Abstract
Suboptimal intrauterine development has been linked to predisposition to cardiovascular disease in adulthood, a concept termed ‘developmental origins of health and disease’. Although the exact mechanisms underlying this developmental programming are unknown, a growing body of evidence supports the involvement of epigenetic regulation. Epigenetic mechanisms such as DNA methylation, histone modifications and micro-RNA confer added levels of gene regulation without altering DNA sequences. These modifications are relatively stable signals, offering possible insight into the mechanisms underlying developmental origins of health and disease. This review will discuss the role of epigenetic mechanisms in heart development as well as aberrant epigenetic regulation contributing to cardiovascular disease. Additionally, we will address recent advances targeting epigenetic mechanisms as potential therapeutic approaches to cardiovascular disease.
Keywords: DNA methylation, histone modifications, micro-RNA, heart development, cardiovascular disease
Developmental origins of health and disease
Since the early studies in the 1980s, factors contributing to an adverse intrauterine environment have been linked to increased risk of disease in adulthood. Initial studies on the developmental origins of adult disease, performed by Barker et al., revealed a possible link between intrauterine adversity and an increased risk of cardiovascular disease in adulthood [1]. Subsequent studies confirmed that intrauterine insults such as undernutrition, hypoxia, microbial toxins and chemical agents increased the risk of cardiovascular and metabolic disorders later in life, including metabolic syndrome, type 2 diabetes mellitus, hypertension and ischemic heart disease [2]. As a result of these studies, Hales and Barker developed the ‘thrifty phenotype hypothesis’ to explain this phenomenon [3]. This hypothesis proposes that, when confronted with a suboptimal intrauterine environment, the developing fetus makes irreversible adaptation to ensure survival in such an environment. Following birth, however, these changes can prove to be maladaptive, rendering the neonate and growing individual susceptible to disease development. Although the exact mechanisms translating unfavorable intrauterine conditions into susceptibility to cardiovascular and metabolic diseases are not yet known, recent studies on the role of epigenetic modifications have provided valuable insights into the developmental origins of health and disease. In this review, we summarize recent progress made in our understanding of major epigenetic mechanisms and their roles in regulating heart development and disease.
Epigenetic mechanisms
Epigenetic regulatory mechanisms are, by definition, capable of modulating gene expression without altering DNA sequences. These mechanisms can act at genomic (DNA methylation) or nucleosomal and chromatin (histone modifications, chromatin remodeling complexes) levels. In addition, a post-transcriptional level of regulation is conferred by small, noncoding RNAs termed micro-RNAs (miRs). Although these regulatory systems have distinct mechanisms, there is considerable functional overlap and crosstalk among them. This review will address the roles of DNA methylation, histone modifications and miRs in heart development and disease. For in-depth reviews of the roles of chromatin remodeling complexes in heart development and disease see [4,5].
DNA methylation
DNA methylation: location and function
DNA methylation is the only known epigenetic regulatory mechanism that affects the DNA molecule directly. It has been implicated in gene imprinting, X-chromosome inactivation, the regulation of tissue-specific gene expression, cell development, genomic stability and modulation of splicing events [6,7]. DNA methylation most often occurs in mammals at the fifth carbon of cytosine nucleotides located within CG dinucleotides, termed CpG sites. These CpG sites are distributed throughout the genome in genic and intergenic regions. Genic regions include the promoter and gene body, whereas intergenic regions include distal regulatory elements as well as repetitive elements [8,9]. Promoter methylation is generally repressive and the extent of promoter methylation is inversely related to the CpG density of the promoter. This observation prompted the classification of promoters into two groups based on CpG density: high CpG density (HCG) and low CpG density (LCG) promoters [10]. HCG promoters make up [s1]72% of promoters in the human genome. These promoters contain clusters of CpG sites termed CpG islands, which are largely devoid of methylation, resulting in overall hypomethylation of HCG promoters. These promoters are associated with genes expressed by cells from all or most tissues (housekeeping genes) [10]. By contrast, LCG promoters do not contain CpG islands and are associated with lineage or tissue-specific gene regulation [10–12]. Furthermore, there is evidence to suggest that methylation of gene bodies could provide fine-tuning of promoter methylation patterns, and the combination of promoter hypomethylation and gene body methylation is positively correlated to gene expression [13]. Methylation of gene bodies, defined as the region of the gene beyond the first exon [14], is associated with a higher level of gene expression in dividing cells [8]. Furthermore, CpG sites are more common in exons than introns [10], and recent studies suggest that gene body methylation might play a part in pre-mRNA splicing [15,16]. CpG methylation in intergenic regions affects distal regulatory elements, such as enhancers [9] and repetitive elements.
Although the exact mechanisms translating the methylation code into gene repression or expression are unknown, two mechanisms have been proposed and well-studied: (i) promoter cytosine methylation prohibits the binding of transcription factors, thus inhibiting transcription of the downstream gene; (ii) cytosine methylation is recognized by DNA methyl-binding proteins (MBPs) that orchestrate chromatin remodeling necessary for gene activation or inhibition. The latter mechanism depends upon the detection of 5-methylcytosine (5mC) by DNA MBPs, of which there are three known families, reviewed in [7]. The methyl-binding domain (MBD) family of MBPs comprises MeCP2 and MBD1–6. Only MBDs1, 2 and 4 have been shown to bind methylated DNA. The set and ring finger associated (SRA) domain family includes ubiquitin PHD[s2] ring finger (UHRF)1 and 2. Of these, UHRF1 is notable for its essential role in DNA methyltransferase (DNMT)1-mediated maintenance of DNA methylation. Finally, zinc finger family proteins Kaiso and zinc finger protein (ZFP)57 are MBPs responsible for maintenance of methylation of imprinted genes [7].
DNA methylation: reprogramming phases
In mammalian development, there are two phases of DNA methylation erasure and reestablishment or reprogramming. The first episode of DNA methylation erasure occurs in the blastocyst following fertilization [17]. The second episode of DNA methylation occurs in primordial germ cells (PGCs) traveling to the gonadal ridge for development into gametocytes [18]. These DNA methylation reprogramming phases are thought to prime cells for lineage specification and differentiation processes, as well as prohibit the transgenerational propagation of disadvantageous epimutations [19]. Certain regions of the genome retain their methylation patterns and escape demethylation during these phases. Imprinting control regions (ICRs) escape DNA methylation erasure during the blastocyst stage, but they are fully reprogrammed during the PGC reprogramming episode. By contrast, transposable elements including intracisternal A-particles (IAPs) escape methylation erasure during both reprogramming phases, highlighting the importance of repression of these elements [17,19]. These crucial phases of DNA methylation reprogramming are dependent on cellular machinery responsible for deletion, reintroduction and maintenance of these methylation marks.
DNA methylation: enzymatic regulation
DNMTs are a family of enzymes responsible for maintaining and/or introducing DNA methylation marks. The DNMT family comprises three main members: DNMT1, DNMT3A and DNMT3B. A fourth member, DNMT3L, lacks catalytic activity and is thought to regulate the activities of DNMT3A and DNMT3B. DNMT1, together with the DNA MBP UHRF1, maintains DNA methylation during replication. DNMT3A and DNMT3B are responsible for de novo DNA methylation during gamete development and during blastocyst implantation, respectively. In addition, these enzymes have been implicated in maintenance of DNA methylation [19,20].
DNA demethylation can occur by passive or active means. Passive DNA demethylation occurs via exclusion of DNMT1 and UHRF1 from the nucleus, resulting in replication-dependent loss of methylation marks. Although no enzyme is yet known that directly converts 5mC to cytosine via active DNA demethylation, indirect demethylation mechanisms have been described, reviewed in [19]. The ten-eleven-translocation (TET) family members TET1–3 oxidize 5mC to 5-hydroxymethylcytosine (5hmC). These TET proteins are also capable of oxidizing 5hmC to 5-formylcytosine (5fC) or 5-carboxylcytosine (5caC). These oxidation products are then diluted by passive demethylation, because they are not detected by DNMT1. Alternatively, they can be actively removed by DNA glycosylases and base excision repair machinery. Other mechanisms of active demethylation have been described, including activation-induced cytidine deaminase (AID) or apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC)-mediated conversion to either 5-hydroxymethyluridine or thymidine, followed by base excision repair [19].
Histone modifications
Histone modification: type and function
Histone proteins (H1, H2A, H2B, H3 and H4) are major organizational and regulatory units of chromatin. DNA is wrapped around histone octamers, composed of two each of histones H2A, H2B, H3 and H4, to form nucleosome cores. Histone H1 facilitates internucleosomal organization, resulting in higher order chromatin packaging [21]. Although histones were initially believed to have only structural and/or mechanical roles in chromatin packaging, it is now known that post-translational modifications to these proteins are a crucial mechanism of gene regulation [22]. These modifications are usually made on amino-terminal histone tails, but they have been reported to occur on the core of the protein as well. Modifications include acetylation [s3]methylation, phosphorylation, sumoylation, ubiquitination, biotinylation and ADP-ribosylation [22,23]. Modification of a histone protein can alter nucleosome structure by: (i) changing the electrostatic charge of the protein and thus its interaction with the DNA; (ii) recruiting ‘reader’ proteins capable of remodeling chromatin; or (iii) directly modulating the organization of higher-order chromatin [24]. Similar to DNA methylation, the location of histone marks greatly influences their regulatory function. In the case of histones, however, the function of the epigenetic mark varies by location on the histone itself, as well as by location relative to the underlying gene structure (promoter, gene body, etc.). In addition, studies suggest that the function of histone modifications is also influenced by the number of marks placed at a particular location (e.g. methylation marks) [24]. In this review, we will focus on two major types of histone modification: acetylation and methylation, and the enzymes that regulate their placement and removal.
Histone modification: enzymatic regulation
Histone acetylation occurs most commonly on lysine residues, and results in neutralization of the positive charge and decreasing the histone affinity for the negatively charged DNA molecule. This process is carried out by a family of enzymes called histone acetyltransferases (HATs), of which there are two main subtypes: type A and type B. Five families are known to belong to the type A HATs: GCN5-related acetyltransferases (GNAT); MOZ, YBF2/SAS3, SAS2 and TIP60 (MYST); CBP/p300; transcription factor III C (TFIIIC) and TAF1; [s4]and nuclear receptor cofactor (NRCF) enzymes. Type A HATs modify histones actively incorporated into nucleosomes, whereas type B HATs typically modify cytosolic histones before their incorporation into the chromatin structure. These HATs include HAT1, HAT2, Rtt109, HatB3.1 and HAT4 [25]. Removal of histone acetylation is accomplished by histone deacetylases (HDACs), which reestablish the positive lysine charge, enhancing histone–DNA binding [26]. HDACs can be classified into three main groups based on sequence similarities, expression, location and enzymatic activity [4]. Class I consists of HDACs 1–3 and 8; class IIa includes HDACs 4, 5, 7 and 9; and class IIb includes HDACs 6 and 10. Classes III and IV also exist for HDACs, with class III being composed of the sirtuin family and class IV including only one HDAC: HDAC 11 [27].
Histone methylation most often occurs at lysine and arginine residues. Because methylation does not alter histone charge, its function is associated with the number of methyl groups placed on a particular histone site [26]. Histone methylation is catalyzed by histone methyltransferases (HMTs) and reversed by histone demethylases, including members of the Jumonji family as well as lysine-specific demethylase (LSD)-1. These enzymes vary in their capacities to place or remove mono-, di- or tri-methyl groups [26].
miRs
miRs are single-stranded noncoding RNAs of approximately 19–25 nucleotides in length. They are capable of complementary base pairing with mRNA sequences, which they repress or target for degradation, depending on the degree of complementarity. Thus, they are classified as a post-transcriptional type of epigenetic regulatory mechanism [28].
miRs: biogenesis
The biogenesis of miRs begins with RNA-polymerase-II-mediated transcription of DNA, resulting in a long (>1 kb) primary transcript, or pri-miR [28]. This process can be guided by miR-specific promoters (intergenic miRs) or by promoters for protein-coding genes (intragenic miRs) [29,30]. The pri-miR folds into a stem loop structure and is then cleaved at the base of the stem loop by the endonuclease Drosha/DGRC8 into a 60–90 nucleotide stem loop intermediate, called the miR precursor, or pre-miR This cleavage reaction results in a 2–3 nucleotide overhang ending in a 5′ phosphate, which is detected by the cytoplasmic endonuclease Dicer following Ran-GTP- and exportin-5-mediated pre-miR export from the nucleus. In the cytoplasm, Dicer proceeds to cleave both strands of the duplex approximately two helical turns away from the base of the stem loop, effectively removing the terminal base pairs and loop of the pre-miR. This process also leaves a 5′ phosphate end and 2–3 nucleotide overhang. The duplex, approximately 20 base pairs long, includes the mature miR and its complement, the miR* sequence. This duplex is then loaded on [s5]the RNA-induced silencing complex (RISC), the helicase and endonuclease components of which un-zipper the duplex and degrade the miR* strand [28].
miRs: mechanism of action
Once properly loaded into the RISC, the miR is responsible for guiding the complex to target mRNA. This is accomplished by the nucleotides at positions 2–8 in the miR, termed the seed region, which interact with the 3′ untranslated region (UTR) of target mRNA [31,32]. The degree of complementarity of the miR to the target mRNA determines the final outcome of the mRNA: degradation or translational inhibition. Perfect complementarity with the target mRNA results in cleavage and degradation, whereas partial complementarity results in translational repression. Notably, the seed region of the miR will always have perfect complementarity with the target mRNA, regardless of outcome based on the remaining miR sequence. Argonaute (Ago)2, a member of the RISC, has important roles in this process. Ago2 is responsible for correct positioning of the miR within the RISC, as well as cleaving the target mRNA and/or recruiting proteins involved in translational repression according to the outcome dictated by the miR [33].
Epigenetics and heart development
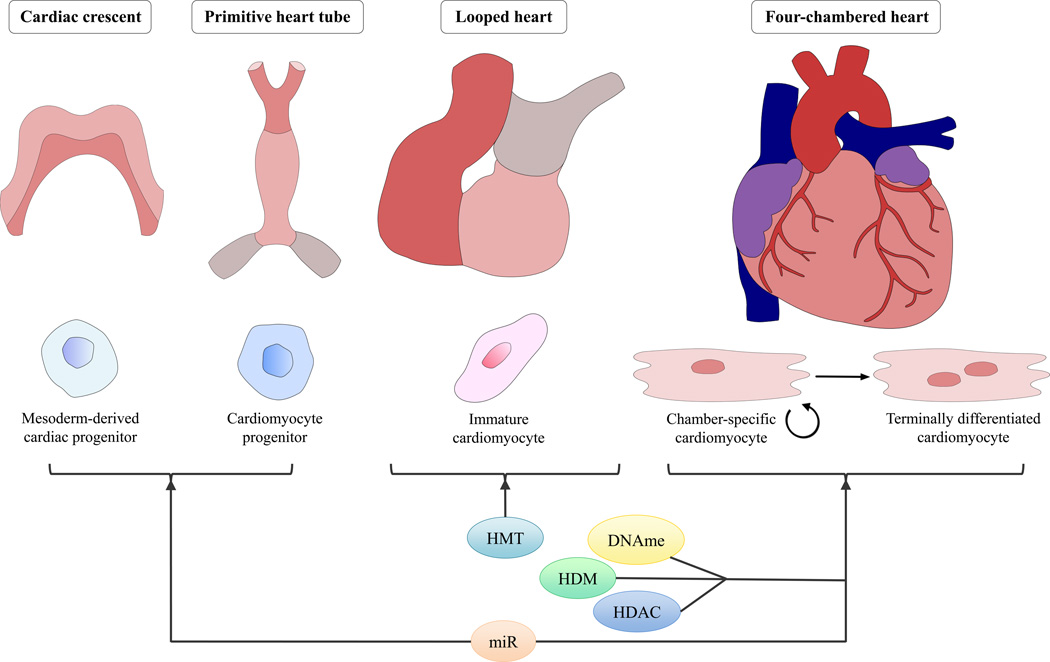
The development of the heart is a complex process involving coordinated cellular proliferation, migration, differentiation and programmed cell death as well as structural remodeling including looping and septation (Figure 1). In rodents, this process begins with the migration of lateral plate mesoderm-derived cardiomyocyte progenitors to the cardiac region on either side of the midline at embryonic day 6.5 (E6.5). The first wave of cardiomyocytes to populate this region is termed the primary heart field; a later group of cardiomyocytes arriving to the region is termed the second heart field. The primary heart field, in addition to its contributions to the left ventricle (LV) and parts of the atria, is thought to provide a scaffold for the development of the second-heart-field-derived right ventricle, outflow tract and significant portions of the atria. Primary heart field cells migrate across the midline at E7.5 and fuse to form the cardiac crescent. This is followed by fusion of the arms of the cardiac crescent on E8.0, forming the primitive heart tube, which is the first functional organ to develop in the embryo. In response to endocardial and epicardial signals, cardiomyocytes proliferate and cause uneven growth and remodeling in the primitive heart tube. Subsequently, rightward looping results in the proper relative orientation of the ventricles, atria and future outflow tract. Cardiac neural crest cells orchestrate the outflow tract septation and patterning of aortic arch arteries. Septation of the ventricles and valve formation form the four-chambered heart by E15 [34,35]. Spatio-temporal gene regulation is paramount to the success of this developmental process. Even subtle alterations in this process can have detrimental effects on the future cardiovascular health of the individual. For example, early terminal differentiation of cardiomyocytes, resulting in nondividing (or very slowly proliferating) cardiomyocytes, reduces total heart cardiomyocyte cell number in adulthood and increases vulnerability to ischemic injury [36]. Proper gene regulation during heart development is accomplished by several key transcription factors, including GATA4, TBX5 and Nkx2.5 [37]. Of importance, epigenetic mechanisms have been implicated in the direct regulation of gene expression patterns as well as indirectly by modulating transcription factor expression and function during heart development [37].
Figure 1. The roles of major epigenetic mechanisms in key stages of heart development.
Cardiac progenitors derived from lateral plate mesoderm migrate to the cardiogenic region to form the cardiac crescent and later the primitive heart tube. Cardiac looping ensures proper orientation of the future atria, ventricles and outflow tract. Finally, development of the conduction and valvular systems, as well as septation of atria, ventricles and outflow tract, results in the four-chambered heart. Cardiomyocyte terminal differentiation, which occurs well after the formation of the four-chambered heart, determines final cardiac contractile endowment. Epigenetic regulatory mechanisms have been described in all stages of heart development. miRs have been shown to regulate cellular differentiation processes in early heart development (miR-1 and miR-499) as well as cardiomyocyte proliferation and terminal differentiation (miR-133). Histone modifications regulate right ventricle development (SMYD1) as well as cardiac septation (Jumonji and LSD-1) and cardiomyocyte function and proliferation (HDACs 1–3). DNA methylation also contributes to outflow tract septation[s14] and cardiomyocyte proliferation and terminal differentiation. Abbreviations: HMT, histone methyltransferases; DNAme, DNA methylation; HDM, histone demethylases; HDAC, histone deacetylases; LSD-1, lysine-specific demethylase; miR, micro-RNA.
DNA methylation and heart development
The importance of DNA methylation in heart development was first suggested by studies involving mice lacking key enzymes regulating DNA methylation and demethylation. These studies demonstrated that the absence of DNMT1 led to global hypomethylation and early embryonic death [38–41]. DNMT3A–null mice survive until birth but die around 4 weeks of age, whereas DNMT3B–null mice die during gestation [42]. These observations are consistent with the studies showing that DNMT3A is abundant in differentiated cells, whereas DNMT3B is present at high levels in early embryos [20,43]. Tet1-null mice are viable [44], although they have reduced body size at birth and are subfertile [45]. Absence of Tet2 has no effects on embryogenesis; however, it has profound effects on hematopoiesis [46–49]. These mice eventually succumb to myeloid leukemia at 4–6 months of age [20]. Inactivation of zygotic Tet3 results in defective DNA methylation erasure and embryonic developmental failure [50]. Together, these results indicate that balance between DNA methylation and demethylation is imperative for proper mammalian development.
DNA methylation patterns and dynamics have been described throughout various developmental processes, including gametogenesis, hematopoiesis and stem cell differentiation, as well as various disease processes [6]. However, the role of DNA methylation in regulating heart development is relatively understudied. Most of what is known about its role in heart development is extrapolated from organism-wide development or from embryonic stem cell (ESC) studies. Although general rules of DNA methylation apply to heart tissues [51], recent studies suggest that other predictions could be limited to certain cardiac cell subsets or spatio-temporal contexts. For instance, it is known that genomic methylation status decreases during cellular development and differentiation [12,13,52,53], with exceptions including pluripotency-associated gene promoters such as those of Oct4 and Nanog [6]. Furthermore, inhibition of DNA methylation in ESCs using 5-azacytidine (5-azaC) has been shown to induce cardiomyocyte differentiation [54]. However, recent studies of murine embryonic heart development have shown that heart development is associated with increases and decreases (greater increases than decreases) in DNA methylation, particularly of cardiac-specific genes [55]. In addition, perturbations in normal developmental conditions, such as hypoxia, have been shown to induce premature terminal differentiation of fetal rat cardiomyocytes by increasing DNA methylation, an effect abolished by administration of 5-azaC [56]. These findings are consistent with studies of postnatal cardiac development in rats, which demonstrate increases in DNA methylation in cardiomyocytes, as well as increased DNMT1, MBD1–3 and MeCP2, over the weeks and months following birth. Furthermore, these studies report an increase in DNA synthesis in cardiomyocytes treated with 5-azaC, corresponding to an earlier stage of development, as confirmed by increased atrial natriuretic peptide (ANP) and β-myosin heavy chain (β-MHC) and reduced α-MHC expression [57]. Recent evidence has also implicated DNA methylation in outflow tract septation through promoter regulation of the VANGL2 gene. VANGL2 is essential for the polarization of cells at the outflow tract, because homozygous mutation of the gene in mice results in a phenotype resembling Tetralogy of Fallot (overriding aorta, double-outlet right ventricle and ventricular septal defects). Indeed, VANGL2 promoter methylation has been found to be elevated in patients with Tetralogy of Fallot compared with healthy controls [58]. The role of DNA methylation in heart development probably varies by genomic location within cells, by cell type within the developing heart, as well as across various stages of development. Further work is needed to elucidate the role of DNA methylation in the regulation of heart development.
Histone modifications and heart development
Recent evidence supports a prominent role for epigenetic histone modifications in normal and aberrant development. Histone acetyltransferase p300 knockout mice exhibit prominent cardiovascular defects and embryonic lethality [59,60]. It has been hypothesized that this could be caused by a loss of transcriptional coactivation of GATA5, an essential transcription factor in heart development [4]. Histone deacetylation has also been studied in heart development. HDAC1 and 2 appear to have redundant roles in cardiac development, because only double knockout mice display cardiac defects. Mice lacking HDAC1 and 2 die early in neonatal life from arrhythmias and dilated cardiomyopathy [61]. Studies have shown that these outcomes are due in part to the loss of inhibition of fetal calcium channel signaling through If and T-type calcium channels, as well as contractile proteins such as α-skeletal actin [61]. HDAC3 has also been linked to heart development, particularly to cardiomyocyte metabolism and proliferation. Loss of HDAC3 results in impairment of glucose metabolism and skewing toward increased lipid metabolism [62]. Overexpression of HDAC3 promotes cardiomyocyte proliferation via reduction of cell cycle inhibitors. Similar to HDAC1 and 2, HDAC5 and 9 are functionally redundant and do not cause notable cardiac defects when deleted individually. However, combined loss of HDAC5 and 9 results in ventricular septal defects and embryonic or neonatal lethality [63]. In addition to histone acetylation and deacetylation, histone methylation status also influences cardiac development. SMYD1, a histone methyltransferase, has been implicated in heart development, because mice lacking SMYD1[s6] exhibit hypoplastic right ventricles and embryonic lethality [64]. Interestingly, Hand2 and Irx4 transcription factors are also reduced in these mice, suggesting that SMYD1-mediated histone methylation is necessary for the expression of these essential cardiac transcription factors [64,65]. Members of the histone demethylase Jumonji family have been implicated in septation, because their deletion results in ventricular septal defects and outflow tract defects [66–68]. In addition, other lysine demethylases such as LSD-1 have also been shown to cause ventricular septal defects [69,70]. Together, these findings illustrate the pervasive roles of histone modifications in heart development.
miR-mediated regulation of cardiomyogenesis
Cardiac development is intricate, involving the differential expression of multiple factors including miRs. This differential regulation, along with spatio-temporal distribution of these miRs, provides insight into their importance during development. In zebrafish, miR expression has been shown to increase during the development in a tissue-specific manner with most expression occurring after organogenesis [71]. miRs are generally conserved sequences directly involved in regulating target mRNA expression. The conservation further highlights the importance in development. In the heart and skeletal muscle, miR-1 and miR-133 expression increases during the development from the embryonic to neonatal stage, albeit to levels still significantly lower than adult tissues [72,73]. In Drosophila, mutations resulting in the loss of miR-1 are lethal and are observed to result in severe defects in muscle genes such as MHC. miR-1 is associated with differentiation, indicated by expression of myogenic markers including myogenin and MHC [72]. The differentiation induced by miR-1 is also associated with a loss of proliferation and a reduction in protein synthesis [74]. Insertion of miR-1 at the single-cell stage results in tissue disorganization, decreased proliferation and the complete absence of cardiac tissue [72]. A number of targets of miR-1 have been identified that are essential to cardiac development including HDAC4, an inhibitor of skeletal muscle gene expression [72], Hand2 [75], GJA1 [76] and KCNJ2 [76]. miR-1 and miR-133 originate from a common bicistronic transcript. miR-133 is also an important regulator of gene expression, with an opposing effect to miR-1, noted to stimulate proliferation and inhibit the expression of myogenin and MHC [72]. Unlike mutations in miR-1, loss of miR-133 does not result in lethality of all effected animals, with about 24% surviving into adulthood [77]. However, the surviving adults display signs of fibrosis, cardiac contractility abnormalities, cardiomyopathy and aberrant proliferation [77]. Cyclin D2 and serum response factor (SRF) have been identified as targets of miR-133 [72]. Knockdown of miR-133 results in increased expression of cyclin D2, providing a mechanism by which miR-133 suppresses proliferation [77]. The knockdown of miR-133 is also associated with the re-induction of fetal genes, further supporting its role as a regulator of maturation and development [74]. Another miR involved in differentiation is miR-499, a cardiac-specific miR that can induce differentiation when in culture with human cardiomyocyte progenitor cells (hCMPCs) [78]. SOX6, a regulator of cell fate during development, is a target of miR-499 resulting in inhibition of its function. Other heart-specific miRs include miR-206 and miR-208. An intriguing observation in animals with loss of miR-208 is the failure to induce cardiac remodeling in response to stress, thus demonstrating the necessity of miR-208 for fibrosis and hypertrophic growth [79]. Together, these findings illustrate the essential roles that miRs have in heart development.
Epigenetics mechanisms in cardiovascular disease
Heart failure (HF) is the final outcome of a variety of cardiovascular pathologies, including hypertension, valvular diseases, cardiomyopathy, arrhythmias, atherosclerosis/coronary artery disease and myocardial infarction [80]. Many of these cardiac stressors trigger similar pathophysiologic events such as cardiomyocyte loss, hypertrophy of remaining cardiomyocytes and proliferation and activation of cardiac fibroblasts resulting in increased extracellular matrix deposition (fibrosis). This process is collectively referred to as cardiac remodeling. This remodeling also alters normal cardiac conduction circuitry, resulting in an increased propensity for arrhythmias. At the molecular level, the development of HF is characterized by changes in gene expression including the reduction of normal adult cardiac muscle proteins such as α-MHC and sarcoplasmic reticulum Ca2+-ATPase (SRCA) and concomitant reactivation of the ‘fetal gene program’ consisting of genes such as atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP) and β-MHC [81]. HF occurs when the heart is incapable of maintaining adequate blood flow to the body owing to impairment of the physico-mechanical pump action of the heart [82].
The overlapping mechanisms of cardiac demise have been proposed to participate in a ‘unifying pathway’ [83], culminating in altered gene expression profiles characteristic of end-stage HF, regardless of the primary contributing pathology. However, recent studies suggest that certain gene regulatory mechanisms, such as epigenetic mechanisms, have distinct activities depending on the specific pathology, despite their common culmination in HF (Figure 2). In other words, whereas pathologies contributing to HF share a variety of cellular processes, epigenetic mechanisms represent disease-specific targets for therapeutic approaches. Here, we address the roles of DNA methylation, histone modifications and miR-mediated gene regulation in key pathologies contributing to HF.
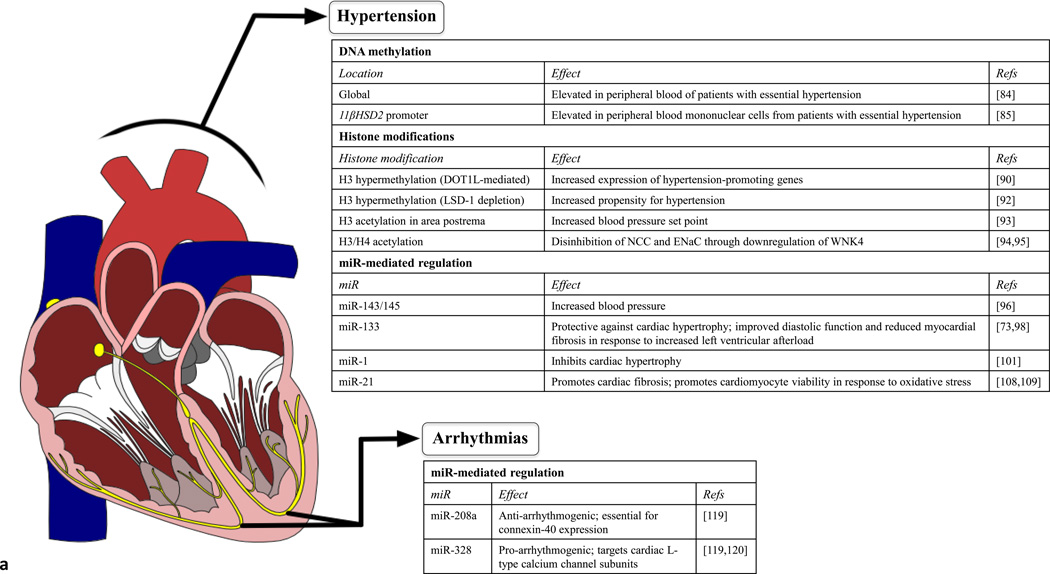
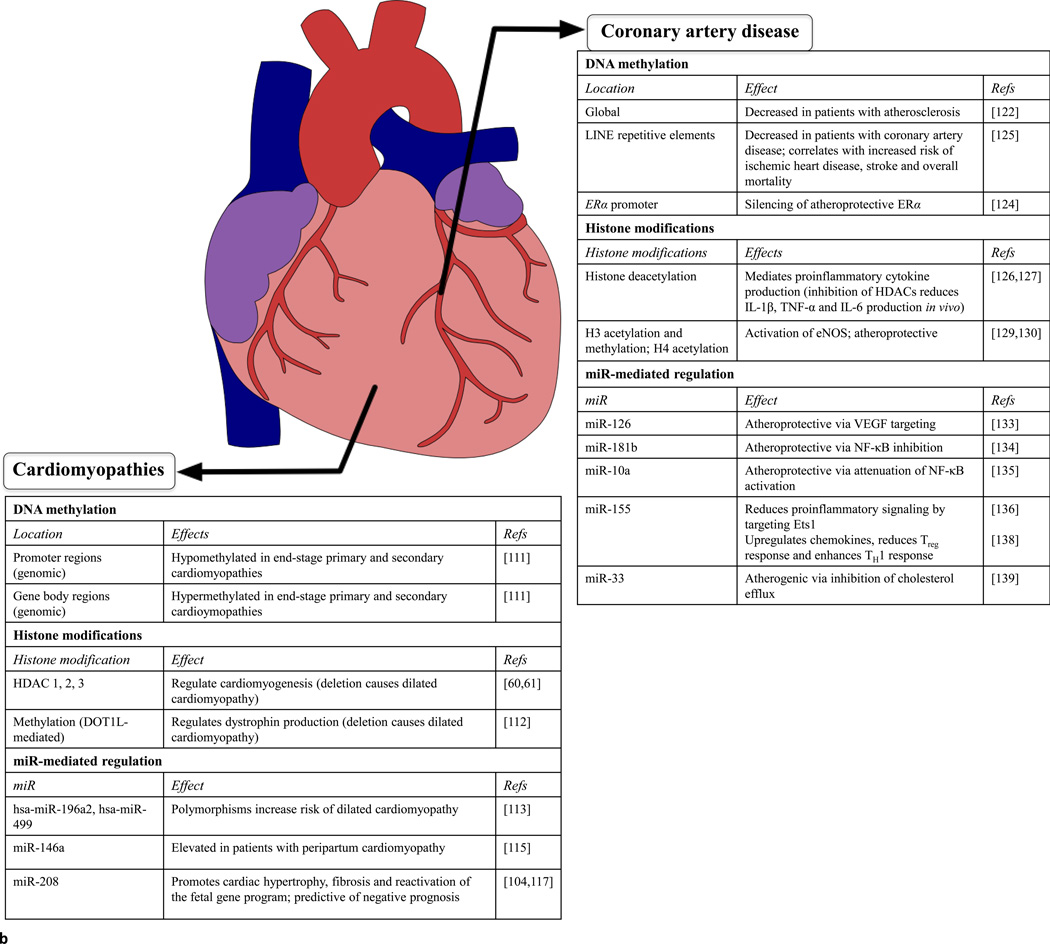
Figure 2. Epigenetic mechanisms in cardiovascular diseases contributing to heart failure.
Regardless of anatomical location, epigenetic mechanisms are emerging as crucial orchestrators of cardiovascular diseases, including previously idiopathic diseases such as essential hypertension or certain cardiomyopathies. (a) Epigenetic mechanisms associated with hypertension and arrhythmias. Differential DNA methylation or expression of miRs detectable in the peripheral blood of patients suffering from left ventricular afterload or conduction disorders can aid in the early detection and diagnosis of disease. In addition, identifying epigenetic mechanisms that contribute to hypertension or arrhythmias (such as methylation of histone H3 or miR-328) or have protective effects (miR-133, miR-1, miR-208a) could aid in the development of therapies for these diseases. (b) Epigenetic mechanisms associated with coronary artery disease and cardiomyopathies. Epigenetic mechanisms can have prognostic value (such as methylation of LINE repetitive elements) or potentially protective effects (H3 acetylation/methylation or H4 acetylation) in coronary artery disease. In addition, DNA methylation and miR expression can serve as diagnostic and prognostic tools for cardiomyopathies.
Hypertension
LV afterload is the pressure that the LV must overcome to eject blood into the aorta. Increased LV afterload can be caused by hemodynamic alterations such as high blood pressure (hypertension) or mechanical obstruction such as aortic stenosis. Under such conditions of chronically increased LV afterload, the heart undergoes remodeling processes, such as dilation and hypertrophy, to ensure adequate blood circulation. Eventually, however, increased cardiomyocyte loss and reduced contractility compromise the function of the organ, leading to HF. Because approximately one-third of the adults in the USA have hypertension [84], the detrimental impact of this disease will become increasingly apparent in the coming years, emphasizing the need for better preventive measures and therapeutic modalities to slow the progression of this disease to HF.
DNA methylation
Patients with essential hypertension were found to have lower levels of global 5mC in peripheral blood cells than healthy control individuals. Furthermore, the levels of 5mC correlated to the stage of hypertension [85]. Interestingly, promoter methylation of 11βHSD2, the gene encoding 11 β-hydroxysteroid dehydrogenase 2 (11βHSD2), has been shown to be elevated in the peripheral blood mononuclear cells of patients with essential hypertension [86]. 11βHSD2 is responsible for the breakdown of cortisol to cortisone that is biologically inactive. Reduced activity of 11βHSD2 leads to cortisol-mediated activation of mineralocorticoid receptors and increased blood pressure [87]. In addition, other key mediators of blood pressure regulation, such as somatic angiogensin-converting enzyme (sACE), are influenced by DNA methylation in human cell lines [88,89]. Taken together, these studies suggest that aberrant DNA methylation could play a causative part in essential hypertension in a gene-specific manner, and that an overall reduction in global methylation is indicative of disease progression.
Histone modifications
A variety of histone modifications have been implicated in the pathophysiology of hypertension. Methylation or acetylation changes are observed in over half of the genes differentially expressed after cardiac pressure overload [90]. Hypermethylation of H3 by disruptor of telomeric silencing-1-like (DOT1L), a methyltransferase, increases expression of hypertension-promoting telomeric genes [91]. However, DOT1L also plays an antihypertensive part by inhibiting amiloride-sensitive renal epithelial sodium channel (ENaC) expression. Aldosterone is capable of inhibiting DOT1L, thus increasing ENaC expression and raising blood pressure [92]. Hypermethylation of H3 caused by a lack of LSD-1 has also been associated with increased propensity for hypertension in animal models [93]. Histone acetylation status also has an impact on blood pressure regulation. For example, H3 acetylation in the area postrema causes upregulation of sympathetic outflow from the rostral ventrolateral medulla and raises blood pressure set point [94]. Additionally, inhibition of HDAC8 via β2-adrenergic receptor stimulation has been shown to increase H3 and H4 acetylation, resulting in glucocorticoid-mediated suppression of the WNK4 gene. WNK4 is a serine/threonine kinase that downregulates expression of sodium chloride transporters (NCC) and ENaCs [95,96].
miR-mediated regulation
miRs are involved in the development of hypertension as well as the cardiac remodeling that occurs as the heart adapts to overcome the high pressure in the vasculature. Certain miRs influence vascular tone, contributing to increased afterload, whereas other miRs promote pathological cardiac remodeling in response to the increased blood pressure. Other miRs contribute to vascular dysregulation as well as cardiac responses to this increased pressure.
In the vasculature, miR-143 and miR-145 play important parts in regulating tone by maintaining the normal phenotype and function of vascular smooth muscle cells (VSMCs) [97]. Under pathological circumstances, VSMCs undergo a phenotypic change from contractile to synthetic/proliferating cells, an event underlying hypertension and atherosclerosis. However, although miR-143 and miR-145 appear to be important mediators of vascular tone, their role in chronic vascular stress such as hypertension is less clear. Deletion or inhibition of miR-143 and miR-145 results in reduced blood pressure, although the mechanism of this hypotensive effect is not understood. Indeed, it would seem that elimination of miR-143 and miR-145 would promote VSMC differentiation to the secretory phenotype, with a concomitant increased risk of atherosclerosis and coronary artery disease [97]. This effect could be counteracted by the differential effects of these miRs on other cell types, such as endothelial cells, within the vasculature. Thus, more work is needed to elucidate how miR-143 and miR-145 contribute to maintenance of blood pressure and to what extent this mechanism can be targeted to treat hypertension. miR-133 is the most highly expressed miR in human cardiac tissue [98]. In the context of heart disease, miR-133 is thought to counteract cardiac hypertrophy, because in vivo antagomir-mediated miR-133 inhibition induces hypertrophy [74]. Approximately half of miR-133a-1 and miR-133a-2 knockout mice die during early development owing to ventricular–septal defects, and surviving double knockout mice eventually develop dilated cardiomyopathy and HF [77]. Interestingly, a study of acute increased LV afterload caused by transverse aortic constriction found that overexpression of miR-133a in vivo improved myocardial fibrosis and diastolic function, but had no effects on hypertrophy or mRNA expression profiles [99]. Thus, the mechanism of miR-133a in cardioprotection under conditions of increased LV afterload remains unclear. The second most abundant miR in the human heart is miR-1, which is transcribed from bicistronic units along with the other members of the miR-133 family (miR-133a, miR-133b and miR-206) [100]. miR-1 has an inhibitory effect on cardiac hypertrophy that is mediated by targeting of the calcineurin/NFAT [101] and insulin-like growth factor (IGF)-1 signaling pathways [102].
Other miRs can contribute to vascular contributions to hypertension as well as cardiac remodeling in response to hypertension. For example, miR-21 is upregulated in vascular neointimal lesions, post-myocardial infarction tissues and hypertrophic hearts [103–108]. Studies have shown that miR-21 has an important role in regulating vascular neointimal lesion formation by promoting proliferation and reducing apoptosis of VSMCs [109]. In addition, miR-21 promotes cardiac remodeling by influencing the extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) pathway in cardiac fibroblasts [108], although the contribution of this pathway is not essential because inhibition or deletion of miR-21 does not completely attenuate stress-dependent cardiac remodeling [110]. Interestingly, miR-21 appears to promote the viability of cardiomyocytes by regulating programmed cell death protein 4 (PDCD4) [109]. These cell-type differential effects could help explain the seeming inconsistencies relating to miR-21 function in cardiac remodeling: while miR-21 promotes fibroblast-mediated remodeling of the heart in response to cardiac stress and presumed increased cell death it also prevents cardiomyocyte cell death, thus altering the dynamics of the remodeling process.
Cardiomyopathies
Cardiomyopathies, or disorders of the myocardial component of the heart, can be either primary (idiopathic) or secondary to ischemia or myocardial infarction. In addition, they can be described on the basis of structural remodeling that occurs as part of the progression to HF, including ventricular dilation and hypertrophy.
DNA methylation
Two studies have recently described DNA methylation patterns in hearts from patients undergoing cardiac transplantation for primary or secondary cardiomyopathies [111,112]. Genomic profiling of DNA methylation in end-stage failing hearts from patients with primary or secondary cardiomyopathies revealed significant hypomethylation at promoter regions and hypermethylation at gene body regions compared with control hearts. Notably, no differences were found in intergenic regions, including enhancer regions [112]. In a second study, Haas et al. identified differential methylation in pathways related to cardiac disease, as well as differential methylation of several key genes, in hearts from patients with primary cardiomyopathy compared with hearts from control individuals [111].
Histone modifications
Histone acetylation and methylation have also been investigated in the context of cardiomyopathies. In particular, studies on the effects of deletion of histone-modifying enzymes have yielded insight into the roles of these epigenetic marks in preventing cardiomyopathies. For instance, deletion of HDAC1, 2 or [s7]3 each resulted in dilated cardiomyopathy [61,62]. Similarly, deletion of histone methyltransferase DOT1L causes dilated cardiomyopathy and systolic dysfunction in humans through reduced dystrophin transcription. Furthermore, restoration of dystrophin transcription via adenovirus-mediated expression alleviates the effects of DOT1L deletion [113].
miR-mediated regulation
A study of genetic polymorphisms in pre-miRs (hsa-miR-196a2, hsa-miR-499 and hsa-miR-146a) found that polymorphisms in hsa-miR-196a2 and hsa-miR-499 were associated with an increased risk of dilated cardiomyopathy [114]. Studies by Matkovich et al. later reported that miR-499 expression is elevated in human and murine cardiac hypertrophy and cardiomyopathy, and that induction of miR-499 expression results in HF and promotes cardiac remodeling in a pressure-overloaded animal model [115]. miR-146a has recently been linked to peripartum cardiomyopathy, acting as an essential mediator of 16 kDa N-terminal prolactin fragment (16KPRL)-driven suppression of NRAS signaling in endothelial cells. In addition, miR-146a can be delivered to cardiomyocyte cells through endothelial-cell-derived exosomes, resulting in impaired metabolism of the recipient cardiomyocyte cell. Interestingly, this study found that miR-146a is elevated in the plasma of patients with peripartum cardiomyopathy, but not patients with dilated cardiomyopathy [116]. Studies from patients with dilated cardiomyopathy have revealed that miRs can be used as clinical diagnostic and prognostic indicators [117]. It has been demonstrated that miR-208 is a predictor of progression to HF and cardiac death [118]. At the cellular level, miR-208 is thought to regulate the expression of β[s8]-MHC, raising the possibility that miR-208 might promote the reactivation of the fetal cardiac gene profile [105,118]. Overexpression of miR-208 has been shown to induce cardiac hypertrophy and fibrosis in response to pressure-overload stress [105,118], suggesting it might have multidimensional roles in the progression of cardiomyopathy to HF.
Arrhythmia
Because the conduction of the heart depends on the propagation of the electrical signal through and between cells, proper structural integrity and ion channel function are essential. Thus, arrhythmias can be caused by disruption of the cardiac conduction system by structural changes (fibrosis) or ion channel dysfunction [119].
In addition to its roles in the development and potential utility in diagnosis of dilated cardiomyopathy, miR-208 can also influence vulnerability to cardiac arrhythmias. miR-208a is required for connexin-40 expression, because miR-208a knockout mice expressed significantly reduced levels of the gap junction protein, resulting in abnormal electrical activation of atrial contraction. Indeed, electrocardiograms from miR-208a–null mice lack P waves preceding QRS complexes, indicating that these mice experience atrial fibrillation [120]. Reports that miR-328 targets cardiac L-type calcium channel subunits α-1c and β-1 also implicated miR-328 in contributing to vulnerability to atrial fibrillation. This was confirmed by studies that overexpressed miR-328 in dogs and mice, inducing atrial fibrillation, and by antagomir-mediated reversal of this atrial fibrillation vulnerability [120,121]. See [119] for an extensive review of other miRs (including miR-1, miR-133 and miR-212) contributions to cardiac arrhythmias.
Atherosclerosis and coronary artery disease
Atherosclerosis, or vascular inflammation, is a chronic disease process that occurs when vascular endothelial cell homeostasis is disrupted (termed endothelial dysfunction) and lipids accumulate within blood vessels, leading to immune-cell-mediated fatty plaque formation [122]. VSMCs also participate in the process of plaque formation by undergoing phenotypic modulation from contractile cells to secretory/proliferative cells responsible for production of extracellular matrix and proinflammatory cytokines. These plaques can occur anywhere in the systemic vasculature and are the primary cause of coronary artery disease (CAD). CAD, in turn, is a major contributor to myocardial ischemia and HF.
DNA methylation
Global DNA methylation is reduced in peripheral blood cells of patients with atherosclerosis [123]. It is also reduced in VSMCs from atherosclerotic lesions and during VSMC phenotypic modulation [124]. However, increased DNA methylation of the promoter region of the atheroprotective estrogen receptor (ER)-α gene, resulting in ER-α silencing, has been observed in VSMC phenotypic modulation [125]. Thus, whereas global DNA methylation is often indicative of disease state, differential DNA methylation patterns occur at gene-specific regions. Regarding CAD, a study by Baccarelli et al. in 2010 reported reduced DNA methylation of long interspersed nucleotide element-1 (LINE-1) repetitive elements in peripheral blood cells of patients with ischemic heart disease. Furthermore, lower LINE-1 methylation levels correlated with increased risk for ischemic heart disease, stroke and overall mortality [126].
Histone modifications
Growing evidence suggests that histone acetylation is correlated to vascular inflammation. Multiple studies have shown that HDAC inhibitors reduce proinflammatory cytokine levels in vivo, including interleukin (IL)-1β , tumor necrosis factor (TNF)-α and IL-6 [127,128]. In addition, various studies have undertaken to address the role of histone modifications in VSMC phenotypic modulation and proliferation in response to these cytokines, as reviewed in [129]. Furthermore, histone modifications have been implicated in endothelial cell function and dysfunction, particularly in the regulation of endothelial nitric oxide synthase (eNOS). Histone acetylation of H3 and H4 as well as methylation of H3 at NOS3, the gene encoding eNOS, promoter regions in endothelial cells results in activation of the gene [130]. However, in VSMCs, which do not normally express eNOS, these activating epigenetic marks are missing [130]. However, under hypoxic conditions, endothelial dysfunction ensues as a result of a loss of activating histone marks at the NOS3 promoter region, resulting in a decline in eNOS production [131]. Thus, histone modifications govern multiple aspects of atherogenesis, including inflammation, VSMC phenotypic modulation and endothelial dysfunction.
miR-mediated regulation
Several miRs have been reported to be involved in the development or control of atherosclerosis. miRs can also be useful in diagnosing CAD, perhaps even facilitating the identification of nonclassically-presenting (atypical) CAD [132]. miR-126 is one of the most highly expressed miRs in endothelial cells [133]. It is thought to have an atheroprotective role in the vasculature by targeting vascular endothelial growth factor (VEGF) signaling [134]. miR-181b has also been shown to oppose the progression of atherosclerosis, but this miR does so by inhibiting nuclear factor (NF)-κB activity. NF-κB is a pivotal transcription factor responsible for regulating many proinflammatory cytokines and adhesion molecules across diverse cell types. miR-181b has been reported to inhibit NF-κB nuclear translocation in endothelial cells, resulting in decreased expression of vascular cell adhesion molecule (VCAM)-1 and E-selectin [135]. Similarly, miR-10a negatively regulates NF-κB activation in endothelial cells by attenuating the MAP3K7 and TAK1-mediated disinhibition of NF-κB [136]. The role of miR-155 in atherosclerosis is less clear. miR-155 has been regulated [s9]by the transcription factor Ets1 in endothelial cells, resulting in reduced proinflammatory signaling molecules such as VCAM-1 and MCP [137]. The antiatherogenic roles of miR-155 are highlighted by the finding that peripheral blood miR-155 levels are lower in patients with CAD than their healthy counterparts [138]. However, atherogenic roles for miR-155 have also been described, including upregulation of chemokines, reduction of anti-inflammatory T regulatory cells and enhanced helper T cell type 1 (TH1) cell response [139]. Other miRs also appear to promote atherogenesis, including miR-33, which promotes dyslipidemia by impairing cholesterol efflux by targeting ATP-binding cassette (ABC) transporters [140]. Inhibition of miR-33 reverses atherosclerotic progression [141].
Therapeutic implications: targeting epigenetic mechanisms to treat cardiovascular disease
DNA methylation
DNA methylation inhibitor 5-azaC has shown significant benefits in the treatment of hematological diseases including malignancies. In cardiovascular research, 5-azaC has received increased attention as a result of findings that reported the DNA methylation inhibitor promotes the differentiation of cardiomyocytes from mesenchymal stem cells [142–144], although at low frequencies [145]. However, the potential benefits of 5-azaC in cardiac regeneration studies are limited by toxicity, including myeloid suppression and induction of cardiomyocyte cell death. Interestingly zebularine, another cytidine analog, has less toxicity compared with 5-azaC, making it a promising candidate for studies on cardiomyocyte differentiation from ESCs. Indeed, elegant work by Horrillo et al. has recently demonstrated that zebularine promotes cardiomyocyte differentiation from ESCs with increased expression of cardiac-specific genes [146]. It remains to be determined whether these findings will translate into advances in the development of stem-cell-based therapy for degenerative cardiac diseases.
Histone modifications
Most of the work done on the therapeutic potential of histone modifiers in cardiovascular disease has focused on HDAC inhibitors. These inhibitors include the commonly used antiepilepsy drug valproic acid (an HDAC6 and HDAC8 inhibitor), the small molecule MPT0E014 (HDAC1, 2 and 6 inhibitor) and HDAC class I and II inhibitors TSA and suberoylanilide[s10] hydroxamic acid (SAHA). These agents have been found to reduce fibrosis and cardiac remodeling in preclinical models of HF, although their mechanisms are not clear [147,148]. In addition, curcumin, a turmeric derivative, exerts antifibrotic and antihypertrophic effects by inhibiting HDAC1, 3 and 8 and suppressing p300 activity, respectively [147].
miR-mediated regulation
Although there is growing evidence of miR expression alterations correlating to cardiovascular diseases [149], several miRs have emerged as potential therapeutic targets. For example, miR-15 inhibition improves cardiac function in animal models of HF by reducing infarct size, fibrosis and remodeling [149]. Also, inhibition of miR-208a has been shown to improve survival and cardiac function in a hypertension-induced HF rat model [150]. In addition, other miRs have been shown to slow or inhibit cellular processes contributing to disease, such as cardiomyocyte apoptosis and fibroblast activation [151]. Future work must be conducted to evaluate the effects of targeting and/or introducing miRs on the functional outcome of cardiovascular diseases.
Concluding remarks
Epigenetic mechanisms of gene regulation represent a breakthrough in modern science and, in our understanding of intra- and inter-cellular processes, contribute to normal development as well in most, if not all, diseases. These mechanisms offer novel perspectives on approaches to therapy for diseases ranging from developmental defects to aging processes to cancer. In this review, we have addressed key roles of DNA methylation, histone acetylation and methylation, and miR-mediated regulation in heart development and disease in an attempt to introduce the reader to the most recent work in this field as well as pivotal findings that continue to cultivate further progress in the prevention and treatment of cardiovascular disease.
Highlights.
Epigenetic mechanisms influence gene expression at key cardiac developmental stages and during major cellular events
Epigenetic mechanisms can directly contribute to the development of cardiovascular diseases
Epigenetic mechanisms represent novel and promising targets for therapeutic development in the treatment of cardiovascular diseases
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL082779 (L.Z.), HL083966 (L.Z.), HL118861 (L.Z.) and HL118861S1 (S.R.M.). We apologize to all authors whose work could not be cited because of space limitations.
Biography

Shannalee Martinez
Shannalee Martinez received her BS degree from Southern Adventist University in 2009 and is currently pursuing her MD/PhD at Loma Linda University. Her graduate studies have been facilitated by the Loma Linda University Initiative for Maximizing Student Development Program through the National Institutes of Health. Her current research is supported by the NIH National Heart Lung and Blood Institute Diversity Supplement Program. Her research focuses on epigenetic mechanisms in heart development and developmental origins of health and disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: Spatio-temporal epigenetic gene regulation is essential for the proper development of the heart, and aberrant epigenetic mechanisms contribute to susceptibility to cardiovascular disease.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Barker DJ, Osmond C. Low birth weight and hypertension. BMJ. 1988;297:134–135. doi: 10.1136/bmj.297.6641.134-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen M, Zhang L. Epigenetic mechanisms in developmental programming of adult disease. Drug Discov. Today. 2011;16:1007–1018. doi: 10.1016/j.drudis.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br. Med. Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- 4.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 5.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 7.Buck-Koehntop BA, Defossez PA. On how mammalian transcription factors recognize methylated DNA. Epigenetics. 2013;8:131–137. doi: 10.4161/epi.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore LD, et al. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxonov S, et al. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazor KL, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boland MJ, et al. Epigenetic regulation of pluripotency and differentiation. Circ. Res. 2014;115:311–324. doi: 10.1161/CIRCRESAHA.115.301517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenet F, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maunakea AK, et al. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seisenberger S, et al. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guibert S, et al. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22:633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auclair G, Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94:2202–2211. doi: 10.1016/j.biochi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Guibert S, Weber M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr. Top. Dev. Biol. 2013;104:47–83. doi: 10.1016/B978-0-12-416027-9.00002-4. [DOI] [PubMed] [Google Scholar]
- 21.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 22.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 23.Vaquero A, et al. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003;2003:Re4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 24.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Dysregulation of histone acetyltransferases and deacetylases in cardiovascular diseases. Oxid. Med. Cell Longev. 2014;2014:641979. doi: 10.1155/2014/641979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dokmanovic M, et al. Histone deacetylase inhibitors: overview and perspectives. Mol. Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Vickers KC, et al. MicroRNAs in the onset and development of cardiovascular disease. Clin. Sci. (Lond.) 2014;126:183–194. doi: 10.1042/CS20130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berillo OA, et al. Characteristics of binding sites of intergenic, intronic and exonic miRNAs with mRNAs of oncogenes coding intronic miRNAs. Afr. J. Biotechnol. 2013;12:1016–1024. [Google Scholar]
- 31.Guo H, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Xin M, et al. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paradis AN, et al. Binucleation of cardiomyocytes: the transition from a proliferative to a terminally differentiated state. Drug Discov. Today. 2014;19:602–609. doi: 10.1016/j.drudis.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuhrenberg T, et al. Epigenetics in cardiac development, function, and disease. Cell Tissue Res. 2014;356:585–600. doi: 10.1007/s00441-014-1887-8. [s11] [DOI] [PubMed] [Google Scholar]
- 38.Arand J, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirasawa R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurihara Y, et al. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev. Biol. 2008;313:335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 41.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 42.Okano M, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 43.Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 44.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi S, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko M, et al. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 51.Choy MK, et al. Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Genomics. 2010;11:519. doi: 10.1186/1471-2164-11-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bibikova M, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abbey D, Seshagiri PB. Aza-induced cardiomyocyte differentiation of P19 EC-cells by epigenetic co-regulation and ERK signaling. Gene. 2013;526:364–373. doi: 10.1016/j.gene.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 55.Chamberlain AA, et al. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J. Am. Heart Assoc. 2014;3:e000976. doi: 10.1161/JAHA.114.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paradis A, et al. Endothelin-1 promotes cardiomyocyte terminal differentiation in the developing heart via heightened DNA methylation. Int. J. Med. Sci. 2014;11:373–380. doi: 10.7150/ijms.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kou CY, et al. Epigenetic regulation of neonatal cardiomyocytes differentiation. Biochem. Biophys. Res. Commun. 2010;400:278–283. doi: 10.1016/j.bbrc.2010.08.064. [DOI] [PubMed] [Google Scholar]
- 58.Yuan Y, et al. Promoter methylation and expression of the VANGL2 gene in the myocardium of pediatric patients with Tetralogy of Fallot. Birth Defects Res. A Clin. Mol. Teratol. 2014 doi: 10.1002/bdra.23291. [DOI] [PubMed] [Google Scholar]
- 59.Yao TP, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 60.Shikama N, et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. Clin. Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gottlieb PD, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava D, et al. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 66.Lee Y, et al. Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- 67.Jung J, et al. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J. Biol. Chem. 2005;280:30916–30923. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- 68.Schneider JE, et al. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev. Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 70.Nicholson TB, et al. A hypomorphic lsd1 allele results in heart development defects in mice. PLoS One. 2013;8:e60913. doi: 10.1371/journal.pone.0060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 72.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rao PK, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 75.Kwon C, et al. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 77.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sluijter JP, et al. MicroRNA-1 and −499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 2010;30:859–868. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 79.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duygu B, et al. Genetics and epigenetics of arrhythmia and heart failure. Front. Genet. 2013;4:219. doi: 10.3389/fgene.2013.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dirkx E, et al. Regulation of fetal gene expression in heart failure. Biochim. Biophys. Acta. 1832:2414–2424. doi: 10.1016/j.bbadis.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 82.Bronze-da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. Biomed. Res. Int. 2014;2014:985408. doi: 10.1155/2014/985408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Creemers EE, et al. Heart failure: advances through genomics. Nat. Rev. Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 84.Go AS, et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smolarek I, et al. Global DNA methylation changes in blood of patients with essential hypertension. Med. Sci. Monit. 2010;16:Cr149–Cr155. [PubMed] [Google Scholar]
- 86.Friso S, et al. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199:323–327. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 87.Stewart PM, et al. Syndrome of apparent mineralocorticoid excess. A defect in the cortisol-cortisone shuttle. J. Clin. Invest. 1988;82:340–349. doi: 10.1172/JCI113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Friso S, et al. Epigenetics and arterial hypertension: the challenge of emerging evidence. Transl. Res. 2015;165:154–165. doi: 10.1016/j.trsl.2014.06.007. [s12] [DOI] [PubMed] [Google Scholar]
- 89.Riviere G, et al. Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics. 2011;6:478–489. doi: 10.4161/epi.6.4.14961. [DOI] [PubMed] [Google Scholar]
- 90.Papait R, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodriguez-Iturbe B. Arteriolar remodeling in essential hypertension: are connective tissue growth factor and transforming growth factor involved? Kidney Int. 2006;69:1104–1105. doi: 10.1038/sj.ki.5000222. [DOI] [PubMed] [Google Scholar]
- 92.Zhang D, et al. Epigenetics and the control of epithelial sodium channel expression in collecting duct. Kidney Int. 2009;75:260–267. doi: 10.1038/ki.2008.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pojoga LH, et al. Histone demethylase LSD1 deficiency during high-salt diet is associated with enhanced vascular contraction, altered NO-cGMP relaxation pathway, and hypertension. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1862–H1871. doi: 10.1152/ajpheart.00513.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Irmak MK, Sizlan A. Essential hypertension seems to result from melatonin-induced epigenetic modifications in area postrema. Med. Hypotheses. 2006;66:1000–1007. doi: 10.1016/j.mehy.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 95.Nguyen Dinh Cat A, et al. Conditional transgenic mice for studying the role of the glucocorticoid receptor in the renal collecting duct. Endocrinology. 2009;150:2202–2210. doi: 10.1210/en.2008-1531. [DOI] [PubMed] [Google Scholar]
- 96.Mu S, et al. Epigenetic modulation of the renal beta-adrenergic-WNK4 pathway in salt-sensitive hypertension. Nat. Med. 2011;17:573–580. doi: 10.1038/nm.2337. [DOI] [PubMed] [Google Scholar]
- 97.Albinsson S, Sward K. Targeting smooth muscle microRNAs for therapeutic benefit in vascular disease. Pharmacol. Res. 2013;75:28–36. doi: 10.1016/j.phrs.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 98.Luo X, et al. Regulation of human cardiac ion channel genes by microRNAs: theoretical perspective and pathophysiological implications. Cell Physiol. Biochem. 2010;25:571–586. doi: 10.1159/000315076. [DOI] [PubMed] [Google Scholar]
- 99.Matkovich SJ, et al. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ. Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Topkara VK, Mann DL. Clinical applications of miRNAs in cardiac remodeling and heart failure. Per. Med. 2010;7:531–548. doi: 10.2217/pme.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ikeda S, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol. Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elia L, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hergenreider E, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 104.Elia L, et al. The knockout of miR-143 and −145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dong S, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheng Y, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J. Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thum T, et al. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc. Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 109.Ji R, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ. Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 110.Patrick DM, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haas J, et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 2013;5:413–429. doi: 10.1002/emmm.201201553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Movassagh M, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nguyen AT, et al. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev. 2011;25:263–274. doi: 10.1101/gad.2018511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou B, et al. Common genetic polymorphisms in pre-microRNAs were associated with increased risk of dilated cardiomyopathy. Clin. Chim. Acta. 2010;411:1287–1290. doi: 10.1016/j.cca.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 115.Matkovich SJ, et al. Direct and indirect involvement of microRNA-499 in clinical and experimental cardiomyopathy. Circ. Res. 2012;111:521–531. doi: 10.1161/CIRCRESAHA.112.265736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Halkein J, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J. Clin. Invest. 2013;123:2143–2154. doi: 10.1172/JCI64365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan KL, et al. Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. 2013;65:12–16. doi: 10.1016/j.ihj.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Satoh M, et al. Expression of microRNA-208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J. Card. Fail. 2010;16:404–410. doi: 10.1016/j.cardfail.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 119.Kim GH. MicroRNA regulation of cardiac conduction and arrhythmias. Transl. Res. 2013;161:381–392. doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Callis TE, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu Y, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 122.Libby P, et al. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 123.Castro R, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin. Chem. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 124.Hiltunen MO, et al. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc. Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 125.Ying AK, et al. Methylation of the estrogen receptor-alpha gene promoter is selectively increased in proliferating human aortic smooth muscle cells. Cardiovasc. Res. 2000;46:172–179. doi: 10.1016/s0008-6363(00)00004-3. [DOI] [PubMed] [Google Scholar]
- 126.Baccarelli A, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–828. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fass DM, et al. Effect of inhibiting histone deacetylase with short-chain carboxylic acids and their hydroxamic acid analogs on vertebrate development and neuronal chromatin. ACS Med. Chem. Lett. 2010;2:39–42. doi: 10.1021/ml1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cardinale JP, et al. HDAC inhibition attenuates inflammatory, hypertrophic, and hypertensive responses in spontaneously hypertensive rats. Hypertension. 2010;56:437–444. doi: 10.1161/HYPERTENSIONAHA.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Findeisen HM, et al. Epigenetic regulation of vascular smooth muscle cell function in atherosclerosis. Curr. Atheroscler. Rep. 2013;15:319. [PubMed] [Google Scholar]
- 130.Yan MS, et al. Epigenetics of the vascular endothelium. J. Appl. Physiol. 2010;109:916–926. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 131.Ho JJ, et al. Nitric oxide signaling in hypoxia. J. Mol. Med. (Berl) 2012;90:217–231. doi: 10.1007/s00109-012-0880-5. [DOI] [PubMed] [Google Scholar]
- 132.Wang J, et al. Altered serum microRNAs as novel diagnostic biomarkers for atypical coronary artery disease. PLoS One. 2014;9:e107012. doi: 10.1371/journal.pone.0107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wei Y, et al. MicroRNA-126, −145, and −155: a therapeutic triad in atherosclerosis? Arterioscler. Thromb. Vasc. Biol. 2013;33:449–454. doi: 10.1161/ATVBAHA.112.300279. [DOI] [PubMed] [Google Scholar]
- 134.Menghini R, et al. MicroRNAs in vascular aging and atherosclerosis. Ageing Res Rev. 2014;17:68–78. doi: 10.1016/j.arr.2014.03.005. [s13] [DOI] [PubMed] [Google Scholar]
- 135.Faraoni I, et al. miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 136.Fang Y, et al. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu N, et al. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis. 2011;215:286–293. doi: 10.1016/j.atherosclerosis.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 138.Fichtlscherer S, et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 139.Ma X, et al. MicroRNA-155 in the pathogenesis of atherosclerosis: a conflicting role? Heart Lung Circ. 2013;22:811–818. doi: 10.1016/j.hlc.2013.05.651. [DOI] [PubMed] [Google Scholar]
- 140.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Naeem N, et al. DNA methylation inhibitors, 5-azacytidine and zebularine potentiate the transdifferentiation of rat bone marrow mesenchymal stem cells into cardiomyocytes. Cardiovasc. Ther. 2013;31:201–209. doi: 10.1111/j.1755-5922.2012.00320.x. [DOI] [PubMed] [Google Scholar]
- 143.Antonitsis P, et al. In vitro cardiomyogenic differentiation of adult human bone marrow mesenchymal stem cells. The role of 5-azacytidine. Interact. Cardiovasc. Thorac. Surg. 2007;6:593–597. doi: 10.1510/icvts.2007.157875. [DOI] [PubMed] [Google Scholar]
- 144.Qian Q, et al. 5-Azacytidine induces cardiac differentiation of human umbilical cord-derived mesenchymal stem cells by activating extracellular regulated kinase. Stem Cells Dev. 2012;21:67–75. doi: 10.1089/scd.2010.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martin-Rendon E, et al. 5-Azacytidine-treated human mesenchymal stem/progenitor cells derived from umbilical cord, cord blood and bone marrow do not generate cardiomyocytes in vitro at high frequencies. Vox. Sang. 2008;95:137–148. doi: 10.1111/j.1423-0410.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 146.Horrillo A, et al. Zebularine regulates early stages of mESC differentiation: effect on cardiac commitment. Cell Death Dis. 2013;4:e570. doi: 10.1038/cddis.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tao H, et al. Histone deacetylases in cardiac fibrosis: current perspectives for therapy. Cell Signal. 2014;26:521–527. doi: 10.1016/j.cellsig.2013.11.037. [DOI] [PubMed] [Google Scholar]
- 148.Schuetze KB, et al. Targeting cardiac fibroblasts to treat fibrosis of the heart: focus on HDACs. J. Mol. Cell. Cardiol. 2014;70:100–107. doi: 10.1016/j.yjmcc.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sayed AS, et al. Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ. 2014;23:503–510. doi: 10.1016/j.hlc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 150.Montgomery RL, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sala V, et al. MicroRNAs in myocardial ischemia: identifying new targets and tools for treating heart disease. New frontiers for miR-medicine. Cell. Mol. Life Sci. 2014;71:1439–1452. doi: 10.1007/s00018-013-1504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]