Abstract
Schizophrenia is associated with alterations in working memory that reflect dysfunction of dorsolateral prefrontal cortex (DLPFC) circuitry. Working memory depends on the activity of excitatory pyramidal cells in DLPFC layer 3, and to a lesser extent in layer 5. Although many studies have profiled gene expression in DLPFC gray matter in schizophrenia, little is known about cell type-specific transcript expression in these two populations of pyramidal cells. We hypothesized that interrogating gene expression specifically in DLPFC layer 3 or 5 pyramidal cells would reveal new and/or more robust schizophrenia-associated differences that would provide new insights into the nature of pyramidal cell dysfunction in the illness. We also sought to determine the impact of other variables, such as a diagnosis of schizoaffective disorder or medication use at time of death, on the patterns of gene expression in pyramidal neurons.
Individual pyramidal cells in DLPFC layers 3 or 5 were captured by laser microdissection from 36 subjects with schizophrenia or schizoaffective disorder and matched normal comparison subjects. The mRNA from cell collections was subjected to transcriptome profiling by microarray followed by qPCR validation.
Expression of genes involved in mitochondrial (MT) or ubiquitin-proteasome system (UPS) functions were markedly down-regulated in the patient group (p values for MT-related and UPS-related pathways were <10−7 and <10−5 respectively). MT-related gene alterations were more prominent in layer 3 pyramidal cells, whereas UPS-related gene alterations were more prominent in layer 5 pyramidal cells. Many of these alterations were not present, or found to a lesser degree, in samples of DLPFC gray matter from the same subjects, suggesting that they are pyramidal cell-specific. Furthermore, these findings principally reflected alterations in the schizophrenia subjects, were not present or present to a lesser degree in the schizoaffective disorder subjects (diagnosis of schizoaffective disorder was the most significant covariate, p<10−6), and were not attributable to factors frequently comorbid with schizophrenia.
In summary, our findings reveal expression deficits in MT- and UPS-related genes specific to layer 3 and/or layer 5 pyramidal cells in the DLPFC of schizophrenia subjects. These cell type-specific transcriptome signatures are not characteristic of schizoaffective disorder, providing a potential molecular-cellular basis of differences in clinical phenotypes.
Introduction
Cognitive deficits, including impairments in working memory, are a core and clinically-critical feature of schizophrenia1. Working memory depends upon the sustained firing of pyramidal cells in layer 3, and to a lesser extent in layer 5, in the dorsolateral prefrontal cortex (DLPFC)2. This persistent activity, that is thought to hold relevant information “on line” in the absence of sensory input, requires recurrent excitatory connections among layer 3 pyramidal cells that terminate on dendritic spines3. Thus, abnormalities in dendritic spines on layer 3 pyramidal cells would be expected to contribute to impairments in working memory.
Consistent with this prediction, postmortem studies have reported lower dendritic spine density, as well as smaller somal volumes and shorter dendritic trees, on DLFPC layer 3 pyramidal cells in subjects with schizophrenia4–8. Similar findings have been reported for layer 5 pyramidal cells, although the studies are less numerous and the findings inconsistent4,7,9. Importantly, dendritic spine abnormalities do not seem to be related to treatment with antipsychotic medications or other potential confounds10 and therefore may reflect the underlying disease. Consistent with this interpretation, transcript levels of certain proteins that regulate spine maintenance and plasticity are altered in the DLPFC of schizophrenia subjects, and the expression levels of these transcripts correlate with spine density on layer 3 pyramidal cells11,12. Furthermore, for some of these proteins, depletion in mouse models triggered PFC spine loss during adolescence and impairments in working memory13.
However, most gene expression studies of the DLPFC in schizophrenia have focused on grey matter samples and have not interrogated the transcriptomes of specific cell types. Cell type- and cortical layer-specific measures of gene expression are particularly important because the cellular heterogeneity present across cortical grey matter is likely to result in a lower sensitivity to detect cell type-specific differences in gene expression14 and to contribute to the variability of findings across studies15. Indeed, transcriptome profiling of laser microdissected samples of layers 2–3 versus layers 5–6 of human DLPFC revealed substantial laminar differences in gene expression in healthy subjects as well as layer-specific alterations in schizophrenia16,17. We hypothesized that interrogating gene expression specifically in DLPFC layer 3 or 5 pyramidal cells would reveal new and/or more robust schizophrenia-associated differences that would provide new insights into the nature of pyramidal cell dysfunction in the illness. In addition, we sought to determine the impact of other variables, such as a diagnosis of schizoaffective disorder or medication use at time of death, on the patterns of gene expression in pyramidal neurons. Consequently, we used laser microdissection to obtain samples of individually-captured pyramidal cells in layers 3 or 5 of the DLPFC from 36 subjects with schizophrenia or schizoaffective disorder and matched comparison subjects and assessed gene expression in these samples using microarrays and quantitative PCR. Similar studies were conducted in DLPFC pyramidal neurons from monkeys exposed chronically to antipsychotic medications.
Methods
Human subjects
Brain specimens (n=72) were obtained during autopsies conducted at the Allegheny County Office of the Medical Examiner (Pittsburgh, PA) after consent was obtained from the next-of-kin. An independent committee of experienced research clinicians made consensus DSM-IV diagnoses for all 72 subjects on the basis of medical records and structured diagnostic interviews conducted with the decedent’s family members6. Each subject with schizophrenia (n=24) or schizoaffective disorder (n=12) was matched with one normal comparison subject for sex and as closely as possible for age; subject groups did not differ in mean age, postmortem interval (PMI), brain pH, RNA integrity number (RIN) or tissue storage time at −80°C (all t71< 3.36; all p>0.07) or in race (Χ2=2.67; p=0.102) (Table 1). Details for each subject, including cause of death, lifetime history of cannabis use, tobacco use at time of death, substance use diagnoses, and psychotropic medication use at time of death are provided in Supplemental Table 1. All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research.
Table 1.
Subject characteristics.
| Comparison | Patients | |
|---|---|---|
| Number | 36 | 36 |
| Sex | 27 M, 9 F | 27 M, 9 F |
| Race | 30 W, 6 B | 24 W, 12 B |
| Age (years) | 48.1 (13.0) | 46.9 (12.4) |
| PMI (h) | 17.6 (6.1) | 18.0 (8.8) |
| Brain pH | 6.7 (0.2) | 6.6 (0.4) |
| RIN | 8.3 (0.6) | 8.2 (0.6) |
| Storage time (months at −80°C) | 122.2 (49.8) | 125.7 (53.1) |
Values are mean (SD). PMI: postmortem interval. RIN: RNA integrity number.
See Supplemental Table 1 for details of individual subjects.
Laser microdissection
The right hemisphere of each brain was blocked coronally, immediately frozen, and stored at −80°C18. For all procedures, samples from each subject in a given pair were processed together in order to control for experimental variance. Tissue sections (12 μm) containing DLPFC area 9 were cut on a cryostat, mounted on glass polyethylene naphthalate membrane slides (Leica Microsystems, Bannockburn, IL) which were blinded as to diagnosis, and stained with thionin for Nissl substance19,20. Using a Leica laser microdissection system (LMD 6500), pyramidal neuron cell bodies with a characteristic triangular shape and prominent apical dendrite were identified and dissected from layers 3 or 5 (Supplemental Figure 1A,B). Dissections were performed in successive collections from two subsets of the 36 subject pairs (cohorts 1 and 2 in Supplemental Table 1). Based on the results of pilot studies (Supplemental Methods), two samples of 100 pyramidal cells were individually dissected from layer 3 and from layer 5 of area 9 from adjacent tissue sections for cohort 1. To prevent potential confounding effects of collection order, pyramidal cells were collected in the opposite order from layers 3 and 5 across the two sections per subject. Preliminary results from cohort 1 transcriptome profiling showed a significantly (p<10−4) higher mean correlation between replicate than between non-replicate (i.e., random pairing) samples; thus, the microarray results from the two cohort 1 samples were averaged for data analysis and one sample of 200 pyramidal cells was collected in each layer from one tissue section for cohort 2.
Microarray and qPCR
For each sample, RNA was extracted using the QIAGEN Micro RNeasy kit Plus. cDNA was synthesized and amplified using the Ovation Pico WTA System, labeled using the Encore Biotin module and loaded on an Affymetrix GeneChip® HT HG-U133+ PM Array Plate which is designed to assess expression levels of transcripts in the human genome. Transcriptome profiling was performed using two successive batches of cDNA synthesized from each cohort.
Differentially expressed transcripts (DETs) detected by microarray were assessed by qPCR as previously described19,21 using the cDNA template obtained from cohort 1 samples. Transcripts selected for validation had a differential expression between subject groups >20% and a high level of expression (Robust Multi-array Average (RMA) normalized value >7). Primer sets were designed outside of the Affymetrix target except when precluded by the small size of some transcripts (Supplemental Table 2). Selected DETs were also assessed by qPCR in grey matter homogenates of DLPFC area 9; for these samples, total RNA was converted to cDNA using the High Capacity cDNA Archive Kit from Applied Biosystems (Foster City, CA). Primer set efficiencies and normalizers for qPCR are described in Supplemental Methods and Supplemental Table 2.
Antipsychotic-treated monkeys
Three groups (n=6 per group) of young adult, male, macaque monkeys (Macaca fascicularis) were exposed for 17 to 27 months to oral haloperidol, olanzapine, or placebo at doses that produced trough serum levels in the therapeutic range for the treatment of schizophrenia22. Animals were euthanized in triads and tissue was processed as described previously23. From each monkey, 200 individually-dissected pyramidal cells were collected from DLPFC layers 3 and 5 from two adjacent tissue sections. Total RNA was extracted (QIAGEN RNeasy microkit Plus), cDNA synthesized (QUANTA BioSciences qScript™ cDNA SuperMix) and qPCR conducted as described above with all samples from a given triad processed together.
Statistical analysis
A detailed description of statistical and pathway analyses is provided in the Supplemental Methods. Briefly, Affymetrix CEL files were normalized and log2 transformed using RMA Express24. Due to the high computational demand of our approach for assessing the impact of covariates (see below) and the expectation that many of the probe sets on the arrays would be expressed at very low levels and/or be non-informative, we used a previously reported approach25 to filter the 54,715 probe sets on the arrays in two steps: 1) Removal of low expression probe sets by filtering out the probe sets with the lowest 40% mean intensities across all samples, and 2) Removal of non-informative probe sets by excluding probe sets with the lowest 40% standard deviation across all samples. This approach (54,715 probe sets X 0.6 × 0.6) resulted in 19,667 probe sets for analysis. The impact of this approach relative to other filtering strategies on the number of differentially expressed probe sets at FDR of 0.01 or 0.05 is shown in Supplemental Table 3. The resulting dataset of 19,697 was then split by layer and the dataset for each layer was analyzed separately using a Random Intercept Model with Bayesian Information Criterion variable selection (RIM-BIC; Supplemental Methods;26). This method provides greater statistical power by accounting, in the present study, for the paired design, sample size, expression effect size and potential confounding variables (see below). An adaptively weighted (AW) Fisher’s method27 was used to combine the differential expression information (i.e., p-values) of both layers for each transcript. Unlike Fisher’s method that assigns equal weight for both studies, the AW method searches for the best 0–1 (0 for absence and 1 for presence of differential expression) adaptive weight for each differentially-expressed probe set (DEP) in order to capture similarities or differences across layers. For example, an adaptive weight of (1,1) indicates the differential expression of a transcript in both layers, (1,0) indicates significant differential expression only in layer 3 and (0,1) only in layer 5. The meta-analyzed p-values from AW were then adjusted for multiple comparisons28 to control false discovery rate (using “p.adjust” function in R). Following meta-analysis, the potential influence of 10 variables (sex, age, schizoaffective diagnosis, suicide, PMI, pH, RIN, benzodiazepine use at time of death (ATOD), antidepressant use ATOD and tobacco use ATOD) on DEP were assessed (Supplemental Methods). For pathway enrichment analysis, a total of 2,092 pathways from the GO, KEGG, Biocarta and Reactome databases were assessed. Human and monkey data from qPCR were analyzed using ANCOVA models (Supplemental Methods).
Results
Differentially expressed genes between subject groups
Findings demonstrating the empirical basis for the number of cells collected per sample, the consistency of findings between the two subject cohorts and the cell type and laminar specificity of the LMD-obtained samples (Supplemental Table 4) are provided in the Supplemental Results. Using a false discovery rate of 5%, we identified 1,783 DEPs in pyramidal cells between the subject groups after controlling for multiple testing (adjusted p value=q value< 0.05). Table 2A lists the 40 DEPs with the smallest overall q values altered in both layers 3 and 5 (full list is available upon request). The majority of the DEPs in layer 3 (71% or 1,265 DEPs) and in layer 5 (70% or 1,250 DEPs) were down-regulated in the patient group. Sixty-five percent (1,158) of the DEPs were differentially expressed in both layers 3 and 5 (Table 2A), 10% (180) were altered only in layer 3 pyramidal cells and 25% (445) only in layer 5 pyramidal cells; accordingly, Table 2B lists the top 10 and 25 DEPs with the lowest q values in layer 3 or in layer 5, respectively.
Table 2A.
List of the 40 DEPs with the most significant q values combining data from pyramidal cells in both layers 3 and 5.*
| Gene Symbol | Layer 3 | Layer 5 | Combined q value | ||
|---|---|---|---|---|---|
| % change | q value | % change | q value | ||
| ZSWIM7 | −46.8 | <0.05 | −54.7 | <10−16 | <10−17 |
| APOA1BP | −37.8 | <0.05 | −42.1 | <0.005 | <10−17 |
| UQCRFS1 | −37.6 | <0.05 | −36.2 | <10−16 | <10−17 |
| TIMM10 | −35.5 | <0.05 | −27.4 | <10−3 | <10−17 |
| BID | −35.2 | <0.005 | −28.8 | <0.05 | <10−17 |
| MRPS6 | −34.2 | <0.01 | −30.1 | <0.005 | <10−17 |
| PTS | −32.2 | <0.05 | −31.5 | <10−3 | <10−17 |
| CMC1 | −31.3 | <0.1 | −42.7 | <10−3 | <10−17 |
| TMEM14B | −30.6 | <0.01 | −36.6 | <0.005 | <10−17 |
| RPS9 | −30.2 | <0.05 | −31.9 | <10−3 | <10−17 |
| SPATA7 | −29.9 | <0.1 | −39.0 | <10−16 | <10−17 |
| DCTN3 | −29.8 | 0.307 | −34.2 | <10−16 | <10−17 |
| COX7A1 | −29.5 | <0.05 | −29.3 | <10−16 | <10−17 |
| LGALS1 | −28.3 | <0.01 | −28.4 | <0.005 | <10−17 |
| PSMC6 | −27.0 | <0.05 | −25.3 | <10−3 | <10−17 |
| UQCRQ | −26.4 | <0.05 | −24.2 | <0.05 | <10−17 |
| NDUFV1 | −26.1 | 0.124 | −28.0 | <10−16 | <10−17 |
| IGFBP6 | −25.8 | <0.1 | −29.8 | <10−3 | <10−17 |
| COX7B | −25.7 | <0.05 | −26.0 | <10−3 | <10−17 |
| ETV1 | −25.3 | <0.05 | −32.6 | <10−3 | <10−17 |
| MGST3 | −24.7 | <0.005 | −24.4 | <10−3 | <10−17 |
| MMGT1 | −24.4 | <0.05 | −34.8 | <10−3 | <10−17 |
| ACTR10 | −24.0 | <0.1 | −28.8 | <10−16 | <10−17 |
| PDZD11 | −24.0 | <0.05 | −30.4 | <10−16 | <10−17 |
| VTA1 | −23.4 | <0.01 | −24.3 | <0.01 | <10−17 |
| NDUFS4 | −23.2 | <0.05 | −22.7 | <0.005 | <10−17 |
| C1orf212 | −23.2 | <0.05 | −27.4 | <10−3 | <10−17 |
| NDUFB9 | −21.7 | <0.05 | −23.3 | <10−3 | <10−17 |
| C3orf14 | −21.6 | <0.05 | −22.7 | <0.005 | <10−17 |
| FAM103A1 | −21.3 | <0.05 | −27.5 | <10−3 | <10−17 |
| C20orf24 | −21.2 | <0.1 | −30.7 | <10−16 | <10−17 |
| PHYH | −20.3 | <0.1 | −25.8 | <10−3 | <10−17 |
| NSDHL | −20.2 | <0.05 | −22.0 | <10−3 | <10−17 |
| NFU1 | −19.0 | <0.05 | −27.4 | <0.005 | <10−17 |
| UBE2E3 | −17.6 | <0.05 | −21.5 | <0.005 | <10−17 |
| ASAP2 | −15.0 | 0.314 | −30.6 | <10−16 | <10−17 |
| MEG3 | 43.4 | <0.05 | 71.2 | <10−3 | <10−17 |
| MEG3 | 54.9 | <0.05 | 44.7 | <10−3 | <10−17 |
| RASEF | 63.5 | <0.01 | 50.9 | <0.005 | <10−17 |
| MEG3 | 76.6 | <0.01 | 61.2 | <10−3 | <10−17 |
List excludes non-annotated probe sets as well as sequences associated with hypothetical proteins. Note that AW-Fisher’s test can attribute significant alterations in both layers (i.e., 1,1 for expression alterations in both layers 3 and 5) even though q exceeds 0.05 in one layer.
Table 2B.
List of top DEPs with significant expression differences in a single layer.*
| Gene Symbol | Layer 3 | Layer 5 | Combined q value | ||
|---|---|---|---|---|---|
| % change | q value | % change | q value | ||
| EMG1 | −30.3 | <10−15 | −19.5 | 0.753 | <10−17 |
| GGH | −37.8 | <0.01 | −39.2 | 0.511 | <10−3 |
| MRPL36 | −25.3 | <0.01 | −10.1 | 0.455 | <10−3 |
| TMEM19 | −22.1 | <0.05 | −12.2 | 0.462 | <0.005 |
| TINF2 | −24.0 | <0.05 | −1.9 | 0.781 | <0.005 |
| BAG1 | −26.2 | <0.05 | −15.2 | 0.475 | <0.005 |
| MRPL48 | −23.5 | <0.05 | −20.5 | 0.366 | <0.005 |
| ERGIC2 | −25.7 | <0.05 | −20.1 | 0.575 | <0.005 |
| SCNM1 | −28.8 | <0.05 | −19.8 | 0.922 | <0.005 |
| DEF8 | −22.6 | <0.05 | −18.1 | 0.897 | <0.005 |
|
| |||||
| PCSK1 | −16.2 | 0.179 | −27.1 | <10−16 | <10−17 |
| PIGH | −22.0 | 0.223 | −31.0 | <10−16 | <10−17 |
| PFDN5 | −25.6 | 0.431 | −35.2 | <10−3 | <10−3 |
| RSL24D1 | −20.5 | 0.799 | −26.6 | <10−3 | <10−3 |
| RPS27A | −28.0 | 0.285 | −26.1 | <10−3 | <10−3 |
| RPS3A | −24.7 | 0.593 | −28.5 | <10−3 | <10−3 |
| AHCY | −22.4 | 0.324 | −25.8 | <10−3 | <10−3 |
| AP1S2 | −22.6 | 0.343 | −34.4 | <10−3 | <10−3 |
| PSMB4 | −25.0 | 0.437 | −29.2 | <10−3 | <10−3 |
| ACTR3 | −19.5 | 0.465 | −23.8 | <10−3 | <10−3 |
| RPS3A | −24.5 | 0.545 | −26.7 | <10−3 | <10−3 |
| NDUFA8 | −23.6 | 0.449 | −26.7 | <10−3 | <10−3 |
| RPS10 | −23.7 | 0.374 | −22.8 | <10−3 | <10−3 |
| DHX29 | −6.1 | 0.791 | −33.3 | <10−3 | <10−3 |
| CAMK2D | −22.6 | 0.409 | −27.0 | <10−3 | <10−3 |
| C9orf64 | −12.2 | 0.713 | −24.2 | <10−3 | <10−3 |
| TOMM20 | −21.6 | 0.386 | −26.8 | <0.005 | <0.005 |
| UBE3B | 2.1 | 0.961 | 39.4 | <0.005 | <0.005 |
| RPS3A | −26.0 | 0.533 | −27.0 | <0.005 | <0.005 |
| TMEM38A | −14.6 | 0.510 | −25.7 | <0.005 | <0.005 |
| FBXO32 | −8.6 | 0.698 | −32.2 | <0.005 | <0.005 |
| WDYHV1 | −22.0 | 0.884 | −40.4 | <0.005 | <0.005 |
| CCT4 | −30.0 | 0.432 | −33.3 | <0.005 | <0.005 |
| GYG1 | −18.9 | 0.342 | −38.2 | <0.005 | <0.005 |
| SH3GL3 | 11.8 | 0.721 | 32.7 | <0.005 | <0.005 |
List of the top 10 DEPs, ranked by layer-specific q values, with significantly different expression in layer 3 pyramidal cells only (above line) and of top 25 DEPs with significantly different expression in layer 5 pyramidal cells (below line). List excludes non-annotated probesets as well as sequences associated with hypothetical proteins.
Pathway analysis
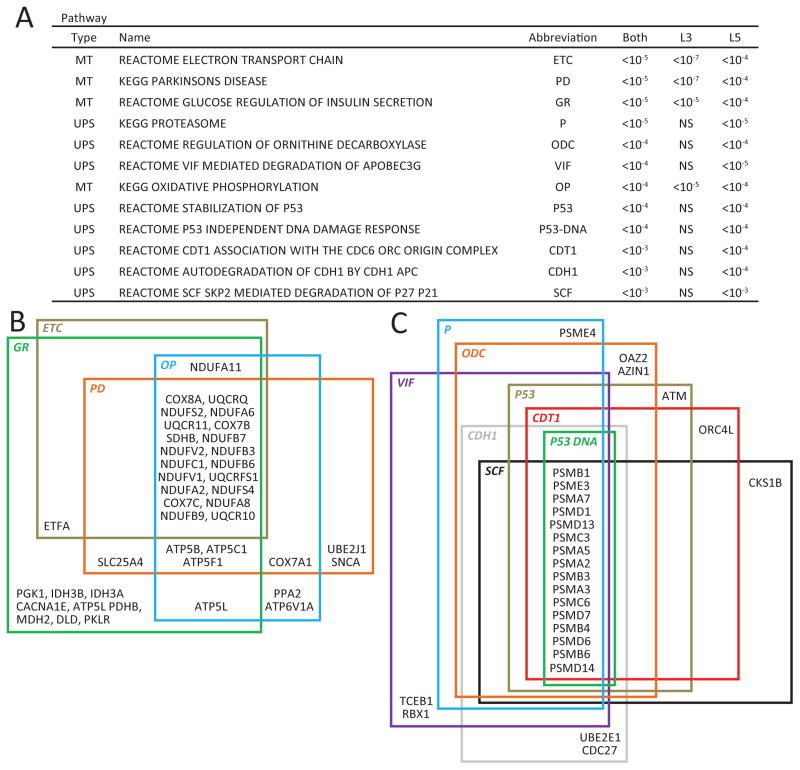
The 1,783 DEPs detected by microarray correspond to 1,420 genes. After selecting pathways containing >10 and <200 genes and using a false discovery rate of 5%, these 1,420 differentially expressed genes were enriched in 68 biological pathways. Of the top 12 affected pathways as ranked by q values (Figure 1A), four were related to mitochondrial (MT) function and eight to ubiquitin proteasome system (UPS) function. The high level of overlap among the DEPs belonging to the MT- or the UPS-related pathways is shown in Figures 1B and 1C, respectively. Furthermore, the three most altered pathways involve MT functions, and of the 40 DETs with the most significant differences (all q<10−17; Table 2) between the subject groups, 11 were related to MT function. All of these transcripts were down-regulated in schizophrenia subjects.
Figure 1. Top 12 gene pathways based on most significant q values altered in pyramidal cells in subjects with schizophrenia.
(A) Twelve most significantly altered gene pathways, based on smallest q values across both layers 3 and 5. Note that MT-related pathways show smaller p values in layer 3 (L3) than layer 5 (L5) pyramidal cells and that UPS-related pathways are not altered in layer 3 pyramidal cells. NS, non-significant. (B) Venn diagram illustrating the overlap in genes among the four most altered MT-related pathways in schizophrenia; (C) Venn diagram illustrating the overlap in genes among the eight most altered UPS-related pathways in schizophrenia.
For all 4 MT pathways, the q values were smaller for layer 3 compared to layer 5. Conversely, for all 8 UPS pathways the q values were significant only in layer 5 (Figure 2A). These differences suggest laminar-specificity in altered pyramidal cell gene expression in schizophrenia, with MT-related alterations more robust in layer 3 and UPS-related alterations restricted to layer 5.
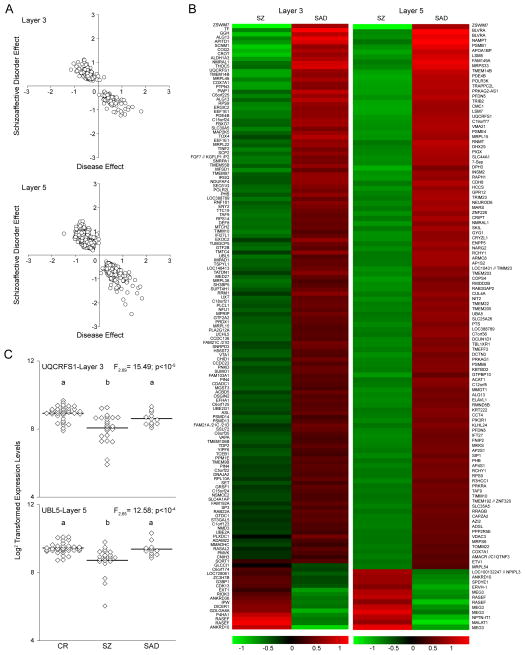
Figure 2. Comparison of the main effect of disease relative to the covariate of schizoaffective disorder.
(A) The effect of disease relative to the effect of the covariate of schizoaffective disorder is shown for each of the 1,000 permutations where a significant effect of schizoaffective disorder was detected in both layers 3 and 5. Due to the filters used for detection of DEPs, there are no data points around a disease effect equal to zero. Note that the main effect of disease and effect of schizoaffective disorder are in the opposite directions in both layers. (B) Heat map illustrating the transcripts that were differentially expressed in pyramidal neurons between subjects with schizophrenia (SZ) and subjects with schizoaffective disorder (SAD) for each layer. Due to the high number of DEPs, only transcripts with a minimum log 2 transformed expression level ≥5 are shown for each layer. In addition, for layer 5 only DEPs with a disease effect greater than +/−0.7 are shown. (C) Log 2 transformed transcript expression levels for UQCRFS1 in layer 3 and UBL5 in layer 5 for comparison (CR), schizophrenia (SZ) and schizoaffective disorder (SAD) subjects. For each transcript, groups not sharing the same letter are significantly different by Tukey post-hoc analysis.
Analysis of potential confounding factors
Of the 10 covariates evaluated (sex, age, schizoaffective diagnosis, suicide, PMI, pH, RIN, benzodiazepine use ATOD, antidepressant use ATOD and tobacco use ATOD), only three achieved overall genome-wide statistical significance (i.e., were confounded with disease effect in more DEPs than expected by chance). The confounding effect of age was significant only in layer 3 and had the same directionality as the disease effect (Supplemental Figure 2B); after controlling for the effect of age, the effect of disease remained significant (p=0.035). The effects of schizoaffective disorder (Figure 2A) and pH (Supplemental Figure 2A) were significant in both layers 3 and 5 (p< 0.001), but were in the opposite direction of the overall disease effect; that is, they blunted the effect present in the subjects with schizophrenia. The genes with the greatest differential expression between subjects with schizophrenia versus schizoaffective disorder are shown in Figure 2B. The effect of schizoaffective disorder versus schizophrenia is illustrated for one MT-related DET in layer 3 (UQCRFS1) and one UPS-related DET in layer 5 (UBL5) in Figure 2C. The mean expression levels across diagnoses were significantly different for UQCRFS1 (F2,69=15.5; p<10−04) and UBL5 (F2,65=12.6; p<10−04), with each transcript lower in the schizophrenia subjects relative to both the schizoaffective and comparison subjects, which did not differ from each other.
The effects of antipsychotic medications were assessed in layer 3 and layer 5 pyramidal cells dissected from the DLPFC of monkeys following long-term exposure to placebo, haloperidol or olanzapine. For each layer, 5 transcripts (ATP5C1, FBXW7, UCHL5, SDHB and UQCRQ for layer 3; ATP5B, COPS4, MRPL33, NDUFB2 and PSMB4 for layer 5) that displayed significantly lower expression in schizophrenia subjects by microarray and by qPCR were selected. Expression of these transcripts did not differ across the three groups of monkeys (Supplemental Table 5). Consistent with these findings, the chlorpromazine equivalent daily dose of antipsychotic medications at time of death (Supplemental Table 1) was not significantly associated (all r<I0.382I, all p>0.198) with the expression level (assessed by either microarray or PCR) of any of these 12 transcripts in layer 3 or layer 5 pyramidal cells in the schizophrenia subjects. Furthermore, alterations in MT and UPS gene expression were not seen in schizoaffective disorder subjects, even though they were treated with antipsychotic medications. Together, these observations suggest that the transcript alterations observed in schizophrenia are not a consequence of antipsychotic medications.
qPCR confirmation of altered gene expression in pyramidal cells
To validate the findings of altered MT and UPS gene expression, 18 DETs related to different functions within these gene groups were assessed by qPCR in cohort 1 pyramidal cell samples from the layer in which the greatest disease-related difference in expression was observed. Consistent with the array results, the mean expression levels of ATP5C1, UQCRQ, COX4I1, IDH3B, SDHB, FBXW7 and UCHL5 in layer 3 pyramidal cells and of ATP5B, NDUFB2, MRPL33, COPS4 and PSMB4 in layer 5 pyramidal cells were significantly lower in the schizophrenia subjects (Table 3); in addition, qPCR and microarray results were significantly correlated across subjects (Table 3). Of the six transcripts that were not significantly different between schizophrenia and control subjects by qPCR, the array and qPCR results were significantly correlated across subjects for VDAC3 and NDFIP2, and nearly so for PSMA2 and CUL3, suggesting that the failure to achieve significance by qPCR might reflect the smaller sample size used in these studies. Only TOMM20 and CUL4A failed to validate by either statistical assessment.
Table 3.
qPCR validation of selected transcripts and correlations with array results across subjects.
| Gene Pathway | Gene Symbol | % change | qPCR* ANCOVA |
Array and qPCR correlation | ||
|---|---|---|---|---|---|---|
| Array | qPCR | r | p | |||
| MT | ATP5C1 | −26.7 | −22.4 | <0.05 | 0.75 | <10−3 |
| MT | UQCRQ | −30.6 | −40.5 | <0.005 | 0.79 | <10−3 |
| MT | COX4I1 | −21.1 | −30.3 | <0.005 | 0.68 | <10−3 |
| MT | IDH3B | −35.1 | −34.4 | <0.05 | 0.41 | <0.05 |
| MT | SDHB | −28.3 | −31.8 | <0.05 | 0.75 | <10−3 |
|
| ||||||
| MT | ATP5B | −38.5 | −22.4 | <0.05 | 0.64 | <10−3 |
| MT | NDUFB2 | −31.8 | −47.5 | <0.005 | 0.74 | <10−3 |
| MT | MRPL33 | −44.1 | −40.2 | <0.05 | 0.68 | <10−3 |
| MT | VDAC3 | −42.8 | −5.3 | 0.750 | 0.57 | <10−3 |
| MT | TOMM20 | −38.7 | 22.1 | 0.220 | −0.15 | 0.392 |
|
| ||||||
| UPS | CUL4A | −42.9 | −12.2 | 0.597 | −0.01 | 0.972 |
| UPS | FBXW7 | −28.8 | −18.2 | <0.05 | 0.56 | <10−3 |
| UPS | UCHL5 | −37.2 | −29.7 | <0.05 | 0.66 | <10−3 |
| UPS | PSMA2 | −38.2 | 6.9 | 0.684 | 0.32 | <0.1 |
|
| ||||||
| UPS | CUL3 | −28.2 | −8.6 | 0.613 | 0.31 | <0.1 |
| UPS | COPS4 | −58.7 | −26.4 | <0.05 | 0.44 | <0.05 |
| UPS | NDFIP2 | −30.9 | −24.8 | 0.220 | 0.43 | <0.05 |
| UPS | PSMB4 | −39.7 | −31.3 | <0.005 | 0.72 | <10−3 |
Nine MT- or UPS-related transcripts were chosen for validation for layer 3 (ATP5C1, UQCRQ, COX4I1, IDH3B, SDHB, CUL4A, FBXW7, UCHL5 and PSMA2) and layer 5 (ATP5B, NDUFB2, MRPL33, VDAC3, TOMM20, CUL3, COPS4, NDFIP2 and PSMB4).
Results of unpaired ANCOVAs excluding outliers. Potential outliers were defined as having an expression ratio more that than three times different from the mean expression ratio of all subjects for that particular transcript. For pair 1, the SZ subject was found to be an outlier for the following transcripts: COX4I1, SDHB, UCHL5 and UQCRQ. For pair 2, the control subject was found to be an outlier for COPS4.
We also sought to determine if cannabis use, factors predictive of illness severity or measures of functional outcome had an influence on the expression levels of these validated transcripts. None of the transcript expression levels differed significantly as a function of a history of cannabis use, age of disease onset, presence of a first-degree relative with schizophrenia, marriage history, socioeconomic status, or independent living status (Supplemental Table 6). The only exception was for marriage history and IDH3B mRNA levels in layer 3 pyramidal cells, but here the individuals with schizophrenia who had never married had higher expression levels than those who had married (Supplemental Table 6).
Cell type specificity of altered gene expression
Many of the DETs observed in pyramidal cells were not previously reported in studies of DLPFC grey matter in schizophrenia. To determine if these DETs reflected pyramidal cell-specific alterations, we conducted qPCR on total area 9 grey matter for nine DETs selected on the basis of a q value <10−4 and differential expression of >20% in both layers 3 and 5 of schizophrenia subjects. Seven of these DETs (TIMM10, LGALS1, UQCRQ, PSMC6, COX7A1, RPS9 and COPS4) were down-regulated and two (MEG3 and CALR) were up-regulated in the schizophrenia subjects. To maximize the detection of differential expression in grey matter, we tested only the 24 subject pairs with schizophrenia. Differential expression by qPCR in grey matter was over 3 times less than that detected in pyramidal cells for CALR, MEG3, NDUFV1, PSMC6; was absent for TIMM10, LGALS1, UQCRQ, COX7A1, and COPS4; and was in the opposite direction for RPS9 (Supplemental Table 7). In concert, these findings suggest that the largest and most robust differences in gene expression between schizophrenia and comparison subjects in layer 3 and layer 5 pyramidal cells may be specific to or enriched in these neurons.
Discussion
In this study, a cell type-specific approach was used to examine gene expression alterations in the DLPFC of subjects with schizophrenia or schizoaffective disorder. Most DETs were altered in the same direction in both layer 3 and 5 pyramidal cells, and most were under-expressed in affected subjects, although a subset was substantially over-expressed. Some of these observed alterations appeared to be specific to, or at least substantially enriched in, pyramidal cells. For most DETs, the disease effect was present in subjects with schizophrenia and was not evident in subjects with schizoaffective disorder. Pathway analysis revealed an overrepresentation of DETs related to MT function in layer 3 and UPS function in layer 5. These alterations in gene expression were not attributable to the effects of antipsychotic medications or other factors frequently co-morbid with schizophrenia.
Cell type-specific transcriptome profiling reveals novel and robust abnormalities in schizophrenia
The cell type-specific gene expression profiling used here carries several important advantages. First, in studies of tissue homogenates, a DET could represent either a difference in relative number of cells expressing that transcript or an altered level of expression per cell. In contrast, our collection of the same number of neurons from every subject indicates that the findings represent group differences in transcript levels per cell. Second, the capture of specific cell types facilitates the detection of gene expression differences that might be obscured in studies of tissue homogenates. For example, studies of tissue homogenates have revealed inconsistent findings regarding alterations in MT and UPS gene expression in schizophrenia29–32,32–37. A recent meta-analysis of grey matter microarray studies38 and next generation sequencing39 did not detect predominant MT or UPS signatures. Indeed, even in our previous transcriptome study of DLPFC layers 2–3 versus layers 5–6 in schizophrenia and comparison subjects, no MT- or UPS-related signatures of schizophrenia were detected, even though marked laminar differences in other transcripts were evident17.
Pyramidal cell gene expression alterations are selective for schizophrenia relative to schizoaffective disorder
A striking finding from this study was the presence of gene expression alterations in schizophrenia that were not evident in schizoaffective disorder; this difference by diagnosis was present for most of the MT- and UPS-related gene alterations. A study of the hippocampal dentate granule cell layer revealed altered expression of transcripts related to MT and UPS in schizophrenia, but not in bipolar or depression subjects40. Other studies also support gene expression differences between subjects with schizophrenia or schizoaffective disorder in the DLPFC. For example, metabotropic glutamate receptor 1 mRNA levels were lower in schizophrenia but higher in schizoaffective disorder41, and lower levels of neuropeptide Y mRNA42 or glutamic acid decarboxylase 65kD43 were detected in schizoaffective disorder but not schizophrenia subjects. On the other hand, alterations in the expression of other transcripts that regulate GABA neurotransmission in the DLPFC did not differ between schizophrenia and schizoaffective disorder subjects19,21,23,44. In concert, these findings suggest that schizophrenia and schizoaffective disorder subjects share a number of disturbances in cortical GABA neurons, but that pyramidal cell alterations in MT and UPS gene expression are distinctive to schizophrenia. Thus, our findings may contribute to the cellular and molecular basis for the observation that working memory, which depends on sustained activity in DLPFC layer 3 pyramidal neurons, is more compromised in schizophrenia than schizoaffective disorder45,46.
Alterations in MT and UPS pathways in DLPFC pyramidal cells in schizophrenia
The finding of MT and UPS alterations raises the question of the extent to which they reflect antemortem effects due to hypoxia or acidosis. In the present study, all subjects died suddenly outside of a hospital setting, with little or no agonal state which can affect brain pH and RNA integrity37,47,48. Indeed, mean brain pH (6.6) and RIN (8.2) were excellent in the schizophrenia subjects and did not differ across subject groups. Furthermore, when evident, the effect of pH on DETs was generally in the opposite direction of the disease effect.
High demand for ATP and for UPS activity at pre- and post- synaptic terminals where protein turnover and vesicle trafficking take place renders the synaptic area particularly sensitive to MT and UPS deficits49. Such deficits could reduce the capacity of pyramidal cells to sustain a normal complement of dendritic spines, leading to the lower DLPFC spine density reported in schizophrenia5,6. Alternatively, the presence of fewer dendritic spines, the principal site of excitatory inputs to pyramidal neurons, would lead to less excitatory drive and a decreased need for ATP production. Dendritic spine deficits are more apparent on DLPFC layer 3 than layer 5 pyramidal cells in schizophrenia4, paralleling the greater deficit in MT pathways in layer 3 than layer 5 pyramidal cells in the present study. The observations that schizophrenia is associated with 1) increased likelihood of de novo mutations in genes encoding proteins that regulate the actin cytoskeleton50 and 2) altered expression of layer 3-specific transcripts that regulate spine stability12 suggest that MT gene expression deficit in layer 3 pyramidal cells may be secondary to dendritic destabilization and a resulting lower number of spines to receive glutamatergic inputs. Whether lower levels of MT transcripts in DLPFC layer 3 pyramidal cells reflect a cell-autonomous disturbance or are secondary to a deficit in excitatory drive, this evidence of a hypotmetabolic state suggests that these neurons are less active in individuals with schizophrenia. Thus, these findings are not consistent with the idea of disinhibited pyramidal cells in schizophrenia51 due to an upstream deficit in inhibition, and may be more consistent with a model in which both excitation and inhibition are lower in DLPFC layer 3 circuitry52. This interpretation is supported by meta-analyses of working memory in schizophrenia that have converged on hypoactivation of the DLPFC as the most common finding53.
What upstream factors might contribute to these gene pathway alterations? Genetic risk factors for schizophrenia could contribute to the MT/UPS deficits. For example, deletions in chromosome 22q11 increase the risk of schizophrenia 30-fold54,55. Transcriptional profiling of the PFC in a mouse model of the 22q11 deletion revealed expression deficits in genes involved in oxidative phosphorylation56, with a substantial overlap in the MT and UPS genes (GEO dataset GSE10784) that are altered in DLPFC pyramidal cells in schizophrenia. Thus, energy and protein metabolism deficits in the PFC are associated with a copy number variant with the largest known effect size in conferring schizophrenia susceptibility; indeed, genes with products involved in mitochondrial function are over-represented among copy number variants in schizophrenia patients57. The apparent segregation of MT/UPS gene decreases to pyramidal cells could be caused by lower levels of factors important to maintaining the homeostasis of this cell type in DLPFC. For instance, the basic helix-loop-helix transcription factor, neurogenic differentiation 6 (NEUROD6), was down-regulated by more than 40% in both layer 3 and 5 pyramidal cells in our dataset. In vitro studies have implicated NEUROD6 in the regulation of MT biogenesis and function during neuronal differentiation58,59, and NEUROD6 over-expression resulted in an up-regulation of molecular chaperones, ETC proteins and proteasomal components60. While no previous gene expression study of the DLPFC reported differences in NEUROD6, a recent study combining multiple datasets reported a decrease in NEUROD615. The robust decrease in NEUROD6 observed in layer 3 and layer 5 pyramidal cells in the present study further highlights the power of the cell type-specific approach.
In summary, using pyramidal cell-specific dissection, we uncovered transcriptome deficits in MT- and UPS-related genes specific to layer 3 and or layer 5 pyramidal cells in the DLPFC of schizophrenia subjects. These cell type-specific transcriptome signatures are not characteristic of schizoaffective disorder, providing a potential molecular-cellular basis of differences in clinical phenotypes.
Supplementary Material
Acknowledgments
The authors thank Mary L. Brady, Carol Sue Johnston, Mary Ann Kelly, Kiley Laing, Amy Truong and Vishal Patel for excellent technical assistance, and Laura English for assistance in preparing the manuscript. David A. Lewis currently receives investigator-initiated research support from Bristol-Myers Squibb and Pfizer and in 2012–2014 served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals, and Sunovion.
Grant Support: This work was supported by a grant from Bristol-Myers Squibb and National Institutes of Health Grants MH043784, MH084053, and MH103204.
Footnotes
Disclosures:
All other authors declare no conflict of interest.
References
- 1.Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70:1107–1112. doi: 10.1001/jamapsychiatry.2013.155. [DOI] [PubMed] [Google Scholar]
- 2.Goldman-Rakic PS. Cellular basis of working memory. Neuron. 1995;14:477–485. doi: 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- 3.Arnsten AF, Wang MJ, Paspalas CD. Neuromodulation of thought: flexibilities and vulnerabilities in prefrontal cortical network synapses. Neuron. 2012;76:223–239. doi: 10.1016/j.neuron.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolluri N, Sun Z, Sampson AR, Lewis DA. Lamina-specific reductions in dendritic spine density in the prefrontal cortex of subjects with schizophrenia. Am J Psychiatry. 2005;162:1200–1202. doi: 10.1176/appi.ajp.162.6.1200. [DOI] [PubMed] [Google Scholar]
- 5.Garey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM, et al. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry. 1998;65:446–453. doi: 10.1136/jnnp.65.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 7.Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry. 1998;55:215–224. doi: 10.1001/archpsyc.55.3.215. [DOI] [PubMed] [Google Scholar]
- 8.Pierri JN, Volk CLE, Auh S, Sampson A, Lewis DA. Decreased somal size of deep layer 3 pyramidal neurons in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2001;58:466–473. doi: 10.1001/archpsyc.58.5.466. [DOI] [PubMed] [Google Scholar]
- 9.Black JE, Kodish IM, Grossman AW, Klintsova AY, Orlovskaya D, Vostrikov V, et al. Pathology of layer V pyramidal neurons in the prefrontal cortex of patients with schizophrenia. Am J Psychiatry. 2004;161:742–744. doi: 10.1176/appi.ajp.161.4.742. [DOI] [PubMed] [Google Scholar]
- 10.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 12.Ide M, Lewis DA. Altered cortical CDC42 signaling pathways in schizophrenia: Implications for dendritic spine deficits. Biol Psychiatry. 2010;68:25–32. doi: 10.1016/j.biopsych.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VanLeeuwen JE, Penzes P. Long-term perturbation of spine plasticity results in distinct impairments of cognitive function. J Neurochem. 2012;123:781–789. doi: 10.1111/j.1471-4159.2012.07899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath S, Mirnics K. Schizophrenia as a disorder of molecular pathways. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.01.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Santiago J, Diez-Alarcia R, Callado LF, Zhang JX, Chana G, White CH, et al. A combined analysis of microarray gene expression studies of the human prefrontal cortex identifies genes implicated in schizophrenia. J Psychiatr Res. 2012;46:1464–1474. doi: 10.1016/j.jpsychires.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Arion D, Unger T, Lewis DA, Mirnics K. Molecular markers distinguishing supragranular and infragranular layers in the human prefrontal cortex. Eur J Neurosci. 2007;25:1843–1854. doi: 10.1111/j.1460-9568.2007.05396.x. [DOI] [PubMed] [Google Scholar]
- 17.Arion D, Horvath S, Lewis DA, Mirnics K. Infragranular gene expression disturbances in the prefrontal cortex in schizophrenia: signature of altered neural development? Neurobiol Dis. 2010;37:738–746. doi: 10.1016/j.nbd.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase 67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 19.Arion D, Lewis DA. Altered expression of regulators of the cortical chloride transporters NKCC1 and KCC2 in schizophrenia. Arch Gen Psychiatry. 2011;68:21–31. doi: 10.1001/archgenpsychiatry.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datta D, Arion D, Lewis DA. Development of GABAA receptor subunits in layer 3 pyramidal cells of monkey prefrontal cortex. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu040. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volk DW, Matsubara T, Li S, Sengupta EJ, Georgiev D, Minabe Y, et al. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169:1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorph-Petersen KA, Pierri JN, Perel JM, Sun Z, Sampson AR, Lewis DA. The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharm. 2005;30:1649–1661. doi: 10.1038/sj.npp.1300710. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Huber W, Irizarry RA, Dudoit S. Bioinformatics and computational biology solutions using R and bioconductor. Springer Science+Business Media, Inc; New York, NY: 2005. [Google Scholar]
- 26.Wang X, Lin Y, Song C, Sibille E, Tseng GC. Detecting disease-associated genes with confounding variable adjustment and the impact on genomic meta-analysis: with application to major depressive disorder. BMC Bioinformatics. 2012;13:52. doi: 10.1186/1471-2105-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Tseng GC. An adaptively weighted statistic for detecting differential gene expression when combing multiple transcriptomic studies. Annals of Applied Statistics. 2011;5:994–1019. [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 29.Kung L, Roberts RC. Mitochondrial pathology in human schizophrenic striatum: a postmortem ultrastructural study. Synapse. 1999;31:67–75. doi: 10.1002/(SICI)1098-2396(199901)31:1<67::AID-SYN9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 30.Maurer I, Zierz S, Moller H. Evidence for a mitochondrial oxidative phosphorylation defect in brains from patients with schizophrenia. Schizophr Res. 2001;48:125–136. doi: 10.1016/s0920-9964(00)00075-x. [DOI] [PubMed] [Google Scholar]
- 31.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 32.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 33.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: A preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 35.Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E, et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:494–502. doi: 10.1002/ajmg.b.31006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 37.Vawter MP, Tomita H, Meng F, Bolstad B, Li J, Evans S, et al. Mitochondrial-related gene expression changes are sensitive to agonal-pH state: implications for brain disorders. Mol Psychiatry. 2006;11:615, 663–615, 679. doi: 10.1038/sj.mp.4001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mistry M, Gillis J, Pavlidis P. Genome-wide expression profiling of schizophrenia using a large combined cohort. Mol Psychiatry. 2013;18:215–225. doi: 10.1038/mp.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fillman SG, Cloonan N, Catts VS, Miller LC, Wong J, McCrossin T, et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2013;18:206–214. doi: 10.1038/mp.2012.110. [DOI] [PubMed] [Google Scholar]
- 40.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Volk DW, Eggan SM, Lewis DA. Alterations in metabotropic glutamate receptor 1alpha and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. Am J Psychiatry. 2010;167:1489–1498. doi: 10.1176/appi.ajp.2010.10030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris HM, Stopczynski RE, Lewis DA. NPY mRNA expression in the prefrontal cortex: Selective reduction in the superficial white matter of subjects with schizoaffective disorder. Schizophr Res. 2009;115:261–269. doi: 10.1016/j.schres.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glausier JR, Kimoto S, Fish KN, Lewis DA. Lower glutamic acid decarboxylase 65kD mRNA and protein levels in the prefrontal cortex in schizoaffective disorder but not schizophrenia. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.05.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldonado-Aviles JG, Curley AA, Hashimoto T, Morrow AL, Ramsey AJ, O’Donnell P, et al. Altered markers of tonic inhibition in the dorsolateral prefrontal cortex of subjects with schizophrenia. Am J Psychiatry. 2009;166:450–459. doi: 10.1176/appi.ajp.2008.08101484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruber O, Gruber E, Falkai P. Articulatory rehearsal in verbal working memory: a possible neurocognitive endophenotype that differentiates between schizophrenia and schizoaffective disorder. Neurosci Lett. 2006;405:24–28. doi: 10.1016/j.neulet.2006.06.062. [DOI] [PubMed] [Google Scholar]
- 46.Correll CU. Understanding schizoaffective disorder: from psychobiology to psychosocial functioning. J Clin Psychiatry. 2010;71 (Suppl 2):8–13. doi: 10.4088/JCP.9096su1cc.02. [DOI] [PubMed] [Google Scholar]
- 47.Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, et al. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13:609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- 48.Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng ZH, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Homayoun H, Moghaddam B. NMDA receptor hypofunction produces opposite effects on prefrontal cortex interneurons and pyramidal neurons. J Neurosci. 2007;27:11496–11500. doi: 10.1523/JNEUROSCI.2213-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kern RS, Horan WP, Barch DM. On altered patterns of brain activation in at-risk adolescents and young adults. Am J Psychiatry. 2013;170:1226–1231. doi: 10.1176/appi.ajp.2013.13081089. [DOI] [PubMed] [Google Scholar]
- 54.Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 57.Szatkiewicz JP, O’Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uittenbogaard M, Baxter KK, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 confers tolerance to oxidative stress by triggering an antioxidant response and sustaining the mitochondrial biomass. ASN Neuro. 2010;2:e00034. doi: 10.1042/AN20100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baxter KK, Uittenbogaard M, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 enhances mitochondrial biogenesis and bioenergetics to confer tolerance of neuronal PC12-NeuroD6 cells to the mitochondrial stressor rotenone. Exp Cell Res. 2012;318:2200–2214. doi: 10.1016/j.yexcr.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uittenbogaard M, Baxter KK, Chiaramello A. NeuroD6 genomic signature bridging neuronal differentiation to survival via the molecular chaperone network. J Neurosci Res. 2010;88:33–54. doi: 10.1002/jnr.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.