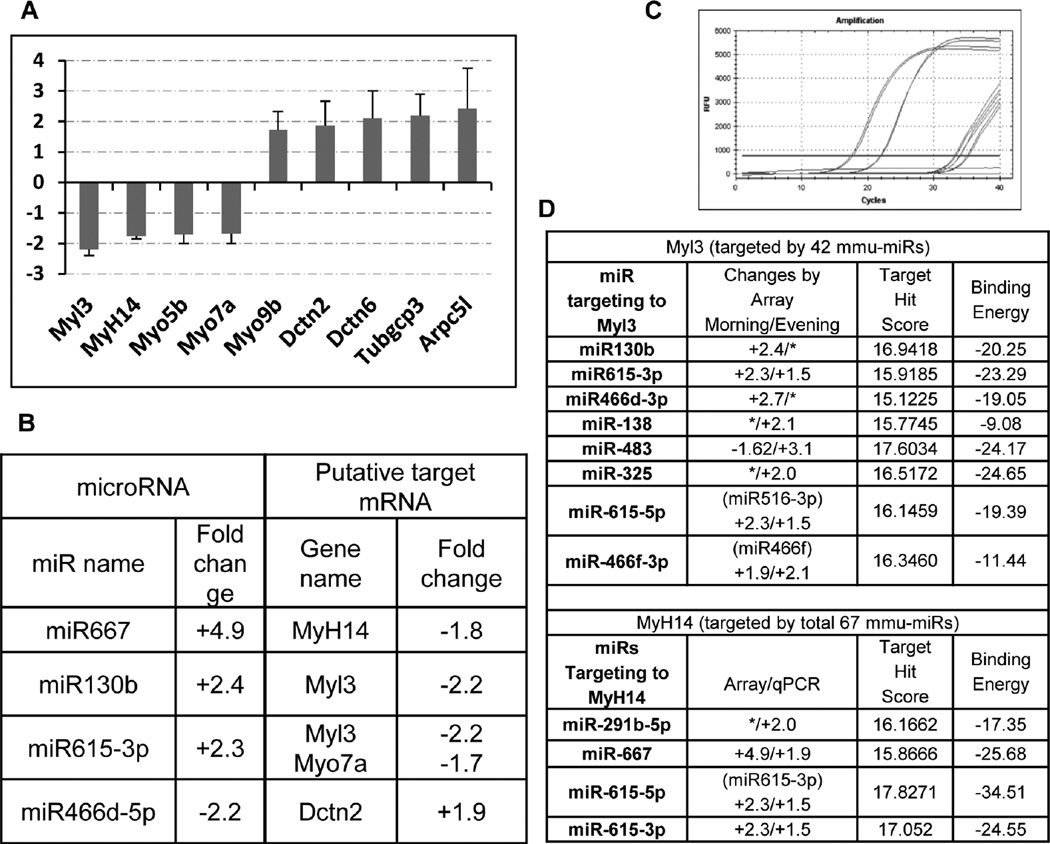
Different algorithms have been developed to identify putative target genes. To identify the potential microRNA targets that are involved in the phagocytic process, we first searched internet-based miRNA target prediction databases including the MicroCosm, miRBase, TargetScan; we then selected those that may participate in cytoskeletal regulation or function. Of the 16 genes studied, nine showed changes of ≥1.5-fold by real time qPCR (Fig. 4A, 4C) with 4 down- and 5 up-regulated. The change tendency of some target mRNAs was inversely correlated with the change trend of the miRNAs as detected by both microarray and real-time PCR (Fig. 4B). Remarkably, all four of the down-regulated target genes are different non-conventional myosin subunits. Among the 21 upregulated miRNAs as shown in figure 2 and table 1, several members, identified through search of the EMBL-EBI MicroCosm database (http://www.ebi.ac.uk/), might collectively contribute to the downregulation of their common putative target genes, especially for the Myh14 and Myl3 (Fig. 4D). Interestingly, consistent with the decreased Myo7a expression pattern we observed from several experiments, real-time PCR quantification of the Myo7a mRNA in the Mertk−/− RPE cells also showed a 1.5-fold decrease. The Myo7a mutation has been shown to cause the Usher syndrome in humans [30], a disease with congenital hearing loss accompanied with a typical presentation of retinitis pigmentosa.
Figure 4. Changes of the putative target genes are correlative with the regulation of miRNAs.
(A) The expression level of selected putative target genes was measured by real-time PCR and normalized to S18 RNA internal control and plotted as fold changes in the Mertk−/− vs. WT RPE cells prepared at 2 hr after the onset of light. Data are expressed as mean of triplicate samples ±SD. Only changes ≥ ±1.5-fold are shown. (B) Shows correlative changes of putative target mRNA with the regulatory miRNAs, § represents data obtained from the morning group. (C) Shows the amplification plot of Myl3 qPCR amplification. Based on the microarray data as shown in Table 1 or Figures 3 and 4 (D) lists all the up-regulated miRNAs (or their precursors) that are predicted to bind to either Mhyl3 or Myh14, along with the calculated target hit score and binding energy. [*] represent fold changes by <± 1.5.
Discussion
Although miRNAs are known to play important roles in a variety of biological processes, little is known about their expression levels or functions in RPE cells. In this study, we uncovered a significant difference in the miRNA expression in both Mertk−/− and WT RPE cells. Mertk regulates RPE phagocytic function, which is essential for timely removal of the shedding photoreceptor OS [31]. Both RPE phagocytic activity and photoreceptor OS shedding are tightly regulated within a diurnal lighting cycle. The phagocytic uptake of OS in mice exhibits as a robust, light-driven event with peak at 2 hr after lighting [14, 15]. The circadian expression of the genes that are important for RPE phagocytosis might be modulated by the daily oscillation of the miRNA system. The Mertk mediated phagocytosis reaches to a peak in the early morning and gradually diminishes in the afternoon, and such inhibition activity might be guided at least in part through a mechanism of gene regulation by miRNAs. When comparing the morning and evening groups, we found that 6 miRNAs were differentially expressed in the WT cells, while more than 32 were altered in the Mertk−/− RPE cells, implying that Mertk mutation caused more dramatic change in miRNA expression. We therefore focused on the miRNAs that were differentially expressed in the Mertk−/− RPE by performing further validation of the 13 miRNAs with real-time qPCR. Eight of the 13 showed correlated changes by both microarray and qPCR.
In searching for the putative miRNA target gene, the upregulated miR617 was putative to bind myosin-VIIa (Myo7a) mRNA. The decreased Myo7a mRNA detected by qPCR is inversely correlated with the increased miR617 expression founded in the Mertk mutant cells. Myosin VIIa is required for melanosome and lysosome movement in RPE cells, its mutation in human causes Usher syndrome type 1B [30], and the patients with this disease develop retinal degeneration and hearing loss as teenagers [32]. Studies on shaker-1 mice deficient in myosin-VIIa suggested that the transport of the ingested OS out of the apical region of the RPE cell was inhibited [33]. In addition, three other non-conventional myosin components, e.g., myosin-Va (Myo5a), myosin heavy chains 14 (MyH14) and myosin regulatory light chain 3 (Myl3), were also detected at lower levels; nevertheless, the myosin-IXa (Myo9a) was found to be increased, a phenomenon that might or might not be regulated by those miRNAs or might be compensatively up-regulated due to the loss of other myosin chains. Interestingly, a few of the upregulated mRNAs identified in this study can act as putative binding partners for those down-regulated subunits. Consistent with upregulation of the candidate miRNAs (including miR667 and miR130b in the Mertk−/− RPE cells), their common putative targets, MyH14 and Myl3, were down-regulated.
Most of genes, including MyH14 and Myl3, are potentially regulated by a group of miRNAs. Remarkably, among the 21 up-regulated miRNAs shown in table 1, many are theoretically able to bind and regulate one or both of the MyH14 and Myl3 genes. The importance of non-conventional myosins in the RPE phagocytosis has been recently demonstrated by selective inhibition of the myosin-II heavy chains with specific inhibitor or siRNA [34]. Myosin II consists of three isoforms of the non-muscle myosin heavy chains (NMHC), II-A, II-B and II-C, which are the products of three different genes, namely MyH9, MyH10 and MyH14 in human, respectively [35]. NMHC-IIA and –IIB isoforms were shown to participate in the Mertk-mediated RPE phagocytosis [34]. Upon binding of the photoreceptor OS to the RPE cells, both NMHC-IIA and –IIB redistribute to the phagosome formation site in the RPE cells. Furthermore, the Mertk is a prerequisite for such movement [34]. This study has identified a simultaneous down-regulation of multiple myosin subunits through up-regulation of their theoretical miRNA regulators in the Mertk−/− RPE cells, which might contribute to impaired phagocytosis by the mutant RPE cells. However, whether or not that those candidate genes are bona fide targets for the differentially regulated miRNA candidates which are potentially regulated in circadian pattern by Mertk and in turn affect RPE cell phagocytosis, are needed to be further validated. In the near future, we can continue identifying the authentic target genes for these differentially expressed miRNAs using commercial available miRNA 3’-UTR target clone libraries; and further verify their phagocytic regulatory functions by inhibition of candidate miRNA using specific inhibitors or by overexpression of these candidate miRNAs in the phagocytosing RPE cells.
Acknowledgement
This paper was supported by NIH grants R01EY018830 and Research to Prevent Blindness.
REFERENCES
- 1.Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaVail MM. Kinetics of rod outer segment renewal in the developing mouse retina. J Cell Biol. 1973;58:650–661. doi: 10.1083/jcb.58.3.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young RW, Bok D. Participation of the retinal pigment epithelium in the rod outer segment renewal process. J Cell Biol. 1969;42:392–403. doi: 10.1083/jcb.42.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards RB, Szamier RB. Defective phagocytosis of isolated rod outer segments by RCS rat retinal pigment epithelium in culture. Science. 1977;197:1001–1003. doi: 10.1126/science.560718. [DOI] [PubMed] [Google Scholar]
- 5.Bok D, Hall MO. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J Cell Biol. 1971;49:664–682. doi: 10.1083/jcb.49.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, et al. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;9:645–651. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- 7.Nandrot E, Dufour EM, Provost AC, Pequignot MO, Bonnel S, et al. Homozygous deletion in the coding sequence of the c-mer gene in RCS rats unravels general mechanisms of physiological cell adhesion and apoptosis. Neurobiol Dis. 2000;7:586–599. doi: 10.1006/nbdi.2000.0328. [DOI] [PubMed] [Google Scholar]
- 8.Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398:723–728. doi: 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- 9.Duncan JL, LaVail MM, Yasumura D, Matthes MT, Yang H, et al. An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest Ophthalmol Vis Sci. 2003;44:826–838. doi: 10.1167/iovs.02-0438. [DOI] [PubMed] [Google Scholar]
- 10.Tschernutter M, Jenkins SA, Waseem NH, Saihan Z, Holder GE, et al. Clinical characterisation of a family with retinal dystrophy caused by mutation in the Mertk gene. Br J Ophthalmol. 2006;90:718–723. doi: 10.1136/bjo.2005.084897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHenry CL, Liu Y, Feng W, Nair AR, Feathers KL, et al. MERTK arginine-844-cysteine in a patient with severe rod-cone dystrophy: loss of mutant protein function in transfected cells. Invest Ophthalmol Vis Sci. 2004;45:1456–1463. doi: 10.1167/iovs.03-0909. [DOI] [PubMed] [Google Scholar]
- 12.Gal A, Li Y, Thompson DA, Weir J, Orth U, et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26:270–271. doi: 10.1038/81555. [DOI] [PubMed] [Google Scholar]
- 13.Thompson DA, McHenry CL, Li Y, Richards JE, Othman MI, et al. Retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. Am J Hum Genet. 2002;70:224–229. doi: 10.1086/338455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaVail MM. Rod outer segment disk shedding in rat retina: relationship to cyclic lighting. Science. 1976;194:1071–1074. doi: 10.1126/science.982063. [DOI] [PubMed] [Google Scholar]
- 15.LaVail MM. Circadian nature of rod outer segment disc shedding in the rat. Invest Ophthalmol Vis Sci. 1980;19:407–411. [PubMed] [Google Scholar]
- 16.Nandrot EF, Anand M, Sircar M, Finnemann SC. Novel role for alphavbeta5-integrin in retinal adhesion and its diurnal peak. Am J Physiol Cell Physiol. 2006;290:C1256–C1262. doi: 10.1152/ajpcell.00480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, et al. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J Exp Med. 2004;200:1539–1545. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robosky LC, Wade K, Woolson D, Baker JD, Manning ML, et al. Quantitative evaluation of sebum lipid components with nuclear magnetic resonance. J Lipid Res. 2008;49:686–692. doi: 10.1194/jlr.D700035-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Lu H, Lu Q, Gaddipati S, Kasetti RB, Wang W, et al. IKK2 inhibition attenuates laser-induced choroidal neovascularization. PLoS One. 2014;9:e87530. doi: 10.1371/journal.pone.0087530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 22.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 23.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 24.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 25.Huang KM, Dentchev T, Stambolian D. MiRNA expression in the eye. Mamm Genome. 2008;19:510–516. doi: 10.1007/s00335-008-9127-8. [DOI] [PubMed] [Google Scholar]
- 26.Pegoraro M, Tauber E. The role of microRNAs (miRNA) in circadian rhythmicity. J Genet. 2008;87:505–511. doi: 10.1007/s12041-008-0073-8. [DOI] [PubMed] [Google Scholar]
- 27.Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, et al. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin Y, Lu Q, Li Q. 14-3-3sigma controls corneal epithelial cell proliferation and differentiation through the Notch signaling pathway. Biochem Biophys Res Commun. 392:593–598. doi: 10.1016/j.bbrc.2010.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandrot EF, Finnemann SC. Altered rhythm of photoreceptor outer segment phagocytosis in beta5 integrin knockout mice. Adv Exp Med Biol. 2006;572:119–123. doi: 10.1007/0-387-32442-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 31.Prasad D, Rothlin CV, Burrola P, Burstyn-Cohen T, Lu Q, et al. TAM receptor function in the retinal pigment epithelium. Mol Cell Neurosci. 2006;33:96–108. doi: 10.1016/j.mcn.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Smith RJ, Berlin CI, Hejtmancik JF, Keats BJ, Kimberling WJ, et al. Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet. 1994;50:32–38. doi: 10.1002/ajmg.1320500107. [DOI] [PubMed] [Google Scholar]
- 33.Gibbs D, Kitamoto J, Williams DS. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc Natl Acad Sci U S A. 2003;100:6481–6486. doi: 10.1073/pnas.1130432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strick DJ, Feng W, Vollrath D. Mertk drives myosin II redistribution during retinal pigment epithelial phagocytosis. Invest Ophthalmol Vis Sci. 2009;50:2427–2435. doi: 10.1167/iovs.08-3058. [DOI] [PubMed] [Google Scholar]
- 35.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]