Abstract
Culex pipiens mosquitoes are important disease vectors inhabiting temperate zones, worldwide. The seasonal reduction in temperature and photoperiod accompanying late summer and early fall prompts female mosquitoes to enter diapause, a stage of developmental arrest and physiological conditioning that enhances survival during the winter months. To investigate the molecular mechanisms underlying diapause induction, we used custom whole-transcriptome microarrays to identify differences in gene expression following exposure to nondiapause (long days, 25°C) and diapause-inducing (short days, 18°C) environmental conditions. Using a two-way ANOVA we identified 1130 genes that were differentially expressed. We used the expression of these genes across three time points to construct a gene co-expression network comprising five modules. Genes in modules 1, 2 and 3 were largely up-regulated, while genes in modules 4 and 5 were down-regulated when compared to nondiapause conditions. Pathway enrichment analysis of the network modules revealed some potential regulatory mechanisms driving diapause induction. Module 1 was enriched for genes in the TGF-β and Wnt signaling pathways; module 2 was enriched for genes involved in insect hormone biosynthesis, specifically, ecdysone synthesis; module 3 was enriched for genes involved in chromatin modification, and module 5 was enriched for genes in the circadian rhythm pathway. Our results suggest that TGF-β signaling and chromatin modification are key drivers for the integration of environmental signals into the diapause induction phase in C. pipiens mosquitoes.
Keywords: house mosquito, gene network, overwintering, seasonality, gene regulation
Introduction
Culex pipiens (L), the northern house mosquito, is a vector of several important human diseases including West Nile virus (Fonseca et al. 2004) and lymphatic filariasis in Egypt (Harb et al. 1993; Abdel-Hamid et al. 2011). They have a broad geographic distribution, inhabiting temperate zones on six continents (Mattingly 1951; Vinogradova 2000). The seasonal changes in day length and temperature accompanying late summer and early fall program C. pipiens females for diapause. Mosquitoes are programmed by the shorter days and lower temperatures only during immature stages, specifically, the late 4th instar and early pupal stages (Spielman and Wong 1972). In C. pipiens, diapause manifests as developmental, behavioral and physiological changes that enhance survival during the unfavorable conditions associated with overwintering in temperate climates. After emergence and before the onset of winter, diapause-destined females mate, accumulate energy reserves and seek shelter. In addition, dehydration resistance and cold hardiness are enhanced (Rhinehart et al. 2006; Benoit and Denlinger 2007), while ovary development is arrested (Eldridge 1968; Spielman and Wong 1973).
One distinct behavioral change associated with diapause is the propensity for host seeking. Non-diapausing females seek a blood meal shortly after emerging, while diapausing females seek only carbohydrate rich diets (Mitchel 1983). This shift from host seeking behavior to sugar gluttony is accompanied by an increase in the expression of genes associated with lipid store accumulation, and a reduction in the expression of genes associated with blood meal digestion (Robich and Denlinger 2005). Consequently, diapausing females enticed to blood feed are unable to effectively digest the blood and will often expel the undigested blood meal (Mitchel 1983).
Several mechanisms regulating the diapause program in C. pipiens have been identified. The shutdown of juvenile hormone (JH) by the corpora allata is a key endocrine regulator in C. pipiens diapause (Spielman 1974), as well as other cases of adult diapause in insects (Denlinger 2002). However, recent studies have revealed a likely role for insulin signaling as an upstream regulator of JH. This was first demonstrated by Sim and Denlinger (2008) when RNAi-mediated knock-down of the insulin receptor (InR) in mosquitoes reared in non-diapause conditions resulted in arrest of ovarian follicle development at stage and size resembling diapause. This effect was reversed by the application of JH. They were also able to show that a shutdown in insulin signaling caused the activation of FOXO (forkhead transcription factor), while the knock-down of FOXO resulted in the reduction of fat stores and a reduced life span in mosquitoes programmed for diapause (Sim and Denlinger 2008). In a follow up study by the same group, the shutdown of insulin-like peptide-1 (ILP-1) was shown to be critical for diapause (Sim and Denlinger 2009).
Although several key components of the diapause program have been elucidated, the regulatory networks leading to these complex changes have yet to be fully described. To help fill some of these gaps, we investigated the molecular events driving diapause induction by measuring complete transcriptome expression at three time points during the early pupal stage in nondiapause (25°C; 16 h light/8 h dark) and diapause-inducing conditions (18°C; 8 h light/16 h dark). We then used a gene co-expression network approach to help identify regulatory networks driving diapause induction.
Materials and methods
Sample collections
The laboratory strain used in this study was established from >4000 larvae collected from a single container in South Bend, IN, USA, during the summer of 2009. Adults were fed a 5% sucrose solution ad libitum, and maintained in 60 × 60 × 60 cm screen cages at 21–23°C with a natural photoperiod for Notre Dame, IN, USA. Mosquitoes were blood fed on anesthetized rats, and egg rafts were laid and allowed to hatch en masse. On the day of hatching, 100 1st instar larvae were placed in 32.5 × 17.6 × 6.5 cm plastic trays (Cambro) containing 1 L of reverse osmosis water. They were fed a slurry of beef liver powder ad libitum and placed in an environmental chamber (Sanyo) at nondiapause conditions (long days, 25°C). In the morning on the day when the majority of females pupated, all pupae were cleared from the trays at 9 AM. Eight hours later female pupae were collected and placed in eight plastic cups containing ~200 ml of deionized water, each with ~20 pupae. Four cups were placed back into the chamber at nondiapause conditions, and four cups were placed in a chamber at diapause-inducing conditions (18°C; 8 h light/16 h dark). The time the lights turned on/off in nondiapause and diapause-inducing conditions was 5 AM/9 PM and 9 AM/5 PM, respectively. Eight, 16 and 24 hours later one cup from each environmental treatment was collected and the pupae stored in RNALater (Ambion). The fourth cup was left in the respective treatment to evaluate the diapause phenotype; at ten days post-emergence, females were collected and stored at −20°C until they could be dissected and the primary ovarian follicles measured (proxy for diapause). Five follicles from each female (~20 females per biological replicate) were measured using an optical micrometer. Total RNA was extracted from pools of 10 pupae using the Qiagen Qiashredder and RNeasy Extraction Kits (Qiagen). Four biological replicates were used in the analysis.
Microarray analysis
Probe sets were designed using the CpipJ1.2 annotation in Vectorbase (Lawson et al. 2009; Arensburger et al. 2010). The probe set represented 18,692 protein coding genes, and each gene was represented by three 60-mer probes per gene and arrayed in duplicate on NimbleGen 12-plex chips (Roche NimbleGen Expr12x135K). The details of the array design, sample description and expression data are available at Gene Expression Omnibus (GEO) under accession number GSE56722. The quality of each RNA sample was assessed using the Bioanalyzer 2100 (Agilent Technologies) and RNA 6000 Nano kit (Agilent Technologies). RNA and cDNA concentrations were measured using the Nanodrop ND-2000 (ThermoScientific), and cDNA libraries were constructed with 10.0 µg total RNA using the SuperScript double-stranded cDNA Synthesis Kit (Invitrogen). Samples were labeled using the NimbleGen One Color labelling kit (Roche NimbleGen). Hybridization and post-hybridization washes were conducted using the NimbleGen Hybridization and Wash Buffer Kits (Roche NimbleGen). Images were acquired using a NimbleGen MS200 Microarray Scanner, at 2 µm resolution.
Microarray validation
Quantitative RT-PCR (qRT-PCR) was used to measure relative expression of five genes in three biological replicates across two time points (16 and 24 hours after transfer to diapause inducing conditions) to evaluate the microarray results. We chose genes in three different network modules so both up-regulated and down-regulated genes were included in the validation. Primers were designed using the Primer3 plus program (Untergasser et al. 2007). Genes and their corresponding primer sequences are listed in Table S1. Total RNA was treated with RQ1 DNase (Promega), and first strand cDNA synthesis was performed using gene specific primers and SuperScript II (Invitrogen) using 200 ng RNA in 10 µl reactions with a final primer concentration of 0.4 µM. Real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the 7500 Fast Real-Time PCR System (Applied Biosystems). A final primer concentration for both the forward and reverse primers was 0.2 µM in a 25 µl reaction. The thermal cycle was the same for each primer set: hold for 2 min. at 50°C, an initial denature at 95°C for 10 min., 40 cycles of 95°C for 15 sec. with annealing at 60.0°C for 1 min., followed by melt curve analysis. Expression values were normalized against ribosomal protein S17 (RpS17) as the reference control (Morlais and Severson 2001). The R statistical environment was used to perform a Spearman’s rank test to measure correlation between microarray and qRT-PCR log2-transformed fold changes between nondiapause and diapause-inducing environmental conditions.
Statistical analysis
NimbleGen array image data were processed using NimbleScan (version 2.5) to extract intensity value for each gene. Raw gene expression data were log2-transformed and normalized using quintile normalization method (Bolstad et al. 2003). A two-way ANOVA was used to identify differentially expressed genes based on time-point, environmental treatment, and their interaction. A false discovery rate (0.05) control was used to correct for multiple comparisons (Benjamini and Hochberg 1995).
Network construction
Network analysis was performed using R package WGCNA (Langfelder and Horvath, 2008) on the expression profiles of the 1130 differentially expressed (DE) genes in diapause-inducing conditions. We applied WGCNA in this study to analyze co-expressed gene networks and to detect co-expressed gene modules, which are defined as group of highly correlated genes using a topological overlap measure (Ravasz et al. 2002). The network and module structures were visualized using Cytoscape (Shannon et al. 2003; Smoot et al. 2011). The Genesis program was used to generate heat maps and expression plots for the network modules (Sturn et al. 2002).
Enrichment analysis
GO term and pathway enrichment analysis was performed using Drosophila melanogaster orthologs predicted by the OrthoDB database (Waterhouse et al. 2013). The web-based gene set analysis toolkit WebGestalt was used to measure GO term enrichment in network modules against a background of genes used in the analysis (Zhang et al. 2005; Wang et al. 2013). Pathway genes and transcription factors were mapped using KEGG annotations (Kanehisa and Goto 2000; Kanehisa et al. 2014). P-values for pathway and GO term enrichment were calculated using a Fisher’s exact test.
Results
Sample collections
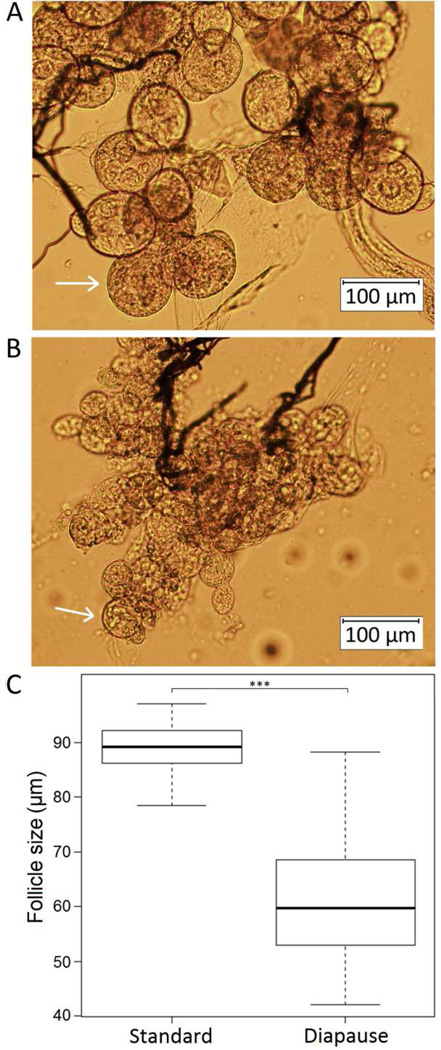
The mean primary follicle size (range) in mosquitoes reared in nondiapause and diapause-inducing conditions was 89.1 (78.4–97.0 µm) (Figure 1A) and 61.0 µm (44.0–88.2 µm) (Figure 1B). The range of follicle sizes in mosquitoes reared in nondiapause conditions was within that expected in non-diapausing mosquitoes, while the size range in mosquitoes reared in diapause-inducing conditions indicated that most females entered diapause (Figure 1C).
Figure 1.
Images of follicles 10 days post-emergence in mosquitoes reared in nondiapause (A) and diapause-inducing (B) conditions. Box plot (C) showing follicle sizes of the combined replicates 10 days post-emergence in nondiapause and diapause-inducing conditions (*** = p-value < 0.001).
Microarray analysis and Network construction
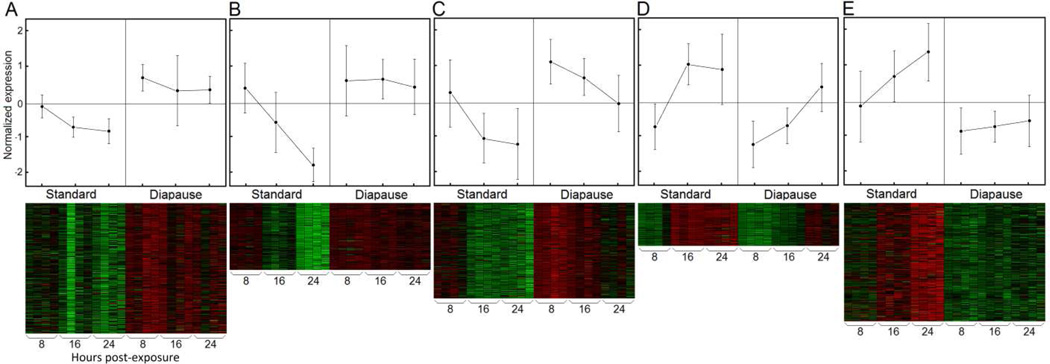
The total number of genes used in the statistical analysis after quality filtering was 16,313. There were 1,130 differentially expressed (DE) genes based on ANOVA following FDR corrections. Fold change between nondiapause-inducing and diapause-inducing conditions of five genes measured using qRT-PCR were highly correlated (rho = 0.818; p = 0.0068) with fold change based on the microarray results (Figure 1S). Expression of the 1,130 DE genes across three time-points (8, 16, and 24 hours) following initial exposure to diapause-inducing conditions was used to construct a gene co-expression network. Gene network analysis revealed two primary gene clusters: one cluster comprised of three modules (1–3) that were up-regulated, and one cluster comprised of two modules (4 and 5) that were down-regulated in diapause-inducing conditions (Figure 2). The number of genes in modules 1–5 was 325, 167, 237, 106, and 295, respectively.
Figure 2.
Gene network based on correlations between 1,130 differentially expressed genes across three time points in the early pupal stage: module 1 (red); module 2(blue); module 3 (green); module 4 (yellow); module 5 (white).
Enrichment analysis and transcription factor mapping
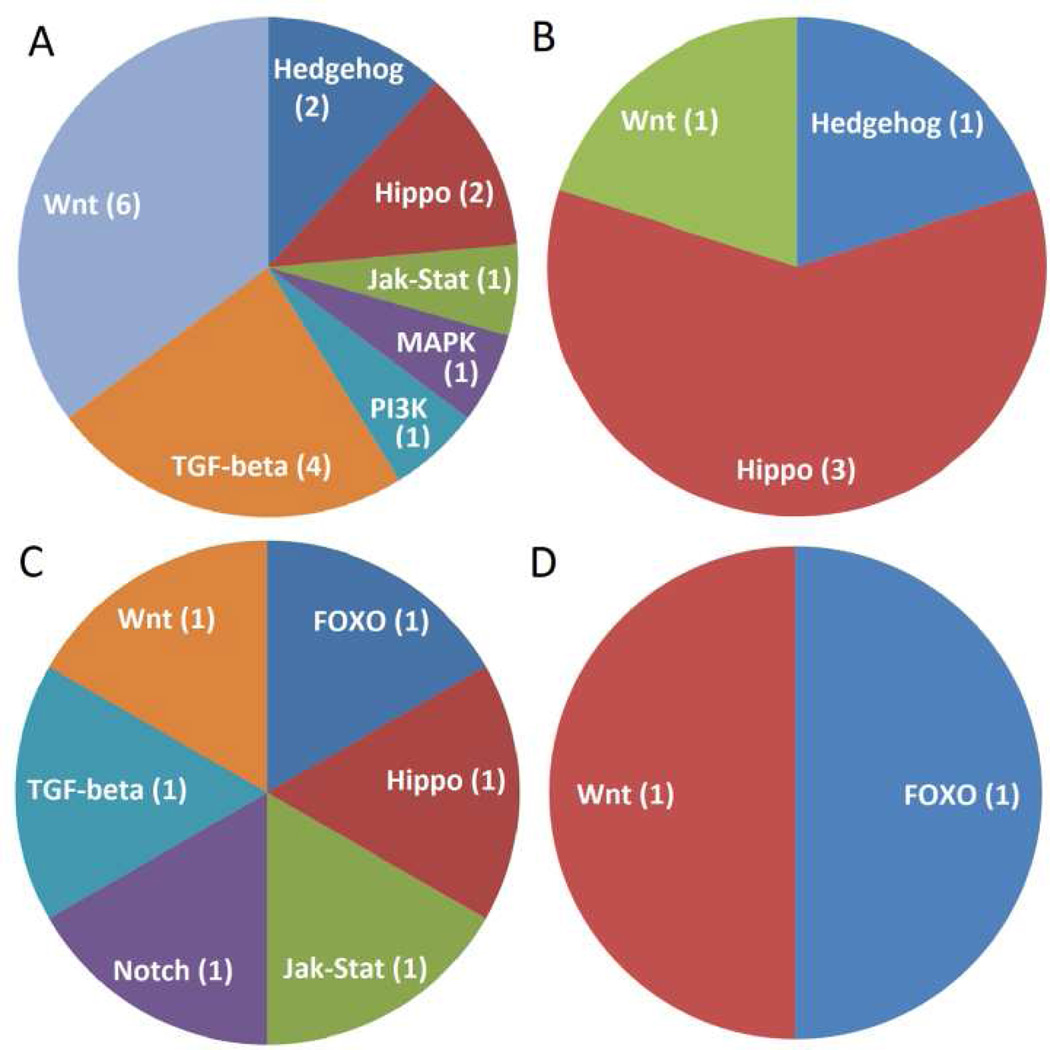
Module 1 contains genes that were up-regulated in pupae following transfer to diapause-inducing conditions (Figure 3). GO term analysis revealed enrichment of genes associated with development, chromatin remodeling, calcium ion binding, and DNA-dependent ATPase activity (Table 1). Module 1 was also enriched for genes in the TGF-β and Wnt signaling pathways, and genes involved in spliceosome function (Table 2). The percentage of transcription factors (Table 3) and pathway genes (Figure 4) mapping to module 1 (8.0%) was greater than any other module (0–5.1%). Module 2 was also up-regulated under diapause-inducing conditions (Figure 3), and was enriched for genes involved in morphogenesis, epidermal cell differentiation, and serine-type endopeptidase activity (Table 1). Genes in the insect hormone biosynthesis, tryptophan metabolism, and phagosome degradation pathways were also enriched (Table 2). The two genes mapping to the insect hormone biosynthesis pathway were spook (CPIJ006322) and disembodied (CPIJ010826), genes involved in ecdysone synthesis. Module 3, another cluster of genes up-regulated under diapause-inducing conditions, was enriched for genes associated with mRNA splicing, gamete generation, snRNA binding, and the Sin3 and NuRD complexes (Table 1). Genes involved in the spliceosome pathway were enriched (Table 2), while six transcription factors (Table 3) and six signal transduction pathway genes (Figure 4) mapped to module 3.
Figure 3.
Expression profiles and heat maps for network modules 1–5, A–E, respectively.
Table 1.
The top five enriched GO terms for each of the five network modules.
| GO Categorya |
GO terms | ratiob | p-value |
|---|---|---|---|
| module 1 | |||
| BP | organ morphogenesis | 32/481 | 8.8E-5 |
| BP | sensory organ development | 23/310 | 2.0E-4 |
| CC | chromatin remodeling complex | 8/68 | 2.2E-3 |
| MF | calcium ion binding | 10/129 | 6.6E-3 |
| MF | DNA-dependent ATPase activity | 4/26 | 7.9E-3 |
| module 2 | |||
| BP | instar larval or pupal morphogenesis | 17/343 | 2.0E-6 |
| BP | epidermal cell differentiation | 6/38 | 9.9E-6 |
| CC | integral to plasma membrane | 9/131 | 2.3E-5 |
| MF | fatty-acyl-CoA reductase (alcohol forming) activity | 2/4 | 1.1E-3 |
| MF | serine-type endopeptidase activity | 5/61 | 1.5E-3 |
| module 3 | |||
| BP | mRNA splicing via spliceosome | 13/169 | 9.9E-6 |
| BP | gamete generation | 19/433 | 2.0E-4 |
| CC | Sin3 complex | 2/2 | 4.0E-4 |
| MF | snRNA binding | 2/5 | 3.6E-3 |
| CC | NuRD complex | 2/5 | 3.6E-3 |
| module 4 | |||
| CC | structural constituent of cuticle | 11/51 | 2.8E-15 |
| MF | retinal binding | 2/3 | 1.0E-4 |
| BP | chitin metabolic process | 3/41 | 2.0E-4 |
| BP | chitin-based cuticle development | 3/49 | 6.0E-4 |
| MF | integral to plasma membrane | 4/131 | 1.8E-3 |
| module 5 | |||
| BP | organonitrogen compound biosynthetic process | 11/187 | 2.0E-4 |
| BP | response to alkaloid | 3/11 | 6.0E-4 |
| MF | alpha-amino acid metabolic process | 5/62 | 2.9E-3 |
| MF | fatty acid biosynthetic process | 3/19 | 3.2E-3 |
| CC | vacuolar proton-transporting V-type ATPase complex | 2/14 | 1.8E-2 |
BP = biological process, MF = molecular function, CC = cellular component
Ratio = the number of module genes in category/the total number of genes in category
Table 2.
Enriched pathways for each module based on KEGG annotations.
| Pathway | # Genes (# Drosophila orthologs) | Ratioa | p-value |
|---|---|---|---|
| Module 1 | 325 (257) | ||
| TGF-β signaling pathway | 4/26 | 0.003 | |
| Wnt signaling pathway | 6/64 | 0.0014 | |
| Spliceosome | 7/106 | 0.03 | |
| Module 2 | 167 (121) | ||
| Insect hormone biosynthesis | 2/13 | 0.002 | |
| Tryptophan metabolism | 2/19 | 0.003 | |
| Phagosome degradation | 3/50 | 0.005 | |
| Module 3 | 237 (180) | ||
| Spliceosome | 9/106 | 0.001 | |
| Module 5 | 295 (200) | ||
| Circadian rhythm | 4/9 | 0.001 | |
| Beta-alanine metabolism | 2/17 | 0.030 | |
Ratio = the number of module genes in category/the total number of genes in category
Table 3.
Transcription factors mapped to the network modules. Gene names are based on D. melanogaster orthologs.
| Gene ID | Gene name | Interpro classification |
|---|---|---|
| Module 1 | ||
| CPIJ006384 | Zn finger homeodomain 2 | Homeobox, zinc finger C2H2 |
| CPIJ008660 | pointed | Ets, pointed, winged helix-turn-helix |
| CPIJ018751 | forkhead domain 96Ca | Forkhead, winged helix-turn-helix |
| CPIJ002027 | CG1965 | Gc-rich sequence DNA-binding factor |
| CPIJ012632 | tailup | Homeobox, zinc finger LIM-type |
| CPIJ005379 | MTA1-like | Zinc finger GATA-type, ELM2, SANT |
| CPIJ019088 | NFAT | Ipt, NFAT, p53-like, rel-homology |
| CPIJ007784 | optomotor-blind-related-gene-1 | T-box, p53-like |
| CPIJ013984 | TfAP-2 | Transcription factor AP-2, C-terminal |
| CPIJ017673 | TfIIFβ | Winged helix-turn-helix |
| CPIJ013972 | trithorax | Zinc finger nuclear hormone receptor-type |
| CPIJ018355 | dSmad2 | SMAD Dwarfin-like |
| Module 2 | ||
| CPIJ017141 | sister of odd and bowl | Zinc finger C2H2 |
| CPIJ004513 | grainy head | CP2 transcription factor |
| CPIJ009088 | E5 | Homeobox |
| Module 3 | ||
| CPIJ003409 | enhancer of split mβ | Orange, Myc-type basic helix-loop-helix |
| CPIJ016941 | vrille | Basic-leucine zipper |
| CPIJ001711 | jumeau | Forkhead, winged helix-turn-helix |
| CPIJ002815 | eyegone | Homeobox, paired, winged helix-turn-helix |
| CPIJ006382 | shaven | Paired, homeo-like, winged helix-turn-helix |
| CPIJ018505 | ovo | Zinc finger C2H2 |
| Module 5 | ||
| CPIJ002146 | clock | PAS, nuclear translocator, PAC motif |
| CPIJ002147 | clock | PAS, nuclear translocator, PAC motif |
| CPIJ014945 | ftz transcription factor 1 | Nuclear hormone receptor ligand binding core |
| CPIJ003767 | kayak | Fos transforming, basic-leucine zipper |
| CPIJ006398 | twin of eyeless | Homeobox, paired, winged helix-turn-helix |
| CPIJ003297 | Max | Myc-type basic helix-loop-helix |
Figure 4.
Network module genes mapping to signal transduction pathways: module 1 (A), module 2 (B), module 3 (C), and module 5 (D). Module 4 did not have any signal transduction pathway genes.
Modules 4 and 5 represented genes that were down-regulated under diapause-inducing conditions when compared to nondiapause conditions. Module 4 was the smallest of the modules with 106 genes, and was enriched for genes associated with the structural constituent of cuticle, retinal binding, chitin metabolism, and genes integral to the plasma membrane (Table 1). No significant enrichment of pathway genes was detected, and no transcription factors mapped to module 4. Expression of genes in module 5 in diapause-inducing conditions was down-regulated early in the time series and increased only marginally compared to nondiapause conditions where expression increased considerably across the time-points (Figure 3). Module 5 was enriched for genes associated with organonitrogen biosynthesis, response to alkaloid, alpha-amino acid metabolism, fatty acid biosynthesis, vacuolar proton-transporting, and the V-type ATPase complex (Table 1). Module 5 was also enriched for genes in the circadian rhythm and beta-alanine metabolism pathways (Table 2), while six transcription factors (Table 3) were mapped to this module. Four circadian rhythm pathway genes in module 5 (clock, CPIJ002146; clock, CPIJ002147; period, CPIJ007193; timeless, CPIJ007082) was reduced and the upward trend over the three time points was dampened considerably when compared to expression in nondiapause conditions (Figure 3).
Discussion
Gene expression analysis of diapausing C. pipiens was previously studied using suppressive subtractive hybridization; however, their focus was on diapause-specific expression in adult females from 7–10 days post-emergence to 56–59 days post-emergence—early to late stage diapause (Robich et al. 2007), periods well after the induction phase. In light of the work by Spielman and Wong (1973), who identified the day of the 4th instar-pupa molt as being the end of the environmentally sensitive period for diapause induction, we reasoned that the gene expression changes observed during the early pupal stage in diapause-inducing conditions could help to identify mechanisms associated with the integration of the environmental cues into the diapause program.
Gene expression profiles for the five modules revealed that most of the differences in expression between nondiapause and diapause-inducing conditions occurred later in the time course (16 or 24 hours) (Figure 3). However, modules 1 and 3 were up-regulated at the earliest time point (8 hours) and include the majority of genes associated with signal transduction pathways (Figure 4), suggesting that genes in these modules likely code for upstream effectors that drive the molecular cascade initiating the diapause program. Module 1 was enriched for genes in the TGF-β signaling pathway, a highly conserved signaling pathway involved in Caenorhabditis elegans dauer formation, a nematode state similar to insect diapause (Inoue and Thomas 2000). Among TGF-β genes associated with dauer formation are the SMADs, intracellular proteins that function in signal transduction following TGF-β ligand binding and receptor activation (Inoue and Thomas 2000). Members of the TGF-β superfamily can be grouped into one of three branches based on the ligand-receptor combinations involved: the TGF-βs, activins, and bone morphogenetic proteins (BMPs) (Nguyen et al. 2000). Membership of the four TGF-β signaling pathway genes in module 1 (maverick, saxophone, thrombospondin, and dSmad2) could be made to both the TGF-β (dSmad2) and the BMP (saxophone) branches, while membership of maverick and thrombospondin to a specific branch remains ambiguous. These results suggest that both the BMP and the classical TGF-βs branches could be involved in the signal transduction events leading to diapause.
A number of genes involved in chromatin modification were differentially expressed following exposure to diapause-inducing conditions. Module 3 was enriched for genes comprising the SIN3 and NuRD complexes, multi-protein histone deacetylase (HDAC) complexes that facilitate changes in chromatin structure which ultimately alter gene expression. Deacetylated histones are generally associated with condensed chromatin and transcriptional repression, while acetylated histones are associated with open chromatin and activation of gene expression. The SIN3 and NuRD complexes have been shown to have an important role in development (Ahringer 2000). The MTA1-like protein, a transcription factor in module 1, is another component of the NuRD complex (Ng and Bird 2000). Module 1 also includes trithorax, a transcriptional activator that functions as an antagonist of the polycomb gene repressor group and helps maintain transcription following activation of target gene expression (Schuettengruber et al. 2011). Interestingly, trithorax has been shown to modulate longevity and the stress response in Drosophila through antagonism of polycomb silencing (Siebold et al. 2009). These data together suggest a possible role for epigenetic regulation as a component of the diapause program.
Nearly 80 years have passed since Bünning (1936) proposed that circadian clock forms the basis for the photoperiodic response driving the induction of diapause, yet a clear connection has yet to be demonstrated. Five of nine circadian rhythm pathway genes (clock, period, timeless, vrille) were differentially expressed in our data set, with four of these being in module 5, a module with expression that apparently was dampened compared to expression in nondiapause conditions (Figure 3). Because of the differences in photoperiod between the two experimental conditions and the cyclic nature of the expression of most clock genes, we cannot conclude that the expression differences are involved with diapause induction.
Previous studies have clearly demonstrated a role for insulin/FOXO (Sim and Denlinger 2008; Sim and Denlinger 2009) and JH signaling (Spielman 1974; Denlinger 2002, Kang et al. 2014) in the maintenance phase of diapause in C. pipiens. However, we found few genes directly related to these pathways that were differentially expressed in our study. ILP-5 (CPIJ001698) and cyclin B3 (CPIJ014315), a target of FOXO, were in module 3, while pepck (CPIJ010515), another FOXO target, was in module 5. Sim and Denlinger (2009) reported a considerable reduction in the expression of ILP-5 in diapausing C. pipiens; however, this was measured one week post-adult eclosion—a point well after the induction phase for C. pipiens diapause (Spielman and Wong 1973).
A study by Kang et al. (2014) suggests that a decrease in the expression of allatotropin, a regulator of JH production in the corpora allata, is an important component of the diapause program in C. pipiens. We did not find significant expression differences in this or other genes regulating the shutdown or degradation of JH (allatotropin, juvenile hormone epoxide hydrolase, juvenile hormone esterase), which was not unexpected since we did not see the changes in insulin signaling that promote the shutdown of JH (Sim and Denlinger 2013). Our results suggest that the major changes in insulin/FOXO signaling that drive the shutdown of JH do not occur during the first hours of diapause induction; therefore, we hypothesize that TGF-β signaling and chromatin modification are key drivers for the integration of environmental signals into the diapause induction phase.
The environmental conditions used in this study (nondiapause: 25°C long days; diapause-inducing: 18°C short days) were following those used by Spielman and Wong (1973) to characterize the environmentally sensitive stage for diapause induction. Two confounding factors make the interpretation of these data challenging. First is the influence of photoperiod on the expression of genes under circadian control. To help reduce the effect of photoperiod on these genes we used a centrally-anchored photoperiod paradigm rather than aligning lights off or lights on. We acknowledge, however, that we may not have completely eliminated the circadian rhythm influence on gene expression. Second, due to the different temperatures, the pupae were not developmentally synchronized. Consequently, some differences in gene expression could be due to insect development alone, and not related to the induction of diapause.
We present here the first whole transcriptome study investigating the gene expression changes during the photosensitive stage for diapause induction in C. pipiens. By measuring gene expression changes across three time points we were able to identify correlations among 1,130 DE genes and construct a gene network representing expression changes during diapause induction.
Supplementary Material
Acknowledgements
We thank Ryan Hemme for collecting the larvae used in establishment of the C. pipiens South Bend strain. We also thank Melissa Stephens in the University of Notre Dame Genomics Core Facility for processing the microarrays. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (RO1-AI079125-A1) to D.W.S.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Benoit JB, Denlinger DL. Suppression of water loss during adult diapause in the northern house mosquito, Culex pipiens. J Exp Biol. 2007;210:217–226. doi: 10.1242/jeb.02630. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bünning E. Die endogene tagesrhythmik als grundlage der photoperiodischen reaktion. Ber Dtsch Bot Ges. 1936;54:590–607. [Google Scholar]
- Denlinger DL. Regulation of diapause. Annu Rev Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- Eldridge BF. The effect of temperature and photoperiod on blood-feeding and ovarian development in mosquitoes of the Culex pipiens complex. Am J Trop Med Hyg. 1968;17:133–140. doi: 10.4269/ajtmh.1968.17.133. [DOI] [PubMed] [Google Scholar]
- Emerson KJ, Bradshaw WE, Holzapfel CM. Complications of complexity: integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 2009;25:217–225. doi: 10.1016/j.tig.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Mogi M, Fleischer RC, Wilkerson RC. Emerging vectors in the Culex pipiens complex. Science. 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]
- Harb M, Faris R, Gad AM, Hafez ON, Ramzy R, Buck AA. The resurgence of lymphatic filariasis in the Nile delta. B World Health Organ. 1993;71:49–54. [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Thomas JH. Targets of TGF-b signaling in Caenorhabditis elegans dauer formation. Dev Biol. 2000;217:192–204. doi: 10.1006/dbio.1999.9545. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DS, Denlinger DL, Sim C. Suppression of allatotropin simulates reproductive diapause in the mosquito Culex pipiens. 2014;64:48–53. doi: 10.1016/j.jinsphys.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialanas E, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:583–587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matingly PF, Rozeboom LE, Knight KL, Laven H, Drummond FH, Christophers SR, Shute PG. The Culex pipiens complex. Trans R Ent Soc Lond. 1951;102:331–382. [Google Scholar]
- Mitchell CJ. Differentiation of host-seeking behavior from blood-feeding behavior in overwintering Culex pipiens (Diptera: Culicidae) and observations on gonotrophic dissociation. J Med Entomol. 1983;20:157–163. doi: 10.1093/jmedent/20.2.157. [DOI] [PubMed] [Google Scholar]
- Ng H-H, Bird A. Histone deacetylases: silencers for hire. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Parker L, Arora K. Identification of maverick a novel member of the TGF-β superfamily in Drosophila. Mech Develop. 2000;95:201–206. doi: 10.1016/s0925-4773(00)00338-5. [DOI] [PubMed] [Google Scholar]
- Oda T. Studies on the follicular development and overwintering of the house mosquito, Culex pipiens pallens, in Nagasaki area. Trop Med. 1968;10:195–216. [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Rhinehart JP, Robich RM, Denlinger DL. Enhanced cold and desiccation tolerance in diapason adults of Culex pipiens and a role for Hsp70 in response to cold shock but not as a component of the diapause program. J Med Entomol. 2006;43:713–722. doi: 10.1603/0022-2585(2006)43[713:ECADTI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Robich RM, Denlinger DL. Diapause in the mosquito Culex pipiens evokes a metabolic switch from blood feeding to sugar gluttony. PNAS. 2005;102:15912–15917. doi: 10.1073/pnas.0507958102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robich RM, Rinehart JP, Kitchen LJ, Denlinger DL. Diapause-specific gene expression in the northern house mosquito, Culex pipiens L., identified by suppressive subtractive hybridization. J Insect Physiol. 2007;53:235–245. doi: 10.1016/j.jinsphys.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez A-M, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Bio. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb repressive complex 2 and trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci. USA. 2009;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. PNAS. 2008;105:6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol Bio. 2009;18:325–332. doi: 10.1111/j.1365-2583.2009.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Denlinger DL. Insulin signaling and the regulation of insect diapause. Front Physiol. 2013;4:189. doi: 10.3389/fphys.2013.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M, Ono K, Johannes Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman A, Wong J. Environmental control of ovarian diapause in Culex pipiens. Ann Entomol Soc Am. 1973;66:905–907. [Google Scholar]
- Spielman A. Effect of synthetic juvenile hormone on ovarian diapause of Culex pipiens mosquitoes. J Med Entomol. 1974;11:223–225. doi: 10.1093/jmedent/11.2.223. [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova EB. Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control. Pensoft, Sofia: 2000. [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov SA, Snoddy JR. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.